Abstract
Among different species or cell types, or during early embryonic cell divisions that occur in the absence of cell growth, the size of subcellular structures, including the nucleus, chromosomes, and mitotic spindle, scale with cell size. Maintaining correct subcellular scales is thought to be important for many cellular processes and, in particular, for mitosis. In this review, we provide an update on nuclear and chromosome scaling mechanisms and their significance in metazoans, with a focus on Caenorhabditis elegans, Xenopus and mammalian systems, for which a common role for the Ran (Ras-related nuclear protein)-dependent nuclear transport system has emerged.
Introduction
Absolute and relative size of biological entities varies widely, both within and among species at all levels of organization above the atomic/molecular: the organism, the cells that make up the organism, and the components of the cells. How does scaling occur so that everything fits and functions properly? Until recently, the control systems that a cell uses to regulate and coordinate the size of its internal structures were virtually unknown. One candidate coordinator is the small GTPase Ran and its downstream transport machinery, which are involved in many cellular processes in both interphase and mitosis, from nucleo-cytoplasmic transport to spindle morphogenesis to nuclear envelope assembly [1,2]. We will start with a brief overview of the Ran pathway and discuss recent work that elucidates mechanisms of subcellular scaling and the potential importance for cell function and division.
The RanGTP pathway and spindle assembly
RanGTP marks the genome in both interphase and mitosis and acts as a molecular switch. In the nucleus, Ran is concentrated in its GTP state due to the chromatin-associated RanGEF (Guanine nucleotide Exchange Factor) RCC1. In the cytoplasm, Ran is found in its GDP form due to the activity of cytoplasmic RanGAP (GTPase Activating Protein). RanGTP binds both importins and exportins, stabilizing the exportin-cargo interaction required for nuclear export, while releasing cargoes from importins. As a result, proteins with an NLS (Nuclear Localization Signal) are transported into the nucleus by importins and accumulate in the nucleus, while NES (Nuclear Export Signal)-containing proteins are transported out of the nucleus (Figure 1A).
Figure 1.
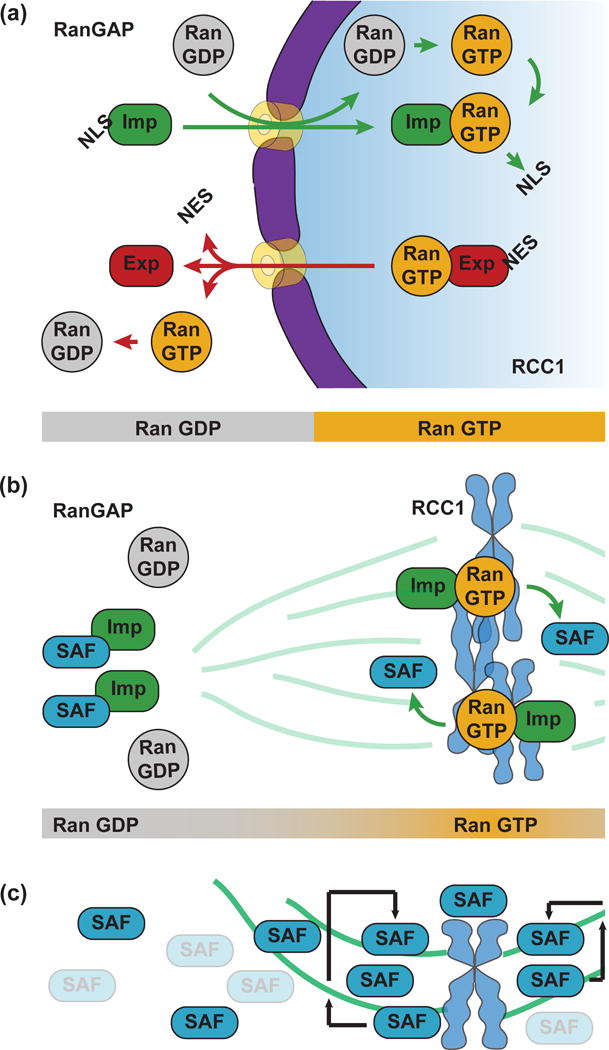
The RanGTP pathway and spindle assembly. (a) In interphase, Ran is GTP-bound in the nucleus due to the chromatin-associated RanGEF, RCC1, and GDP-bound in the cytoplasm due to cytoplasmic RanGAP. Proteins harboring an NLS are imported into the nucleus by importins and released when importins interact with RanGTP. Proteins containing an NES are exported out of the nucleus by RanGTP-bound exportins and released by GTP hydrolysis. (b) In mitosis, chromosome-bound RCC1 creates a Ran-GTP gradient near the chromosomes where NLS-containing SAFs are released from importins, promoting microtubule nucleation and stabilization. (c) Following microtubule nucleation by SAFs, the interaction between SAFs and microtubules leads to a feedback that further enriches SAFs on microtubules.
