Abstract
DNA replication proceeds along spatially and temporally coordinated patterns within the nucleus, thus protecting the genome during the synthesis of new genetic material. While we have been able to visualize replication patterns on DNA fibers for 50 years, recent developments and discoveries have provided a greater insight into how DNA replication is controlled. In this review, we highlight many of these discoveries. Of great interest are the physiological role of the replication timing program, cis and trans-acting factors that modulate replication timing and the effects of chromatin structure on the replication timing program. We also discuss future directions in the study of replication timing.
Introduction
Duplication of the genetic material prior to each cell division requires stepwise detangling, unwinding, synthesis and reassembly of the entire length of nuclear DNA. Because this process necessarily endangers genomic stability, replication is highly coordinated spatially and temporally within the nucleus. Replication patterns of single DNA fibers were first visualized in mammalian cells half a century ago [1] and follow-up experiments detected DNA replication events as foci within interphase nuclei [2–4]. Both visualization approaches clearly indicate that DNA synthesis proceeds along temporal and spatial patterns that are correlated with nuclear structure. Advances in whole-genome analyses of replicating DNA continue to support the notion that nuclear DNA replicates with a distinct, consistent and often tissue-specific order (“replication timing”) that is strongly associated with spatial patterns of chromatin organization ([5,6]; for a recent review, see [7]).
In mammals, DNA replication starts at many sites (replication origins) on each chromosome. Those origins often cluster into larger domains called replication timing domains, each containing multiple origins that replicate concomitantly or within a short time window [8,9]. These domains vary in length from hundreds of kilobases to megabases [8–14]. Replicon clusters can be visualized as discrete foci of pulse-labelled DNA (replication foci) whose density, size and distribution vary as cells progress from early to late synthesis (S) phase of the cell cycle. Sequential pulse-chase experiments have demonstrated that the DNA sequences replicated at individual sites are consistent temporally and spatially as the cell progresses through the cell cycle and into subsequent generations [15].
Early replicating DNA often consists of regions of open chromatin that contain transcribed regions. As S-phase proceeds, early replicating domains complete replication, and DNA synthesis transitions gradually to replicate more condensed, less transcriptionally active chromatin in mid S-phase. Condensed, heterochromatic DNA often replicates in late S-phase (Figure 1). For example, centromeric cores, which contain centromeric heterochromatin, replicate in mid S-phase, before heavily compacted classical heterochromatin [16]. Genomic analysis of replication timing domains demonstrate that these domains are conserved within cells of any particular tissue type or developmental stage, and domain boundary analysis showed that replication domains highly colocalize with topologically associating domains (TADs), which are substructures located within chromatin compartments identified by high-throughput chromosome conformation capture (Hi-C) [12,17–20]. These studies imply that the order of DNA replication is associated with the same spatial features that constrain the distribution of chromatin in the interphase nucleus.
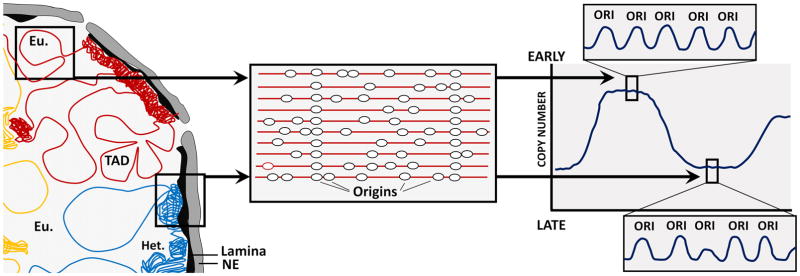
Figure 1.
Replication timing and nuclear structure: To co-ordinate DNA replication with transcription and chromatin assembly, chromatin inside the nucleus is organized into distinct replication timing domains. Replication timing domains are highly colocalized with topologically associating domains (TADs). Some domains contain euchromatin with actively transcribed genes and usually replicate at early S phase; some domains contain heterochromatin with silenced genes and replicate at late S phase. Inside each replication timing domain, many origins cluster together and initiate at similar times. Replication timing can be quantified by measuring relative copy number in proliferating cells. Replication timing domains can be detected as magabase-sized peaks when relative copy number is plotted, and replication origins can be detected as small peaks (ripples) within replication timing domains. Eu: euchromatin. Het: heterchromatin. Ori: origins. NE: nuclear envelope.
Because cells of different tissues of origin and developmental stages require specialized transcription programs, the ability to alter the order of DNA replication might confer an evolutionary advantage by facilitating a rapid adaptation to a changing chromatin landscape during differentiation. In support of this hypothesis, the order of replication changes in a large fraction of the human genome during nuclear reorganization processes associated with differentiation and development [21,22]. In this review, we will discuss the possible role of a replication timing program in metazoans and explore the known determinants of the order of replication in somatic cells with a special emphasis on how the replication program relates to the determinants of chromatin organization in the nucleus.
Physiological role of the replication timing program
The need for a consistent, developmentally and tissue-specific temporal order for eukaryotic genome duplication is not immediately evident. Entire genomes (for example, during the early embryonic stages in flies and frogs) can be assembled and rapidly duplicated without an apparently consistent order [23]. Consistent replication timing programs, however, appear to be ubiquitous in differentiated somatic cells and in embryonic cells after the activation of zygotic transcription, suggesting that the consistent order of DNA replication might facilitate genome integrity by coordinating replication with transcription and chromatin assembly on the shared chromatin template.
