Abstract
Polyamines such as putrescine, spermidine, and spermine are small aliphatic cations that serve myriad biological functions in all forms of life. While polyamine biosynthesis and cellular trafficking pathways are generally well defined, it is only recently that the molecular basis of reversible polyamine acetylation has been established. In particular, enzymes that catalyze polyamine deacetylation reactions have been identified and structurally characterized: histone deacetylase 10 (HDAC10) from Homo sapiens and Danio rerio (zebrafish) is a highly specific N8-acetylspermidine deacetylase, and its prokaryotic counterpart, acetylpolyamine amidohydrolase (APAH) from Mycoplana ramosa, is a broad-specificity polyamine deacetylase. Similar to the greater family of HDACs, which mainly serve as lysine deacetylases, both enzymes adopt the characteristic arginase-deacetylase fold and employ a Zn2+-activated water molecule for catalysis. In contrast with HDACs, however, the active sites of HDAC10 and APAH are sterically constricted to enforce specificity for long, slender polyamine substrates and exclude bulky peptides and proteins containing acetyl-L-lysine. Crystal structures of APAH and D. rerio HDAC10 reveal that quaternary structure, i.e., dimer assembly, provides the steric constriction that directs the polyamine substrate specificity of APAH, whereas tertiary structure – a unique 310 helix defined by the P(E,A)CE motif – provides the steric constriction that directs the polyamine substrate specificity of HDAC10. Given the recent identification of HDAC10 and spermidine as mediators of autophagy, HDAC10 is rapidly emerging as a biomarker and target for the design of isozyme-selective inhibitors that will suppress autophagic responses to cancer chemotherapy, thereby rendering cancer cells more susceptible to cytotoxic drugs.
Graphical abstract

Introduction
Polyamines comprise a diverse array of small aliphatic polycations essential for function in all domains of life,1-4 and common polyamines such as putrescine, spermidine, and spermine derive from amino acid catabolism (Figure 1).5-8 The 4-carbon diamine putrescine is generated through the decarboxylation of L-ornithine, as catalyzed by ornithine decarboxylase; L-ornithine is generated through the hydrolysis of L-arginine by the urea cycle enzyme arginase. Alternatively, L-arginine can be converted into agmatine via decarboxylation by arginine decarboxylase, after which agmatine is hydrolyzed by agmatinase to form putrescine. Until somewhat recently, this alternative pathway was thought to exist only in bacteria, plants, and certain lower species;1,9 however, recent studies show this pathway also exists in certain mammalian cell types.4,10-12
Figure 1. Eukaryotic and prokaryotic polyamine metabolism.

ADC, arginine decarboxylase; APAO, N1-acetylpolyamine oxidase; APAH, acetylpolyamine amidohydrolase; HDAC10, histone deacetylase 10; ODC, ornithine decarboxylase; P/CAF, P300/CBP associated factor; SMOX, spermine oxidase; SMS, spermine synthase; SRM, spermidine synthase; SSAT, spermidine/spermine acetyltransferase.
Spermidine synthase catalyzes the reaction of putrescine with the 3-carbon aminopropyl group of S-adenosylmethioninamine (generated by S-adenosylmethionine decarboxylase) to form the asymmetric triamine spermidine. In turn, spermidine acquires a second aminopropyl group to form spermine in a reaction catalyzed by spermine synthase. A notable difference between polyamine metabolism in prokaryotes and eukaryotes is that spermine is absent or rare in prokaryotes.13-15 Also notable is that the biosynthetic intermediate N8-acetylspermidine is generated in the nucleus of eukaryotic cells,16 and that the histone acetyltransferase P/CAF is responsible for this activity.17 Once generated, N8-acetylspermidine is exported to the cytoplasm, where it can be deacetylated and recycled by transport back into the nucleus.16,18,19
Typically present at millimolar concentrations in the cell, polyamines serve myriad chemical and biological functions. For example, polyamines influence the stabilization, condensation, and packaging of DNA,20-23 the stabilization and modulation of RNA structure,24-27 transcriptional and translational regulation,28-30 posttranscriptional regulation,31,32 and cell cycle progression.2,30,33 Because polyamines are positively charged at physiological pH, they often associate with negatively charged sites on proteins, phospholipids, and nucleic acids to exert their various functions.
Structure and catalytic mechanism of polyamine deacetylases
Many enzymes of prokaryotic and eukaryotic polyamine metabolism are well characterized in terms of their structure and function. For example, crystal structures of arginase,34-36 ornithine decarboxylase,37-39 spermidine synthase,40,41 spermine synthase,42 a broad-specificity plant polyamine oxidase,43 and a mammalian polyamine oxidase similar in activity to spermidine oxidase44 have illuminated molecular details of catalysis. Among the enzymes of prokaryotic and eukaryotic metabolism, however, structure-function relationships for polyamine deacetylases have remained enigmatic until recent years. The broad-specificity prokaryotic enzyme, acetylpolyamine amidohydrolase (APAH) from Mycoplana ramosa, was first discovered 30 years ago,45,46 but its crystal structure was not reported until 2011 by Lombardi and colleagues.47 Eukaryotic polyamine deacetylase activity was discovered 40 years ago18,19 and was characterized by strict substrate specificity for N8-acetylspermidine hydrolysis.48 However, the enzyme responsible for this activity was not identified. This deacetylase activity was distinct from that of eukaryotic histone deacetylases (HDACs) that catalyze the deacetylation of acetyl-L-lysine residues in histone proteins.49 In 2011, Lombardi and colleagues47 predicted that one of the two cytosolic class IIb HDACs, HDAC6 or HDAC10, might be responsible for the cytosolic N8-acetylspermidine deacetylation activity observed several decades earlier.18,19 In 2017, the N8-acetylspermidine deacetylase was identified as HDAC10 based on structural and functional studies of HDAC10 from Homo sapiens (human) and Danio rerio (zebrafish) reported by Hai and colleagues.50
The crystal structure of APAH47 revealed an α/β fold identical to that of histone deacetylases, which in turn resembled the unique α/β fold first identified in rat liver arginase34 and subsequently observed in a bacterial histone deacetylase-like protein (Figure 2).51 Accordingly, this α/β fold is designated the arginase-deacetylase fold. Even though the amino acid sequences of arginases and metal-dependent deacetylases have diverged to identities generally less than 20%,50 the binding site of the catalytic Zn2+ ion in the deacetylases is conserved as the Mn2+B binding site in the binuclear manganese cluster of the arginases.52-54
Figure 2. The arginase-deacetylase fold.