During mitosis, RCC1 remains associated with the chromosomes following nuclear envelope breakdown, enriching RanGTP in the zone where the spindle will assemble. As RanGTP diffuses away from the chromatin, RanGAP in the cytoplasm converts it to RanGDP, creating a RanGTP gradient. Numerous NLS-containing SAFs (Spindle Assembly Factors) are released within this gradient where they contribute to spindle assembly by nucleating and organizing microtubules [2] (Figure 1B). A recent study in cultured cells has revealed an interesting feedback mechanism that results from the binding of RanGTP-activated SAFs to microtubules [3]**. Microtubule binding serves to concentrate microtubule nucleators on the forming spindle, amplifying microtubule polymerization and rendering the length of the spindle insensitive to the size of the Ran gradient (Figure 1C). This microtubule-dependent amplification mechanism would explain why increasing the amount of chromatin in the spindle, and therefore the amount of RCC1 and RanGTP, does not increase spindle length in Xenopus egg extracts [4], while increasing the amount of microtubule polymer by addition of a drug dramatically increases spindle size [5]. Importantly, however, NLS-containing SAFs that regulate microtubule nucleation and dynamics downstream of RanGTP, including TPX2 and kif2a, have been shown to act as scaling factors for centrosomes and/or spindles and whose activities are regulated by importin α [6–8]. Thus, the emerging concept is that while RanGTP acts as a trigger for spindle assembly, the complex interplay between SAFs, microtubules and importins contributes to spindle scaling. Mechanisms of spindle size regulation have been elucidated in a variety of systems, particularly Xenopus, and are discussed in detail elsewhere [2,9].
Mechanisms of nuclear scaling
In contrast to the apparent independence of spindle size from the RanGTP gradient, strong evidence has accumulated that nucleocytoplasmic transport via the Ran pathway regulates nuclear scaling through the import of nuclear lamins. The nuclear lamina, which forms an intermediate filament meshwork underlying the inner nuclear membrane, is a major regulator of nuclear morphology in metazoans [10,11]. In Xenopus, lamin B3, the major lamin isoform in the egg, was found to be a key cargo regulating nuclear size differences between two different-sized frog species, X. laevis and X. tropicalis, which scale at organismal, cellular and subcellular levels [12,13]. Slower nuclear import rates and reduced accumulation of lamin B3 in egg extracts of the smaller X. tropicalis was shown to be due to differences in the levels of two nuclear transport factors, importin α and Ntf2 [13]. In contrast to importin α that promotes lamin import and nuclear growth, Ntf2 negatively regulates import of large cargoes such as lamin oligomers, and its expression decreases lamin import and nuclear size [14]*. In C. elegans, nuclear transport and lamin levels were also demonstrated to regulate nuclear size [15,16]. Thus, it appears that the more import of lamin into the nucleus, the larger it gets (Figure 2A).
Figure 2.
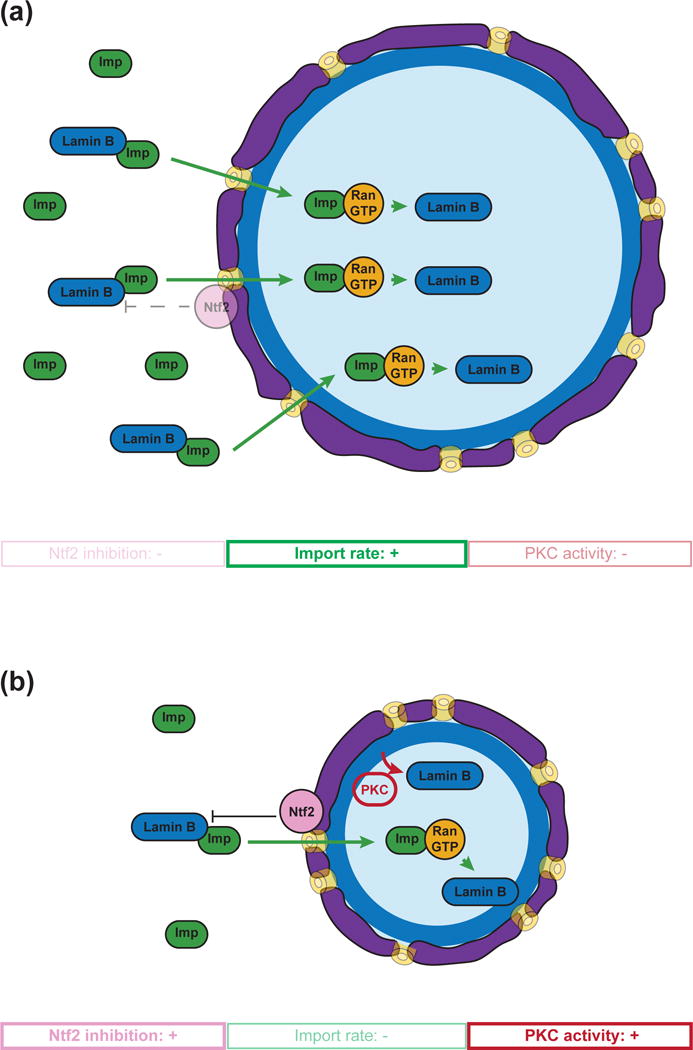
Mechanisms of nuclear scaling. (a) Nuclear import of lamins promotes nuclear growth. (b) Reduced import rate of lamins and PKC-driven lamin removal scale nuclei smaller. Moreover, Ran-dependent association of Ntf2 to the nuclear pore also affects nuclear size by inhibiting lamin import, perhaps by reducing the diameter of the nuclear pore complex.
