Abstract
During female meiosis, only one of four meiotic products is retained in the egg. It was previously proposed that chromosomes might compete for inclusion in the egg via their centromere ‘strength’. Recent findings have revealed the primary requirements for such ‘centromere drive’. First, CDC42 signaling from the oocyte cortex renders the meiotic I spindle asymmetric. Second, ‘stronger’ centromeres preferentially detach from microtubules in cortical proximity, making them more likely to orient away from the cortex, and be included in the egg. Third, centromeric satellite DNA expansions result in greater recruitment of centromeric proteins. Despite these mechanistic insights, it is still unclear if centromere drive elicits rapid evolution of centromeric proteins, thereby driving cellular incompatibilities and wreaking havoc on centromere stability.
Introduction
Centromeres are chromosomal regions responsible for proper chromosome segregation in eukaryotes. Centromeres recruit kinetochore proteins, which attach to the spindle, allowing chromosomes to be pulled apart in an orchestrated manner during cell division. Proper centromere function is essential for eukaryotic life. Given this vital role, centromeric DNA and proteins are expected to evolve under strict evolutionary constraint. In contrast to this prediction, centromeric DNA and proteins evolve rapidly across diverse eukaryotic taxa. This evolutionary enigma is known as the centromere paradox [1]. The centromere drive model provides a plausible explanation for the centromere paradox. The model posits that centromeric DNA evolves rapidly because centromeres are engaged in a genetic conflict, in which stronger centromeres can bias their own transmission to the egg rather than perishing in polar bodies during female meiosis [1,2].
Centromere drive has been observed in Mimulus guttatus monkeyflower populations [3]. More recently, quantitative measurements of satellite DNA also discovered inheritance patterns consistent with centromere drive in Drosophila melanogaster [4]. Although these and other systems have contributed much insight into to the evolution and ecology of centromere drive, the key cell biological system to study centromere drive has been female meiosis in mice. Recent studies have combined previous genetic findings together with advances in cellular manipulation and imaging to bring centromere drive into sharp focus.
The cell biology of centromere drive
Centromere drive is hypothesized to occur in the first meiotic cell division (meiosis I, or MI). Meiosis I is a reductional division in which homologous chromosomes separate from one another, but sister chromatids are held together at their centromeres. In contrast, during the second, equational meiotic division (Meiosis II), the two sister chromatids segregate away from each other.
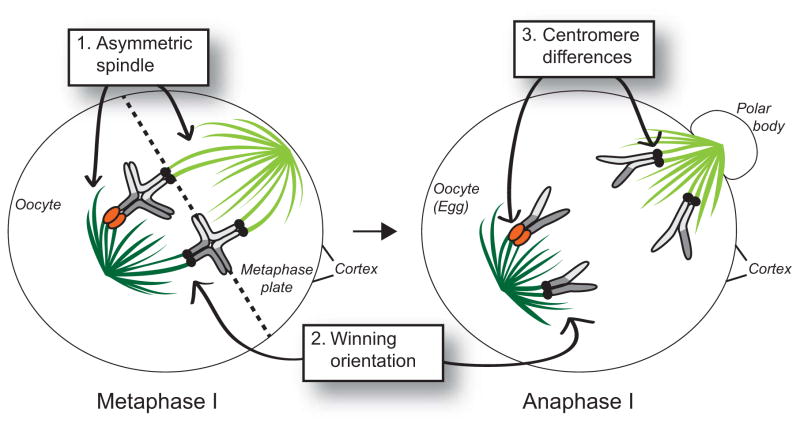
There are three primary requirements for centromere drive (Fig 1). First, centromere drive relies on asymmetry of the MI spindle. The asymmetric MI spindle enables centromeres to orient themselves relative to each other and to the cortex of the oocyte. Second, a preferred centromere position on the asymmetric MI spindle must predictably dictate chromosomal fates i.e., whether chromosomes are retained in the egg or degraded in polar bodies. The third requirement is centromere heterozygosity, in which homologous chromosomes have different propensities to exploit the asymmetry of the female meiotic spindle.
Figure 1. The three cellular requirements of centromere drive.
In order for centromere drive to occur, three basic requirements must be met. First, the meiotic I spindle must be asymmetric (dark green vs light green microtubules). Second, there must be a preferred (winning) orientation that dictates which chromosomes will segregate to the egg instead of the polar body. In mouse oocytes, chromosomes positioned toward the center of the oocyte nucleus will end up in the egg while chromosomes positioned near the oocyte cortex will segregate to the polar body. Third, there must be centromeric heterozygosity (one homolog has large, orange, centromeres, while the other three have small black centromeres). Chromosomes drawn here are acrocentric (have their centromeres on one end) and are paired with one crossover each in late metaphase I. They segregate to opposite poles in anaphase I. DNA is in shades of grey.