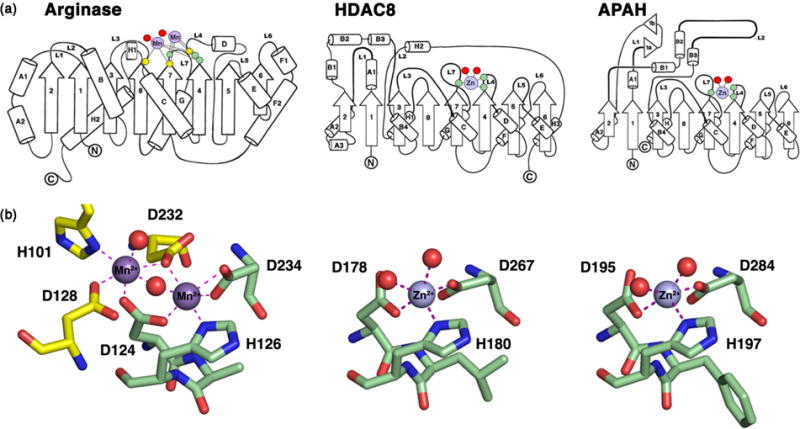
(a) Arginase, HDAC8, and APAH share a conserved α/β fold consisting of a central 8-stranded parallel β-sheet flanked by a-helices. Residues that coordinate to catalytic Mn2+ ions in arginase are located on loops L3, L4, and L7 (yellow and green circles), and residues that coordinate to the catalytic Zn2+ ion in HDAC8 and APAH are conserved on loops L4 and L7 (green circles). Red circles signify metal-bound water molecules or hydroxamate inhibitor atoms. (b) The Mn2+B site of arginase is conserved in HDACs and APAH as D(A,V,L,F)HX~100D (boldface indicates metal ligands). The Mn2+A site of arginase is not conserved in the deacetylase family. Reprinted from ref. 54, copyright 2011, with permission from Elsevier.
In addition to the histidine and two aspartate residues that coordinate to the catalytic Zn2+ ion, key residues conserved in the active sites of prokaryotic APAH and eukaryotic HDAC10 include tandem histidine residues and a tyrosine residue. These residues are also conserved in the active site of the related class I deacetylase HDAC8 (Figure 2), which is perhaps the most-studied deacetylase in terms of structure-mechanism relationships. It is likely that the chemical mechanism of amide bond hydrolysis is generally identical for HDAC8, APAH, and HDAC10. Interestingly, HDAC8 can utilize either Zn2+ or Fe2+ for catalysis,55 so it is possible that the polyamine deacetylases can similarly function with alternative metal ions. To this end, maximal catalytic activity is measured for APAH with Mn2+, perhaps as a vestige of its evolutionary relationship with the Mn2+-dependent arginases.46
In the catalytic mechanism of HDAC8, APAH, and HDAC10 (Figure 3), the scissile carbonyl of the substrate coordinates to Zn2+ and also accepts a hydrogen bond from the catalytic tyrosine in the precatalytic enzyme-substrate complex. Structural evidence for this binding mode was first provided by crystal structures of the catalytically inactive Y306F and H143A HDAC8 variants complexed with an intact substrate.56,57 Similarly, the amide carbonyl of N8-acetylspermidine coordinates to Zn2+ and accepts a hydrogen bond from Y323 in the active site of H159A APAH (Figure 4a).47 Thus, both Zn2+ coordination and a hydrogen bond with the catalytic tyrosine are required to polarize the scissile carbonyl for nucleophilic attack by a Zn2+-bound water molecule in metal-dependent deacetylases.
Figure 3. Proposed mechanism of acetylpolyamine hydrolysis.
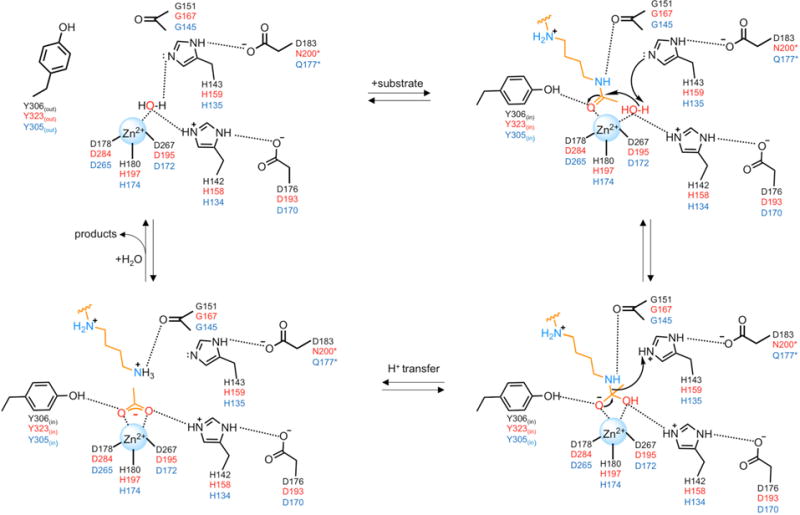
Residue labels for HDAC8, APAH, and HDAC10 are color-coded black, red, and blue, respectively. Note that for APAH and HDAC10, the substrate is N8-acetylspermidine as shown; for HDAC8, the substrate would be an acetyl-L-lysine residue on a peptide or protein substrate (not shown). Also note that the residue that hydrogen bonds with the second of the tandem histidine residues differs in each of these enzymes. This feature may influence the chemical function of this histidine residue in general base-general acid catalysis.
Figure 4. Stereoviews of polyamine deacetylase complexes.

(a) Structure of the H159A APAH-N8-acetylspermidine complex reveals that the substrate carbonyl group coordinates to Zn2+ and accepts a hydrogen bond from Y323in (the Y323out conformer is not shown for clarity). Reprinted from Ref. 63. Copyright 2015 American Chemical Society. (b) Structure of the APAH-AAT complex reveals the binding of the trifluoromethylketone as a gem-diolate that mimics the tetrahedral intermediate and flanking transition states for the hydrolysis of N8-acetylspermidine. Reprinted from Ref. 63. Copyright 2015 American Chemical Society. (c) Structure of the HDAC10-AAT complex similarly reveals the binding of the trifluoromethylketone as a gem-diolate transition state analogue. E274 plays a critical electrostatic role in enzyme-substrate recognition, and the P(E,A)CE motif (purple) sterically constricts the active site to favor the binding of the slender acetylpolyamine substrate. Reprinted from ref. 50.
Interestingly, the catalytic tyrosine adopts two different conformations designated “in” and “out” in the APAH-substrate complex: Y323in hydrogen bonds with the substrate and Y323out is oriented toward solvent.47 This suggests the possibility of induced-fit substrate binding in the deacetylase active site. Consistent with this possibility, the catalytic tyrosine is preceded by a glycine-rich segment, conserved in deacetylases as GGY or GGGGY, that may facilitate conformational flexibility. Conformational mobility is observed for this segment in molecular dynamics simulations of HDAC8; moreover, the substitution of alanine residues for these glycine residues, which would hinder conformational mobility, compromises catalytic activity.58
Nucleophilic attack of Zn2+-bound water at the amide carbonyl group polarized by Zn2+ and the catalytic tyrosine is facilitated by a general base (Figure 3). In HDAC8, H143 is a single general acid-general base, and H142 serves as an electrostatic catalyst that stabilizes the resulting tetrahedral intermediate.59,60 However, structural studies of the related isozyme HDAC6, in which the intact substrate and tetrahedral intermediate are trapped in the active sites of the Y745F and H574A variants, respectively, leave open the possibility that the first histidine of the tandem pair (H573) serves as a general base and the second histidine (H574) serves as a general acid.61 Possibly, the function of the second histidine in the tandem pair is modulated by different hydrogen bond partners in different deacetylases (Figure 3).