Two lines of evidence suggest that a consistent replication timing program plays a role in the maintenance of genomic stability by preventing conflicts between genome duplication and other enzymatic processes occurring concomitantly on chromatin, such as transcription. First, changes in replication timing are hallmarks of certain cancers [24–26] and also accompany pathological chromosomal rearrangements in several rare genetic disorders [27,28]. Second, a distinct order of DNA replication is primarily a property of the euchromatin component of nuclear DNA. In fact, the order of replication within extended heterochromatin domains does not seem to be consistent. For example, an allele-specific chromosomally phased study of the replication timing program along the active and inactive chromosomes revealed that replication proceeded with a consistent order in the active X chromosome, whereas the order of replication within the inactive X varied among individuals and resembled a random, unstructured process [8,29]. Late-replicating, transcriptionally inactive regions on the autosomes also replicated in a similar manner, suggesting that in mammalian cells, inactive and condensed chromatin regions replicate rapidly in an apparently random order [29]. Furthermore, late replication of heterochromatin may preserve the structural integrity of the nucleus by preventing rapid, massive chromatin decondensation and re-condensation at the early stages of genome duplication [30]. Taken together with evidence for stochastic replication in cellular systems that do not activate transcription, such as during early embryogenesis [23], these observations imply that the temporal order of DNA replication facilitates coordination of replication with gene expression.
An additional role of the replication timing program, in particular the early replication of highly expressed genomic regions, might be to provide the necessary templates for transcription of genes that are typically expressed during S-phase. For example, transcription of the histone genes, which in most cells replicate early during S-phase, was halved when the replication timing of histone gene regions was delayed [31]. In addition, studies in induced human embryonic stem cell reveal that during differentiation, almost all the expressed genes, whether constitutive or differentiation-dependent, replicate at early S-phase. Only a very small fraction of expressed genes, which typically exhibit low levels of expression, replicate late [32]. These data suggest that DNA replication in early S-phase contributes towards maintaining high expression levels of genes actively transcribed during S-phase.
The mutation landscape of the genome is also influenced by replication timing. This influence is particularly apparent during carcinogenesis and genome evolution. Sections of DNA that replicate at different times during S-phase tend to have different mutation patterns. Point mutations and copy number losses correlate with late replication, whereas copy number gains and other rearrangements correlate with early replication (for a review, see [33]). Early replicating regions exhibit lower point mutation rates than regions replicating late, possibly because mismatch repair (MMR) efficiency is higher during early replication [34–36]. Higher MMR efficiency during early S-phase might therefor select for the replication of actively transcribed genes in early S-phase. Deregulated initiation of DNA replication on transcribed DNA increases the number of head-on collisions between DNA replication and transcription machinery. The collisions lead to R-loops and genome instability [37] that demonstrate the importance of coordinating replication timing and transcription.
Cis-elements as determinants of replication timing
Mechanistically, replication timing domains that replicate in a consistent order require the activation of groups of replication origins at distinct times during S-phase. The sequential activation of replication origins is evident within replication timing domains. For example, in human basal erythroblasts, small highly reproducible peaks (Figure 1) are detectable within the well-characterized, megabase-sized replication timing domains correlate with clusters of replication origins detected by nascent strand analyses [8]. Experiments correlating the frequency of replication initiation events as measured by the direction of Okazaki fragments with large replication timing domains also support this conclusion [38].
Although epigenetic properties, such as histone modifications, play an important role in determining initiation frequency [10,39,40], replication origin activity is determined in part by the primary DNA sequence, as origin sequences can initiate DNA replication at ectopic sites [41–43]. Primary DNA sequence properties also correlate in part with replication timing. For example, early-replicating regions correspond to gene-rich, high-GC domains, whereas late-replicating regions correspond to gene-poor, low-GC domains [44]. However, replication timing does not correlate with either GC or repeat content in zebrafish [45], suggesting a link between DNA replication and functional aspects of the genome rather than sequence composition.
Replication origins may regulate chromatin and replication timing via specific DNA sequences that are easily unwound or by recruiting sequence-specific proteins that modify local and distal interactions on chromatin. Consistent with this suggestion, inherited variants that affect replication origin activity and replication timing have been identified in two experiments: analyses of replication timing in 161 samples of proliferating cells [46] and analyses of phased genomes, which permit identification of paternal or maternal origins to characterize the effects of specific sequence variations on origin activity [8,9]. In parallel, replication origins may also interact directly or indirectly with local or long distance cis-elements to modulate replication (see [47] for details). For example, an interaction between the replication origin and the locus control region at the human β-globin locus [48] affects the time of replication and is essential for both replication initiation and prevention of gene silencing [41,42,49].
In addition to sequences at replication origins, cis-acting sequences that modulate large chromatin domains can also affect the order of replication. For example, heterochromatin formation and replication delay can be modulated via non-coding RNAs such as XIST for the X chromosome [29,47] and ASAR6 and ASAR15 for the autosomal chromosome 6 and 15, respectively, in cancer cells [50,51]. These long non-coding RNAs, which remain associated with the chromosome territories from which they are transcribed, are implicated in the control of monoallelic gene expression but may be involved more broadly in chromosomal maintenance throughout the cell cycle [50,51]. Replication associated with ASAR6 transgenes requires an anti-sense transcript of an L1 retrotransposon, implicating L1 antisense RNA as a player in the regulation of chromosome-wide replication timing [51]. Cis-acting genetic elements at TADs can also determine, at least in part, the locations of megabase-scale replication timing domains. In cancer cells, the replication of entire chromosomes may be delayed in a sequence-specific manner, and interactions with long non-coding RNAs could alter the timing of replication for entire chromosomes [50].
Chromatin structure
The partial dependence on the primary sequence does not necessarily imply that specific sequences solely determine the timing of DNA replication, either directly or indirectly. In fact, recent studies using a combination of mathematical modelling and high-throughput genomics provide evidence to the contrary, suggesting that the distribution of replication origins by itself is sufficient to determine the order of replication initiation, and hence the replication timing program. For example, a mathematical model assuming a single rate-limiting factor for DNA replication and relying solely on DNase hypersensitivity data and the relative efficiencies of initiation at specific loci [52] accurately recapitulated cell-specific DNA replication timing patterns, including abnormal timing in cancer cells. This observation implies that it is possible to predict replication timing with high accuracy in human cells without assuming any “replication timing factor”. A second model [53], assuming spontaneous stochastic initiation within euchromatin and facultative heterochromatin, successfully predicted the three-dimensional spatial and temporal organization of replication events as well as the timing of replication based on higher chromatin organization. The density of the mini-chromosome maintenance (MCM) replicative helicase, assuming a high level of MCMs at early origins [54], was also able to predict the replication timing program, suggesting that the spatial distribution of replication origins determines the temporal organization of the replication process.
