Abstract
Malfunction of nuclear-cytoplasmic transport contributes to many diseases including cancer. Defective nuclear transport leads to changes in both the physiological levels and temporal-spatial location of tumor suppressors, proto-oncogenes and other macromolecules that in turn affect the tumorigenesis process and drug sensitivity of cancer cells. In addition to their nuclear transport functions in interphase, Karyopherin nuclear transport receptors also have important roles in mitosis and chromosomal integrity. Therefore, alterations in the expressions or regular functions of Karyopherins may have substantial effects on the course and outcome of diseases.
Introduction
Trafficking of macromolecules across the nuclear envelope is essential to signal transduction, in order to regulate and finely tune a multitude of biological pathways. Proper temporal-spatial localization of macromolecules is regulated in a bidirectional manner through the highly selective nuclear pore complex (NPC). While small molecules (such as ATP) and solutes travel through the NPC via passive diffusion, this mode of transport is not feasible with the increased molecular mass of macromolecules [1] Therefore, to achieve nuclear-cytoplasmic transport of macromolecules in physiologically relevant time scales, they are transported through the NPC in transport receptor- and energy-dependent manners. Members of the Karyopherin-β (Kap) family of nuclear transport receptors are responsible for the majority of the shuttling of cargo proteins from cytoplasm to nucleus (β-Importins) and from nucleus to cytoplasm (Exportins)[2–4]. β-Importins and Exportins recognize specific signals within the cargo proteins termed nuclear localization signal (NLS) and the nuclear export signal (NES), respectively. At this time, a few more than 20 Kaps have been reported in human cells (Table 1). β-Importins and Exportins are each composed of ~20 consecutive HEAT repeats (each composed of a pair of antiparallel α-helices) that are arranged to form super-helical or ring-shaped proteins.
Table 1.
Human importins and exportins
| Karyopherin-β proteins in Nuclear Export | |||
|---|---|---|---|
| Human Protein/Gene Name | Aliases* | Example of cargos** | Implicated in cancer*** |
| Exportin 1 (XPO1) | CRM1; exp1; emb | ad1, Rio2, CDC7, CPEB4 SNUPN, X11L2, PKIα, p73, STAT-1-3, MEK1, c-Abl, Paxillin, ADAR1, HPV16 E7, APC2, mdm2 | Lymphomas, gynecological malignancies glioblastoma, head & neck squamous cell carcinoma, liposarcoma, multiple myeloma, lung, prostate, hepatocellular, cervical cancer |
| Cellular apoptosis susceptibility (CAS) | CAS; CSE1; CSEL1 XPO2 | Impα1, Impα3, Impα4, Impα5, Impα6, Impα7, Impα8 | Bladder cancer, osteosarcoma, melanoma, leukemia, breast cancer, hepatocellular carcinoma, gastric cancer, ovarian cancer, colorectal cancer, thyroid cancer |
| Exportin for tRNA (XPOT) | XPO3 | aminoacylated tRNAs | Breast cancer, ovarian cancer, mesothelioma |
| Exportin 5 (XPO5) | exp5 | Jaz, pre-microRNA | Colorectal cancer, breast cancer, bladder, thyroid cancer, melanoma, thyroid, liver cancer, larynx cancer, small-cell lung cancer, gastric cancer, renal cell carcinoma, esophageal cancer |
| Exportin 6 (XPO6) | EXP6; RANBP20 | Nuclear actin | prostate cancer, breast cancer |
| Exportin 7 (XPO7) | EXP7; RANBP16 | p50RhoGAP, 14-3-3, STRAD | non-small lung cancer, prostate cancer, ovarian cancer oligodendrogliomas |
| Karyopherin-β proteins in Nuclear Import | |||
| Importin subunit beta 1 (KPNB1) | Impβ; MB1; IPO1; IPOB; Impnb; NTF97 | Snurportin-1, cyclin B1, SREPB2, CREB | cervical cancer, gastric cancer, breast cancer, hepatocellular cancer, diffuse large B-cell lymphoma, multiple myeloma |
| Transportin 1 (TNPO1) | Kapβ2; MIP; TRN; IPO2; MIP1; KPNB2 | FUS, EWS, hnRNA-A1,2,3, -D,-G-H-M, NFX1 | n/a |
| Transportin 2 (TNPO2) | IPO3; TRN2; KPNB2B | n/a | n/a |
| Transportin 3 (TNPO3) | TRN-SR; TRN-SR2; IPO12; TRNSR; LGMD1F; MTR10A; | SRSF1, ASF/SF2, SC35HIV integrase | n/a |
| Importin 4 (IPO4) | Imp4 | TP2, Vitamin D receptor | n/a |
| Importin 5 (IPO5) | IMB3; Pse1; imp5; KPNB3; RANBP5 | HPV-16-E5(16E2), p60TRP, Rag-2, Apolipoprotein A-I PGC7/Stella | cervical cancer, Kaposi’s sarcoma |
| Importin 7 (IPO7) | Imp7; RANBP7 | EZI, ERK2, SMAD3, RPL23A, RPS7 and RPL5 | colorectal cancer, prostate cancer, lung cancer, ependymoma |
| Importin 8 (IPO8) | RANBP8 | cap-free eIF4E, SMAD4, | acute myeloid leukemia |
| Importin 9 (IPO9) | Imp9 | nuclear actin and cofilin. | n/a |
| Importin 11 (IPO11) | RanBP11 | UbcM2, Ube2e3, Ub- primed PTEN | bladder cancers, lung cancer, squamous cell carcinoma |
| Nuclear Import Adaptors: Importin-α Proteins | |||
| Human Impα5/Karyopherin subunit alpha 1 (KPNA1) | RCH2; SRP1; IPOA5; NPI-1 | ADAR2, LSD1, Arx (NLS1), NF-κB (p50/p65) | n/a |
| Human Impα1/Karyopherin subunit alpha 2 (KPNA2) | QIP2; RCH1; IPOA1; SRP1alpha; SRP1-alpha | BRCA1, NBS1 RAD51 E2F1, | gastric cancer, colon cancer, endometrial cancer, prostate cancer, CRC, bladder cancer, non-small-cell lung cancer and breast cancer |
| Human Impα4/Karyopherin subunit alpha 3 (KPNA3) | SRP1; SRP4; IPOA4; hSRP1; SRP1gamma | RRC1, RanBP3, XPA, NF-κB (p50/p65) | chronic lymphocytic leukemia and mantle cell lymphoma |
| Human Impα3/Karyopherin subunit alpha 4 (KPNA4) | QIP1; SRP3; IPOA3 | RRC1, RanBP3, hMSH2, p53, NF-κB (p50/p65) | breast cancer, prostate cancer, glioblastoma |
| Human Impα6/Karyopherin subunit alpha 5 (KPNA5) | SRP6; IPOA6 | ARHI (DIRAS3), BRMS1, NF-κB (p50/p65) | colorectal cancer, breast cancers |
| Human Impα7/Karyopherin subunit alpha 6 (KPNA6) | IPOA7; KPNA7 | ARHI (DIRAS3), Keap1, pSTAT1 | smooth muscle neoplasm, chronic myeloid leukemia |
| Human Impα8/Karyopherin subunit alpha 7 (KPNA7) | IPOA8 | n/a | pancreatic cancer |
| Bidirectional transporter/Karyopherin beta | |||
| Exportin 4 (XPO4) | exp4 | Import cargo: Sox2, SRY Export cargo: eIF5A1, eIF5A2, Smad3 |
hepatocellular carcinoma, breast cancer |
| Importin 13 (IPO13) | IMP13; LGL2; KAP13; RANBP13 | Import cargo: Mago-Y14, Ubc9 Export cargo: eIF1A |
endometriosis and endometrial carcinoma |
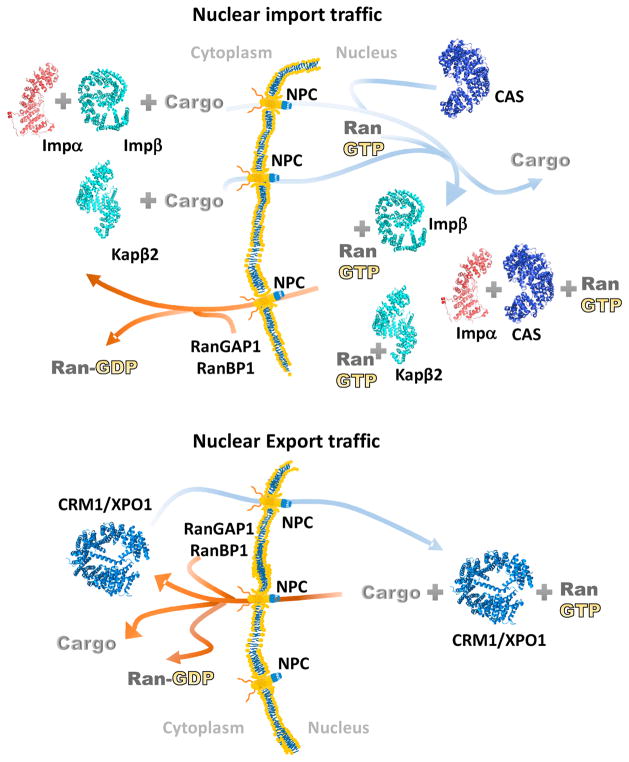
Kap-mediated active nuclear transport is regulated by the small Ras related GTPase, Ran, which controls assembly and disassembly of Kap-cargo complexes [5,6]. The direction of nuclear transport is determined by the asymmetric concentrations of the GTP- versus GDP-bound forms of Ran in the nucleus and the cytoplasm, respectively. RanGTP and nuclear export cargos bind with positive cooperativity to Exportins leading to formation of ternary Exportin-RanGTP-cargo complexes in the nucleus to begin the nuclear export process [7,8]. Upon translocation to the cytoplasm, RanGTP is hydrolyzed to RanGDP by the actions of RanGAP1 and RanBP1/RanBP2 causing the trimeric complexes to dissociate. The opposite Kap-cargo-Ran reactions occur in nuclear import. NLS-containing cargos and RanGTP bind Importins with negative cooperativity [9,10]. Importins will only bind their cargos in the cytoplasm where RanGTP is absent (due to the actions of RanGAP1 and RanBP1/RanBP2). Once Importin-cargo complexes enter the nucleus, RanGTP binds with high affinity to Importins causing cargo release (Figure 1).
Figure 1. Kap-mediated nuclear import and export.