The oxyanion of the resulting tetrahedral intermediate is stabilized by coordination to Zn2+ and hydrogen bonding with the catalytic tyrosine. The trifluoromethylketone analogue of N8-acetylspermidine, 7-[(3-aminopropyl)amino]-1,1,1-trifluoroheptan-2-one (AAT),62 binds as a tetrahedral gem-diolate to both APAH and HDAC10, thereby mimicking the tetrahedral intermediate and its flanking transition states in catalysis (Figure 4b,c).50,63 These crystal structures show that the Zn2+ ion, tyrosine, and tandem histidine residues contribute to transition state stabilization in each deacetylase. Finally, collapse of the tetrahedral intermediate requires a proton donor, and the second histidine of the tandem pair must serve as the general acid due to its proximity to the leaving amino group (Figure 3).
Structural determinants of polyamine substrate specificity
APAH functions as homodimer with broad substrate specificity toward both small and large acetylpolyamines.45,46 Crystal structures of the intact substrates acetylspermine or N8-acetylspermidine bound in the active site of catalytically inactive H159A APAH reveal key intermolecular interactions with substrate amino groups that account for the molecular recognition of polyamine substrates (the structure of the complex with N8-acetylspermidine is shown in Figure 4a).47 In addition to the hydrogen bond between the side chain of catalytic tyrosine Y323 and the scissile amide carbonyl described in the previous section, the amide NH group of the polyamine substrate donates a hydrogen bond to the backbone carbonyl group of G167. The N4 amino group engages in a cation-π interaction with the aromatic side chain of F225 and forms a hydrogen bond (acetylspermine) or electrostatic interaction (N8-acetylspermidine) with E117. Additionally, the N1 amino group of N8-acetylspermidine donates a hydrogen bond to the carboxylate group of E106 in the adjacent monomer of the APAH dimer. A comparable interaction is observed for the N9 amino group of acetylspermine. Both E106 and E117 undergo substantial conformational changes to accommodate substrate binding. Finally, the N12 amino group of acetylspermine donates a hydrogen bond to the side chain of Y19 in the complex with H159A APAH.
In contrast with APAH, HDAC10 functions as a monomer with much narrower substrate specificity (Figure 5a,b).50 HDAC10 preferentially catalyzes the deacetylation of N8-acetylspermidine in addition to the smaller substrates acetylputrescine and acetylcadaverine; HDAC10 does not appreciably catalyze the deacetylation of N1-acetylspermidine or acetylspermine. The crystal structure of D. rerio (zebrafish) Y307F HDAC10 complexed with the trifluoromethylketone AAT reveals critical insight regarding strict substrate specificity for N8-acetylspermidine (Figure 4c).50 First, in contrast with lysine deacetylases such as the related class II isozyme HDAC6, the active site of HDAC10 is surrounded by a surface of negative electrostatic potential resulting from several aspartate and glutamate residues highly conserved among HDAC10 orthologs. This feature likely facilitates the recruitment of cationic acetylpolyamine substrates. However, the electrostatic surface ultimately becomes positively charged at the base of the active site, closer to the catalytic Zn2+ ion. This could disfavor the binding of N1-acetylspermidine relative to N8-acetylspermidine, since the positively charged N4 amino group of N1-acetylspermidine would be closer to this cationic surface.
Figure 5. Catalytic activity of human HDAC10 (hHDAC10) and zebrafish HDAC10 (zHDAC10).

(a,b) Steady-state kinetics measured using acetylpolyamines and acetyl-L-lysine (K(Ac)) peptides reveal a clear preference for N8-acetylspermidine and acetylputrescine hydrolysis, with little to no acetyl-L-lysine peptide hydrolysis. Abbreviations: AcPUT, acetylputrescine; N8-AcSPD, N8-acetylspermidine; AcCAD, acetylcadaverine; AcDAO, N-(aminooctyl)acetamide; N1,N8-diAcSPD, N1,N8 –diacetylspermidine; N1-AcSPD, N1-acetylspermidine; N1-AcSPM, N1-acetylspermine; AcPAD, N-(3-aminopropyl)acetamide; BTA, N-butylacetamide. (c) Ratio of catalytic efficiencies (kcat/KM) for polyamine deacetylase (PDAC) activity measured with N8-acetylspermidine and acetyl-L-lysine deacetylase (HDAC) activity measured with RGK(ac)-AMC (AMC = aminomethylcoumarin). APAH, hHDAC10, and zHDAC10 exhibit a clear catalytic preference for PDAC activity. The E274L mutation converts zHDAC10 from a PDAC into an HDAC, and the zHDAC10 ΔηA2 mutant is a bifunctional PDAC-HDAC. Abbreviations: zHDAC10Δ, proteolytically nicked zHDAC10 used for crystal structure determination; hHDAC6 CD12, human HDAC6 construct containing both catalytic domains; zHDAC6 CD1 or CD2, zebrafish HDAC6 catalytic domain 1 or catalytic domain 2. Reprinted from ref. 50.
Second, the side chain of E274 serves as a gatekeeper in the HDAC10 active site. This residue is strictly conserved in HDAC10 orthologues; within the greater HDAC family, this residue appears mainly as a leucine. In HDAC10, the negatively charged carboxylate of E274 confers specificity toward positively charged polyamine substrates. While the E274 carboxylate is not sufficiently close to form a hydrogen bond with the N4 amino group of AAT (3.7 Å; Figure 4c), it nonetheless makes a favorable electrostatic interaction. Moreover, the E274L substitution converts HDAC10 from an N8-acetylspermidine deacetylase into a lysine deacetylase (Figure 5c), so this electrostatic interaction is critical for substrate specificity.50
Finally, both APAH and HDAC10 share key active site residues found in lysine deacetylases (i.e., HDACs) required for the chemistry of amide hydrolysis. However, neither APAH nor HDAC10 exhibit appreciable lysine deacetylase activity with peptide substrates (Figure 5c).47,50 Crystal structures reveal that the active sites of APAH and HDAC10 are sterically constricted so as to favor the binding of long, slender acetylpolyamine substrates, thereby disfavoring the binding of sterically bulky acetyl-L-lysine peptides. The steric constriction of the APAH active site results from dimer assembly, such that the approach to the active site is made through a narrow “L-shaped” tunnel (Figure 6a). In contrast, the steric constriction of the HDAC10 active site results from the unique 310-helix ηA2. Compared with the CD2 domain of HDAC6, the ηA2 helix of HDAC10 is formed by a two-residue insertion and a two-residue substitution in the L1 loop, conserved in HDAC10 orthologs as P(E,A)CE (Figure 4c, Figure 6b). Deletion of this segment diminishes polyamine deacetylase activity and enhances lysine deacetylase activity in the zHDAC10 ΔηA2 construct (Figure 5c).50 Thus, the strict substrate specificity exhibited by HDAC10 for N8-acetylspermidine that excludes acetyl-L-lysine peptide substrates is rooted in the electrostatic effects mediated by gatekeeper E274 as well as the steric constriction of the active site introduced by the P(E,A)CE motif. As APAH and HDAC10 are compared, it is interesting to note that the steric constriction of their respective active sites is mediated by quaternary structure in the prokaryotic enzyme and by tertiary structure in the eukaryotic enzyme.