But is the situation reversed when nuclei scale smaller? During both early Xenopus and C. elegans embryonic development, the size of the nucleus decreases with decreasing cell size [13,17,18], which reduces the amount of cytoplasm surrounding the nucleus and the materials (like lamins) necessary for nuclear growth. Interestingly, microfluidic encapsulation of Xenopus egg extracts also revealed a role for microtubules in determining the speed of nuclear expansion through dynein-mediated membrane transport [19]. In vivo studies revealed that prior to the mid-blastula transition (MBT) and the onset of zygotic transcription, the reduction in nuclear size in Xenopus correlates with reduced import rates and levels of cytoplasmic importin α. Ectopic importin α expression was sufficient to increase nuclear size in pre-MBT embryos [17]. Although lamin isoform expression changes during Xenopus development, nuclear scaling was sensitive to the total lamin concentration, and was not altered by specific lamin isotypes [10]. After MBT, but prior to gastrulation, nuclear size reduction was shown to depend on protein kinase C (cPKC) activity, which correlated with the removal of lamins from the nuclear envelope [20]. Moreover, PKC-mediated phosphorylation of lamins in interphase contributed to reductions in nuclear size in both Xenopus and mammalian cells [21] (Figure 2B). However, increasing lamin levels in post-MBT embryos or mammalian tissue culture cells also decreased nuclear size [10]. A possible explanation for this paradox is that lamin expression is precisely tuned in transcriptionally active cells, and their incorporation above a certain threshold alters mechanical properties of the lamina and, as a consequence, distorts nuclear size and shape. Consistent with this idea, a recent biophysical study revealed that lamin levels control nuclear stiffness in response to large deformations in mammalian cells [22]*.
What are the functional consequences of nuclear scaling? As well as nuclear mechanics, nuclear size is thought to impact chromatin organization and gene expression, and defects in nuclear size are associated with disease [23]. In Xenopus, altering the nucleo-cytoplasmic ratio by modulating either DNA content (ploidy) or nuclear scaling factors affected MBT timing and the onset of zygotic gene expression [17,24]. A recent study in embryonic stem cells demonstrated that changing nuclear size and shape by altering nuclear envelope components impacted gene regulation and lineage differentiation [25]. Interestingly, however, manipulations of nuclear scaling factors in frog or mouse did not negatively affect embryonic development, and the functional significance of nuclear size remains to be elucidated.
Mechanisms of chromosome scaling
A current limit to our understanding of mitotic chromosome scaling is the fact that chromosome architecture itself is poorly understood. Recently, a technical breakthrough that allows much improved visibility of the DNA by electron tomography showed definitively that rather than orderly packaging, chromatin exists as a disordered granular chain with a diameter of 5 to 24 nm. These chains are packed at variable concentration densities inside the nucleus, and at even higher densities in mitotic chromosomes [26]**. This apparent disorder allows flexible bending, enabling high packing densities of DNA. Thus, rather than higher order folding of a nucleosome fiber, mitotic chromosomes contain the same 5 to 24 nm chromatin chains as in interphase, but packed at a higher concentration density.
Thus, a candidate mechanism for how mitotic chromosomes scale with cell size is through changes in chromatin packing density. Consistent with this idea, mitotic chromosome size decreased with nuclear size during both Xenopus and C. elegans embryogenesis [15,16,27]. Furthermore, intra-nuclear DNA density was shown to correlate with the packing ratio of mitotic chromosomes across a variety of species [28]. Interestingly, when nuclei from small embryonic cells were allowed to expand in an interphase Xenopus egg extract prior to inducing chromosome condensation, small mitotic chromosomes were still produced, arguing against a relationship between nuclear size and chromosome size. However, progression through a full cell cycle in egg extract re-established characteristic chromosome lengths [27]. In contrast, artificially reducing nuclear size by blocking nuclear transport or adding an inhibitory lamin antibody to egg extracts, or by reducing RanGTP levels or nuclear import in C. elegans, led to the formation of smaller mitotic chromosomes [15,16]. These studies link mitotic chromosome size scaling with the Ran pathway and nuclear import and are consistent with a model in which chromatin has a “memory” of how compact it was in the interphase nucleus. In this model, scaling factors would be imported or exported from the nucleus during interphase and loaded on to chromatin during DNA replication, thereby setting chromosome size (Figure 3).