Nuclear import and export of macromolecules occur through the nuclear pore complex (NPC) and are mediated by the Karyopherin family of nuclear transport receptors. Kap-mediated nuclear transport is regulated by the small GTPase Ran, which controls assembly and disassembly of Kap-cargo complexes. The Importins Impβ and Kapβ2 are in cyan, the adaptor protein Impα is in red and the Exportins CRM1/XPO1 and CAS are in blue and dark blue, respectively.
Most Kaps bind directly to their cargo proteins in order to translocate through the NPC. The amphipathic HEAT repeats of Kaps provide multiple hydrophobic patches on their outer surfaces to bind dynamically to Phe-Gly (FG) repeats found in many nucleoporins of the NPC. The highly dynamic and intrinsically disordered FG repeats in FG-nucleoporins form the permeability barrier in the center of NPC, which prevents passage of unaccompanied macromolecules while promoting the selective and efficient transport of Kap-cargo complexes [11–14].
In addition to the β-Importins, adaptor proteins named Importin-αs (Impα, Karyopherin-α) also play important roles in nuclear import. Impα binds directly to both the classical-NLS (cNLS) in cargo proteins and to Impβ [15–17]. Subsequently, Impβ interacts with the NPC to carry the Impβ-Impα-cargo complex into the nucleus. Seven different Impα proteins have been identified in human cells [18]. All Impαs share a highly conserved protein structure of a flexible N-terminal Impβ binding (IBB) domain followed by a central ARM domain (contains 10 ARM repeats) and a short C-terminal disordered tail. The ARM domain of Impα binds cNLS and its IBB domain binds Impβ [19].
The spectrum of cellular functions for Kap cargos is huge and proper nuclear-cytoplasmic localization of these cargos is critical to their abilities to execute normal cellular functions (Table 1). Many nuclear transport cargos have been implicated in enabling characteristics and hallmarks of cancer [20]. Thus, aberrant temporal-spatial localization of these cargos, due to altered behavior and/or expression of Karyopherins, are likely to impact the biology of cancer cells (Figure 2). Aberrant nuclear-cytoplasmic shuttling of key cellular regulator macromolecules, such as oncogenes and tumor suppressor genes, has been reported in variety of cancers and viral infections. The majority of published studies have focused on pathogenic mutations in the cargo proteins rather than on the Kaps that transport them. Although mutational analyses of cargo proteins have helped identification of many NLSs and NESs, the importance of the Karyopherins in cancer causation, progression and treatment has only begun to be highlighted in the past decade. Here, we compile and review studies in cancer biology where Karyopherins were shown to be deregulated and/or mutated.
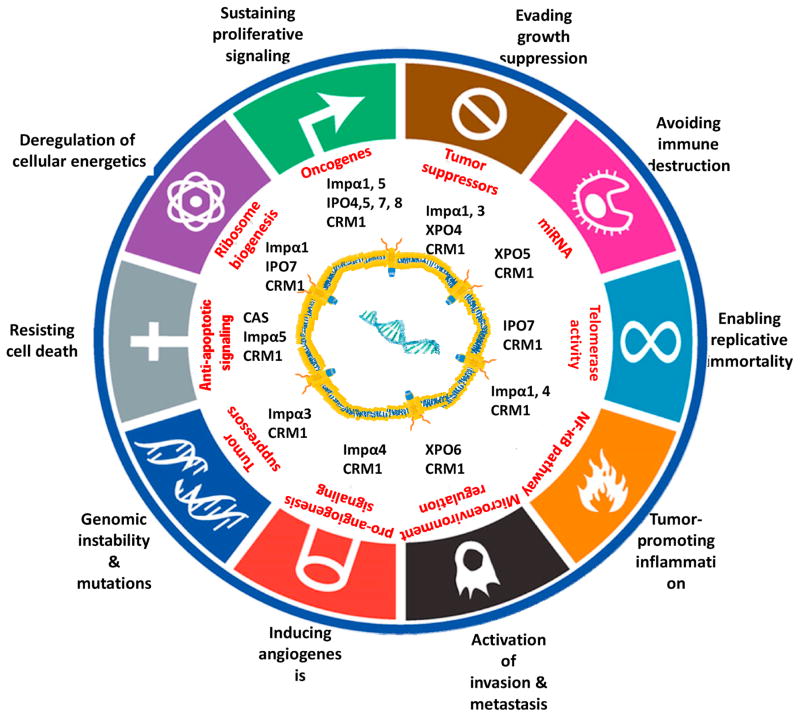
Figure 2. Kap-mediated nuclear transport systems and their impact on the hallmarks of cancer.
The ten hallmarks of cancer are shown with associated Kaps that were reported to be involved in cancer onset and progression. The cancer hallmarks diagram is adapted from [20].
Kap-mediated nuclear export and cancer
CRM1/Exportin 1 (XPO1)
CRM1/XPO1 is the Exportin that is responsible for nuclear export of hundreds to more than a thousand NES-containing proteins and RNAs. Many protein cargos of CRM1/XPO1 are tumor suppressors and/or regulators of cell proliferation [21]. CRM1/XPO1 has been shown to display high mRNA and protein levels in many types of cancers [22]. Although the molecular mechanisms that lead to CRM1/XPO1 overexpression remain mostly unknown, CRM1/XPO1 gene copy number gain (chromosome 2p16.1-2p15 locus) has been reported in hematological malignancies [23].
Recently, somatic missense mutations were found in codon 571 of CRM1/XPO1, which results in a mutation of residue Glu571 of CRM1/XPO1 to either a glycine or a lysine (p.E571K/G) [23,24]. Glu571 lies within the hydrophobic NES-binding groove of CRM1/XPO1. Recurrent patterns of E571K/G mutations have been reported in both solid and hematologic cancers [23,24]. However, the molecular consequences of these CRM1/XPO1 mutations and how they lead to cancer, remain to be elucidated.
Two recent studies on functional genomic and pharmacologic profiling of rare sarcoma and KRAS-mutant non-small cell lung cancer identified and evaluated CRM1/XPO1 as a native and generic potential therapeutic target [25,26]. In these independent studies, CRM1/XPO1 was shown to be a context-independent cancer dependent or synthetic lethality gene in a number of well-annotated primary cell-lines that were used to represent heterogeneity of tumors.
Atomic level knowledge of how CRM1/XPO1 recognizes the NES [27] allowed in silico small molecule docking studies by Karyopharm Therapeutics Inc. (Newton, MA) to develop compounds named Selective inhibitors of Nuclear Export (KPT-SINEs) into potent CRM1/XPO1 inhibitors. KPT-SINEs are orally bioavailable inhibitors that bind covalently, but in a slowly reversible fashion, to the Cys528 residue that is located in the CRM1/XPO1 NES-binding groove. Binding of KPT-SINE to CRM1 blocks binding of NESs of CRM1/XPO1 cargos and therefore efflux of the cargos into the cytoplasm. This ends up concentrating the cargos in the nucleus where they can potentially mediate apoptosis in response to DNA damage, to the cell’s microenvironment or to chemotherapy [28,29]. More than 40 pre-clinical studies, reporting on the application and efficacy of KPT-SINE compounds in hematopoietic malignancies and solid tumors were published from 2012 till the present. Broad antitumor activity of the SINE compounds relies on nuclear retention of a variety of CRM1/XPO1 cargos (such as p53, IκB, NPM1, PAR-4, FOXO, p73, p27, topoisomerase IIα, MDM2, mTOR) in a cell context dependent manner. Retention of these regulators in the nucleus led to subsequent activation of cell-cycle arrest, apoptotic-, anti-inflammatory- and stress-related gene expression [30,31]. Three KTP-SINEs, namely Selinexor (KPT-330) (https://ClinicalTrials.gov, NCT01607892, NCT01607905, NCT02025985, NCT01896505, NCT02025985, NCT02606461, NCT02250885, NCT02606461) [32], Verdinexor (KPT-335) (https://ClinicalTrials.gov, NCT02431364) and Eltanexor (KPT-8602) (https://ClinicalTrials.gov, NCT02649790) are being investigated in over fifty clinical trials for wide varieties of both solid and hematologic cancers. The clinical-trials in human patients include both single agent trials and trials that combine nuclear export inhibition by Selinexor with cell cycle or proteasome inhibitors or monoclonal antibodies for further multi-pathway/multi-dimensional targeting of critical signaling pathways [33–36].
Cellular apoptosis susceptibility (CAS)/Exportin 2 (XPO2)
CAS is the Exportin for the Impα subunits of the classical Impα/β nuclear import system, and is responsible for recycling Impα back to the cytoplasm for additional rounds of nuclear import [37]. CAS was identified as a putative oncogene. Its overexpression was reported in several tumor types and correlated with cancer grades, cancer stages, and poor outcomes of cancer patients [38]. Recently, additional studies have identified a variety of unexpected links between CAS and different non-Impα proteins suggesting that diverse and context-specific functions of CAS that are still incompletely understood [38,39].
Exportin for tRNA (XPO-t)/Exportin 3 (XPO3)
The Exportin for tRNAs or XPO-t is the major nuclear exporter of aminoacylated tRNAs [40,41]. Recently, in an attempt to identify new prognostic markers through comparative expression pattern analysis among tumor tissues and cell-lines, XPO-t overexpression was shown to correlate with poor prognosis in breast, ovarian cancers and mesothelioma [42,43]. These correlations are reasonable considering the findings that tRNA-derived fragments are dysregulated in several cancers [44]. Other than these preliminary correlative observations, the tumorigenic consequence of XPO-t overexpression and the mechanism of how that influences cancer progression remain to be understood.