Figure 6. Structural determinants of polyamine substrate specificity.

(a) Assembly of the APAH dimer constricts the approach to the active site through an “L”-shaped tunnel indicated by red dotted line. Reprinted from ref. 47. Copyright 2011 American Chemical Society. (b) Stereoview showing the superposition of the polyamine deacetylase domain of HDAC10 with HDAC6 catalytic domains CD1 and CD2. The P(E,A)CE motif (purple) is conserved in HDAC10 orthologs and constricts the approach to the active site, as indicated by the binding of the transition state analogue AAT (stick figure). A close-up view of the P(E,A)CE motif is also visible in Figure 4c. Reprinted from Ref. 50.
Structural aspects of domain assembly in class IIb HDACs
Both HDAC10 and HDAC6 are unique among the greater family of metal-dependent HDACs in that they are the only isozymes that contain two deacetylase domains. In HDAC6, catalytic domains 1 and 2 (CD1 and CD2) are catalytically active, although they exhibit different substrate specificities.61 In HDAC10, only one domain is active as a polyamine deacetylase (PDAC); although the second domain adopts the characteristic α/β arginase-deacetylase fold, it is approximately 100 residues smaller and lacks all residues important for metal binding and catalysis.50 This catalytically inactive domain is thus designated as a pseudo-deacetylase domain (ΨDAC). The YDAC domain exhibits greater sequence divergence across HDAC10 isozymes from different species, and it is implicated in directing the cytoplasmic enrichment of this isozyme.64
Comparisons of the crystal structures of an intact CD1-CD2 construct of zebrafish HDAC6 and a proteolytically nicked yet fully assembled PDAC-YDAC construct of zebrafish HDAC10 reveal a common mode of domain-domain assembly (Figure 7).50,65 The CD1 and CD2 domains of HDAC6 assemble with butterfly-like architecture such that their active sites are oriented away from the domain-domain interface and separated by approximately 50 Å.65 The central feature of domain-domain assembly is a 4-helix bundle involving H13/14 and H15 of CD1, and H32/33 and H34 of CD2; these helices are perpendicular to a pseudo-twofold symmetry axis between CD1 and CD2. Additionally contributing to the domain-domain interface are H17, H18, and the H17-H18 loop of CD1, which interact with the corresponding segments of CD2 (H36, H37, and the H36-H37 loop). The CD1-CD2 interdomain linker and the C-terminus of CD2 also contribute to the domain-domain interface. In total, approximately 2,100 Å2 protein surface area is buried at the domain-domain interface, and this interface is defined by van der Waals interactions between nonpolar residues as well as residues that form inter-domain hydrogen bonds.
Figure 7. Domain architecture of class IIb HDACs.

(a) The HDAC6 structure reported by Miyake and colleagues65 (PDB 5G0J) consists of two catalytically active domains, CD1 (cyan) and CD2 (mauve), connected by an interdomain linker (green). Active sites are indicated by red arrows. Helices that mediate domain-domain association are indicated. (b) The HDAC10 structure reported by Hai and colleagues50 (PDB 5TD7) consists of a catalytically active polyamine deacetylase domain (PDAC, blue) and a catalytically inactive pseudo-deacetylase domain (ΨDAC, green). The active site in the PDAC domain is indicated by the binding of transition state analogue AAT (stick figure). The interdomain linker is proteolytically nicked and is not observed in the crystal structure.
Similar to the CD1 and CD2 domains of HDAC6, the PDAC and YDAC domains of HDAC10 assemble with overall butterfly-like architecture, burying approximately 1,700 Å2 surface area; a pseudo-twofold symmetry axis runs perpendicular to a central 4-helix bundle defined by helices HaF and HaG of the PDAC domain and the corresponding helices of the ΨDAC domain.50 These helices are topologically identical to those that define the central 4-helix bundle at the CD1-CD2 interface in HDAC6. Intriguingly, the conserved architecture observed for CD1-CD2 assembly in HDAC6 and PDAC-ΨDAC assembly in HDAC10 may originate from an ancestral HDAC, as exemplified by the recent crystal structure determination of homodimeric Clr3 HDAC from Schizosaccharomyces pombe.66 The interdomain linker is dispensable for the assembly and catalytic activity of HDAC10 in vitro, since the proteolytically nicked enzyme is fully active (Figure 5c).65
HDAC10 as a target for cancer chemotherapy
Neuroblastoma is the most common childhood extracranial tumor and is a major cause of cancer-related deaths in children.67 Of the 11 metal-dependent HDACs, only HDAC10 expression levels significantly correlate with poor overall survival in neuroblastoma patients, and HDAC10 expression levels can also be used to predict recurrence and survival in medulloblastoma patients.68 Thus, HDAC10 may be useful as a biomarker to predict clinical outcomes for advanced-stage cancer patients diagnosed with these highly malignant tumors. Further experiments show that HDAC10 promotes autophagy-mediated cell survival in cultured neuroblastoma cells treated with the cytotoxic chemotherapy drug Doxorubicin (Figure 8a), possibly through a mechanism involving Hsp70 family proteins.68 Additionally, HDAC10 has been shown to promote angiogenesis, a crucial physiological process that supports tumor growth.69 Genetic knockdown or chemical inhibition of HDAC10 disrupts HDAC10-mediated autophagy and angiogenesis,68,69 so this HDAC isozyme plays a critical regulatory role in these cellular functions.
Figure 8. HDAC10 as a mediator of autophagy.

(a) As reported by Oehme and colleagues,68 BE(2)-C human neuroblastoma cells were stably transfected with short hairpin RNAs targeting HDAC10 (shR-1, -2, -3, -4) or a negative control (shR-NC). Cells were treated with 0.05 μg/mL doxorubicin (a cytotoxic cancer chemotherapy drug) or normal culture medium. After 10 days, colonies were stained and results were quantified (bar diagram; shR-NC, open bars; shR-HDAC10, filled bars). The Western Blot shows HDAC10 expression in stably transfected cells, and β-actin was used as a loading control. Knockdown of HDAC10 clearly enhances the cytotoxicity of doxorubicin relative to the negative control treated with shR-NC. Reprinted with permission from Ref. 68. (b) Summary of proposed roles of HDAC10 and N8-acetylspermidine in promoting autophagy. A selective inhibitor of HDAC10 (HDACi) will blunt the autophagic response to cytotoxic cancer drugs such as doxorubicin and thus render cancer cells more susceptible to chemotherapy.