Figure 3.
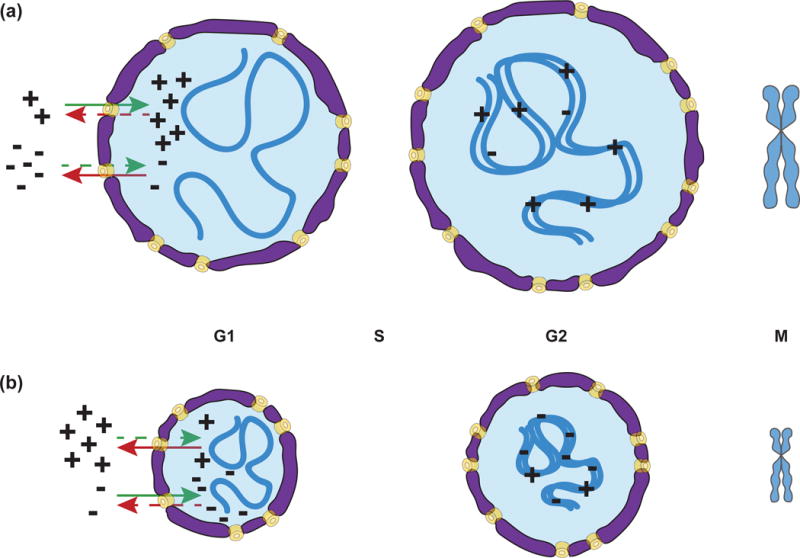
A possible model for chromosome scaling. Scaling factors that increase (+) or decrease (−) chromosome size are differentially imported/exported in interphase leading to more (+) factors and/or less (−) factors in large nuclei (a) and vice-versa in small nuclei (b). Scaling factors are then loaded during DNA replication and thus set chromosome size for mitosis.
Even if chromosome scaling were simply due to physical effects of nuclear scaling, factors likely act to maintain higher levels of chromatin compaction in smaller cells to facilitate mitotic chromosome scaling. A screen for proteins essential for embryogenesis in a C. elegans strain harboring a long chromosome identified two potential mitotic chromosome scaling factors as topoisomerase II (topo-II) and the centromere-specific histone H3 variant CENP-A [29]**. Since C. elegans chromosomes are holocentric, CENP-A is found periodically all along the length of mitotic chromosomes, forming a platform for kinetochore assembly and spindle microtubule attachment [30]. Interestingly, CENP-A levels and chromosome staining decrease during development, and depletion of CENP-A, or reduction of its nuclear import, further reduced chromosome length. These findings implicate CENP-A as one nuclear cargo that could act at the chromosome surface, perhaps by organizing chromatin domains whose abundance correlates with chromosome size. While a CENP-A-driven mechanism of chromosome scaling would be limited to holocentric chromosomes, it suggests where and how a vertebrate scaling factor could operate. However, recent functional studies of proteins at the periphery of human mitotic chromosomes, including Ki-67 that act as a surfactant to disperse mitotic chromosomes [31] and the BAF protein that clusters mitotic chromosomes together [32] did not reveal any role in setting or maintaining their size.
In contrast to CENP-A, partial depletion of topo-II from C. elegans embryos was found to increase chromosome length [29]**. Topo-II levels or staining was unchanged during development, however, and the effects of its depletion may reflect a more general role in establishing chromosome architecture. Indeed, depleting or interfering with chromosome structural proteins often results in chromosomes with improper length, shape or compaction; these include condensin and cohesin [33], as well as linker [34] and core histones [35]**. It is thus plausible that chromosome scaling factors include chromatin structural proteins with known functions in compaction and organization. However, elucidating scaling roles for one or more of these factors may prove challenging considering the complex relationships among them, such as the interplay between condensin, cohesin and topo-II [33,36], and post-translational modifications that alter their activity and distribution [37,38].