Exportin 5 (XPO5)
Xpo5 is the Exportin that transports precursor micro-RNAs (pre-miRNAs) from the nucleus to the cytoplasm [45], a key step in miRNA biogenesis [46]. Both overexpression and decreased expression of XPO5 have been observed in cancer. XPO5 overexpression was shown in colorectal, breast, bladder, thyroid carcinomas and melanomas [47,48]. The molecular basis for XPO5 overexpression and for the resulting tumorigenic activity is not yet known, but XPO5 overexpression originating from 6p polyploidy was shown to be associated with gastric cancer [49]. In contrast, lower expression levels of XPO5 due to the recurrent rs11077 single-nucleotide polymorphism (SNP) located in the 3′ UTR of XPO5, was shown to closely associate with thyroid cancer, liver cancer, larynx cancer, colorectal cancer and leukoplakia [50,51]. SNPs related to the miRNA pathway, known as miR-SNPs [52], can influence miRNA functions either by directly perturbing miRNA expression levels or by perturbing miRNA binding sequence in target genes. miR-SNPs can affect cancer development and prognosis by changing the global miRNA profile of cells. The recently identified TG variant (carries T or G allele) of the rs11077 miR-SNPs, which leads to XPO5 downregulation, correlates with distant metastasis and lymphatic invasion in thyroid cancer [50]. On the other hand, the more frequent heterozygous AC genotype of the miR-SNPs rs11077, in which a genotype from one parent carries a specific A mutation and the genotype from the other parent carries a C mutation, is associated with improved chemotherapy response in non-small cell lung cancer and with longer overall survival in chemosensitive multiple myeloma and non-small cell lung cancer patients [53,54]. The AC genotype is also associated with increased risk in esophageal and renal cell cancers [55,56]. XPO5 downregulation can also result from a loss of function mutation, which generates an inactive C-terminally truncated XPO5 protein. This mutation was shown to be present in 11.6% of the primary malignancies with microsatellite instability including colorectal cancer, gastric and endometrial tumors [57]. Besides altered expression level and gene mutations of XPO5, extracellular-signal-regulated kinase (ERK)-driven XPO5 phosphorylation significantly reduces nuclear efflux of pre-miRNA, and was recently shown to correlate with poor prognosis in liver cancer patients [58]. ERK-mediated XPO5 suppression of miR-122 in liver cells leads to increased microtubule dynamics resulting in tumorigenesis along with drug resistance. Although miR-SNP studies of XPO5 are currently still at early stages, further studies in identification of miRNA dysregulation affected by XPO5 mutations should lead to a better understanding of chemotherapy sensitivity, cancer survival and epidemiology of a given cancer type.
Exportin 6 (XPO6) and Exportin 7 (XPO7)
Exportin 6 is key for the nuclear export of nuclear-actin [59], which was proposed to mediate both growth and quiescence in mouse epithelial cells acting though the laminin-111 (LN1)/PI3K/XPO6/N-actin pathway [60]. In the context of normal mammary basal membrane, LN1 impedes PI3K-induced XPO6 activation thus affecting nuclear export nuclear-actin, which is crucial for cells to become quiescent. This proposed mechanism along with statistical analysis of clinical datasets, led to the proposal that XPO6 expression correlates with poor survival in breast cancer patients. On the other hand, gene expression analysis of samples from prostate cancer patients showed that elevated levels of XPO6 can be used a prognostic marker of reoccurrence [61,62].
XPO7 mediates nuclear export of various cytoplasmic cargo proteins such as 14-3-3, p50RhoGAP and STRAD [63]. Proper cytoplasmic localization of these cargo proteins is important for their normal cellular functions, such as 14-3-3-dependent tuning of apoptosis and cell-cycle checkpoint. Thus, it is not surprising that irregularly elevated cytoplasmic levels of XPO7 is strongly associated with poor overall survival in epithelial ovarian cancer patients [64]. A point mutation where residue Asp237 of XPO7 is mutated to asparagine (p.D237N) was recently identified in oligodendrogliomas tumor samples [65]. We performed homology modeling and sequence alignment of XPO7 with crystal structures of CRM1/XPO1 and XPO4, which placed Asp237 in the putative loop between HEAT repeats 4 and 5 in XPO7 (unpublished data). Increased proliferation observed in an overexpression study indicated that D237N mutation affects the functional property of XPO7 in oligodendroglial and HEK293 cell lines, but further studies along with a broader range of samples are needed for proper characterization of the mutation [65].
Nuclear import and cancer
The classical Importin-α•Importin-β system
Importin-β (KPNB1)
Importin-β (Impβ) is part of the heterodimeric Impα/β nuclear importer complex for cNLS-harboring proteins [66]. Impβ also regulates mitotic progression subsequent to nuclear envelope disintegration [67]. Impβ, often coupled with Impα, sequesters many spindle assembly factors (SAFs) in areas distant from the chromatin and then releases them in a RanGTP-dependent manner near the mitotic chromosomes/spindles to execute the proper order of events in cell division. These SAF proteins include, but are not limited to, tumor suppressor proteins, Ran-dependent microtubule stabilizers, and a subset of nucleoporins [68]. Impβ was reported to be overexpressed in cervical cancer, gastric cancer [69], breast cancer [70], hepatocellular cancer [71], diffuse large B-cell lymphoma [72] and multiple myeloma [73]. Impβ was also implicated in interference with cell survival and proliferation [69,72,73]. Recent cell culture studies identified two small molecule Impβ inhibitors named the inhibitor of Nuclear Import-43 (INI-43) [74] and its 2-aminothiazole derivative 1 [75], both with therapeutic potential for cancer treatment. Both compounds, at nanomolar concentrations, elicited G2-M cell-cycle arrest in cancer cells and induced an intrinsic apoptotic pathway but did not display adverse effects on normal in vitro cell culture models [74,75]. INI-43 also showed substantial inhibitory effect on the growth of esophageal and cervical tumor cells in subcutaneously xenografted models [74]. Impβ plays central roles in both cell cycle regulation and Impα-dependent nuclear import. Since many cancer-sustaining pathways share these fundamental cellular processes, selective inhibition of Impβ represents a powerful approach for anticancer therapeutics.
Adaptor proteins for Importin-β: the Importin-α (Impα) proteins
There are a few examples of the involvement of human Impα proteins in malignancies and in cancer biology. In the following sections we compile knowledge on the different Impα proteins that are grouped into three subfamilies defined by their amino acid similarities and evolutional conservations: 1) the α1 subfamily: Impα1 and Impα8, 2) the α2 subfamily: Impα3 and Impα4 and 3) the α3 subfamily: Impα5, Impα6 and Impα7 [66].
The α1 subfamily: Human Impα1 (KNPA2) and Impα8 (KNPA7)
Impα1, one of seven known Impα adaptor proteins of Impβ, binds directly to many cNLS-containing protein cargos including cancer associated proteins such as BRCA1, NBS1 and RAD51 and E2F1 [76–78] and the DNA double-strand break repair complex MRN [79]. Elevated levels of Impα1 protein was reported in gastric cancer, colon cancer, endometrial cancer, prostate cancer, colorectal cancer, bladder cancer, non-small cell lung cancer and breast cancer, and were associated with poor prognosis [80–82]. Because of the aberrant and cancer-related high levels of Imp-α1, the protein was recently identified as one of the target oncogenes for the tumor suppressor microRNA miR-26b [83,84]. In these studies, downregulation of miR-26b was highly associated with upregulation of Impα1 mRNA/protein levels and with unfavorable prognosis in gastric and ovarian cancer patients. The cancer cell secretome, which is the collection of extracellular proteins secreted by cancer cells, provides a novel approach for cancer biomarker identification. Surprisingly, in addition to intracellular overexpression of Impα1, elevated levels of serum Impα1 were detected in lung cancer, colorectal cancer and esophageal squamous cell carcinoma [85,86], making the protein a potential cancer biomarker.
Another Impα subtype, Impα8, is virtually absent in most adult tissues. Impα8 is found in very low levels in oocytes, dendric cells and the small intestine [87]. Its expression is also very tightly regulated during development. Overexpression of Imp-α8 has been shown to promote malignant properties of pancreatic cancer [88].
The α2 subfamily: Human Impα3 (KNPA4) and Impα4 (KPNA3)
The evolutionary relationship between Impα3 and Impα4 partitions the two proteins into the same Impα subfamily [66]. They share several cargo proteins that are specific to this subfamily, such as RCC1 and RanBP3 [66]. Other than common specialized cargos, both Impα3 and Impα4 have been shown to be required for proper nuclear influx of some important cancer related gene products. While Impα3 recognizes and rapidly transfers both hMSH2 and p53 under stress conditions [89,90], Impα4 facilitates UV-induced nuclear accumulation of a critical sensor of S-phase DNA damage, the Xeroderma pigmentosum Group A (XPA) protein [91].
Impα3 was also identified as a direct target for tumor suppressor micro-RNAs miR-708, miR-181b and Hsa-miR-567 [92–94]. Downregulation of miR-708 and miR-181b levels results in Imp-α3 overexpression associated with skeletal metastasis of prostate cancer and glioblastoma with poor overall survival, respectively. Impα3 overexpression was also reported in the highly aggressive MDA-MB-231 breast cancer cell line and in samples from patients with poor prognosis [93]. Furthermore, ectopic expression of Hsa-miR-567 in a breast cancer cell line strongly inhibits cell proliferation and migration in vitro and in mouse xenografts.
Impα4 expression is down-regulated by miR-223, leading to the inhibition of NF-κB signaling in glomerular endothelial cells [95]. Impα4-dependent NF-κB signaling attenuation was also reported in chronic lymphocytic leukemia and mantle cell lymphoma with loss of 13q14.3. The latter chromosomal aberration leads to Impα4 haploinsufficiency (down-regulation), which in turn may associate with tumorigenesis [96].
The α3 subfamily: Human Impα6 (KPNA5) and Impα7 (KPNA6)
There are a few reports of involvement of α3 subfamily members in cancer. Through genome-wide somatic mutation analysis, Impα6 was found to be recurrently mutated in breast cancer tumor samples and thus considered to be a candidate cancer gene [97,98]. These recurrent mutations include the p.F48L mutation within its IBB motif, the p.L179V mutation in its 2nd ARM domain and the p.R319S mutation in its 5th ARM domain. Imp-α7 was identified in a study to evaluate differential gene expression in chronic myeloid leukemia (CML) versus healthy volunteers, as one of the overexpressed genes that may be involved in this disease [99]. In addition to overexpression, recurrent Imp-α7 haploinsufficiency due to regional loss of chromosome 1p was reported in smooth muscle neoplasm of the uterus [100]. However, no clear evidence supporting the importance of the loss of Imp-α7 has been published.
Impα proteins are involved in many cellular processes that range from embryonic stem cell fate to normal neuronal function. Therefore, further research is needed to gain a better understanding of how expression patterns and Importin-cargo specificity for the each Impα subtype affect normal and disease states of cells.