The original centromere drive model [1] proposed that stronger centromeres (that are more successful in segregating to the egg during female meiosis) recruit more centromeric proteins, compared to their homologous centromere competitors. This hypothesis was elegantly demonstrated in crosses between mouse strains possessing homologous chromosomes with differential recruitment of centromere proteins [5]. For example, in heterozygous CHPO/CF-1 mice, particular CF-1 chromosomes are preferentially transmitted through female meiosis. The ability of CF-1 chromosomes to preferentially transmit was directly correlated with the increased amount of inner and outer kinetochore proteins recruited to CF-1 versus CHPO centromeres [5]. Moreover, chromosomes that were subject to meiotic drive were positioned off-center at the MI metaphase plate (Figure 1). This study established that increased levels of centromere proteins correlate with increased likelihood of transmission to the egg in female meiosis. These experiments helped establish a powerful model system that enabled further mechanistic studies of the meiotic spindle, chromosome behavior on the spindle and the molecular basis for centromere asymmetry.
Molecular basis of meiotic spindle asymmetry
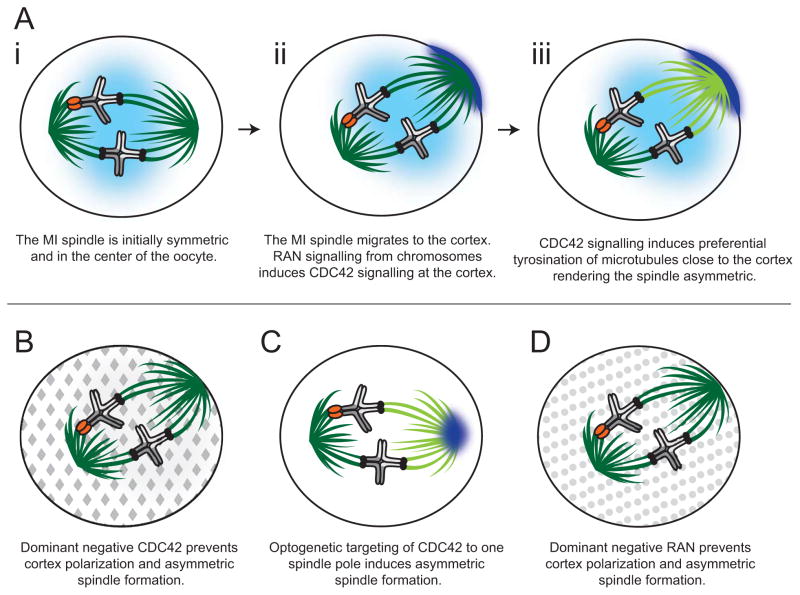
The meiotic spindle in oocytes is different from mitotic spindles in that it lacks microtubule-organizing centers called centrosomes. Instead, meiotic chromosomes organize the microtubules of the MI spindle, which forms first in the center of the oocyte but then moves in an actin-dependent manner perpendicularly towards the oocyte cortex (Figure 2Ai–Aiii).
Figure 2. Steps to spindle polarization in mouse oocyte meiosis I.
(a) The meiotic I (MI) spindle is initially symmetric and is located in the center of the oocyte (i). The spindle migrates toward the cell cortex where a RAN-GTP gradient (light blue) emanates from the chromosomes and induces CDC42 signaling (dark blue), creating a polarized cortex (ii). CDC42 signaling from the polarized cortex induces tyrosination of α-tubulin (light green microtubules) on the side of the spindle closest to the cortex. The microtubules emanating from the spindle pole oriented toward the center of the oocyte remain primarily de-tyrosinated (dark green microtubules) (iii). (b) Global expression of a dominant negative CDC42 (grey diamonds) prevents cortex polarization and prevents the MI spindle from becoming asymmetrically tyrosinated (note: all microtubules are de-tyrosinated, dark green). (c) Optogenetic targeting of active CDC42 (dark blue) to one spindle pole induces asymmetric spindle tyrosination (dark vs. light green microtubules) even before spindle migration towards the cortex. (d) Global expression of a dominant negative RAN (grey circles) prevents cortex polarization and prevents the spindle from becoming asymmetrically tyrosinated.
A recent study investigated meiotic spindle asymmetry by examining post-translational modifications of microtubules [6]. It found that the MI spindle is preferentially enriched for α-tubulin tyrosination on its cortical side (which would result in polar body inclusion) but depleted on the egg side (Figure 2Aiii). α-tubulin tyrosination results in decreased microtubule stability. Thus, the cortical side microtubules are more dynamic than those oriented towards the egg side. Intriguingly, this asymmetry is not evident in early meiosis I when symmetric spindles are formed in the center of mouse oocytes (Figure 2Ai), but is established upon spindle positioning at the cortex.