High-resolution whole-genome analyses have revealed that replication timing domains often reflect chromatin modifications [11,21]. Early replicating regions associate with transcriptionally active topological domains, whereas late replicating origins often associate with heterochromatin. The establishment of the replication-timing program occurs during the early G1 phase of the cell cycle, around the same time as TADs have been shown to assemble and become compartmentalized in mouse cells. The association between replication timing domains and TADs suggests that the timing of activation of replication origins reflects a fundamental structural property of the nucleus [17,19,21,55]. Replication origins are known to associate with nuclear structural features such as matrix attachment sites, scaffold attachment sites and stabilizing anti repressor elements [47,56] as well as with lamins and cohesins [39,47]. Replication origins also associate with chromatin modulators, such as a phosphorylated from of the histone deacetylase SIRT1 that prevents the initiation of DNA replication from a group of “dormant” origins [57].
The replication timing of specific genomic loci can vary with tissue type and differentiation status. For example, the human beta-globin locus replicates early during S-phase in erythroleukaemia cells whereas in non-erythroid cells it replicates later in S-phase [58], suggesting that other than sequence, chromatin environments also affect replication timing. Replication timing and initiation patterns tend to depend on cellular lineage rather than on cancer status in cancer cells [59]. Early replicating regions associate with open chromatin features, such as acetylation of H3K9, H3K18 and H3K27, methylation H3K4me and H3K36me3 [59]. Transcriptional activity coordinates with the replication timing program [32] and replication delays often accompany gene silencing. Origins that associate with open chromatin are activated in many cell types, and are enriched in moderately active transcription start sites [13,60]. In contrast, late replicating regions associate with closed chromatin features, like hypoacetylation of H3 and H4, methylation H3K9 and H3K27, and they often initiate replication in a cell type specific manner [39,59].
Trans-acting factors
The binding patterns of pre-replication complexes do not provide clues to the principles of origin choice because the origin recognition complex, which anchors the pre-replication complexes to chromatin, does not bind specific DNA sequences [61]. Interactions between replication origins and components of pre-replication complexes, therefore, are essential for initiation but cannot provide a simple mechanistic explanation for the consistent replication patterns observed in most mitotic cell cycles.
The transcription factors Forkhead 1 (Fkh1) and Forkhead 2 (Fkh2) are required for clustering a subset of replication origins during G1 phase and for early initiation of these origins in S-phase in S. cerevisiae [62,63]. Fkh1 and Fkh2 promote early replication by recruiting replication factors. Fkh1 and Fkh2 regulate origin timing and establish changes in timing during the late G1 phase of the cell cycle [64], indicating that replication timing can be reset after origin licensing.
Distal DNA sequences modify transcriptional activity and origin activity through long-distance interactions [65–67]. Such interactions can be facilitated by chromatin remodelling factors, transcriptional activators and other proteins that bind enhancers and locus control regions [40,42,68]. For example, RepID, a protein that interacts with a group of replication origins, is associated with an origin-activating chromatin loop between the replication origin and the locus control region that facilitates early replication at the human beta-globin locus in erythroid cells [43].
Rif1, a shelterin component, is involved in regulating replication-timing in all eukaryotes studied so far by modulating the three-dimensional organization of the genome and recruiting protein phosphatase 1 (PP1). Rif1 interacts with G-quadruplex structures and recruits PP1 to modulate the chromatin binding of pre-initiation complex components [69–71] in genomic regions that undergo late replication. These actions might delay replication by preventing the essential replication kinase, DDK, from phosphorylating the replicating helicase (MCM2-7) while associating with nuclear architectural structures that anchor heterochromatin. Recent papers confirmed a direct interaction of Rif1 and PP1 in both human and mouse cells [72–74]. In fission yeast, a second Shelterin component, Taz1, binds along with Rif1 to heterochromatin-euchromatin boundaries to form heterchromatin compartments and regulate gene expression and replication timing [75,76].
The ORC-associated protein (ORCA/LRWD1), which stabilizes ORC on chromatin [77], temporally associates with late replication origins marked with H3K9me3 and methylated CpGs during G1 phase. In ORCA-depleted cells, levels of H3K9me3 and DNA methylation were altered at ORCA binding sites [78]. H4K20 tri-methylation mediated by Suv4-20h is necessary for the licensing and activity of some ORCA/LRWD1-associated origins. These origins ensure that late-replicating heterochromatin domains are replicated at the correct time [79].
Perspective and future directions: the role of the replication timing program
We have known for half a century that replication order follows a strict temporal and spatial organization, and recent studies on the whole-genome level have confirmed that replication patterns are tightly linked to nuclear architectural features. The challenge remains to tease out the determinants of replication timing, distinguish cause from effect, and address the biological questions associated with the replication timing program. An understanding of the association between late-replicating genomic regions and higher mutation rates would be particularly rewarding, as many genomic regions with recurrent mutations in cancer replicate late [46,80]. Lastly, the mechanism by which replication timing affects human genome sequence composition via GC-biased substitutions and gene conversions is an interesting subject of exploration [81–83].
Most studies measuring replication timing have been performed with cells growing in culture, in which the order of replication might reflect selective pressure for rapid proliferation. To understand the dynamics of replication timing more completely, especially the effects of changes in timing in cancer cells, replication timing will need to be investigated in patient samples. Such cells are hard to grow in vitro, but the development of xenografts in immunodeficient mice [84] has opened a novel avenue of research.