Other β-Importin systems: Kapβ2/TNPO1, TNPO3, IPO4, IPO5, IPO7, IPO8, IPO9, IPO11
Karyopherin-β2/Transportin-1, Transportin-3/Transportin-SR, Importin-4 and Importin-9
Many studies of Karyopherin-β2 (Kapβ2; also known as Transportin-1 or TNPO1), Transportin 3 (TNPO3; also known as Transportin-SR), Importin 4 (IPO4) and Importin 9 (IPO9) have identified nuclear import cargos for each of the Importins [3,4]. Kapβ2/TNPO1 is one of the major players of the nuclear import system and shown to mediate nuclear import of > 30 of cargos harboring the well characterized PY(proline/tyrosine)-NLS [2]. Many of these cargos, such as hnRNP A1, hnRNP M and FUS, are classified as RNA processing proteins [101]. TNPO3 was initially identified as a novel importer for conserved SR-proteins that are involved in RNA splicing family (e.g. ASF/SF2, SC35) [102], but was later shown to be required for several lentiviruses infection [102]. Kapβ2/TNPO1 and TNPO3 have been shown to play important roles in the pathogenesis of diseases such as familial FUS amyotrophic lateral sclerosis [103] and Limb-girdle muscular dystrophy 1F [104], respectively. However, at this time, none of the four Importins (Kapβ2, TNPO3, IPO4 and IPO9) have reported roles in cancer etiology.
Importin 5 (IPO5)
IPO5 imports a variety of functionally diverse cargos by binding to IK-NLSs in their polypeptide chains [2,105]. Among its cargos are several viral proteins. IPO5 binds directly to a human papilloma virus (HPV) protein named HPV-16-E5(16E2), which is an important mediator of oncogenic transformation [106]. Interestingly, HPV-16-E5(16E2) does not have an NLS and is not found in the nucleus, thus it is unclear what role IPO5 assumes in this interaction. In a more conventional role, IPO5 imports other HPV proteins such as the NLS-containing L2 proteins of HPV-11 and HPV-16, into the nucleus [107]. Finally, the IPO5 mRNA was found to be the target of a miRNA produced by the human herpesvirus 8 (HHV-8; an oncogenic virus associated with Kaposi sarcoma and primary effusion lymphoma) [108]. In all the examples above, interactions between IPO5 and viral oncogenes were documented but the pathological significance of these interactions as they pertain to cancer needs further investigation.
Importin 7 (IPO7)
There is limited information about IPO7 cellular function and its cargo proteins. IPO7 was shown to directly interact with ribosomal proteins RPL23A, RPS7 and RPL5 in order to facilitate their nuclear import [109]. Overexpression of IPO7 was reported and implicated as a notable factor in colorectal cancer, prostate and lung cancers [110–112]. In many cases, IPO7 upregulation is induced at the transcriptional level either by the c-myc oncogene, which is commonly overexpressed in colorectal cancer [109] or by promoter hypomethylation in pediatric malignancy [113]. While being positively upregulated by c-myc, both mRNA and protein levels of IPO7 were shown to be downregulated by p53 [109]. Interestingly, in the same study, it has been proposed that dysregulation of ribosomal biogenesis by IPO7 deletion results in a p53-driven cell growth arrest. This sequence of events hints that IPO7 may be a therapeutic target, a notion that is further supported by the antitumor effect of IPO7 knockdown in a mouse lung cancer model [110].
Importin 8 (IPO8)
Few nuclear import cargos are known for IPO8. One of them is the protooncogene eIF4E, a translation initiation factor, which binds the 5′ cap of mRNAs to direct them for translation [114]. IPO8 binds only the unliganded (5′cap-mRNAs-free) form of eIF4E, and IPO8-dependent nuclear import of eIF4E is important for both tumor formation and metastasis. At this time, the connection between IPO8 and cancer is limited to the study of eIF4E [114]. Elevated levels of IPO8 were found in acute myeloid leukemia but reports on cancer gene expression studies described IPO8 as a housekeeping gene that displays similar levels of expression among tissue types and treatment conditions [115,116].
Importin 11 (IPO11)
Importin 11 mediates the import of ubiquitinated protein cargos into the nucleus [117]. It is also the Importin for the E2 ubiquitin-conjugating enzyme UBE2E3 [118]. At this time, only a few studies have indicated links between IPO11 and cancer. In non-muscle-invasive bladder cancer, IPO11 overexpression that results from Chromosome 5 aneuploidy is strongly associated with poor prognosis [119]. IPO11 was also proposed to be a potential biomarker for bladder cancer. In addition, a recent publication revealed that IPO11 selectively binds to the monoubiquinated form of both PTEN (tumor suppressor protein) and UBE2E1 (E2 ubiquitin–conjugating enzyme) to mediate their nuclear import. IPO11-driven removal of PTEN and UBE2E1 from the cytoplasm prevents degradation of PTEN in the cytoplasm. The role of IPO11 in regulation of PTEN tumor suppressor activity is further supported by identification of frequent occurrence of IPO11 loss of heterozygosity along with reduced level of PTEN in many cancer genomic datasets [120].
Bidirectional nuclear transporters and cancer
Bidirectional Kaps display dual functionality – they mediate nuclear import of certain cargos and nuclear export of other cargos. There are only two known human bidirectional Kaps, Exportin 4 (XPO4) and Importin 13 (IPO13).
Exportin 4 mediates nuclear export of eIF5A and SMAD3, and nuclear import of the SOX2 and SRY proteins [121]. XPO4 was identified as a putative tumor suppressor gene in a hepatocellular carcinoma model [122]. XPO4 loss, commonly observed in human hepatocellular carcinoma specimens, contributes to oncogenesis by promoting aberrant nuclear accumulation of it exports cargos eIF5A1, eIF5A2 and SMAD3 [123,124]. It has also been demonstrated that reintroduction of XPO4 into XPO4-deficient tumor cells selectively suppressed tumorigenesis. Similarly, low XPO4 expression correlates with poor prognosis in hepatocellular carcinoma and breast cancer patients.
Importin 13 is another a bidirectional transporter that is responsible for nuclear import of the Mago-Y14 and Ubc9 proteins while facilitating nuclear export of eIF1A [125]. Clinical relevance of IPO13 has been documented in few human disease cases such as asthma and pterygium (corneal abnormality) [126,127]. There is no clear link between IPO13 and cancer other than a correlation study reporting elevated IPO13 mRNA and protein levels in endometriosis and endometrial carcinoma specimens [128].
Summary and perspective
Many studies have connected altered Karyopherin expression, mainly elevated protein levels with occasional lower expression, to cell transformation in many different types of cancer cells. Most studies of aberrant Karyopherin levels in cancer seem to focus on identification of novel diagnostic and prognostic factors for a given cancer. Studies to understand the origin of dysregulated Karyopherin expression/protein levels and the resulting tumorigenic mechanism are rare and badly needed.
Inhibitors are available for only four of the >20 Kap systems: CRM1/XPO1, Kapβ2/TNPO1, the Impα/β heterodimer and Impβ alone [129]. Kap-specific nuclear transport inhibitors are not only beneficial for identification of novel cargos and to gain better understanding for cargo specificity and cellular mechanisms of Kap-dependent nuclear transport systems, but they can also be developed into much needed targeted therapeutic agents. At present, there are very few initial studies focused on small-molecule inhibitors of nuclear import proteins. The only two reported examples are development of INI-43 [74] and its 2-aminothiazole derivative 1 [75] as potential Impβ inhibitors, and aerosol administration of shRNAs for IPO7 knockdown [110]. Specific and well-characterized small-molecule inhibitors of CRM1/XPO1, like the KPT-SINEs, so far are the most successful and potent chemotropic agent that are being tested in clinical trials for a large spectrum of malignancies (https://ClinicalTrials.gov). Evidence of single-agent anti-cancer activity of KPT-SINEs, particularly Selinexor/KPT-330, has been documented in more than 2000 patients across more than 29 privately- and investigator-sponsored clinical trials.
Frequently observed cases of elevated levels of Karyopherins (different molecular origins) suggest that cancer cells may have developed dependence and addiction to the nuclear transport machinery in order to sustain their tumorigenic and increased metabolic needs [130]. This hypothesis is also supported by the synthetic lethality phenotype of CRM1/XPO1 observed in heterogenic tumors and cell lines [26]. Therefore, as more genome-wide cell-based chemical/genetic screens are advanced, it will be important to search for other components of the nuclear transport machinery and cargos that are aberrantly expressed in cancer cells in order to identify novel targets that may be exploited in the development of diagnostic, prognostic markers and therapeutic agents for cancer and other diseases.
Acknowledgments
We apologize to all our colleagues whose work could not be properly reviewed and cited here because of space limitation.