These observations suggest that some cortical signaling likely induces the asymmetry of an otherwise symmetric MI spindle. Further experiments showed that cortical signaling by CDC42, a plasma membrane-associated small GTPase involved in a variety of cell polarization processes, is required for the asymmetric tyrosination (Fig 2Aiii) [6]. Expression of a dominant-negative (Fig 2B) or a constitutively active CDC42 decreases or increases α-tubulin tyrosination respectively. Moreover, through clever optogenetic experiments, targeting CDC42 to one pole of a symmetric meiotic spindle induces asymmetric α-tubulin tyrosination even without cortical proximity (Fig 2C). The CDC42 signaling is established by a chromatin-based gradient of RAN, a small GTPase with well-established roles in microtubule dynamics. RAN signaling helps to activate CDC42 and polarize the oocyte cortex. Abrogation of this chromosome-directed RAN signaling is sufficient to eliminate the spindle asymmetry and disrupt biased chromosome orientation required for centromere drive in spite of the proximity of the spindle to the oocyte cortex (Fig 2D).
How do strong centromeres exploit the asymmetry of the meiotic spindle?
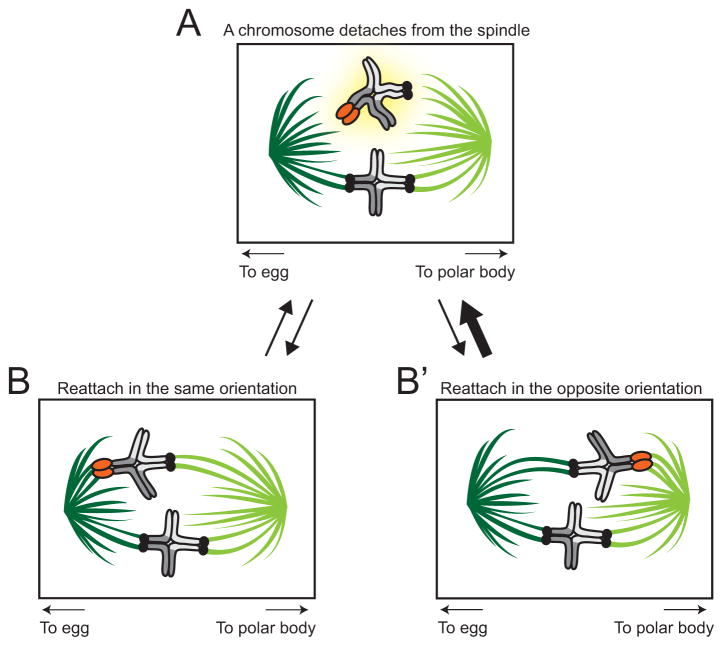
In order to undergo preferential segregation to the egg, chromosomes with stronger centromeres are predicted to preferentially orient on the MI spindle by exploiting its inherent asymmetry. Two recent studies show how strong centromeres achieve this winning orientation. The first of these studies showed that all centromeres (strong or weak) are detached from microtubules in proximity to the meiotic spindle poles [7]. This detachment requires the action of Aurora A kinase, which primarily localizes to the spindle poles. A second study [6] showed that stronger centromeres are more likely to detach than weaker centromeres, and do so at higher frequency on the cortical (‘wrong’) side, likely related to the inherently lower stability conferred by their α-tubulin tyrosination (Fig 3). These properties give stronger centromeres a higher chance of orienting toward the egg pole, thereby achieving preferential transmission into the next generation: centromere drive. This is the key cellular process by which stronger centromeres win [6].
Figure 3. How strong centromeres drive.
(a) When a chromosome detaches from the meiotic I spindle (highlighted in yellow), it has equal likelihood of reattaching in the same orientation (b) or opposite orientation (b′) (equal weight arrows connect (a) to (b) and (a) to (b′). However, a driving centromere is more likely to detach from the cortical spindle (light green microtubules oriented toward polar body, thick arrow from (b) to (a) represents higher likelihood) than from the spindle oriented toward the egg (dark green microtubules, thin arrow from (b′) to (a) represents lower likelihood).
Exactly how stronger centromeres preferentially detach remains unknown. It is possible that centromeres recruit factors like Aurora kinases that can destabilize microtubule-kinetochore attachments in a stoichiometric manner, such that stronger centromeres recruit more kinase, and their attachments to spindle microtubule are, therefore, more likely to be destabilized. If this were the case, proteins that enhance the recruitment or retention of such factors would be expected to directly enhance success in female meiosis, much like kinetochore protein recruitment is predicted to do under the original centromere drive model [1,2,8].
What is the molecular basis of centromere strength?
Earlier work using immunofluorescence supported the model that stronger centromeres recruit more inner and outer kinetochore proteins [5]. However, it remained unclear how chromosomes achieve increased recruitment of centromeric proteins. One hypothesis is that increased recruitment is linked to the underlying centromeric DNA. A recent study compared stronger and weaker centromeres in mouse strains that have identical sequences of both repetitive centromeric satellite DNA (called the minor satellite) and the centromeric histone CENP-A [9]. Closer investigation revealed that the stronger centromeres, from mouse strains CF-1 or C57BL/6J, possessed 6–10 times more copies of the minor satellite repeats than the weaker centromeres from the CHPO strain. These results establish that more centromeric DNA can recruit more centromeric proteins, which can explain the differences in centromere strength that underlie centromere drive [9].