Importance of subcellular scaling in mitosis
Spindle and nuclear scaling with cell size is conserved across metazoans [23,39]. Furthermore, scaling factors have evolved to adapt nuclear and spindle size to cell size in different Xenopus species [13,40], and scaling mechanisms operate during development [7,20]. One would therefore expect these mechanisms to be important for cell and organism viability. However, since few scaling factors have been identified, functional data are limited. As discussed above, modest changes in nuclear scaling affect developmental timing and gene expression, but not embryo viability [17,24]. Decreasing spindle size during the early cleavage divisions of Xenopus embryos by increasing levels of the microtubule depolymerizing spindle scaling factor kif2a caused metaphase spindle alignment defects, but cleavage plane positioning was corrected by interactions of astral microtubules with the cell cortex, resulting in normal development [7]. We can think of two reasons why scaling functions in cell division might be difficult to disrupt. First, in addition to molecular scaling factors, an intrinsic physical mechanism based on cell volume and limiting components plays an important role in subcellular scaling [41,42]. Measurements comparing size variants within and between closely related nematode species indicated that natural selection acts predominantly on cell/embryo size, which then indirectly influences the spindle size [43]. Thus, cell size itself contributes to spindle size and the fidelity of cell division. Second, multiple mechanisms operate across a wide range of cell sizes to facilitate cell division. For example, microtubule amplification and trigger waves function to spatially and temporally coordinate chromosome segregation and cytokinesis in large cells [44,45].
Scaling of chromosome length to anaphase spindle length, which scales to cell length, is obviously crucial for proper chromosome segregation. Landmark studies in plants showed that artificially lengthened chromosomes fail to clear the spindle mid-zone and lead to the formation of micronuclei [46,47]. In animal cells, Aurora B kinase at the spindle mid-zone causes hypercondensation of chromosomes [48,49], which helps to avoid such defects. However, gross inhibition of chromosome condensation by depletion of linker histone H1, for instance, prevents chromosome alignment and segregation [34]. Due to the intimate relationship between chromosome size and architecture, and compensatory mitotic mechanisms, demonstrating a function for mitotic chromosome scaling is a current challenge in the field.
Studies analyzing the effects of variation in size relationships have revealed their relevance to spindle function and accurate chromosome segregation. Manipulating nuclear-cytoplasmic volume ratio by halving or fusing mouse oocytes affected meiotic spindle architecture, assembly kinetics, and chromosome alignment [50]**. Large cytoplasmic volume limited the spindle’s capacity to prevent anaphase entry with misaligned chromosomes, consistent with a study showing that cell size determines the strength of the spindle assembly checkpoint during C. elegans development [51]*. Modeling also predicts that checkpoint silencing entails proper size scaling of the spindle [52]*. Thus, although cells possess robust systems to ensure accurate chromosome transmission, cell size and scaling relationships impact the fidelity of cell division.
Conclusions
The role of the Ran pathway in spindle assembly and nuclear scaling has been studied for quite some time, but its implications for mitotic chromosome scaling are only starting to be appreciated. Although the RanGTP gradient itself does not appear to set spindle size, the transport machinery, particularly importin α, regulates known spindle scaling factors. Lamin import, which is also regulated by importin α, scales nuclear size to cell size during development. In turn, nuclear size affects DNA density and correlates with mitotic chromosome length, though the factors that scale chromosome condensation to cell size are still poorly understood. A mechanistic link between importins and coordinated spindle, nuclear, and mitotic chromosome scaling seems likely, but remains to be elucidated. Although proper scaling relationships are central to the fidelity of chromosome segregation, they have proven difficult to manipulate experimentally and compensating mechanisms operate to facilitate the vitally fundamental process of cell division.