It is intriguing to consider that the molecular function of HDAC10 as an N8-acetylspermidine deacetylase in vitro might be linked to its biological role as a mediator of autophagy in vivo. Notably, increased cellular spermidine levels induce autophagy and extend the lifespan of a variety of cell types.70,71 We speculate that the spermidine generated by HDAC10 may be implicated in this process, although it is presently unclear as to how HDAC10-derived spermidine would be trafficked so as to influence the autophagy program. Even so, it is interesting to note that inhibition of ornithine decarboxylase (Figure 1) with 2-difluoromethylornithine (DFMO) reduces cellular polyamine levels and suppresses autophagy, additionally implicating a connection between polyamine metabolism and autophagy.72 Indeed, DFMO is in clinical trials for the treatment of neuroblastoma.73-76 HDAC10 is also implicated in cell cycle progression,77 DNA mismatch repair,78,79 homologous recombination,80 and melanogenesis,81 so its function as a polyamine deacetylase and a mediator of autophagy may be linked to multiple biological functions.
Concluding Remarks
Recent studies have expanded our understanding of the structural biology and chemistry of polyamine metabolism in prokaryotes and eukaryotes. While reversible polyamine acetylation has remained the most enigmatic leg of eukaryotic polyamine metabolism for many years, the identification of P/CAF as an N8-acetylspermidine acetyltransferase13 and HDAC10 as an N8-acetylspermidine deacetylase50 completes the picture of eukaryotic polyamine metabolism as summarized in Figure 1. The strict substrate specificity of HDAC10 for N8-acetylspermidine contrasts with the broad polyamine substrate specificity of the prokaryotic deacetylase APAH, and the structural basis for polyamine substrate specificity appears to be rooted in hydrogen bond and/or electrostatic interactions in each enzyme active site. Neither APAH nor HDAC10 appreciably catalyze the deacetylation of canonical acetyl-L-lysine peptides, and structural analysis reveals that the active site of each enzyme is sterically constricted so as to favor the binding of long, slender polyamine substrates instead of bulkier peptide substrates. This steric constriction is achieved by quaternary structure in APAH and by tertiary structure in HDAC10.
It is interesting to note that myriad histone and non-histone protein substrates have been identified for HDACs in recent years, including substrates with various covalent modifications to lysine, e.g., fatty acid acylation, crotonylation, etc.82-84 Furthermore, this enzyme family has evolved with impressively varied specificity for protein and non-protein amide substrates. Accordingly, this diverse family of metal-dependent deacetylases is better described more generally as the amidohydrolase family. HDAC10 and APAH are remarkable members of this family in that they catalyze the deacetylation of non-lysine, non-protein small molecule substrates.
HDAC10 is implicated in multiple cellular processes and promotes cancer cell survival, as recently reported for ovarian cancer,85 and the cellular basis of this activity likely resides in the function of HDAC10 as an mediator of autophagy (Figure 8b).68 Thus, HDAC10 is an emerging new target for drug design, since inhibition or knockdown renders cancer cells more susceptible to cytotoxic chemotherapy drugs.68,85 It is perhaps not a coincidence that the product of the HDAC10-catalyzed reaction, spermidine, is also implicated as an inducer of autophagy and is also being studied as a caloric restriction mimetic in the treatment of age-associated disorders.86 With the crystal structure of HDAC10 now available,50 the structure-based design of isozyme-specific inhibitors promises to yield valuable new leads for HDAC10-targeted therapeutics.
Acknowledgments
This work was supported by NIH grant GM49758 to D.W.C. and by NIH grant F32GM125141 to S.A.S.
References
- 1.Tabor CW, Tabor H. Polyamines. Annu Rev Biochem. 1984;53:749–790. doi: 10.1146/annurev.bi.53.070184.003533. [DOI] [PubMed] [Google Scholar]
- 2.Thomas T, Thomas TJ. Polyamines in cell growth and cell death: molecular mechanisms and therapeutic applications. Cell Mol Life Sci. 2001;58:244–258. doi: 10.1007/PL00000852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Casero RA, Jr, Pegg AE. Polyamine catabolism and disease. Biochem J. 2009;421:323–338. doi: 10.1042/BJ20090598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Michael AJ. Polyamines in eukaryotes, bacteria, and archaea. J Biol Chem. 2016;291:14896–14903. doi: 10.1074/jbc.R116.734780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Michael AJ. Evolution of biosynthetic diversity. Biochem J. 2017;474:2277–2299. doi: 10.1042/BCJ20160823. [DOI] [PubMed] [Google Scholar]
- 6.Miller-Fleming L, Olin-Sandoval V, Campbell K, Ralser M. Remaining mysteries of molecular biology: the role of polyamines in the cell. J Mol Biol. 2015;427:3389–3406. doi: 10.1016/j.jmb.2015.06.020. [DOI] [PubMed] [Google Scholar]
- 7.Kahana C. Protein degradation, the main hub in the regulation of cellular polyamines. Biochem J. 2016;473:4551–4558. doi: 10.1042/BCJ20160519C. [DOI] [PubMed] [Google Scholar]
- 8.Michael AJ. Biosynthesis of polyamines and polyamine-containing molecules. Biochem J. 2016;473:2315–2329. doi: 10.1042/BCJ20160185. [DOI] [PubMed] [Google Scholar]
- 9.Satishchandran C, Boyle SM. Purification and properties of agmatine ureohydrolyase, a putrescine biosynthetic enzyme in Escherichia coli. J Bacteriol. 1986;165:843–848. doi: 10.1128/jb.165.3.843-848.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horyn O, Luhovyy B, Lazarow A, Daikhin Y, Nissim I, Yudkoff M, Nissim I. Biosynthesis of agmatine in isolated mitochondria and perfused rat liver: studies with 15N-labelled arginine. Biochem J. 2005;388:419–425. doi: 10.1042/BJ20041260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iyo AH, Zhu MY, Ordway GA, Regunathan S. Expression of arginine decarboxylase in brain regions and neuronal cells. J Neurochem. 2006;96:1042–1050. doi: 10.1111/j.1471-4159.2005.03544.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang X, Ying W, Dunlap KA, Lin G, Satterfield MC, Burghardt RC, Wu G, Bazer FW. Arginine decarboxylase and agmatinase: an alternative pathway for de novo biosynthesis of polyamines for development of mammalian conceptuses. Biol Reprod. 2014;90:84. doi: 10.1095/biolreprod.113.114637. [DOI] [PubMed] [Google Scholar]