These findings challenge the idea that centromeres in animal and plant species are solely epigenetically defined. Despite the fact that mouse centromeres are flanked by large amounts of major satellites (the pericentric satellite repeat), there were very few CENP-A nucleosomes packaged on major satellites, even in weaker centromeres. Therefore, the major satellite is a poor substrate for CENP-A nucleosome assembly whereas minor satellites are preferred. What genetic differences could distinguish the two satellites at mouse centromeres?
The differences between minor and major satellites could be attributed to the only centromeric protein with DNA-binding specificity: CENP-B. Recent studies have shown that CENP-B binding can increase the fidelity of centromere function in humans [10]. Indeed, CENP-B activity can even render the otherwise essential CENP-A dispensable for mitotic centromere function in human cells [11]. Even in human centromeres, the youngest, most abundant dimeric alpha-satellite centromeric repeat units possess CENP-B binding motifs [12] as do alpha-satellite arrays that can assemble centromeres de novo in stably transmitted human artificial chromosomes [13]. Interestingly, minor, but not major, satellites possess binding sites for CENP-B in mouse centromeres. This suggests that the CENP-B binding site is likely an important genetic determinant of centromere function, at least in humans and mouse.
An alternative, and not mutually exclusive, possibility is that satellite specificity could be RNA-mediated. Recent findings have identified centromeric satellite transcripts in complex with centromeric proteins like CENP-A and CENP-C [14]. Under this scenario, the ability to produce more minor satellite RNA by the stronger centromere may underlie its increased recruitment of CENP-A and CENP-C proteins. Conversely, major satellite-derived RNA may preferentially recruit heterochromatic, rather than centromeric, proteins [15–17].
Cellular consequences of centromere drive
Accelerated evolution of centromeric satellite DNA (in size and sequence) is only the first prediction of the centromere drive model [1,2,8]. The second prediction is that centromere drive must have deleterious fitness consequences, either directly as a result of expanded or mismatched centromeric strengths or indirectly due to the hitchhiking of deleterious alleles with driving centromeres. Finally, the third prediction of centromere drive is that centromeric proteins must co-evolve with centromeric DNA in order to suppress the deleterious consequences of centromere drive.
Recent studies have attempted to test both these predictions, starting with the question of whether centromere drive has deleterious consequences. Early work based on human carriers of Robertsonian fusions (in which two acrocentric chromosomes fuse their centromeres to create one metacentric chromosome) suggested that these expanded centromeres cause no somatic (mitotic) defects but result in lower male fertility in the heterozygous state [18,19]. This led to the model that the primary deleterious effects of centromere drive must be in male meiosis or gametogenesis. Supporting this model, taxa lacking male meiosis appear to have not undergone rapid evolution of their centromeric histones suggesting they are not subject to deleterious effects despite apparently undergoing centromere drive [20,21]. For instance, the centromeric satellites of haplodiploid fire ants (males are haploid and do not undergo meiosis) cover nearly a third of the length of their chromosomes [22]. However, recent findings have challenged the hypothesis that the primary deleterious consequence of centromere drive is in male fertility. Recent work in the Mimulus system has shown that driving centromeres cause a myriad of deleterious effects including reduced male and female viability [23] in individuals homozygous for driving centromeres. However, it is likely that the effects of deleterious alleles, which have accumulated in centromere-linked heterochromatin blocks, may have obscured the true deleterious effects of centromere drive. Thus, there is still no robust evidence for the deleterious effects of centromere drive.
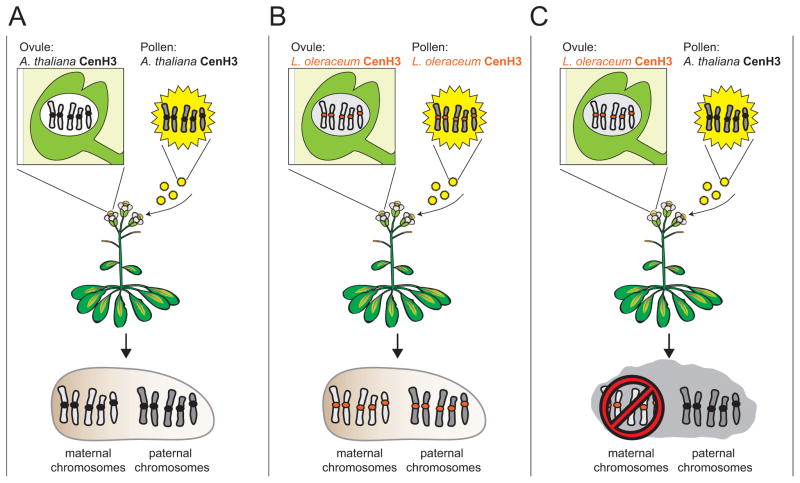
Do centromeric proteins co-evolve with rapidly evolving centromeric DNA? If centromeric proteins evolve rapidly in concert with centromeric DNA, they should be specifically adapted to their own genome. The centromeric histone, CenH3 (CENP-A in humans), is a prime candidate for this co-evolution because it has been shown to evolve rapidly in plants and animals [8], and it directly interacts with centromeric DNA [24–26]. Recently, researchers addressed this question by investigating whether divergent CenH3 orthologs can complement loss of the endogenous CenH3 allele in Arabidopsis thaliana [27]. They found that untagged CenH3 orthologs from Lepidium oleraceum and Zea mays are surprisingly capable of supporting both mitotic and meiotic function in A. thaliana. Heterologous CenH3 bearing plants are fully fertile and yield viable seeds when selfed, comparable to wild-type crosses (Fig 4A, 4B). This would suggest that A. thaliana CenH3 has not specifically adapted to A. thaliana centromeres, even though A. thaliana CenH3 has been shown to evolve rapidly [28]
Figure 4. Competition between CenH3 orthologs induces aneuploidy in Arabidopsis.