Acknowledgments
We thank the members of the Heald lab for suggestions. We thank Dan Levy, Mingxuan Sun, Christopher Brownlee and Coral Zhou for critical reading of the manuscript, and apologize to all those whose work could not be cited due to space limitations. RH was supported by NIH R35 GM118183 and the Flora Lamson Hewlett Chair. RG was supported by an HFSP long term fellowship LT 0004252014-L.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Forbes DJ, Travesa A, Nord MS, Bernis C. Nuclear transport factors: global regulation of mitosis. Curr Opin Cell Biol. 2015;35:78–90. doi: 10.1016/j.ceb.2015.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cavazza T, Vernos I. The RanGTP Pathway: From Nucleo-Cytoplasmic Transport to Spindle Assembly and Beyond. Front Cell Dev Biol. 2016;3 doi: 10.3389/fcell.2015.00082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3**.Oh D, Yu C-H, Needleman DJ. Spatial organization of the Ran pathway by microtubules in mitosis. Proc Natl Acad Sci. 2016;113:8729–8734. doi: 10.1073/pnas.1607498113. Using modeling and by manipulating cultured cells, Oh et al. revealed that the interaction between SAFs and microtubules leads to a feedback mechanism that spatially amplifies spindle assembly factors to render spindle size independent of the RanGTP gradient. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grenfell AW, Strzelecka M, Crowder ME, Helmke KJ, Schlaitz AL, Heald R. A versatile multivariate image analysis pipeline reveals features of Xenopus extract spindles. J Cell Biol. 2016;213:127–136. doi: 10.1083/jcb.201509079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mitchison TJ, Maddox P, Gaetz J, Groen A, Shirasu M, Desai A, Salmon ED, Kapoor TM. Roles of polymerization dynamics, opposed motors, and a tensile element in governing the length of Xenopus extract meiotic spindles. Mol Biol Cell. 2005;16:3064–76. doi: 10.1091/mbc.E05-02-0174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Greenan G, Brangwynne CP, Jaensch S, Gharakhani J, Jülicher F, Hyman AA. Centrosome size sets mitotic spindle length in Caenorhabditis elegans embryos. Curr Biol. 2010;20:353–8. doi: 10.1016/j.cub.2009.12.050. [DOI] [PubMed] [Google Scholar]
- 7.Wilbur JD, Heald R. Mitotic spindle scaling during Xenopus development by kif2a and importin α. Elife. 2013;2:e00290. doi: 10.7554/eLife.00290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Helmke KJ, Heald R. TPX2 levels modulate meiotic spindle size and architecture in Xenopus egg extracts. J Cell Biol. 2014;206:385–93. doi: 10.1083/jcb.201401014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reber S, Goehring NW. Intracellular scaling mechanisms. Cold Spring Harb Perspect Biol. 2015;7 doi: 10.1101/cshperspect.a019067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jevtić P, Edens LJ, Li X, Nguyen T, Chen P, Levy DL. Concentration-dependent effects of nuclear lamins on nuclear size in xenopus and mammalian cells. J Biol Chem. 2015;290:27557–27571. doi: 10.1074/jbc.M115.673798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dechat T, Adam SA, Taimen P, Shimi T, Goldman RD. Nuclear lamins. Cold Spring Harb Perspect Biol. 2010;2:1–23. doi: 10.1101/cshperspect.a000547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brown KS, Blower MD, Maresca TJ, Grammer TC, Harland RM, Heald R. Xenopus tropicalis egg extracts provide insight into scaling of the mitotic spindle. J Cell Biol. 2007;176:765–70. doi: 10.1083/jcb.200610043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Levy DL, Heald R. Nuclear size is regulated by importin α and Ntf2 in Xenopus. Cell. 2010;143:288–98. doi: 10.1016/j.cell.2010.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14*.Vukovic LD, Jevtic P, Zhang Z, Stohr BA, Levy DL. Nuclear size is sensitive to NTF2 protein levels in a manner dependent on Ran binding. J Cell Sci. 2016;129:1115–1127. doi: 10.1242/jcs.181263. Levy and colleagues revealed that NTF2 negatively regulates nuclear size by reducing nuclear pore complex diameter and large lamin cargo import in Xenopus embryos and mammalian tissue culture cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hara Y, Iwabuchi M, Ohsumi K, Kimura A. Intranuclear DNA density affects chromosome condensation in metazoans. Mol Biol Cell. 2013;24:2442–53. doi: 10.1091/mbc.E13-01-0043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ladouceur AM, Dorn JF, Maddox PS. Mitotic chromosome length scales in response to both cell and nuclear size. J Cell Biol. 2015;209:645–652. doi: 10.1083/jcb.201502092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jevtic P, Levy DL. Nuclear Size Scaling during Xenopus Early Development Contributes to Midblastula Transition Timing. Curr Biol. 2014;25:45–52. doi: 10.1016/j.cub.2014.10.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Uppaluri S, Weber SC, Brangwynne CP. Hierarchical Size Scaling during Multicellular Growth and Development. Cell Rep. 2016;17:345–352. doi: 10.1016/j.celrep.2016.09.007. [DOI] [PubMed] [Google Scholar]