This work is funded by Cancer Prevention Research Institute of Texas (CPRIT) Grants RP150053 and RP170170 (YMC), R01 GM069909 (YMC), the Welch Foundation Grant I-1532 (YMC) and the University of Texas Southwestern Endowed Scholars Program (Y.M.C.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Timney BL, Raveh B, Mironska R, Trivedi JM, Kim SJ, Russel D, Wente SR, Sali A, Rout MP. Simple rules for passive diffusion through the nuclear pore complex. Journal of Cell Biology. 2016;215:57–76. doi: 10.1083/jcb.201601004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Soniat M, Chook YM. Nuclear localization signals for four distinct karyopherin-beta nuclear import systems. Biochem J. 2015;468:353–362. doi: 10.1042/BJ20150368. [DOI] [PubMed] [Google Scholar]
- 3.Kimura M, Imamoto N. Biological significance of the importin-beta family-dependent nucleocytoplasmic transport pathways. Traffic. 2014;15:727–748. doi: 10.1111/tra.12174. [DOI] [PubMed] [Google Scholar]
- 4.Chook YM, Suel KE. Nuclear import by karyopherin-betas: recognition and inhibition. Biochim Biophys Acta. 2011;1813:1593–1606. doi: 10.1016/j.bbamcr.2010.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gorlich D, Mattaj IW. Nucleocytoplasmic transport. Science. 1996;271:1513–1518. doi: 10.1126/science.271.5255.1513. [DOI] [PubMed] [Google Scholar]
- 6.Moore MS, Blobel G. A G protein involved in nucleocytoplasmic transport: the role of Ran. Trends Biochem Sci. 1994;19:211–216. doi: 10.1016/0968-0004(94)90024-8. [DOI] [PubMed] [Google Scholar]
- 7.Matsuura Y. Mechanistic Insights from Structural Analyses of Ran-GTPase-Driven Nuclear Export of Proteins and RNAs. J Mol Biol. 2016;428:2025–2039. doi: 10.1016/j.jmb.2015.09.025. [DOI] [PubMed] [Google Scholar]
- 8.Guttler T, Gorlich D. Ran-dependent nuclear export mediators: a structural perspective. EMBO J. 2011;30:3457–3474. doi: 10.1038/emboj.2011.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cavazza T, Vernos I. The RanGTP Pathway: From Nucleo-Cytoplasmic Transport to Spindle Assembly and Beyond. Front Cell Dev Biol. 2015;3:82. doi: 10.3389/fcell.2015.00082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moroianu J, Blobel G, Radu A. Nuclear protein import: Ran-GTP dissociates the karyopherin alpha beta heterodimer by displacing alpha from an overlapping binding site on beta. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:7059–7062. doi: 10.1073/pnas.93.14.7059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Milles S, Mercadante D, Aramburu IV, Jensen MR, Banterle N, Koehler C, Tyagi S, Clarke J, Shammas SL, Blackledge M, et al. Plasticity of an ultrafast interaction between nucleoporins and nuclear transport receptors. Cell. 2015;163:734–745. doi: 10.1016/j.cell.2015.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hough LE, Dutta K, Sparks S, Temel DB, Kamal A, Tetenbaum-Novatt J, Rout MP, Cowburn D. The molecular mechanism of nuclear transport revealed by atomic-scale measurements. Elife. 2015:4. doi: 10.7554/eLife.10027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Radu A, Moore MS, Blobel G. The peptide repeat domain of nucleoporin Nup98 functions as a docking site in transport across the nuclear pore complex. Cell. 1995;81:215–222. doi: 10.1016/0092-8674(95)90331-3. [DOI] [PubMed] [Google Scholar]
- 14.Rout MP, Wente SR. Pores for thought: nuclear pore complex proteins. Trends Cell Biol. 1994;4:357–365. doi: 10.1016/0962-8924(94)90085-x. [DOI] [PubMed] [Google Scholar]
- 15.Conti E, Uy M, Leighton L, Blobel G, Kuriyan J. Crystallographic analysis of the recognition of a nuclear localization signal by the nuclear import factor karyopherin alpha. Cell. 1998;94:193–204. doi: 10.1016/s0092-8674(00)81419-1. [DOI] [PubMed] [Google Scholar]
- 16.Moroianu J, Blobel G, Radu A. Previously identified protein of uncertain function is karyopherin alpha and together with karyopherin beta docks import substrate at nuclear pore complexes. Proc Natl Acad Sci U S A. 1995;92:2008–2011. doi: 10.1073/pnas.92.6.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Enenkel C, Blobel G, Rexach M. Identification of a yeast karyopherin heterodimer that targets import substrate to mammalian nuclear pore complexes. J Biol Chem. 1995;270:16499–16502. doi: 10.1074/jbc.270.28.16499. [DOI] [PubMed] [Google Scholar]
- 18.Goldfarb DS, Corbett AH, Mason DA, Harreman MT, Adam SA. Importin alpha: a multipurpose nuclear-transport receptor. Trends Cell Biol. 2004;14:505–514. doi: 10.1016/j.tcb.2004.07.016. [DOI] [PubMed] [Google Scholar]
- 19.Cingolani G, Petosa C, Weis K, Muller CW. Structure of importin-beta bound to the IBB domain of importin-alpha. Nature. 1999;399:221–229. doi: 10.1038/20367. [DOI] [PubMed] [Google Scholar]
- 20.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 21.Xu DR, Grishin NV, Chook YM. NESdb: a database of NES-containing CRM1 cargoes. Molecular Biology of the Cell. 2012;23:3673–3676. doi: 10.1091/mbc.E12-01-0045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Duijvesz D, Burnum-Johnson KE, Gritsenko MA, Hoogland AM, Vredenbregt-van den Berg MS, Willemsen R, Luider T, Pasa-Tolic L, Jenster G. Proteomic profiling of exosomes leads to the identification of novel biomarkers for prostate cancer. PLoS One. 2013;8:e82589. doi: 10.1371/journal.pone.0082589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jardin F, Pujals A, Pelletier L, Bohers E, Camus V, Mareschal S, Dubois S, Sola B, Ochmann M, Lemonnier F, et al. Recurrent mutations of the exportin 1 gene (XPO1) and their impact on selective inhibitor of nuclear export compounds sensitivity in primary mediastinal B-cell lymphoma. Am J Hematol. 2016;91:923–930. doi: 10.1002/ajh.24451. [DOI] [PubMed] [Google Scholar]
- 24.Xie QL, Liu Y, Zhu Y. Chromosome region maintenance 1 expression and its association with clinical pathological features in primary carcinoma of the liver. Exp Ther Med. 2016;12:59–68. doi: 10.3892/etm.2016.3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25•.Hong AL, Tseng YY, Cowley GS, Jonas O, Cheah JH, Kynnap BD, Doshi MB, Oh C, Meyer SC, Church AJ, et al. Integrated genetic and pharmacologic interrogation of rare cancers. Nat Commun. 2016;7:11987. doi: 10.1038/ncomms11987. Hong et al. presented a very elaborate high throughput screening (HTS) utilizing triple complementary techniques, pooled CRISPR-Cas9 and RNAi loss-of-function screens and a small-molecule screen to develop an unbiased approach to identify potential therapeutic targets in rare cancers. CRM1/XPO1 and CDK4 were identified as novel and probably broad-spectrum cancer-dependency genes even though neither of the genes seem to be mutated or altered in this model system. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26•.Kim J, McMillan E, Kim HS, Venkateswaran N, Makkar G, Rodriguez-Canales J, Villalobos P, Neggers JE, Mendiratta S, Wei S, et al. XPO1-dependent nuclear export is a druggable vulnerability in KRAS-mutant lung cancer. Nature. 2016;538:114–117. doi: 10.1038/nature19771. Kim et al. used well-characterized KRAS mutant repertoire of more than 100 non-small cell lung cancer (NSCLC)–derived cell lines and an HTS siRNA screen. They identified CRM1/XPO1 and other nuclear transport components as synthetic lethal pharmacological targets. By targeting CRM1/XPO1 genetically and pharmacologically, they demonstrated synthetic lethally of CRM1/XPO1 in cell lines and xenografts. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fung HY, Chook YM. Atomic basis of CRM1-cargo recognition, release and inhibition. Semin Cancer Biol. 2014;27:52–61. doi: 10.1016/j.semcancer.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lapalombella R, Sun Q, Williams K, Tangeman L, Jha S, Zhong Y, Goettl V, Mahoney E, Berglund C, Gupta S, et al. Selective inhibitors of nuclear export show that CRM1/XPO1 is a target in chronic lymphocytic leukemia. Blood. 2012;120:4621–4634. doi: 10.1182/blood-2012-05-429506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kandarpa M, Kraftson SJ, Maxwell SP, McCauley D, Shacham S, Kauffman M, Jakubowiak AJ. CRM1 Is Highly Expressed in Myeloma Plasma Cells and Its Inhibition by KPT-SINE Induces Cytotoxicity by Increasing p53 in the Nucleus of Multiple Myeloma (MM) Cells. Blood. 2011;118:806–806. [Google Scholar]
- 30.Etchin J, Kentsis A, Sanda T, Kung AL, Stone RM, McCauley D, Kauffman M, Shacham S, Look T. KPT-SINE, a Potent, Small Molecule Inhibitor of CRM1-Dependent Nuclear-Cytoplasmic Shuttling, with Potent Activity Against T-ALL and AML. Blood. 2011;118:1126–1126. [Google Scholar]
- 31.Haines JD, Herbin O, de la Hera B, Vidaurre OG, Moy GA, Sun QX, Fung HYJ, Albrecht S, Alexandropoulos K, McCauley D, et al. Nuclear export inhibitors avert progression in preclinical models of inflammatory demyelination. Nature Neuroscience. 2015;18:511. doi: 10.1038/nn.3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garzon R, Savona M, Baz R, Andreeff M, Gabrail N, Gutierrez M, Savoie L, Mau-Sorensen PM, Wagner-Johnston N, Yee K, et al. A phase 1 clinical trial of single-agent selinexor in acute myeloid leukemia. Blood. 2017;129:3165–3174. doi: 10.1182/blood-2016-11-750158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Farren MR, Hennessey RC, Shakya R, Elnaggar O, Young G, Kendra K, Landesman Y, Elloul S, Crochiere M, Klebanov B, et al. The Exportin-1 Inhibitor Selinexor Exerts Superior Antitumor Activity when Combined with T-Cell Checkpoint Inhibitors. Mol Cancer Ther. 2017;16:417–427. doi: 10.1158/1535-7163.MCT-16-0498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rosebeck S, Alonge MM, Kandarpa M, Mayampurath A, Volchenboum SL, Jasielec J, Dytfeld D, Maxwell SP, Kraftson SJ, McCauley D, et al. Synergistic Myeloma Cell Death via Novel Intracellular Activation of Caspase-10-Dependent Apoptosis by Carfilzomib and Selinexor. Mol Cancer Ther. 2016;15:60–71. doi: 10.1158/1535-7163.MCT-15-0488. [DOI] [PubMed] [Google Scholar]