A CenH3 null mutant Arabidopsis thaliana plant can be fully rescued by an A. thaliana CenH3 transgene (black centromeres) (a) A self cross between pollen and ovules from an A. thaliana CenH3 transgene plant results in healthy seeds that develop into phenotypically wild-type, fertile plants. (b) A CenH3 null mutant A. thaliana plant can also be rescued by an orthologous CenH3 transgene from L. oleraceum (orange centromeres). A self cross between pollen and ovules from an L. oleraceum CenH3 transgene plant results in healthy seeds that develop into phenotypically wild-type, fertile plants. (c) However, when pollen from an A. thaliana CenH3 transgenic plant is crossed to ovules from a L. oleraceum CenH3 transgenic plant, the resulting progeny have high rates of aneuploidy which is entirely attributable to defects in the maternal, L. oleraceum-CenH3-packaged, genome. Note: all plants have an A. thaliana genetic background.
How do we reconcile the full functionality of CenH3 orthologs, with the evolutionary signature of their rapid evolution? A partial answer is revealed in crosses using pollen from A. thaliana plants encoding A. thaliana CenH3 and ovules from A. thaliana plants encoding a divergent L. oleraceum CenH3 ortholog (Fig 4C). In this cross, although fertilization proceeds normally, the maternal chromosomes, whose centromeres are packaged in L. oleraceum CenH3, undergo dramatic chromosome segregation defects [27]. In contrast, the paternal chromosomes, whose centromeres are packaged in A. thaliana CenH3, undergo proper chromosome segregation. As a result, many progeny plants are aneuploid or haploid, containing solely paternal chromosomes.
What is the nature of the developmental defect induced when genomes packaged by heterologous CenH3s are outcrossed to wild-type A. thaliana? One possibility is that L. oleraceum CenH3 recognizes a different set of satellites than A. thaliana CenH3. However, ChIP-seq experiments with A. thaliana and L. oleraceum CenH3 proteins revealed no significant differences satellite sequence binding in A. thaliana genomes [29]. Although these experiments cannot rule out the possibility that orthologous CenH3 proteins have lower stability than A. thaliana CenH3 despite correct localization, they do weaken the possibility that DNA sequence-binding preferences can fully explain functional differences between CenH3 orthologs [29,30].
In Drosophila species, rapid, divergent evolution of centromeric proteins can also lead to incompatibilities in protein-protein interactions. For instance, the D. melanogaster CenH3 chaperone Cal1 is incompatible with the D. bipectinata CenH3 protein, leading to the mislocalization of exogenous D. bipectinata CenH3 in D. melanogaster cells [31]. Co-expression of the D. bipectinata Cal1 protein can alleviate this mislocalization. However, the finding that plant CenH3 orthologs are intrinsically capable of performing meiotic and mitotic functions in A. thaliana (Figure 4B) weakens the likelihood that protein-protein incompatibilities can explain the Arabidopsis aneuploidy-induction phenotype [27]. Thus, although these studies do find evidence in support of the coevolution of centromeric proteins, they leave mechanistic basis of centromeric incompatibilities unresolved [30].
The perplexing diversity of centromeric protein repertoires
Despite performing similar essential functions across eukaryotes, centromeric and kinetochore protein repertoires are remarkably varied [32]. While we could attribute the rapid changes in essential proteins like CenH3 to centromere drive, other changes are likely to be driven by other differences in chromatin architecture or constraints. One dramatic type of centromere change is the transition from monocentric to holocentric chromosomes [33], which has been suggested to be an evolutionary means to curb centromere drive [8]. Indeed, recent results suggest that centromeric proteins do not evolve rapidly in the case of holocentric plants [34], although this possibility is not fully explored. In another dramatic example, four independent transitions to holocentricity in insect lineages have been accompanied by loss of the otherwise essential CenH3 protein [35]. Trypanosomes also lack CenH3 [36,37]; it remains unclear whether these centromeres represents an ancestral CenH3-less state or whether they secondarily lost CenH3 like holocentric insects.