- 19.Hara Y, Merten CA. Dynein-Based Accumulation of Membranes Regulates Nuclear Expansion in Xenopus laevis Egg Extracts. Dev Cell. 2015;33:562–575. doi: 10.1016/j.devcel.2015.04.016. [DOI] [PubMed] [Google Scholar]
- 20.Edens LJ, Levy DL. cPKC regulates interphase nuclear size during xenopus development. J Cell Biol. 2014;206:473–483. doi: 10.1083/jcb.201406004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Edens LJ, Dilsaver MR, Levy DL. PKC-mediated phosphorylation of nuclear lamins at a single serine residue regulates interphase nuclear size in Xenopus and mammalian cells. Mol Biol Cell. 2017;28:1389–1399. doi: 10.1091/mbc.E16-11-0786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22*.Stephens AD, Banigan EJ, Adam SA, Goldman RD, Marko JF. Chromatin and lamin A determine two different mechanical response regimes of the cell nucleus. Mol Biol Cell. 2017;28:1984–1996. doi: 10.1091/mbc.E16-09-0653. Using novel micromanipulation methods, Stephens et al. identified the differential contributions of chromatin and lamin A/C to cell nuclear mechanical response. They showed that while chromatin governs resistance to small nuclear deformations, lamins provide robust mechanical response to greater nuclear deformations. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vuković LD, Jevtić P, Edens LJ, Levy DL. New Insights into Mechanisms and Functions of Nuclear Size Regulation. Int Rev Cell Mol Biol. 2016;322:1–59. doi: 10.1016/bs.ircmb.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 24.Jevtić P, Levy DL. Both Nuclear Size and DNA Amount Contribute to Midblastula Transition Timing in Xenopus laevis. Sci Rep. 2017;7:7908. doi: 10.1038/s41598-017-08243-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith ER, Meng Y, Moore R, Tse JD, Xu AG, Xu X-X. Nuclear envelope structural proteins facilitate nuclear shape changes accompanying embryonic differentiation and fidelity of gene expression. BMC Cell Biol. 2017;18:8. doi: 10.1186/s12860-017-0125-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26**.Ou HD, Phan S, Deerinck TJ, Thor A, Ellisman MH, O’Shea CC. ChromEMT: Visualizing 3D chromatin structure and compaction in interphase and mitotic cells. Science. 2017;357 doi: 10.1126/science.aag0025. The authors developed an electron tomography technique that enabled the 3D visualization of chromatin structure and compaction within cells. They showed that chromatin exists as flexible and disordered granular chains with a diameter of 5 to 24 nm packed together at different concentration densities in interphase nuclei and mitotic chromosomes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kieserman E, Heald R. Mitotic chromosome size scaling in Xenopus. Cell Cycle. 2011;10:3863–70. doi: 10.4161/cc.10.22.17975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hara Y, Adachi K, Kagohashi S, Yamagata K, Tanabe H, Kikuchi S, Okumura S-I, Kimura A. Scaling relationship between intra-nuclear DNA density and chromosomal condensation in metazoan and plant. Chromosom Sci. 2016;19:43–49. [Google Scholar]
- 29**.Ladouceur AM, Ranjan R, Smith L, Fadero T, Heppert J, Goldstein B, Maddox AS, Maddox PS. CENP-A and topoisomerase-II antagonistically affect chromosome length. J Cell Biol. 2017;216:2645–2655. doi: 10.1083/jcb.201608084. Generating a C. elegans strain with an exceptionally long chromosome, the authors screened for proteins whose nuclear localization is critical for mitotic chromosome scaling and identified CENP-A and topoisomerase-II as two potential holocentric chromosome scaling factors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maddox PS, Oegema K, Desai A, Cheeseman IM. “Holo”er than thou: chromosome segregation and kinetochore function in C. elegans. Chromosome Res. 2004;12:641–53. doi: 10.1023/B:CHRO.0000036588.42225.2f. [DOI] [PubMed] [Google Scholar]
- 31.Cuylen S, Blaukopf C, Politi AZ, Müller-Reichert T, Neumann B, Poser I, Ellenberg J, Hyman AA, Gerlich DW. Ki-67 acts as a biological surfactant to disperse mitotic chromosomes. Nature. 2016;535:308–312. doi: 10.1038/nature18610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Samwer M, Schneider MWG, Hoefler R, Schmalhorst PS, Jude JG, Zuber J, Gerlich DW. DNA Cross-Bridging Shapes a Single Nucleus from a Set of Mitotic Chromosomes. Cell. 2017;170:956–972.e23. doi: 10.1016/j.cell.2017.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shintomi K, Hirano T. The relative ratio of condensin I to II determines chromosome shapes service The relative ratio of condensin I to II determines chromosome shapes. Genes Dev. 2011;25:1464–1469. doi: 10.1101/gad.2060311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maresca TJ, Freedman BS, Heald R. Histone H1 is essential for mitotic chromosome architecture and segregation in Xenopus laevis egg extracts. J Cell Biol. 2005;169:859–869. doi: 10.1083/jcb.200503031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35**.Shintomi K, Inoue F, Watanabe H, Ohsumi K, Ohsugi M, Hirano T. Mitotic chromosome assembly despite nucleosome depletion in Xenopus egg extracts. Science. 2017;356:1284–1287. doi: 10.1126/science.aam9702. This work demonstrated that chromatid-like structures could assemble even in the absence of nucleosomes, suggesting that nucleosome assembly is not as essential as expected for mitotic chromosome assembly. [DOI] [PubMed] [Google Scholar]