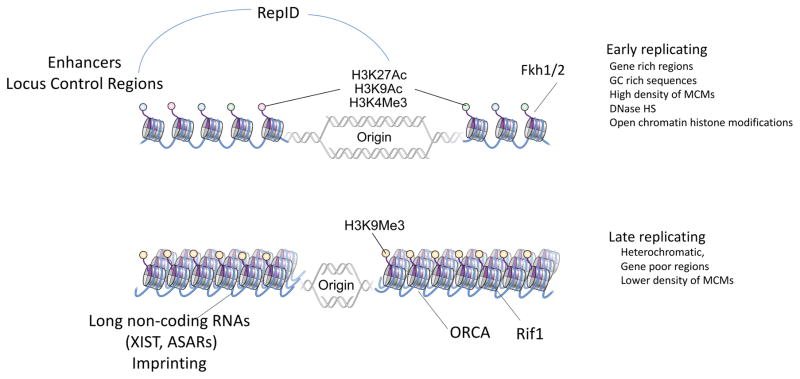
Figure 2.
Cis- and trans-factors establish early DNA replication domains with open chromatin and late DNA replication domains with heterochromatin. In addition to replicators and replication initiation complexes, other cis- and trans-factors affect nuclear structures and DNA replication timing. For example, RepID regulates DNA replication initiation by forming loops linking enhancer and/or locus control regions with replication origins; Fkh1/2 cluster groups of origins in the G1 phase for early replication in S phase; Rif1 and ORCA prevent late replicating regions from replicating earlier. Long non-coding RNAs can also affect chromatin structure and replication timing locally and mediate the action of distal regulators.
Acknowledgments
This study was supported by the Intramural Research Program of the NIH, Center for Cancer Research, National Cancer Institute. The authors wish to thank Dr. Christophe E. Redon for model design and helpful discussion.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Huberman JA, Riggs AD. On the mechanism of DNA replication in mammalian chromosomes. J Mol Biol. 1968;32:327–341. doi: 10.1016/0022-2836(68)90013-2. [DOI] [PubMed] [Google Scholar]
- 2.Jackson DA, Pombo A. Replicon clusters are stable units of chromosome structure: evidence that nuclear organization contributes to the efficient activation and propagation of S-phase in human cells. J Cell Biol. 1998;140:1285–1295. doi: 10.1083/jcb.140.6.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nakamura H, Morita T, Sato C. Structural organizations of replicon domains during DNA synthetic phase in the mammalian nucleus. Exp Cell Res. 1986;165:291–297. doi: 10.1016/0014-4827(86)90583-5. [DOI] [PubMed] [Google Scholar]
- 4.Nakayasu H, Berezney R. Mapping replicational sites in the eucaryotic cell nucleus. J Cell Biol. 1989;108:1–11. doi: 10.1083/jcb.108.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hiratani I, Ryba T, Itoh M, Yokochi T, Schwaiger M, Chang CW, Lyou Y, Townes TM, Schubeler D, Gilbert DM. Global reorganization of replication domains during embryonic stem cell differentiation. PLoS Biol. 2008;6:e245. doi: 10.1371/journal.pbio.0060245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hansen RS, Thomas S, Sandstrom R, Canfield TK, Thurman RE, Weaver M, Dorschner MO, Gartler SM, Stamatoyannopoulos JA. Sequencing newly replicated DNA reveals widespread plasticity in human replication timing. Proc Natl Acad Sci U S A. 2010;107:139–144. doi: 10.1073/pnas.0912402107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rivera-Mulia JC, Gilbert DM. Replicating Large Genomes: Divide and Conquer. Mol Cell. 2016;62:756–765. doi: 10.1016/j.molcel.2016.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mukhopadhyay R, Lajugie J, Fourel N, Selzer A, Schizas M, Bartholdy B, Mar J, Lin CM, Martin MM, Ryan M, et al. Allele-specific genome-wide profiling in human primary erythroblasts reveal replication program organization. PLoS Genet. 2014;10:e1004319. doi: 10.1371/journal.pgen.1004319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bartholdy B, Mukhopadhyay R, Lajugie J, Aladjem MI, Bouhassira EE. Allele-specific analysis of DNA replication origins in mammalian cells. Nat Commun. 2015;6:7051. doi: 10.1038/ncomms8051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aladjem MI, Redon CE. Order from clutter: selective interactions at mammalian replication origins. Nat Rev Genet. 2017;18:101–116. doi: 10.1038/nrg.2016.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dileep V, Ay F, Sima J, Vera DL, Noble WS, Gilbert DM. Topologically associating domains and their long-range contacts are established during early G1 coincident with the establishment of the replication-timing program. Genome Res. 2015;25:1104–1113. doi: 10.1101/gr.183699.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gilbert DM, Takebayashi SI, Ryba T, Lu J, Pope BD, Wilson KA, Hiratani I. Space and time in the nucleus: developmental control of replication timing and chromosome architecture. Cold Spring Harb Symp Quant Biol. 2010;75:143–153. doi: 10.1101/sqb.2010.75.011. [DOI] [PubMed] [Google Scholar]
- 13.Martin MM, Ryan M, Kim R, Zakas AL, Fu H, Lin CM, Reinhold WC, Davis SR, Bilke S, Liu H, et al. Genome-wide depletion of replication initiation events in highly transcribed regions. Genome Res. 2011;21:1822–1832. doi: 10.1101/gr.124644.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Renard-Guillet C, Kanoh Y, Shirahige K, Masai H. Temporal and spatial regulation of eukaryotic DNA replication: from regulated initiation to genome-scale timing program. Semin Cell Dev Biol. 2014;30:110–120. doi: 10.1016/j.semcdb.2014.04.014. [DOI] [PubMed] [Google Scholar]