Other dramatic changes in kinetochore proteins still defy explanation. For instance, the Dam1 outer kinetochore protein complex of budding yeast and the Ska complex in humans may carry out analogous functions in linking kinetochores to spindle microtubules. A recent phylogenomic analysis made a strong case for multiple, independent replacements of the ancestral Ska complex by the Dam1 complex [38]. However, it is unclear why the Dam1 complex was preferred in some lineages and the Ska complex preferred in others. Other recent studies revealed that CenH3 genes in Drosophila and plants have undergone independent duplication events, where they may have subfunctionalized for different centromeric roles [39,40]. These are only a few examples of the gene loss, duplication, and evolutionary innovation that are pervasive across the kinetochores of eukaryotes [41].
Future directions
The identification of specific molecular features such as α-tubulin tyrosination, RAN and CDC42 signaling and greater CENP-A recruitment which help define the asymmetry of the female meiotic spindle in mouse oocytes will facilitate parallel studies in other model systems to ascertain whether similar mechanisms are used across animals and plants. Given the diversity of kinetochore repertoires and the independent evolutionary origins of asymmetric female meiosis (in plants and animals, for instance), one may not be surprised to find diversity of mechanisms underlying the cellular basis of the asymmetric spindle and of centromere drive. Although centromere drive must occur in the first meiotic division, it will be interesting to assess whether similar determinants of spindle asymmetry underlie drive that occurs in the second meiotic division in both plants and animals [42,43].
Highlights.
Cortical CDC42 signaling leads to asymmetric tyrosination of meiosis I spindle microtubules.
Centromeric DNA expansion recruits more CenH3, increases centromere strength.
Stronger centromeres achieve a preferential position during meiosis.
Plant CenH3 ortholog replacements retain functionality, but lack stability.
Acknowledgments
We thank Lukas Chmatal, Luca Comai, Michelle Hays, Rini Kasinathan, Michael Lampson, and Tera Levin for their comments on this review. We are supported by funding from the National Institutes of Health training grants T32 HG000035 and T32 GM007270 (to L.E.K.) and R01 GM074108 (to H.S.M.). H.S.M. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Henikoff S, Ahmad K, Malik HS. The centromere paradox: stable inheritance with rapidly evolving DNA. Science. 2001;293:1098–1102. doi: 10.1126/science.1062939. [DOI] [PubMed] [Google Scholar]
- 2.Malik HS. The centromere-drive hypothesis: a simple basis for centromere complexity. Prog Mol Subcell Biol. 2009;48:33–52. doi: 10.1007/978-3-642-00182-6_2. [DOI] [PubMed] [Google Scholar]
- 3.Fishman L, Saunders A. Centromere-associated female meiotic drive entails male fitness costs in monkeyflowers. Science. 2008;322:1559–1562. doi: 10.1126/science.1161406. [DOI] [PubMed] [Google Scholar]
- 4*.Wei KH, Reddy HM, Rathnam C, Lee J, Lin D, Ji S, Mason JM, Clark AG, Barbash DA. A Pooled Sequencing Approach Identifies a Candidate Meiotic Driver in Drosophila. Genetics. 2017;206:451–465. doi: 10.1534/genetics.116.197335. This paper unveils a clever pooled sequencing strategy that can detect meiotic drive by a patterns of SNP frequencies in driver versus distal loci. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chmatal L, Gabriel SI, Mitsainas GP, Martinez-Vargas J, Ventura J, Searle JB, Schultz RM, Lampson MA. Centromere strength provides the cell biological basis for meiotic drive and karyotype evolution in mice. Curr Biol. 2014;24:2295–2300. doi: 10.1016/j.cub.2014.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6**.Akera T, Chmátal L, Trimm E, Yang K, Aonbangkhen C, Chenoweth DM, Janke C, Schultz RM, Lampson MA. Spindle asymmetry drives non-Mendelian chromosome segregation. Science. 2017;358:668–672. doi: 10.1126/science.aan0092. This groundbreaking paper establishes the cell biological basis for the asymmetry of the female meiotic spindle and its exploitation by ‘stronger’ centromeres. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7*.Chmatal L, Yang K, Schultz RM, Lampson MA. Spatial Regulation of Kinetochore Microtubule Attachments by Destabilization at Spindle Poles in Meiosis I. Curr Biol. 2015;25:1835–1841. doi: 10.1016/j.cub.2015.05.013. This paper first reveals kinetochore detachment from microtubules via Aurora A kinases, a possible spatial regulation of centromere strength. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Malik HS, Henikoff S. Major evolutionary transitions in centromere complexity. Cell. 2009;138:1067–1082. doi: 10.1016/j.cell.2009.08.036. [DOI] [PubMed] [Google Scholar]