- 13.Bachrach U. Metabolism and function of spermine and related polyamines. Annu Rev Microbiol. 1970;24:109–134. doi: 10.1146/annurev.mi.24.100170.000545. [DOI] [PubMed] [Google Scholar]
- 14.Tabor CW, Tabor H. Polyamines in microorganisms. Microbiol Rev. 1985;49:81–99. doi: 10.1128/mr.49.1.81-99.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shah P, Swiatlo E. A multifaceted role for polyamines in bacterial pathogens. Mol Microbiol. 2008;68:4–16. doi: 10.1111/j.1365-2958.2008.06126.x. [DOI] [PubMed] [Google Scholar]
- 16.Libby PR. Rat liver nuclear N-acetyltransferases: separation of two enzymes with both histone and spermidine acetyltransferase activity. Arch Biochem Biophys. 1980;203:384–389. doi: 10.1016/0003-9861(80)90190-3. [DOI] [PubMed] [Google Scholar]
- 17.Burgio G, Corona DF, Nicotra CM, Carruba G, Taibi G. P/CAF-mediated spermidine acetylation regulates histone acetyltransferase activity. J Enzyme Inhib Med Chem. 2016;31:75–82. doi: 10.1080/14756366.2016.1205045. [DOI] [PubMed] [Google Scholar]
- 18.Blankenship J. Deacetylation of N8-acetylspermidine by subcellular fractions of rat tissue. Arch Biochem Biophys. 1978;189:20–27. doi: 10.1016/0003-9861(78)90109-1. [DOI] [PubMed] [Google Scholar]
- 19.Libby PR. Properties of an acetylspermidine deacetylase from rat liver. Arch Biochem Biophys. 1978;188:360–363. doi: 10.1016/s0003-9861(78)80020-4. [DOI] [PubMed] [Google Scholar]
- 20.Thomas TJ, Thomas T. Collapse of DNA in packaging and cellular transport. Int J Biol Macromol. 2017;109:36–48. doi: 10.1016/j.ijbiomac.2017.12.076. [DOI] [PubMed] [Google Scholar]
- 21.Iacomino G, Picariello G, D’Agostino L. DNA and nuclear aggregates of polyamines. Biochim Biophys Acta. 2012;1823:1745–1755. doi: 10.1016/j.bbamcr.2012.05.033. [DOI] [PubMed] [Google Scholar]
- 22.Bloomfield VA. DNA condensation by multivalent cations. Biopolymers. 1997;44:269–282. doi: 10.1002/(SICI)1097-0282(1997)44:3<269::AID-BIP6>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 23.Yoo J, Aksimentiev A. The structure and intermolecular forces of DNA condensates. Nucleic Acids Res. 2016;44:2036–2046. doi: 10.1093/nar/gkw081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trachman RJ, III, Draper DE. Divalent ion competition reveals reorganization of an RNA ion atmosphere upon folding. Nucleic Acids Res. 2017;45:4733–4742. doi: 10.1093/nar/gkw1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Igarashi K, Kashiwagi K. Polyamine modulon in Escherichia coli: genes involved in the stimulation of cell growth by polyamines. J Biochem. 2006;139:11–16. doi: 10.1093/jb/mvj020. [DOI] [PubMed] [Google Scholar]
- 26.Katz AM, Tolokh IS, Pabit SA, Baker N, Onufriev AV, Pollack L. Spermine condenses DNA, but not RNA duplexes. Biophys J. 2017;112:22–30. doi: 10.1016/j.bpj.2016.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oguro A, Yanagida A, Fujieda Y, Amano R, Otsu M, Sakamoto T, Kawai G, Matsufuji S. Two stems with different characteristics and an internal loop in an RNA aptamer contribute to spermine-binding. J Biochem. 2017;161:197–206. doi: 10.1093/jb/mvw062. [DOI] [PubMed] [Google Scholar]
- 28.Childs AC, Mehta DJ, Gerner EW. Polyamine-dependent gene expression. Cell Mol Life Sci. 2003;60:1394–1406. doi: 10.1007/s00018-003-2332-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ivanov IP, Atkins JF, Michael AJ. A profusion of upstream open reading frame mechanisms in polyamine-responsive translational regulation. Nucleic Acids Res. 2010;38:353–359. doi: 10.1093/nar/gkp1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Igarashi K, Kashiwagi K. Modulation of protein synthesis by polyamines. IUBMB Life. 2015;67:160–169. doi: 10.1002/iub.1363. [DOI] [PubMed] [Google Scholar]
- 31.Wang PY, Rao JN, Zou T, Liu L, Xiao L, Yu TX, Turner DJ, Gorospe M, Wang JY. Post-transcriptional regulation of MEK-1 by polyamines through the RNA-binding protein HuR modulating intestinal epithelial apoptosis. Biochem J. 2010;426:293–306. doi: 10.1042/BJ20091459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wallace HM, Fraser AV, Hughes A. A perspective of polyamine metabolism. Biochem J. 2003;376:1–14. doi: 10.1042/BJ20031327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oredsson SM. Polyamine dependence of normal cell-cycle progression. Biochem Soc Trans. 2003;31:366–370. doi: 10.1042/bst0310366. [DOI] [PubMed] [Google Scholar]
- 34.Kanyo ZF, Scolnick LR, Ash DE, Christianson DW. Structure of a unique binuclear manganese cluster in arginase. Nature. 1996;383:554–557. doi: 10.1038/383554a0. [DOI] [PubMed] [Google Scholar]
- 35.Bewley MC, Jeffrey PD, Patchett ML, Kanyo ZF, Baker EN. Crystal structures of Bacillus caldovelox arginase in complex with substrate and inhibitors reveal new insights into activation, inhibition and catalysis in the arginase superfamily. Structure. 1999;7:435–448. doi: 10.1016/s0969-2126(99)80056-2. [DOI] [PubMed] [Google Scholar]
- 36.Di Costanzo L, Sabio G, Mora A, Rodriguez PC, Ochoa AC, Centeno F, Christianson DW. Crystal structure of human arginase I at 1.29-Å resolution and exploration of inhibition in the immune response. Proc Natl Acad Sci USA. 2005;102:13058–13063. doi: 10.1073/pnas.0504027102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kern AD, Oliveira MA, Coffino P, Hackert ML. Structure of mammalian ornithine decarboxylase at 1.6 Å resolution: stereochemical implications of PLP-dependent amino acid decarboxylases. Structure. 1999;7:567–581. doi: 10.1016/s0969-2126(99)80073-2. [DOI] [PubMed] [Google Scholar]
- 38.Grishin NV, Osterman AL, Brooks HB, Phillips MA, Goldsmith EJ. X-ray structure of ornithine decarboxylase from Trypanosoma brucei: the native structure and the structure in complex with a-difluoromethylornithine. Biochemistry. 1999;38:15174–15184. doi: 10.1021/bi9915115. [DOI] [PubMed] [Google Scholar]
- 39.Almrud JJ, Oliveira MA, Kern AD, Grishin NV, Phillips MA, Hackert ML. Crystal structure of human ornithine decarboxylase at 2.1 Å resolution: structural insights to antizyme binding. J Mol Biol. 2000;295:7–16. doi: 10.1006/jmbi.1999.3331. [DOI] [PubMed] [Google Scholar]