- 15.Ma H, Samarabandu J, Devdhar RS, Acharya R, Cheng PC, Meng C, Berezney R. Spatial and temporal dynamics of DNA replication sites in mammalian cells. J Cell Biol. 1998;143:1415–1425. doi: 10.1083/jcb.143.6.1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16*.Wear EE, Song J, Zynda G, LeBlanc C, Lee TJ, Mickelson-Young L, Concia L, Mulvaney P, Szymanski ES, Allen GC, et al. Genomic analysis of the DNA replication timing program during mitotic S-phase in maize (Zea mays L.) root tips. Plant Cell. 2017 doi: 10.1105/tpc.17.00037. The first reported whole-genome replication timing analysis in plants using a novel adaptation of the “Repli-seq” assay for use in intact root tips of maize. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yaffe E, Farkash-Amar S, Polten A, Yakhini Z, Tanay A, Simon I. Comparative analysis of DNA replication timing reveals conserved large-scale chromosomal architecture. PLoS Genet. 2010;6:e1001011. doi: 10.1371/journal.pgen.1001011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ryba T, Hiratani I, Lu J, Itoh M, Kulik M, Zhang J, Schulz TC, Robins AJ, Dalton S, Gilbert DM. Evolutionarily conserved replication timing profiles predict long-range chromatin interactions and distinguish closely related cell types. Genome Res. 2010;20:761–770. doi: 10.1101/gr.099655.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hiratani I, Ryba T, Itoh M, Rathjen J, Kulik M, Papp B, Fussner E, Bazett-Jones DP, Plath K, Dalton S, et al. Genome-wide dynamics of replication timing revealed by in vitro models of mouse embryogenesis. Genome Res. 2010;20:155–169. doi: 10.1101/gr.099796.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weddington N, Stuy A, Hiratani I, Ryba T, Yokochi T, Gilbert DM. ReplicationDomain: a visualization tool and comparative database for genome-wide replication timing data. BMC Bioinformatics. 2008;9:530. doi: 10.1186/1471-2105-9-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pope BD, Ryba T, Dileep V, Yue F, Wu W, Denas O, Vera DL, Wang Y, Hansen RS, Canfield TK, et al. Topologically associating domains are stable units of replication-timing regulation. Nature. 2014;515:402–405. doi: 10.1038/nature13986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rivera-Mulia JC, Gilbert DM. Replication timing and transcriptional control: beyond cause and effect-part III. Curr Opin Cell Biol. 2016;40:168–178. doi: 10.1016/j.ceb.2016.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mechali M, Kearsey S. Lack of specific sequence requirement for DNA replication in Xenopus eggs compared with high sequence specificity in yeast. Cell. 1984;38:55–64. doi: 10.1016/0092-8674(84)90526-9. [DOI] [PubMed] [Google Scholar]
- 24.Chang BH, Smith L, Huang J, Thayer M. Chromosomes with delayed replication timing lead to checkpoint activation, delayed recruitment of Aurora B and chromosome instability. Oncogene. 2007;26:1852–1861. doi: 10.1038/sj.onc.1209995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Amiel A, Litmanovitch T, Lishner M, Mor A, Gaber E, Tangi I, Fejgin M, Avivi L. Temporal differences in replication timing of homologous loci in malignant cells derived from CML and lymphoma patients. Genes Chromosomes Cancer. 1998;22:225–231. doi: 10.1002/(sici)1098-2264(199807)22:3<225::aid-gcc8>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 26.Bras A, Cotrim CZ, Vasconcelos I, Mexia J, Leonard A, Sanzhar I, Akhmatullina N, Rueff J. Asynchronous DNA replication detected by fluorescence in situ hybridisation as a possible indicator of genetic damage in human lymphocytes. Oncol Rep. 2008;19:369–375. doi: 10.3892/or.19.2.369. [DOI] [PubMed] [Google Scholar]
- 27.D’Antoni S, Mattina T, Di Mare P, Federico C, Motta S, Saccone S. Altered replication timing of the HIRA/Tuple1 locus in the DiGeorge and Velocardiofacial syndromes. Gene. 2004;333:111–119. doi: 10.1016/j.gene.2004.02.029. [DOI] [PubMed] [Google Scholar]
- 28.State MW, Greally JM, Cuker A, Bowers PN, Henegariu O, Morgan TM, Gunel M, DiLuna M, King RA, Nelson C, et al. Epigenetic abnormalities associated with a chromosome 18(q21–q22) inversion and a Gilles de la Tourette syndrome phenotype. Proc Natl Acad Sci U S A. 2003;100:4684–4689. doi: 10.1073/pnas.0730775100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koren A, McCarroll SA. Random replication of the inactive X chromosome. Genome Res. 2014;24:64–69. doi: 10.1101/gr.161828.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bustin M, Misteli T. Nongenetic functions of the genome. Science. 2016;352:aad6933. doi: 10.1126/science.aad6933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31**.Muller CA, Nieduszynski CA. DNA replication timing influences gene expression level. J Cell Biol. 2017;216:1907–1914. doi: 10.1083/jcb.201701061. Whole genome replication timing analysis of 7 phylogenetically diverse yeast identified genomic features with conserved replication timing and demonstrated that early replication timing might facilitate maximal expression of genes expressed in S-phase, like histone genes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rivera-Mulia JC, Buckley Q, Sasaki T, Zimmerman J, Didier RA, Nazor K, Loring JF, Lian Z, Weissman S, Robins AJ, et al. Dynamic changes in replication timing and gene expression during lineage specification of human pluripotent stem cells. Genome Res. 2015;25:1091–1103. doi: 10.1101/gr.187989.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sima J, Gilbert DM. Complex correlations: replication timing and mutational landscapes during cancer and genome evolution. Curr Opin Genet Dev. 2014;25:93–100. doi: 10.1016/j.gde.2013.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Supek F, Lehner B. Differential DNA mismatch repair underlies mutation rate variation across the human genome. Nature. 