- 9**.Iwata-Otsubo A, Dawicki-McKenna JM, Akera T, Falk SJ, Chmatal L, Yang K, Sullivan BA, Schultz RM, Lampson MA, Black BE. Expanded Satellite Repeats Amplify a Discrete CENP-A Nucleosome Assembly Site on Chromosomes that Drive in Female Meiosis. Curr Biol. 2017;27:2365–2373. e2368. doi: 10.1016/j.cub.2017.06.069. This paper describes that stronger centromeres recruit more CENP-A via minor satellite expansions. It further shows how sequence preferences preclude pericentric major satellites from becoming centromeric even in weaker centromeres. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10*.Fachinetti D, Han JS, McMahon MA, Ly P, Abdullah A, Wong AJ, Cleveland DW. DNA Sequence-Specific Binding of CENP-B Enhances the Fidelity of Human Centromere Function. Dev Cell. 2015;33:314–327. doi: 10.1016/j.devcel.2015.03.020. This paper establishes the important role of CENP-B proteins in maintaining centromere-kinetochore fidelity of function in human cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoffmann S, Dumont M, Barra V, Ly P, Nechemia-Arbely Y, McMahon MA, Herve S, Cleveland DW, Fachinetti D. CENP-A Is Dispensable for Mitotic Centromere Function after Initial Centromere/Kinetochore Assembly. Cell Rep. 2016;17:2394–2404. doi: 10.1016/j.celrep.2016.10.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12*.Henikoff JG, Thakur J, Kasinathan S, Henikoff S. A unique chromatin complex occupies young alpha-satellite arrays of human centromeres. Sci Adv. 2015:1. doi: 10.1126/sciadv.1400234. This study identifies a recently homogenized α-satellite dimeric unit, occupied by two CENP-A nucleosomes flanking a CENP-B/CENP-C linker, as the dominant repeat of human centromeres. [DOI] [PMC free article] [PubMed]
- 13.Ohzeki J, Nakano M, Okada T, Masumoto H. CENP-B box is required for de novo centromere chromatin assembly on human alphoid DNA. J Cell Biol. 2002;159:765–775. doi: 10.1083/jcb.200207112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14*.McNulty SM, Sullivan LL, Sullivan BA. Human Centromeres Produce Chromosome-Specific and Array-Specific Alpha Satellite Transcripts that Are Complexed with CENP-A and CENP-C. Dev Cell. 2017;42:226–240. e226. doi: 10.1016/j.devcel.2017.07.001. This study adds to a growing body of evidence that centromeric satellite transcripts may play key roles in centromere function and kinetochore assembly. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson WL, Yewdell WT, Bell JC, McNulty SM, Duda Z, O’Neill RJ, Sullivan BA, Straight AF. RNA-dependent stabilization of SUV39H1 at constitutive heterochromatin. Elife. 2017:6. doi: 10.7554/eLife.25299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shirai A, Kawaguchi T, Shimojo H, Muramatsu D, Ishida-Yonetani M, Nishimura Y, Kimura H, Nakayama JI, Shinkai Y. Impact of nucleic acid and methylated H3K9 binding activities of Suv39h1 on its heterochromatin assembly. Elife. 2017:6. doi: 10.7554/eLife.25317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Velazquez Camacho O, Galan C, Swist-Rosowska K, Ching R, Gamalinda M, Karabiber F, De La Rosa-Velazquez I, Engist B, Koschorz B, Shukeir N, et al. Major satellite repeat RNA stabilize heterochromatin retention of Suv39h enzymes by RNA-nucleosome association and RNA:DNA hybrid formation. Elife. 2017:6. doi: 10.7554/eLife.25293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Daniel A. Distortion of female meiotic segregation and reduced male fertility in human Robertsonian translocations: consistent with the centromere model of co-evolving centromere DNA/centromeric histone (CENP-A) Am J Med Genet. 2002;111:450–452. doi: 10.1002/ajmg.10618. [DOI] [PubMed] [Google Scholar]
- 19.Wallace BM, Searle JB, Everett CA. The effect of multiple simple Robertsonian heterozygosity on chromosome pairing and fertility of wild-stock house mice (Mus musculus domesticus) Cytogenet Genome Res. 2002;96:276–286. doi: 10.1159/000063054. [DOI] [PubMed] [Google Scholar]
- 20.Elde NC, Roach KC, Yao MC, Malik HS. Absence of positive selection on centromeric histones in Tetrahymena suggests unsuppressed centromere: drive in lineages lacking male meiosis. J Mol Evol. 2011;72:510–520. doi: 10.1007/s00239-011-9449-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zedek F, Bures P. CenH3 evolution reflects meiotic symmetry as predicted by the centromere drive model. Sci Rep. 2016;6:33308. doi: 10.1038/srep33308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang YC, Lee CC, Kao CY, Chang NC, Lin CC, Shoemaker D, Wang J. Evolution of long centromeres in fire ants. BMC Evol Biol. 2016;16:189. doi: 10.1186/s12862-016-0760-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fishman L, Kelly JK. Centromere-associated meiotic drive and female fitness variation in Mimulus. Evolution. 2015;69:1208–1218. doi: 10.1111/evo.12661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barrey EJ, Heun P. Artificial Chromosomes and Strategies to Initiate Epigenetic Centromere Establishment. Prog Mol Subcell Biol. 2017;56:193–212. doi: 10.1007/978-3-319-58592-5_8. [DOI] [PubMed] [Google Scholar]