- 40.Korolev S, Ikeguchi Y, Skarina T, Beasley S, Arrowsmith C, Edwards A, Joachimiak A, Pegg AE, Savchenko A. The crystal structure of spermidine synthase with a multisubstrate adduct inhibitor. Nat Struct Biol. 2002;9:27–31. doi: 10.1038/nsb737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu H, Min J, Ikeguchi Y, Zeng H, Dong A, Loppnau P, Pegg AE, Plotnikov AN. Structure and mechanism of spermidine synthases. Biochemistry. 2007;46:8331–8339. doi: 10.1021/bi602498k. [DOI] [PubMed] [Google Scholar]
- 42.Wu H, Min J, Zeng H, McCloskey DE, Ikeguchi Y, Loppnau P, Michael AJ, Pegg AE, Plotnikov AN. Crystal structure of human spermine synthase: implications of substrate binding and catalytic mechanism. J Biol Chem. 2008;283:16135–16146. doi: 10.1074/jbc.M710323200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Binda C, Coda A, Angelini R, Federico R, Ascenzi P, Mattevi A. A 30 Å long U-shaped catalytic tunnel in the crystal structure of polyamine oxidase. Structure. 1999;7:265–276. doi: 10.1016/s0969-2126(99)80037-9. [DOI] [PubMed] [Google Scholar]
- 44.Sjögren T, Wassvik CM, Snijder A, Aagaard A, Kumanomidou T, Barlind L, Kaminski TP, Kashima A, Yokota T, Fjellström O. The structure of murine N1-acetylspermine oxidase reveals molecular details of vertebrate polyamine catabolism. Biochemistry. 2017;56:458–467. doi: 10.1021/acs.biochem.6b01140. [DOI] [PubMed] [Google Scholar]
- 45.Fujishiro K, Ando M, Uwajima T. Crystallization and some properties of acetylpolyamine amidohydrolase from Mycoplana bullata. Biochem Biophys Res Commun. 1988;157:1169–1174. doi: 10.1016/s0006-291x(88)80997-5. [DOI] [PubMed] [Google Scholar]
- 46.Sakurada K, Ohta T, Fujishiro K, Hasegawa M, Aisaka K. Acetylpolyamine amidohydrolase from Mycoplana ramosa: gene cloning and characterization of the metal-substituted enzyme. J Bacteriol. 1996;178:5781–5786. doi: 10.1128/jb.178.19.5781-5786.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lombardi PM, Angell HD, Whittington DA, Flynn EF, Rajashankar KR, Christianson DW. Structure of prokaryotic polyamine deacetylase reveals evolutionary functional relationships with eukaryotic histone deacetylases. Biochemistry. 2011;50:1808–1817. doi: 10.1021/bi101859k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marchant P, Manneh VA, Blankenship J. N1-Acetylspermidine is not a substrate for N-acetylspermidine deacetylase. Biochim Biophys Acta. 1986;881:297–299. doi: 10.1016/0304-4165(86)90017-6. [DOI] [PubMed] [Google Scholar]
- 49.Marchant P, Dredar S, Manneh V, Alshabanah O, Matthews H, Fries D, Blankenship J. A selective inhibitor of N8-acetylspermidine deacetylation in mice and HeLa cells without effects on histone deacetylation. Arch Biochem Biophys. 1989;273:128–136. doi: 10.1016/0003-9861(89)90170-7. [DOI] [PubMed] [Google Scholar]
- 50.Hai Y, Shinsky SA, Porter NJ, Christianson DW. Histone deacetylase 10 structure and molecular function as a polyamine deacetylase. Nat Commun. 2017;8:15368. doi: 10.1038/ncomms15368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Finnin MS, Donigan JR, Cohen A, Richon VM, Rifkind RA, Marks PA, Breslow R, Pavletich NP. Structures of a histone deacetylase homologue bound to the TSA and SAHA inhibitors. Nature. 1999;401:188–193. doi: 10.1038/43710. [DOI] [PubMed] [Google Scholar]
- 52.Ash DE, Cox JD, Christianson DW. Arginase: a binuclear manganese metalloenzyme. Metal Ions Biol Syst. 2000;37:407–428. [PubMed] [Google Scholar]
- 53.Christianson DW. Arginase: structure, mechanism, and physiological role in male and female sexual arousal. Acc Chem Res. 2005;38:191–201. doi: 10.1021/ar040183k. [DOI] [PubMed] [Google Scholar]
- 54.Lombardi PM, Cole KE, Dowling DP, Christianson DW. Structure, mechanism, and inhibition of histone deacetylases and related metalloenzymes. Curr Opin Struct Biol. 2011;21:735–743. doi: 10.1016/j.sbi.2011.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gantt SL, Gattis SG, Fierke CA. Catalytic activity and inhibition of human histone deacetylase 8 is dependent on the identity of the active site metal ion. Biochemistry. 2006;45:6170–6178. doi: 10.1021/bi060212u. [DOI] [PubMed] [Google Scholar]
- 56.Vannini A, Volpari C, Gallinari P, Jones P, Mattu M, Carfí A, De Francesco R, Steinkühler C, Di Marco S. Substrate binding to histone deacetylases as shown by the crystal structure of the HDAC8-substrate complex. EMBO Rep. 2007;8:879–884. doi: 10.1038/sj.embor.7401047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dowling DP, Gantt SL, Gattis SG, Fierke CA, Christianson DW. Structural studies of human histone deacetylase 8 and its site-specific variants complexed with substrate and inhibitors. Biochemistry. 2008;47:13554–13563. doi: 10.1021/bi801610c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Porter NJ, Christianson NH, Decroos C, Christianson DW. Structural and functional influence of the glycine-rich loop G302GGGY on the catalytic tyrosine of histone deacetylase 8. Biochemistry. 2016;55:6718–6729. doi: 10.1021/acs.biochem.6b01014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gantt SL, Joseph CG, Fierke CA. Activation and inhibition of histone deacetylase 8 by monovalent cations. J Biol Chem. 2010;285:6036–6043. doi: 10.1074/jbc.M109.033399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gantt SML, Decroos C, Lee MS, Gullet LE, Bowman CM, Christianson DW, Fierke CA. General base-general acid catalysis in human histone deacetylase 8. Biochemistry. 2016;55:820–832. doi: 10.1021/acs.biochem.5b01327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hai Y, Christianson DW. Histone deacetylase 6 structure and molecular basis of catalysis and inhibition. Nat Chem Biol. 2016;12:741–747. doi: 10.1038/nchembio.2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Decroos C, Bowman CM, Christianson DW. Synthesis and evaluation of N8-acetylspermidine analogues as inhibitors of bacterial acetylpolyamine amidohydrolase. Bioorg Med Chem. 2013;21:4530–4540. doi: 10.1016/j.bmc.2013.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Decroos C, Christianson DW. Design, synthesis, and evaluation of polyamine deacetylase inhibitors, and high-resolution crystal structures of their complexes with acetylpolyamine amidohydrolase. Biochemistry. 2015;54:4692–4703. doi: 10.1021/acs.biochem.5b00536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tong JJ, Liu J, Bertos NR, Yang XJ. Identification of HDAC10, a novel class II human histone deacetylase containing a leucine-rich domain. Nucleic Acids Res. 2002;30:1114–1123. doi: 10.1093/nar/30.5.1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Miyake Y, Keusch JJ, Wang L, Saito M, Hess D, Wang X, Melancon BJ, Helquist P, Gut H, Matthias P. Structural insights into HDAC6 tubulin deacetylation and its selective inhibition. Nat Chem Biol. 2016;12:748–754. doi: 10.1038/nchembio.2140. [DOI] [PubMed] [Google Scholar]