2015;521:81–84. doi: 10.1038/nature14173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35*.Poulos RC, Olivier J, Wong JWH. The interaction between cytosine methylation and processes of DNA replication and repair shape the mutational landscape of cancer genomes. Nucleic Acids Res. 2017;45:7786–7795. doi: 10.1093/nar/gkx463. Late replication associated with higher mutation rates, especially at methylated CpGs, in colorectal cancer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lujan SA, Clausen AR, Clark AB, MacAlpine HK, MacAlpine DM, Malc EP, Mieczkowski PA, Burkholder AB, Fargo DC, Gordenin DA, et al. Heterogeneous polymerase fidelity and mismatch repair bias genome variation and composition. Genome Res. 2014;24:1751–1764. doi: 10.1101/gr.178335.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hamperl S, Bocek MJ, Saldivar JC, Swigut T, Cimprich KA. Transcription-Replication Conflict Orientation Modulates R-Loop Levels and Activates Distinct DNA Damage Responses. Cell. 2017;170:774–786. e719. doi: 10.1016/j.cell.2017.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Petryk N, Kahli M, d’Aubenton-Carafa Y, Jaszczyszyn Y, Shen Y, Silvain M, Thermes C, Chen CL, Hyrien O. Replication landscape of the human genome. Nat Commun. 2016;7:10208. doi: 10.1038/ncomms10208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cayrou C, Ballester B, Peiffer I, Fenouil R, Coulombe P, Andrau JC, van Helden J, Mechali M. The chromatin environment shapes DNA replication origin organization and defines origin classes. Genome Res. 2015;25:1873–1885. doi: 10.1101/gr.192799.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fragkos M, Ganier O, Coulombe P, Mechali M. DNA replication origin activation in space and time. Nat Rev Mol Cell Biol. 2015;16:360–374. doi: 10.1038/nrm4002. [DOI] [PubMed] [Google Scholar]
- 41.Fu H, Wang L, Lin CM, Singhania S, Bouhassira EE, Aladjem MI. Preventing gene silencing with human replicators. Nat Biotechnol. 2006;24:572–576. doi: 10.1038/nbt1202. [DOI] [PubMed] [Google Scholar]
- 42.Huang L, Fu H, Lin CM, Conner AL, Zhang Y, Aladjem MI. Prevention of transcriptional silencing by a replicator-binding complex consisting of SWI/SNF, MeCP1, and hnRNP C1/C2. Mol Cell Biol. 2011;31:3472–3484. doi: 10.1128/MCB.05587-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Y, Huang L, Fu H, Smith OK, Lin CM, Utani K, Rao M, Reinhold WC, Redon CE, Ryan M, et al. A replicator-specific binding protein essential for site-specific initiation of DNA replication in mammalian cells. Nat Commun. 2016;7:11748. doi: 10.1038/ncomms11748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Julienne H, Zoufir A, Audit B, Arneodo A. Human genome replication proceeds through four chromatin states. PLoS Comput Biol. 2013;9:e1003233. doi: 10.1371/journal.pcbi.1003233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45**.Siefert JC, Georgescu C, Wren JD, Koren A, Sansam CL. DNA replication timing during development anticipates transcriptional programs and parallels enhancer activation. Genome Res. 2017;27:1406–1416. doi: 10.1101/gr.218602.116. High-resolution whole genome sequencing analysis throughout different stages of Zebrafish development showed dynamic changes in replication timing, transcription and enhancer activity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koren A, Handsaker RE, Kamitaki N, Karlic R, Ghosh S, Polak P, Eggan K, McCarroll SA. Genetic variation in human DNA replication timing. Cell. 2014;159:1015–1026. doi: 10.1016/j.cell.2014.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smith OK, Aladjem MI. Chromatin structure and replication origins: determinants of chromosome replication and nuclear organization. J Mol Biol. 2014;426:3330–3341. doi: 10.1016/j.jmb.2014.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aladjem MI, Groudine M, Brody LL, Dieken ES, Fournier RE, Wahl GM, Epner EM. Participation of the human beta-globin locus control region in initiation of DNA replication. Science. 1995;270:815–819. doi: 10.1126/science.270.5237.815. [DOI] [PubMed] [Google Scholar]
- 49.Lin CM, Fu H, Martinovsky M, Bouhassira E, Aladjem MI. Dynamic alterations of replication timing in mammalian cells. Curr Biol. 2003;13:1019–1028. doi: 10.1016/s0960-9822(03)00382-8. [DOI] [PubMed] [Google Scholar]
- 50.Donley N, Smith L, Thayer MJ. ASAR15, A cis-acting locus that controls chromosome-wide replication timing and stability of human chromosome 15. PLoS Genet. 2015;11:e1004923. doi: 10.1371/journal.pgen.1004923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51*.Platt EJ, Smith L, Thayer MJ. L1 retrotransposon antisense RNA within ASAR lncRNAs controls chromosome-wide replication timing. J Cell Biol. 2017 doi: 10.1083/jcb.201707082. Epub ahead of print Refs. 50 and 51 demonstrate that long non-coding RNAs can regulate the replication timing of entire chromosomes. Ref. 51 suggest that an L1-retrotransposon anti-sense RNA plays a role in regulating replication timing. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gindin Y, Valenzuela MS, Aladjem MI, Meltzer PS, Bilke S. A chromatin structure-based model accurately predicts DNA replication timing in human cells. Mol Syst Biol. 2014;10:722. doi: 10.1002/msb.134859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lob D, Lengert N, Chagin VO, Reinhart M, Casas-Delucchi CS, Cardoso MC, Drossel B. 3D replicon distributions arise from stochastic initiation and domino-like DNA replication progression. Nat Commun. 2016;7:11207. doi: 10.1038/ncomms11207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Das SP, Borrman T, Liu VW, Yang SC, Bechhoefer J, Rhind N. Replication timing is regulated by the number of MCMs loaded at origins. Genome Res. 2015;25:1886–1892. doi: 10.1101/gr.195305.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Moindrot B, Audit B, Klous P, Baker A, Thermes C, de Laat W, Bouvet P, Mongelard F, Arneodo A. 3D chromatin conformation correlates with replication timing and is conserved in resting cells. Nucleic Acids Res. 2012;40:9470–9481. doi: 10.1093/nar/gks736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mechali M, Yoshida K, Coulombe P, Pasero P. Genetic and epigenetic determinants of DNA replication origins, position and activation. Curr Opin Genet Dev. 2013;23:124–131. doi: 10.1016/j.gde.2013.02.010. [DOI] [PubMed] [Google Scholar]