- 25.Rosin LF, Mellone BG. Centromeres Drive a Hard Bargain. Trends Genet. 2017;33:101–117. doi: 10.1016/j.tig.2016.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schalch T, Steiner FA. Structure of centromere chromatin: from nucleosome to chromosomal architecture. Chromosoma. 2017;126:443–455. doi: 10.1007/s00412-016-0620-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27**.Maheshwari S, Tan EH, West A, Franklin FC, Comai L, Chan SW. Naturally occurring differences in CENH3 affect chromosome segregation in zygotic mitosis of hybrids. PLoS Genet. 2015;11:e1004970. doi: 10.1371/journal.pgen.1004970. This study if the first to describe consequences of CenH3 ortholog evolution on function in A. thaliana. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Talbert PB, Masuelli R, Tyagi AP, Comai L, Henikoff S. Centromeric localization and adaptive evolution of an Arabidopsis histone H3 variant. Plant Cell. 2002;14:1053–1066. doi: 10.1105/tpc.010425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maheshwari S, Ishii T, Brown CT, Houben A, Comai L. Centromere location in Arabidopsis is unaltered by extreme divergence in CENH3 protein sequence. Genome Res. 2017;27:471–478. doi: 10.1101/gr.214619.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Comai L, Maheshwari S, Marimuthu MPA. Plant centromeres. Curr Opin Plant Biol. 2017;36:158–167. doi: 10.1016/j.pbi.2017.03.003. [DOI] [PubMed] [Google Scholar]
- 31*.Rosin L, Mellone BG. Co-evolving CENP-A and CAL1 Domains Mediate Centromeric CENP-A Deposition across Drosophila Species. Dev Cell. 2016;37:136–147. doi: 10.1016/j.devcel.2016.03.021. Elegant demonstration of a protein-protein incompatibility between CenH3 and its chaperone Cal1 in two species of Drosophila. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Drinnenberg IA, Henikoff S, Malik HS. Evolutionary Turnover of Kinetochore Proteins: A Ship of Theseus? Trends Cell Biol. 2016;26:498–510. doi: 10.1016/j.tcb.2016.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Melters DP, Paliulis LV, Korf IF, Chan SW. Holocentric chromosomes: convergent evolution, meiotic adaptations, and genomic analysis. Chromosome Res. 2012;20:579–593. doi: 10.1007/s10577-012-9292-1. [DOI] [PubMed] [Google Scholar]
- 34.Zedek F, Bures P. Absence of positive selection on CenH3 in Luzula suggests that holokinetic chromosomes may suppress centromere drive. Ann Bot. 2016;118:1347–1352. doi: 10.1093/aob/mcw186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Drinnenberg IA, deYoung D, Henikoff S, Malik HS. Recurrent loss of CenH3 is associated with independent transitions to holocentricity in insects. Elife. 2014:3. doi: 10.7554/eLife.03676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Akiyoshi B, Gull K. Evolutionary cell biology of chromosome segregation: insights from trypanosomes. Open Biol. 2013;3:130023. doi: 10.1098/rsob.130023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Akiyoshi B, Gull K. Discovery of unconventional kinetochores in kinetoplastids. Cell. 2014;156:1247–1258. doi: 10.1016/j.cell.2014.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van Hooff JJE, Snel B, Kops G. Unique Phylogenetic Distributions of the Ska and Dam1 Complexes Support Functional Analogy and Suggest Multiple Parallel Displacements of Ska by Dam1. Genome Biol Evol. 2017;9:1295–1303. doi: 10.1093/gbe/evx088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Finseth FR, Dong Y, Saunders A, Fishman L. Duplication and Adaptive Evolution of a Key Centromeric Protein in Mimulus, a Genus with Female Meiotic Drive. Mol Biol Evol. 2015;32:2694–2706. doi: 10.1093/molbev/msv145. [DOI] [PubMed] [Google Scholar]
- 40.Kursel LE, Malik HS. Recurrent Gene Duplication Leads to Diverse Repertoires of Centromeric Histones in Drosophila Species. Mol Biol Evol. 2017;34:1445–1462. doi: 10.1093/molbev/msx091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41**.van Hooff JJ, Tromer E, van Wijk LM, Snel B, Kops GJ. Evolutionary dynamics of the kinetochore network in eukaryotes as revealed by comparative genomics. EMBO Rep. 2017;18:1559–1571. doi: 10.15252/embr.201744102. A tour-de-force phylogenomic study recounting the dynamic changes in kintochore protein repertoires across eukaryotic evolution. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu G, Hao L, Han Z, Gao S, Latham KE, de Villena FP, Sapienza C. Maternal transmission ratio distortion at the mouse Om locus results from meiotic drive at the second meiotic division. Genetics. 2005;170:327–334. doi: 10.1534/genetics.104.039479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kanizay LB, Albert PS, Birchler JA, Dawe RK. Intragenomic conflict between the two major knob repeats of maize. Genetics. 2013;194:81–89. doi: 10.1534/genetics.112.148882. [DOI] [PMC free article] [PubMed] [Google Scholar]