- 35.Ranganathan P, Kashyap T, Yu X, Meng X, Lai T-H, McNeil B, Bhatnagar B, Shacham S, Kauffman M, Dorrance AM, et al. XPO1 Inhibition using Selinexor Synergizes with Chemotherapy in Acute Myeloid Leukemia by Targeting DNA Repair and Restoring Topoisomerase IIalpha to the Nucleus. Clinical cancer research: an official journal of the American Association for Cancer Research. 2016;22:6142–6152. doi: 10.1158/1078-0432.CCR-15-2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muqbil I, Aboukameel A, Elloul S, Carlson R, Senapedis W, Baloglu E, Kauffman M, Shacham S, Bhutani D, Zonder J, et al. Anti-tumor activity of selective inhibitor of nuclear export (SINE) compounds, is enhanced in non-Hodgkin lymphoma through combination with mTOR inhibitor and dexamethasone. Cancer Lett. 2016;383:309–317. doi: 10.1016/j.canlet.2016.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kutay U, Bischoff FR, Kostka S, Kraft R, Gorlich D. Export of importin alpha from the nucleus is mediated by a specific nuclear transport factor. Cell. 1997;90:1061–1071. doi: 10.1016/s0092-8674(00)80372-4. [DOI] [PubMed] [Google Scholar]
- 38.Jiang MC. CAS (CSE1L) signaling pathway in tumor progression and its potential as a biomarker and target for targeted therapy. Tumor Biology. 2016;37:13077–13090. doi: 10.1007/s13277-016-5301-x. [DOI] [PubMed] [Google Scholar]
- 39.Cheng DD, Lin HC, Li SJ, Yao M, Yang QC, Fan CY. CSE1L interaction with MSH6 promotes osteosarcoma progression and predicts poor patient survival. Scientific Reports. 2017:7. doi: 10.1038/srep46238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kutay U, Lipowsky G, Izaurralde E, Bischoff FR, Schwarzmaier P, Hartmann E, Gorlich D. Identification of a tRNA-specific nuclear export receptor. Mol Cell. 1998;1:359–369. doi: 10.1016/s1097-2765(00)80036-2. [DOI] [PubMed] [Google Scholar]
- 41.Arts GJ, Fornerod M, Mattaj IW. Identification of a nuclear export receptor for tRNA. Curr Biol. 1998;8:305–314. doi: 10.1016/s0960-9822(98)70130-7. [DOI] [PubMed] [Google Scholar]
- 42.Vaidyanathan S, Thangavelu PU, Duijf PHG. Overexpression of Ran GTPase Components Regulating Nuclear Export, but not Mitotic Spindle Assembly, Marks Chromosome Instability and Poor Prognosis in Breast Cancer. Targeted Oncology. 2016;11:677–686. doi: 10.1007/s11523-016-0432-y. [DOI] [PubMed] [Google Scholar]
- 43.Melaiu O, Melissari E, Mutti L, Bracci E, De Santi C, Iofrida C, Di Russo M, Cristaudo A, Bonotti A, Cipollini M, et al. Expression status of candidate genes in mesothelioma tissues and cell lines. Mutat Res. 2015;771:6–12. doi: 10.1016/j.mrfmmm.2014.11.002. [DOI] [PubMed] [Google Scholar]
- 44.Sun C, Fu Z, Wang S, Li J, Li Y, Zhang Y, Yang F, Chu J, Wu H, Huang X, et al. Roles of tRNA-derived fragments in human cancers. Cancer Lett. 2017;414:16–25. doi: 10.1016/j.canlet.2017.10.031. [DOI] [PubMed] [Google Scholar]
- 45.Bohnsack MT, Czaplinski K, Gorlich D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA. 2004;10:185–191. doi: 10.1261/rna.5167604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Berardino BG, Fesser EA, Canepa ET. Perinatal protein malnutrition alters expression of miRNA biogenesis genes Xpo5 and Ago2 in mice brain. Neurosci Lett. 2017;647:38–44. doi: 10.1016/j.neulet.2017.03.012. [DOI] [PubMed] [Google Scholar]
- 47.Shigeyasu K, Okugawa Y, Toden S, Boland CR, Goel A. Exportin-5 Functions as an Oncogene and a Potential Therapeutic Target in Colorectal Cancer. Clin Cancer Res. 2017;23:1312–1322. doi: 10.1158/1078-0432.CCR-16-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ott CA, Linck L, Kremmer E, Meister G, Bosserhoff AK. Induction of exportin-5 expression during melanoma development supports the cellular behavior of human malignant melanoma cells. Oncotarget. 2016;7:62292–62304. doi: 10.18632/oncotarget.11410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cheng L, Wang P, Yang S, Yang Y, Zhang Q, Zhang W, Xiao H, Gao H, Zhang Q. Identification of genes with a correlation between copy number and expression in gastric cancer. BMC Med Genomics. 2012;5:14. doi: 10.1186/1755-8794-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wen J, Gao Q, Wang N, Zhang W, Cao K, Zhang Q, Chen S, Shi L. Association of microRNA-related gene XPO5 rs11077 polymorphism with susceptibility to thyroid cancer. Medicine (Baltimore) 2017;96:e6351. doi: 10.1097/MD.0000000000006351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Osuch-Wojcikiewicz E, Bruzgielewicz A, Niemczyk K, Sieniawska-Buccella O, Nowak A, Walczak A, Majsterek I. Association of Polymorphic Variants of miRNA Processing Genes with Larynx Cancer Risk in a Polish Population. Biomed Res Int. 2015;2015:298378. doi: 10.1155/2015/298378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mishra PJ, Mishra PJ, Banerjee D, Bertino JR. MiRSNPs or MiR-polymorphisms, new players in microRNA mediated regulation of the cell: Introducing microRNA pharmacogenomics. Cell Cycle. 2008;7:853–858. doi: 10.4161/cc.7.7.5666. [DOI] [PubMed] [Google Scholar]
- 53.de Larrea CF, Navarro A, Tejero R, Tovar N, Diaz T, Cibeira MT, Rosinol L, Ferrer G, Rovira M, Rozman M, et al. Impact of MiRSNPs on survival and progression in patients with multiple myeloma undergoing autologous stem cell transplantation. Clin Cancer Res. 2012;18:3697–3704. doi: 10.1158/1078-0432.CCR-12-0191. [DOI] [PubMed] [Google Scholar]
- 54.Geng JQ, Wang XC, Li LF, Zhao J, Wu S, Yu GP, Zhu KJ. MicroRNA-related single-nucleotide polymorphism of XPO5 is strongly correlated with the prognosis and chemotherapy response in advanced non-small-cell lung cancer patients. Tumour Biol. 2016;37:2257–2265. doi: 10.1007/s13277-015-3980-3. [DOI] [PubMed] [Google Scholar]
- 55.Horikawa Y, Wood CG, Yang H, Zhao H, Ye Y, Gu J, Lin J, Habuchi T, Wu X. Single nucleotide polymorphisms of microRNA machinery genes modify the risk of renal cell carcinoma. Clin Cancer Res. 2008;14:7956–7962. doi: 10.1158/1078-0432.CCR-08-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ye Y, Wang KK, Gu J, Yang H, Lin J, Ajani JA, Wu X. Genetic variations in microRNA-related genes are novel susceptibility loci for esophageal cancer risk. Cancer Prev Res (Phila) 2008;1:460–469. doi: 10.1158/1940-6207.CAPR-08-0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Melo SA, Moutinho C, Ropero S, Calin GA, Rossi S, Spizzo R, Fernandez AF, Davalos V, Villanueva A, Montoya G, et al. A genetic defect in exportin-5 traps precursor microRNAs in the nucleus of cancer cells. Cancer Cell. 2010;18:303–315. doi: 10.1016/j.ccr.2010.09.007. [DOI] [PubMed] [Google Scholar]
- 58.Sun HL, Cui R, Zhou J, Teng KY, Hsiao YH, Nakanishi K, Fassan M, Luo Z, Shi G, Tili E, et al. ERK Activation Globally Downregulates miRNAs through Phosphorylating Exportin-5. Cancer Cell. 2016;30:723–736. doi: 10.1016/j.ccell.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stuven T, Hartmann E, Gorlich D. Exportin 6: a novel nuclear export receptor that is specific for profilin. actin complexes. EMBO J. 2003;22:5928–5940. doi: 10.1093/emboj/cdg565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fiore APZP, Spencer VA, Mori H, Carvalho HF, Bissell MJ, Bruni-Cardoso A. Laminin-111 and the Level of Nuclear Actin Regulate Epithelial Quiescence via Exportin-6. Cell Reports. 2017;19:2102–2115. doi: 10.1016/j.celrep.2017.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hao J, Chiang YT, Gout PW, Wang Y. Elevated XPO6 expression as a potential prognostic biomarker for prostate cancer recurrence. Front Biosci (Schol Ed) 2016;8:44–55. doi: 10.2741/s445. [DOI] [PubMed] [Google Scholar]
- 62.Knyazev EN, Samatov TR, Fomicheva KA, Nyushko KM, Alekseev BY, Shkurnikov MY. MicroRNA hsa-miR-4674 in Hemolysis-Free Blood Plasma Is Associated with Distant Metastases of Prostatic Cancer. Bull Exp Biol Med. 2016;161:112–115. doi: 10.1007/s10517-016-3358-6. [DOI] [PubMed] [Google Scholar]
- 63.Dorfman J, Macara IG. STRAD alpha regulates LKB1 localization by blocking access to importin-alpha, and by association with Crm1 and exportin-7. Molecular Biology of the Cell. 2008;19:1614–1626. doi: 10.1091/mbc.E07-05-0454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Caceres-Gorriti KY, Carmona E, Barres V, Rahimi K, Letourneau IJ, Tonin PN, Provencher D, Mes-Masson AM. RAN nucleo-cytoplasmic transport and mitotic spindle assembly partners XPO7 and TPX2 are new prognostic biomarkers in serous epithelial ovarian cancer. PLoS One. 2014;9:e91000. doi: 10.1371/journal.pone.0091000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Erdem-Eraslan L, Heijsman D, de Wit M, Kremer A, Sacchetti A, van der Spek PJ, Sillevis Smitt PA, French PJ. Tumor-specific mutations in low-frequency genes affect their functional properties. J Neurooncol. 2015;122:461–470. doi: 10.1007/s11060-015-1741-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pumroy RA, Cingolani G. Diversification of importin-alpha isoforms in cellular trafficking and disease states. Biochem J. 2015;466:13–28. doi: 10.1042/BJ20141186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Guttinger S, Laurell E, Kutay U. Orchestrating nuclear envelope disassembly and reassembly during mitosis. Nat Rev Mol Cell Biol. 2009;10:178–191. doi: 10.1038/nrm2641. [DOI] [PubMed] [Google Scholar]