- 57.Utani K, Fu H, Jang S, Marks AB, Redon CE, Smith OK, Zhang Y, Shimizu N, Aladjem MI. Phosphorylated SIRT1 associates with replication origins to prevent excess replication initiation and preserve genomic stability. Nucleic Acids Research. 2017;45:7807–7824. doi: 10.1093/nar/gkx468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Forrester WC, Epner E, Driscoll MC, Enver T, Brice M, Papayannopoulou T, Groudine M. A deletion of the human beta-globin locus activation region causes a major alteration in chromatin structure and replication across the entire beta-globin locus. Genes Dev. 1990;4:1637–1649. doi: 10.1101/gad.4.10.1637. [DOI] [PubMed] [Google Scholar]
- 59.Smith OK, Kim R, Fu H, Martin MM, Lin CM, Utani K, Zhang Y, Marks AB, Lalande M, Chamberlain S, et al. Distinct epigenetic features of differentiation-regulated replication origins. Epigenetics Chromatin. 2016;9:18. doi: 10.1186/s13072-016-0067-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Besnard E, Babled A, Lapasset L, Milhavet O, Parrinello H, Dantec C, Marin JM, Lemaitre JM. Unraveling cell type-specific and reprogrammable human replication origin signatures associated with G-quadruplex consensus motifs. Nat Struct Mol Biol. 2012;19:837–844. doi: 10.1038/nsmb.2339. [DOI] [PubMed] [Google Scholar]
- 61*.Miotto B, Ji Z, Struhl K. Selectivity of ORC binding sites and the relation to replication timing, fragile sites, and deletions in cancers. Proc Natl Acad Sci U S A. 2016;113:E4810–4819. doi: 10.1073/pnas.1609060113. ORC2 binds to open chromatin regions (DNase I-hypersensitive, H3 acetylation and H3K4 methylation) in a non-specific manner and ORC binding density correlates with replication timing. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Knott SR, Peace JM, Ostrow AZ, Gan Y, Rex AE, Viggiani CJ, Tavare S, Aparicio OM. Forkhead transcription factors establish origin timing and long-range clustering in S. cerevisiae. Cell. 2012;148:99–111. doi: 10.1016/j.cell.2011.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63*.Ostrow AZ, Kalhor R, Gan Y, Villwock SK, Linke C, Barberis M, Chen L, Aparicio OM. Conserved forkhead dimerization motif controls DNA replication timing and spatial organization of chromosomes in S. cerevisiae. Proc Natl Acad Sci U S A. 2017;114:E2411–E2419. doi: 10.1073/pnas.1612422114. The transcription factors Fkh1/2 may regulate replication timing by clustering Fkh1/2-bound origins via their conserved forkhead dimerization domains. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Peace JM, Villwock SK, Zeytounian JL, Gan Y, Aparicio OM. Quantitative BrdU immunoprecipitation method demonstrates that Fkh1 and Fkh2 are rate-limiting activators of replication origins that reprogram replication timing in G1 phase. Genome Res. 2016;26:365–375. doi: 10.1101/gr.196857.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Aladjem MI, Rodewald LW, Kolman JL, Wahl GM. Genetic dissection of a mammalian replicator in the human beta-globin locus. Science. 1998;281:1005–1009. doi: 10.1126/science.281.5379.1005. [DOI] [PubMed] [Google Scholar]
- 66.Norio P, Kosiyatrakul S, Yang Q, Guan Z, Brown NM, Thomas S, Riblet R, Schildkraut CL. Progressive activation of DNA replication initiation in large domains of the immunoglobulin heavy chain locus during B cell development. Mol Cell. 2005;20:575–587. doi: 10.1016/j.molcel.2005.10.029. [DOI] [PubMed] [Google Scholar]
- 67.Gerhardt J, Tomishima MJ, Zaninovic N, Colak D, Yan Z, Zhan Q, Rosenwaks Z, Jaffrey SR, Schildkraut CL. The DNA replication program is altered at the FMR1 locus in fragile X embryonic stem cells. Mol Cell. 2014;53:19–31. doi: 10.1016/j.molcel.2013.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Aladjem MI. Replication in context: dynamic regulation of DNA replication patterns in metazoans. Nat Rev Genet. 2007;8:588–600. doi: 10.1038/nrg2143. [DOI] [PubMed] [Google Scholar]
- 69.Hiraga S, Alvino GM, Chang F, Lian HY, Sridhar A, Kubota T, Brewer BJ, Weinreich M, Raghuraman MK, Donaldson AD. Rif1 controls DNA replication by directing Protein Phosphatase 1 to reverse Cdc7-mediated phosphorylation of the MCM complex. Genes Dev. 2014;28:372–383. doi: 10.1101/gad.231258.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70**.Foti R, Gnan S, Cornacchia D, Dileep V, Bulut-Karslioglu A, Diehl S, Buness A, Klein FA, Huber W, Johnstone E, et al. Nuclear Architecture Organized by Rif1 Underpins the Replication-Timing Program. Mol Cell. 2016;61:260–273. doi: 10.1016/j.molcel.2015.12.001. Rif1 coats late-replicating chromatin to define and restrict interactions among replication domains, suggesting a function of Rif1 as a chromatin organizer that sets late DNA replicating and heterochromatin genomic regions. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kanoh Y, Matsumoto S, Fukatsu R, Kakusho N, Kono N, Renard-Guillet C, Masuda K, Iida K, Nagasawa K, Shirahige K, et al. Rif1 binds to G quadruplexes and suppresses replication over long distances. Nat Struct Mol Biol. 2015;22:889–897. doi: 10.1038/nsmb.3102. [DOI] [PubMed] [Google Scholar]