- 66.Job G, Brugger C, Xu T, Lowe BR, Pfister Y, Qu C, Shanker S, Baños Sanz JI, Partridge JF, Schalch T. SHREC silences heterochromatin via distinct remodeling and deacetylation modules. Mol Cell. 2016;62:207–221. doi: 10.1016/j.molcel.2016.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Maris JM. Recent advances in neuroblastoma. N Engl J Med. 2010;362:2202–2211. doi: 10.1056/NEJMra0804577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Oehme I, Linke JP, Böck BC, Milde T, Lodrini M, Hartenstein B, Wiegand I, Eckert C, Roth W, Kool M, Kaden S, Gröne HJ, Schulte JH, Lindner S, Hamacher-Brady A, Brady NR, Deubzer HE, Witt O. Histone deacetylase 10 promotes autophagy-mediated cell survival. Proc Nat Acad Sci USA. 2013;110:E2592–E2601. doi: 10.1073/pnas.1300113110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Duan B, Ye D, Zhu S, Jia W, Lu C, Wang G, Guo X, Yu Y, Wu C, Kang J. HDAC10 promotes angiogenesis in endothelial cells through the PTPN22/ERK axis. Oncotarget. 2017;8:61338–61349. doi: 10.18632/oncotarget.18130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Eisenberg T, Knauer H, Schauer A, Büttner S, Ruckenstuhl C, Carmona-Gutierrez D, Ring J, Schroeder S, Magnes C, Antonacci L, Fussi H, Deszcz L, Hartl R, Schraml E, Criollo A, Megalou E, Weiskopf D, Laun P, Heeren G, Breitenbach M, Grubeck-Loebenstein B, Herker E, Fahrenkrog B, Fröhlich KU, Sinner F, Tavernarakis N, Minois N, Kroemer G, Madeo F. Induction of autophagy by spermidine promotes longevity. Nat Cell Biol. 2009;11:1305–1314. doi: 10.1038/ncb1975. [DOI] [PubMed] [Google Scholar]
- 71.Minois N. Molecular basis of the “anti-aging” effect of spermidine and other natural polyamines – a mini-review. Gerontology. 2014;60:319–326. doi: 10.1159/000356748. [DOI] [PubMed] [Google Scholar]
- 72.Vanrell MC, Cueto JA, Barclay JJ, Carrillo C, Colombo MI, Gottlieb RA, Romano PS. Polyamine depletion inhibits the autophagic response modulating Trypanosoma cruzi infectivity. Autophagy. 2013;7:1080–1093. doi: 10.4161/auto.24709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Samal K, Zhao P, Kendzicky A, Yco LP, McClung H, Gerner E, Burns M, Bachmann AS, Sholler G. AMXT-1501, a novel polyamine transport inhibitor, synergizes with DFMO in inhibiting neuroblastoma cell proliferation by targeting both ornithine decarboxylase and polyamine transport. Int J Cancer. 2013;133:1323–1334. doi: 10.1002/ijc.28139. [DOI] [PubMed] [Google Scholar]
- 74.Lozier AM, Rich ME, Grawe AP, Peck AS, Zhao P, Chang ATT, Bond JP, Sholler GS. Targeting ornithine decarboxylase reverses the LIN28/Let-7 axis and inhibits glycolytic metabolism in neuroblastoma. Oncotarget. 2014;6:196–206. doi: 10.18632/oncotarget.2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sholler GLS, Gerner EW, Bergendahl G, MacArthur RB, VanderWerff A, Ashikaga T, Bond JP, Ferguson W, Roberts W, Wada RK, Eslin D, Kraveka JM, Kaplan J, Mitchell D, Parikh NS, Neville K, Sender L, Higgins T, Kawakita M, Hiramatsu K, Moriya S, Bachmann AS. A phase I trial of DFMO targeting polyamine addiction in patients with relapsed/refractory neuroblastoma. PLOS ONE. 2015;10:e0127246. doi: 10.1371/journal.pone.0127246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bassiri H, Benavides A, Haber M, Gilmour SK, Norris MD, Hogarty MD. Translational development of difluoromethylornithine (DFMO) for the treatment of neuroblastoma. Transl Pediatr. 2015;4:226–238. doi: 10.3978/j.issn.2224-4336.2015.04.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li Y, Peng L, Seto E. Histone deacetylase 10 regulates the cell cycle G2/M phase transition via a novel let-7-HMGA2-cyclin A2 pathway. Mol Cell Biol. 2015;35:3547–3565. doi: 10.1128/MCB.00400-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Radhakrishnan R, Li Y, Xiang S, Yuan F, Yuan Z, Telles E, Fang J, Coppola D, Shibata D, Lane WS, Zhang Y, Zhang X, Seto E. Histone deacetylase 10 regulates DNA mismatch repair and may involve the deacetylation of MutS homolog 2. J Biol Chem. 2015;290:22795–22804. doi: 10.1074/jbc.M114.612945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tao X, Yan Y, Lu L, Chen B. HDAC10 expression is associated with DNA mismatch repair gene and is a predictor of good prognosis in colon carcinoma. Oncol Lett. 2017;14:4923–4929. doi: 10.3892/ol.2017.6818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kotian S, Liyanarachchi S, Zelent A, Parvin JD. Histone deacetylases 9 and 10 are required for homologous recombination. J Biol Chem. 2011;286:7722–7726. doi: 10.1074/jbc.C110.194233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lai IL, Lin TP, Yao YL, Lin CY, Hsieh MJ, Yang WM. Histone deacetylase 10 relieves repression on the melanogenic program by maintaining the deacetylation status of repressors. J Biol Chem. 2010;285:7178–7196. doi: 10.1074/jbc.M109.061861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Aramsangtienchai P, Spiegelman NA, He B, Miller SP, Dai L, Zhao Y, Lin H. HDAC8 catalyzes the hydrolysis of long chain fatty acyl lysine. ACS Chem Biol. 2016;11:2685–2692. doi: 10.1021/acschembio.6b00396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Andrews FH, Shinsky SA, Shanle EK, Bridgers JB, Gest A, Tsun IK, Krajewski K, Shi X, Strahl BD, Kutateladze TG. The Taf14 YEATS domain is a reader of histone crotonylation. Nat Chem Biol. 2016;12:396–398. doi: 10.1038/nchembio.2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kutil Z, Novakova Z, Meleshin M, Mikesova J, Schutkowski M, Barinka C. Histone deacetylase 11 is a fatty-acid deacylase. ACS Chem Biol. 2018 doi: 10.1021/acschembio.7b00942. in press. [DOI] [PubMed] [Google Scholar]
- 85.Islam MM, Banerjee T, Packard CZ, Kotian S, Selvendiran K, Cohn DE, Parvin JD. HDAC10 as a potential therapeutic target in ovarian cancer. Gynecol Oncol. 2017;144:613–620. doi: 10.1016/j.ygyno.2017.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Madeo F, Eisenberg T, Pietrocola F, Kroemer G. Spermidine in health and disease. Science. 2018;359 doi: 10.1126/science.aan2788. [DOI] [PubMed] [Google Scholar]