- 68.Roscioli E, Di Francesco L, Bolognesi A, Giubettini M, Orlando S, Harel A, Schinina ME, Lavia P. Importin-beta negatively regulates multiple aspects of mitosis including RANGAP1 recruitment to kinetochores. Journal of Cell Biology. 2012;196:435–450. doi: 10.1083/jcb.201109104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stelma T, Leaner VD. KPNB1-mediated nuclear import is required for motility and inflammatory transcription factor activity in cervical cancer cells. Oncotarget. 2017 doi: 10.18632/oncotarget.15834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nordgard SH, Johansen FE, Alnaes GI, Bucher E, Syvanen AC, Naume B, Borresen-Dale AL, Kristensen VN. Genome-wide analysis identifies 16q deletion associated with survival, molecular subtypes, mRNA expression, and germline haplotypes in breast cancer patients. Genes Chromosomes Cancer. 2008;47:680–696. doi: 10.1002/gcc.20569. [DOI] [PubMed] [Google Scholar]
- 71.Yang L, Hu B, Zhang Y, Qiang S, Cai J, Huang W, Gong C, Zhang T, Zhang S, Xu P, et al. Suppression of the nuclear transporter-KPNbeta1 expression inhibits tumor proliferation in hepatocellular carcinoma. Med Oncol. 2015;32:128. doi: 10.1007/s12032-015-0559-1. [DOI] [PubMed] [Google Scholar]
- 72.He S, Miao X, Wu Y, Zhu X, Miao X, Yin H, He Y, Li C, Liu Y, Lu X, et al. Upregulation of nuclear transporter, Kpnbeta1, contributes to accelerated cell proliferation- and cell adhesion-mediated drug resistance (CAM-DR) in diffuse large B-cell lymphoma. J Cancer Res Clin Oncol. 2016;142:561–572. doi: 10.1007/s00432-015-2057-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yan W, Li R, He J, Du J, Hou J. Importin beta1 mediates nuclear factor-kappaB signal transduction into the nuclei of myeloma cells and affects their proliferation and apoptosis. Cell Signal. 2015;27:851–859. doi: 10.1016/j.cellsig.2015.01.013. [DOI] [PubMed] [Google Scholar]
- 74.van der Watt PJ, Chi A, Stelma T, Stowell C, Strydom E, Carden S, Angus L, Hadley K, Lang D, Wei W, et al. Targeting the Nuclear Import Receptor Kpnbeta1 as an Anticancer Therapeutic. Mol Cancer Ther. 2016;15:560–573. doi: 10.1158/1535-7163.MCT-15-0052. [DOI] [PubMed] [Google Scholar]
- 75.Ha S, Choi J, Min NY, Lee KH, Ham SW. Inhibition of Importin beta1 With a 2-Aminothiazole Derivative Resulted in G2/M Cell-cycle Arrest and Apoptosis. Anticancer Res. 2017;37:2373–2379. doi: 10.21873/anticanres.11575. [DOI] [PubMed] [Google Scholar]
- 76.Wang CI, Chien KY, Wang CL, Liu HP, Cheng CC, Chang YS, Yu JS, Yu CJ. Quantitative proteomics reveals regulation of karyopherin subunit alpha-2 (KPNA2) and its potential novel cargo proteins in nonsmall cell lung cancer. Mol Cell Proteomics. 2012;11:1105–1122. doi: 10.1074/mcp.M111.016592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Clark SL, Rodriguez AM, Snyder RR, Hankins GDV, Boehning D. Structure-Function Of The Tumor Suppressor BRCA1. Computational and structural biotechnology journal. 2012:1. doi: 10.5936/csbj.201204005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tseng SF, Chang CY, Wu KJ, Teng SC. Importin KPNA2 is required for proper nuclear localization and multiple functions of NBS1. J Biol Chem. 2005;280:39594–39600. doi: 10.1074/jbc.M508425200. [DOI] [PubMed] [Google Scholar]
- 79.Erben PB, Brunner K, Hecht M, Haderlein M, Buttner-Herold M, Agaimy A, Fietkau R, Hartmann A, Distel LV. Low cytoplasmic and nuclear KPNA2 expression in radiotherapy-treated head and neck squamous cell cancer is associated with an adverse outcome. Int J Clin Exp Pathol. 2015;8:15814–15824. [PMC free article] [PubMed] [Google Scholar]
- 80.Ikenberg K, Valtcheva N, Brandt S, Zhong Q, Wong CE, Noske A, Rechsteiner M, Rueschoff JH, Caduff R, Dellas A, et al. KPNA2 is overexpressed in human and mouse endometrial cancers and promotes cellular proliferation. J Pathol. 2014;234:239–252. doi: 10.1002/path.4390. [DOI] [PubMed] [Google Scholar]
- 81.Alshareeda AT, Negm OH, Green AR, Nolan CC, Tighe P, Albarakati N, Sultana R, Madhusudan S, Ellis IO, Rakha EA. KPNA2 is a nuclear export protein that contributes to aberrant localisation of key proteins and poor prognosis of breast cancer. Br J Cancer. 2015;112:1929–1937. doi: 10.1038/bjc.2015.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhou J, Dong D, Cheng R, Wang Y, Jiang S, Zhu Y, Fan L, Mao X, Gui Y, Li Z, et al. Aberrant expression of KPNA2 is associated with a poor prognosis and contributes to OCT4 nuclear transportation in bladder cancer. Oncotarget. 2016;7:72767–72776. doi: 10.18632/oncotarget.11889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tsai MM, Huang HW, Wang CS, Lee KF, Tsai CY, Lu PH, Chi HC, Lin YH, Kuo LM, Lin KH. MicroRNA-26b inhibits tumor metastasis by targeting the KPNA2/c-jun pathway in human gastric cancer. Oncotarget. 2016;7:39511–39526. doi: 10.18632/oncotarget.8629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lin J, Zhang L, Huang H, Huang Y, Huang L, Wang J, Huang S, He L, Zhou Y, Jia W, et al. MiR-26b/KPNA2 axis inhibits epithelial ovarian carcinoma proliferation and metastasis through downregulating OCT4. Oncotarget. 2015;6:23793–23806. doi: 10.18632/oncotarget.4363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang CI, Wang CL, Wang CW, Chen CD, Wu CC, Liang Y, Tsai YH, Chang YS, Yu JS, Yu CJ. Importin subunit alpha-2 is identified as a potential biomarker for non-small cell lung cancer by integration of the cancer cell secretome and tissue transcriptome. Int J Cancer. 2011;128:2364–2372. doi: 10.1002/ijc.25568. [DOI] [PubMed] [Google Scholar]
- 86.Yu L, Wang G, Zhang Q, Gao L, Huang R, Chen Y, Tang Q, Liu J, Liu C, Wang H, et al. Karyopherin alpha 2 expression is a novel diagnostic and prognostic factor for colorectal cancer. Oncol Lett. 2017;13:1194–1200. doi: 10.3892/ol.2017.5579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Laurila E, Vuorinen E, Savinainen K, Rauhala H, Kallioniemi A. KPNA7, a nuclear transport receptor, promotes malignant properties of pancreatic cancer cells in vitro. Exp Cell Res. 2014;322:159–167. doi: 10.1016/j.yexcr.2013.11.014. [DOI] [PubMed] [Google Scholar]
- 88.Vuorinen EM, Rajala N, Rauhala HE, Kallioniemi A. KPNA7 nuclear import protein - a critical regulator of cancer cell growth. Cancer Research. 2016:76. [Google Scholar]
- 89.Knudsen NO, Nielsen FC, Vinther L, Bertelsen R, Holten-Andersen S, Liberti SE, Hofstra R, Kooi K, Rasmussen LJ. Nuclear localization of human DNA mismatch repair protein exonuclease 1 (hEXO1) Nucleic acids research. 2007;35:2609–2619. doi: 10.1093/nar/gkl1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Marchenko ND, Hanel W, Li D, Becker K, Reich N, Moll UM. Stress-mediated nuclear stabilization of p53 is regulated by ubiquitination and importin-alpha3 binding. Cell Death Differ. 2010;17:255–267. doi: 10.1038/cdd.2009.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Li Z, Musich PR, Cartwright BM, Wang H, Zou Y. UV-induced nuclear import of XPA is mediated by importin-alpha4 in an ATR-dependent manner. PLoS One. 2013;8:e68297. doi: 10.1371/journal.pone.0068297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yang J, Lu C, Wei J, Guo Y, Liu W, Luo L, Fisch G, Li X. Inhibition of KPNA4 attenuates prostate cancer metastasis. Oncogene. 2017;36:2868–2878. doi: 10.1038/onc.2016.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bertoli G, Cava C, Diceglie C, Martelli C, Rizzo G, Piccotti F, Ottobrini L, Castiglioni I. MicroRNA-567 dysregulation contributes to carcinogenesis of breast cancer, targeting tumor cell proliferation, and migration. Breast Cancer Res Treat. 2017;161:605–616. doi: 10.1007/s10549-016-4079-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wang H, Tao T, Yan W, Feng Y, Wang Y, Cai J, You Y, Jiang T, Jiang C. Upregulation of miR-181s reverses mesenchymal transition by targeting KPNA4 in glioblastoma. Sci Rep. 2015;5:13072. doi: 10.1038/srep13072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bao H, Chen H, Zhu X, Zhang M, Yao G, Yu Y, Qin W, Zeng C, Zen K, Liu Z. MiR-223 downregulation promotes glomerular endothelial cell activation by upregulating importin alpha4 and alpha5 in IgA nephropathy. Kidney Int. 2014;85:624–635. doi: 10.1038/ki.2013.469. [DOI] [PubMed] [Google Scholar]
- 96.Garding A, Bhattacharya N, Claus R, Ruppel M, Tschuch C, Filarsky K, Idler I, Zucknick M, Caudron-Herger M, Oakes C, et al. Epigenetic Upregulation of IncRNAs at 13q14.3 in Leukemia Is Linked to the In Cis Downregulation of a Gene Cluster That Targets NF-kB. Plos Genetics. 2013:9. doi: 10.1371/journal.pgen.1003373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Jiao X, Wood LD, Lindman M, Jones S, Buckhaults P, Polyak K, Sukumar S, Carter H, Kim D, Karchin R, et al. Somatic mutations in the Notch, NF-KB, PIK3CA, and Hedgehog pathways in human breast cancers. Genes Chromosomes Cancer. 2012;51:480–489. doi: 10.1002/gcc.21935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sjoblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, Mandelker D, Leary RJ, Ptak J, Silliman N, et al. The consensus coding sequences of human breast and colorectal cancers. Science (New York, N Y ) 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 99.Mascarenhas CdC, Ferreira da Cunha A, Brugnerotto AF, Gambero S, de Almeida MH, Carazzolle MF, Pagnano KBB, Traina F, Costa FFd, de Souza CA. Identification of target genes using gene expression profile of granulocytes from patients with chronic myeloid leukemia treated with tyrosine kinase inhibitors. Leukemia & lymphoma. 2014;55:1861–1869. doi: 10.3109/10428194.2013.855311. [DOI] [PubMed] [Google Scholar]