- 36.Piskadlo E, Tavares A, Oliveira RA. Metaphase chromosome structure is dynamically maintained by condensin I-directed DNA (de)catenation. Elife. 2017;6:1–22. doi: 10.7554/eLife.26120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cowell IG, Papageorgiou N, Padget K, Watters GP, Austin Ca. Histone deacetylase inhibition redistributes topoisomerase IIβ from heterochromatin to euchromatin. Nucleus. 2011;2:61–71. doi: 10.4161/nucl.2.1.14194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mukhopadhyay D, Dasso M. The SUMO Pathway in Mitosis. Adv Exp Med Biol. 2017;963:171–184. doi: 10.1007/978-3-319-50044-7_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Crowder ME, Strzelecka M, Wilbur JD, Good MC, Von Dassow G, Heald R. A comparative analysis of spindle morphometrics across metazoans. Curr Biol. 2015;25:1542–1550. doi: 10.1016/j.cub.2015.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Loughlin R, Wilbur JD, McNally FJ, Nédélec FJ, Heald R. Katanin contributes to interspecies spindle length scaling in Xenopus. Cell. 2011;147:1397–407. doi: 10.1016/j.cell.2011.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Good MC, Vahey MD, Skandarajah A, Fletcher Da, Heald R. Cytoplasmic volume modulates spindle size during embryogenesis. Science. 2013;342:856–60. doi: 10.1126/science.1243147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hazel J, Krutkramelis K, Mooney P, Tomschik M, Gerow K, Oakey J, Gatlin JC. Changes in cytoplasmic volume are sufficient to drive spindle scaling. Science. 2013;342:853–6. doi: 10.1126/science.1243110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Farhadifar R, Baer CF, Valfort AC, Andersen EC, Müller-Reichert T, Delattre M, Needleman DJ. Scaling, selection, and evolutionary dynamics of the mitotic spindle. Curr Biol. 2015;25:732–740. doi: 10.1016/j.cub.2014.12.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chang JB, Ferrell JE. Mitotic trigger waves and the spatial coordination of the Xenopus cell cycle. Nature. 2013;500:603–7. doi: 10.1038/nature12321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mitchison TJ, Ishihara K, Nguyen P, Wühr M. Size scaling of microtubule assemblies in early xenopus embryos. Cold Spring Harb Perspect Biol. 2015;7 doi: 10.1101/cshperspect.a019182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schubert I, Oud JL. There is an upper limit of chromosome size for normal development of an organism. Cell. 1997;88:515–20. doi: 10.1016/s0092-8674(00)81891-7. [DOI] [PubMed] [Google Scholar]
- 47.Hudakova S, Künzel G, Endo TR, Schubert I. Barley chromosome arms longer than half of the spindle axis interfere with nuclear divisions. Cytogenet Genome Res. 2002;98:101–7. doi: 10.1159/000068530. [DOI] [PubMed] [Google Scholar]
- 48.Mora-Bermúdez F, Gerlich D, Ellenberg J. Maximal chromosome compaction occurs by axial shortening in anaphase and depends on Aurora kinase. Nat Cell Biol. 2007;9:822–831. doi: 10.1038/ncb1606. [DOI] [PubMed] [Google Scholar]
- 49.Lipp JJ, Hirota T, Poser I, Peters J-M. Aurora B controls the association of condensin I but not condensin II with mitotic chromosomes. J Cell Sci. 2007;120:1245–1255. doi: 10.1242/jcs.03425. [DOI] [PubMed] [Google Scholar]
- 50**.Kyogoku H, Kitajima TS. Large Cytoplasm Is Linked to the Error-Prone Nature of Oocytes. Dev Cell. 2017;41:287–298.e4. doi: 10.1016/j.devcel.2017.04.009. This study showed that cytoplasmic size affects the function of the spindle and the stringency of the spindle assembly checkpoint in mouse oocytes. Greater cytoplasmic volume increases chromosome segregation errors and aneuploidy in female meiosis. [DOI] [PubMed] [Google Scholar]
- 51*.Galli M, Morgan DO. Cell Size Determines the Strength of the Spindle Assembly Checkpoint during Embryonic Development. Dev Cell. 2016;36:344–52. doi: 10.1016/j.devcel.2016.01.003. Galli and Morgan showed that the kinetochore-to-cytoplasm ratio determines spindle assembly checkpoint strength during C. elegans embryogenesis. After each round of division, the checkpoint strength increases due to decreasing cell size. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52*.Chen J, Liu J. Spindle Size Scaling Contributes to Robust Silencing of Mitotic Spindle Assembly Checkpoint. Biophys J. 2016;111:1064–1077. doi: 10.1016/j.bpj.2016.07.039. This work used mathematical modelling to demonstrate that robust and timely spindle assembly checkpoint silencing entails proper size scaling of mitotic spindle. [DOI] [PMC free article] [PubMed] [Google Scholar]