- 72.Sukackaite R, Cornacchia D, Jensen MR, Mas PJ, Blackledge M, Enervald E, Duan G, Auchynnikava T, Kohn M, Hart DJ, et al. Mouse Rif1 is a regulatory subunit of protein phosphatase 1 (PP1) Sci Rep. 2017;7:2119. doi: 10.1038/s41598-017-01910-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hiraga SI, Ly T, Garzon J, Horejsi Z, Ohkubo YN, Endo A, Obuse C, Boulton SJ, Lamond AI, Donaldson AD. Human RIF1 and protein phosphatase 1 stimulate DNA replication origin licensing but suppress origin activation. EMBO Rep. 2017;18:403–419. doi: 10.15252/embr.201641983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74**.Alver RC, Chadha GS, Gillespie PJ, Blow JJ. Reversal of DDK-Mediated MCM Phosphorylation by Rif1-PP1 Regulates Replication Initiation and Replisome Stability Independently of ATR/Chk1. Cell Rep. 2017;18:2508–2520. doi: 10.1016/j.celrep.2017.02.042. References 72–74 provide direct evidence that RIF1 regulates origin activitiation by conteracting DDK/CDK mediated MCM phospharylation via recuiting PP1 in mammalian cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tazumi A, Fukuura M, Nakato R, Kishimoto A, Takenaka T, Ogawa S, Song JH, Takahashi TS, Nakagawa T, Shirahige K, et al. Telomere-binding protein Taz1 controls global replication timing through its localization near late replication origins in fission yeast. Genes Dev. 2012;26:2050–2062. doi: 10.1101/gad.194282.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76*.Toteva T, Mason B, Kanoh Y, Brogger P, Green D, Verhein-Hansen J, Masai H, Thon G. Establishment of expression-state boundaries by Rif1 and Taz1 in fission yeast. Proc Natl Acad Sci U S A. 2017;114:1093–1098. doi: 10.1073/pnas.1614837114. Rif1 and Taz1 can bind to small cis-acting elements to establish heterochromatin–euchromatin boundaries that could simultaneously regulate gene expression and DNA replication timing. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shen Z, Sathyan KM, Geng Y, Zheng R, Chakraborty A, Freeman B, Wang F, Prasanth KV, Prasanth SG. A WD-repeat protein stabilizes ORC binding to chromatin. Mol Cell. 2010;40:99–111. doi: 10.1016/j.molcel.2010.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78*.Wang Y, Khan A, Marks AB, Smith OK, Giri S, Lin YC, Creager R, MacAlpine DM, Prasanth KV, Aladjem MI, et al. Temporal association of ORCA/LRWD1 to late-firing origins during G1 dictates heterochromatin replication and organization. Nucleic Acids Res. 2017;45:2490–2502. doi: 10.1093/nar/gkw1211. References 43, 57 and 78 demonstrate the involvement of trans-acting facors in regulating initiation frequency and replication timing in groups of replication origins. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79**.Brustel J, Kirstein N, Izard F, Grimaud C, Prorok P, Cayrou C, Schotta G, Abdelsamie AF, Dejardin J, Mechali M, et al. Histone H4K20 tri-methylation at late-firing origins ensures timely heterochromatin replication. EMBO J. 2017;36:2726–2741. doi: 10.15252/embj.201796541. Suv4-20h mediated H4K20 tri-methylation is required to sustain the licensing and activity of a subset of ORCA/LRWD1-associated origins to ensure late replication of heterochromatin. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Polak P, Karlic R, Koren A, Thurman R, Sandstrom R, Lawrence M, Reynolds A, Rynes E, Vlahovicek K, Stamatoyannopoulos JA, et al. Cell-of-origin chromatin organization shapes the mutational landscape of cancer. Nature. 2015;518:360–364. doi: 10.1038/nature14221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Stamatoyannopoulos JA, Adzhubei I, Thurman RE, Kryukov GV, Mirkin SM, Sunyaev SR. Human mutation rate associated with DNA replication timing. Nat Genet. 2009;41:393–395. doi: 10.1038/ng.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kenigsberg E, Yehuda Y, Marjavaara L, Keszthelyi A, Chabes A, Tanay A, Simon I. The mutation spectrum in genomic late replication domains shapes mammalian GC content. Nucleic Acids Res. 2016;44:4222–4232. doi: 10.1093/nar/gkw268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chen CL, Rappailles A, Duquenne L, Huvet M, Guilbaud G, Farinelli L, Audit B, d’Aubenton-Carafa Y, Arneodo A, Hyrien O, et al. Impact of replication timing on non-CpG and CpG substitution rates in mammalian genomes. Genome Res. 2010;20:447–457. doi: 10.1101/gr.098947.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84*.Sasaki T, Rivera-Mulia JC, Vera D, Zimmerman J, Das S, Padget M, Nakamichi N, Chang BH, Tyner J, Druker BJ, et al. Stability of patient-specific features of altered DNA replication timing in xenografts of primary human acute lymphoblastic leukemia. Exp Hematol. 2017;51:71–82. e73. doi: 10.1016/j.exphem.2017.04.004. This paper opens an opportunity to study replication timing with patient samples by growing patient cancer cells in immundificient mice. [DOI] [PMC free article] [PubMed] [Google Scholar]