- 100.Buza N, Xu F, Wu W, Carr RJ, Li P, Hui P. Recurrent chromosomal aberrations in intravenous leiomyomatosis of the uterus: high-resolution array comparative genomic hybridization study. Hum Pathol. 2014;45:1885–1892. doi: 10.1016/j.humpath.2014.05.010. [DOI] [PubMed] [Google Scholar]
- 101.Twyffels L, Gueydan C, Kruys V. Transportin-1 and Transportin-2: protein nuclear import and beyond. FEBS Lett. 2014;588:1857–1868. doi: 10.1016/j.febslet.2014.04.023. [DOI] [PubMed] [Google Scholar]
- 102.Maertens GN, Cook NJ, Wang W, Hare S, Gupta SS, Oztop I, Lee K, Pye VE, Cosnefroy O, Snijders AP, et al. Structural basis for nuclear import of splicing factors by human Transportin 3. Proc Natl Acad Sci U S A. 2014;111:2728–2733. doi: 10.1073/pnas.1320755111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhang ZC, Chook YM. Structural and energetic basis of ALS-causing mutations in the atypical proline-tyrosine nuclear localization signal of the Fused in Sarcoma protein (FUS) Proceedings of the National Academy of Sciences of the United States of America. 2012;109:12017–12021. doi: 10.1073/pnas.1207247109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Fanin M, Peterle E, Fritegotto C, Nascimbeni AC, Tasca E, Torella A, Nigro V, Angelini C. Incomplete penetrance in limb-girdle muscular dystrophy type 1F. Muscle Nerve. 2015;52:305–306. doi: 10.1002/mus.24539. [DOI] [PubMed] [Google Scholar]
- 105.Kobayashi J, Matsuura Y. Structural basis for cell-cycle-dependent nuclear import mediated by the karyopherin Kap121p. J Mol Biol. 2013;425:1852–1868. doi: 10.1016/j.jmb.2013.02.035. [DOI] [PubMed] [Google Scholar]
- 106.Krawczyk E, Hanover JA, Schlegel R, Suprynowicz FA. Karyopherin beta3: a new cellular target for the HPV-16 E5 oncoprotein. Biochem Biophys Res Commun. 2008;371:684–688. doi: 10.1016/j.bbrc.2008.04.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bordeaux J, Forte S, Harding E, Darshan MS, Klucevsek K, Moroianu J. The l2 minor capsid protein of low-risk human papillomavirus type 11 interacts with host nuclear import receptors and viral DNA. J Virol. 2006;80:8259–8262. doi: 10.1128/JVI.00776-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Quan L, Qiu T, Liang J, Li M, Zhang Y, Tao K. Identification of Target Genes Regulated by KSHV miRNAs in KSHV-Infected Lymphoma Cells. Pathol Oncol Res. 2015;21:875–880. doi: 10.1007/s12253-015-9902-2. [DOI] [PubMed] [Google Scholar]
- 109.Golomb L, Bublik DR, Wilder S, Nevo R, Kiss V, Grabusic K, Volarevic S, Oren M. Importin 7 and exportin 1 link c-Myc and p53 to regulation of ribosomal biogenesis. Mol Cell. 2012;45:222–232. doi: 10.1016/j.molcel.2011.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lee AY, Kim S, Lee S, Jiang H-L, Kim S-B, Hong S-H, Cho M-H. Knockdown of Importin 7 Inhibits Lung Tumorigenesis in K-rasLA1 Lung Cancer Mice. Anticancer research. 2017;37:2381–2386. doi: 10.21873/anticanres.11576. [DOI] [PubMed] [Google Scholar]
- 111.Szczyrba J, Nolte E, Hart M, Doll C, Wach S, Taubert H, Keck B, Kremmer E, Stohr R, Hartmann A, et al. Identification of ZNF217, hnRNP-K, VEGF-A and IPO7 as targets for microRNAs that are downregulated in prostate carcinoma. Int J Cancer. 2013;132:775–784. doi: 10.1002/ijc.27731. [DOI] [PubMed] [Google Scholar]
- 112.Smith ER, Cai KQ, Smedberg JL, Ribeiro MM, Rula ME, Slater C, Godwin AK, Xu X-X. Nuclear entry of activated MAPK is restricted in primary ovarian and mammary epithelial cells. PloS one. 2010;5:e9295. doi: 10.1371/journal.pone.0009295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Perez-Ramirez M, Hernandez-Jimenez AJ, Guerrero-Guerrero A, Benadon-Darszon E, Perezpena-Diazconti M, Siordia-Reyes AG, Garcia-Mendez A, de Leon FC, Salamanca-Gomez FA, Garcia-Hernandez N. Genomics and epigenetics: A study of ependymomas in pediatric patients. Clin Neurol Neurosurg. 2016;144:53–58. doi: 10.1016/j.clineuro.2016.02.041. [DOI] [PubMed] [Google Scholar]
- 114.Volpon L, Culjkovic-Kraljacic B, Osborne MJ, Ramteke A, Sun Q, Niesman A, Chook YM, Borden KLB. Importin 8 mediates m7G cap-sensitive nuclear import of the eukaryotic translation initiation factor eIF4E. Proceedings of the National Academy of Sciences of the United States of America. 2016;113:5263–5268. doi: 10.1073/pnas.1524291113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Iyer G, Wang AR, Brennan SR, Bourgeois S, Armstrong E, Shah P, Harari PM. Identification of stable housekeeping genes in response to ionizing radiation in cancer research. Sci Rep. 2017;7:43763. doi: 10.1038/srep43763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ayakannu T, Taylor AH, Willets JM, Brown L, Lambert DG, McDonald J, Davies Q, Moss EL, Konje JC. Validation of endogenous control reference genes for normalizing gene expression studies in endometrial carcinoma. Mol Hum Reprod. 2015;21:723–735. doi: 10.1093/molehr/gav033. [DOI] [PubMed] [Google Scholar]
- 117.Plafker SM, Plafker KS, Weissman AM, Macara IG. Ubiquitin charging of human class III ubiquitin-conjugating enzymes triggers their nuclear import. J Cell Biol. 2004;167:649–659. doi: 10.1083/jcb.200406001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Plafker KS, Plafker SM. The ubiquitin-conjugating enzyme UBE2E3 and its import receptor importin-11 regulate the localization and activity of the antioxidant transcription factor NRF2. Mol Biol Cell. 2015;26:327–338. doi: 10.1091/mbc.E14-06-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zhao JJ, Xu WD, He MH, Zhang ZS, Zeng SX, Ma C, Sun YH, Xu CL. Whole-exome sequencing of muscle-invasive bladder cancer identifies recurrent copy number variation in IPO11 and prognostic significance of importin-11 overexpression on poor survival. Oncotarget. 2016;7:75648–75658. doi: 10.18632/oncotarget.12315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120•.Chen M, Nowak DG, Narula N, Robinson B, Watrud K, Ambrico A, Herzka TM, Zeeman ME, Minderer M, Zheng W, et al. The nuclear transport receptor Importin-11 is a tumor suppressor that maintains PTEN protein. J Cell Biol. 2017;216:641–656. doi: 10.1083/jcb.201604025. Post-translationally regulated PTEN nuclear-cytoplasmic shuttling appears to be necessary for its tumor suppression functions upon receipt of apoptotic or cell growth inhibitory triggers. Addition of a single ubiquitin to PTEN was shown to promote its nuclear localization, in contract to poly-ubiquitination leading to cytosolic PTEN degradation and clearance. This reference provides strong evidence that IPO11 exclusively binds to monoubiquitinated PTEN and transports it into the nucleus. Ipo11 mediated nuclear localization keeps PTEN away from the players of the cytoplasmic ubiquitination machinery, specifically from the E3 ubiquitin ligase, NEDD4 and the Nedd4 family interacting protein, NDFIP which facilitates PTEN poly-ubiquitination and degradation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Aksu M, Trakhanov S, Gorlich D. Structure of the exportin Xpo4 in complex with RanGTP and the hypusine-containing translation factor eIF5A. Nat Commun. 2016;7:11952. doi: 10.1038/ncomms11952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zender L, Xue W, Zuber J, Semighini CP, Krasnitz A, Ma B, Zender P, Kubicka S, Luk JM, Schirmacher P, et al. An oncogenomics-based in vivo RNAi screen identifies tumor suppressors in liver cancer. Cell. 2008;135:852–864. doi: 10.1016/j.cell.2008.09.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Zhang H, Wei S, Ning S, Jie Y, Ru Y, Gu Y. Evaluation of TGFbeta, XPO4, elF5A2 and ANGPTL4 as biomarkers in HCC. Exp Ther Med. 2013;5:119–127. doi: 10.3892/etm.2012.750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Liang XT, Pan K, Chen MS, Li JJ, Wang H, Zhao JJ, Sun JC, Chen YB, Ma HQ, Wang QJ, et al. Decreased expression of XPO4 is associated with poor prognosis in hepatocellular carcinoma. J Gastroenterol Hepatol. 2011;26:544–549. doi: 10.1111/j.1440-1746.2010.06434.x. [DOI] [PubMed] [Google Scholar]
- 125.Grunwald M, Lazzaretti D, Bono F. Structural basis for the nuclear export activity of Importin13. EMBO J. 2013;32:899–913. doi: 10.1038/emboj.2013.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Raby BA, Van Steen K, Lasky-Su J, Tantisira K, Kaplan F, Weiss ST. Importin-13 genetic variation is associated with improved airway responsiveness in childhood asthma. Respir Res. 2009;10:67. doi: 10.1186/1465-9921-10-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Wang H, Tao T, Tang J, Mao YH, Li W, Peng J, Tan G, Zhou YP, Zhong JX, Tseng SCG, et al. Importin 13 Serves as a Potential Marker for Corneal Epithelial Progenitor Cells. Stem Cells. 2009;27:2516–2526. doi: 10.1002/stem.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zeng B, Hu J, Yuan R, Hu L, Zhong L, Kang K. Increased expression of importin13 in endometriosis and endometrial carcinoma. Medical science monitor: international medical journal of experimental and clinical research. 2012;18:CR361–367. doi: 10.12659/MSM.882879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Stelma T, Chi A, van der Watt PJ, Verrico A, Lavia P, Leaner VD. Targeting nuclear transporters in cancer: Diagnostic, prognostic and therapeutic potential. IUBMB Life. 2016;68:268–280. doi: 10.1002/iub.1484. [DOI] [PubMed] [Google Scholar]
- 130.DeBerardinis RJ, Chandel NS. Fundamentals of cancer metabolism. Sci Adv. 2016;2:e1600200. doi: 10.1126/sciadv.1600200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kimura M, Morinaka Y, Imai K, Kose S, Horton P, Imamoto N. Extensive cargo identification reveals distinct biological roles of the 12 importin pathways. Elife. 2017:6. doi: 10.7554/eLife.21184. [DOI] [PMC free article] [PubMed] [Google Scholar]