Abstract
Chemotherapy-induced peripheral neuropathy (CIPN) is a painful and debilitating side effect of cancer chemotherapy with an unclear pathogenesis. Consequently, the available therapies for this neuropathic pain syndrome are inadequate, leading to a significantly reduced quality of life in many patients. Complement, a key component of the innate immune system, has been associated with neuro-inflammation, a potentially important trigger of some types of neuropathic pain. However, the role of complement in CIPN remains unclear. To address this issue, we developed a complement C3 knockout (KO) rat model and induced CIPN in these KO rats and wild-type (WT) littermates via the intraperitoneal administration of paclitaxel, a chemotherapeutic agent associated with CIPN. We then compared the severity of mechanical allodynia, complement activation, and intradermal nerve fiber loss between the groups. We found that 1) intraperitoneal paclitaxel administration activated complement in WT rats, 2) paclitaxel-induced mechanical allodynia was significantly reduced in C3 KO rats, and 3) the paclitaxel-induced loss of intradermal nerve fibers was markedly attenuated in C3 KO rats. In in vitro studies, we found that paclitaxel-treated rat neuronal cells activated complement, leading to cellular injury. Our findings demonstrate a previously unknown but pivotal role of complement in CIPN, and suggest that complement may be a new target for the development of novel therapeutics to manage this painful disease.
Keywords: Chemotherapy-induced neuropathy, Complement, Inflammation, Pain
Introduction
Chemotherapy-induced peripheral neuropathy (CIPN) is the most common neurological complication of cancer chemotherapy, which often limits the effectiveness of cancer treatment(1). Current management of CIPN is far from satisfactory largely due to an inadequate understanding of the pathophysiology underlying CIPN. Emerging evidence suggests that the immune system and immune-mediated neuro-inflammation are important in the development of CIPN (2–4). The conventional understanding of paclitaxel-induced CIPN is that paclitaxel binds to microtubule, leading to structural and functional disruption of neurons, resulting in peripheral neuropathy(5).
Complement is an important part of the innate immune system, which plays a primary role in protecting the host from infections(6). Once activated by one of the three main pathways (i.e., the classical, lectin, or alternative pathway), C3, the central component of the complement system, is cleaved by C3 convertases to release C3a and to deposit C3b on the surface of the complement activation site. The deposited C3b facilitates phagocytosis (opsonization) of the target cells and the formation of C5 convertase, which activates C5 into C5a and C5b. The generation of C5b initiates the terminal pathway of complement activation to assemble the membrane attack complex (MAC, a.k.a., C5b-9) that inserts “holes” in cells, causing direct cellular injury or lysis. An increasing body of evidence has suggested that complement is integrally involved in certain types of neuropathic pain (7–9), but whether complement plays any role in the pathogenesis of CIPN including paclitaxel-induced CIPN remains unclear. An earlier study (10) reported that Cremophor EL, the commonly-used solvent for paclitaxel, but not the paclitaxel itself, activates complement in vitro after incubating with serum. However, whether complement is activated in vivo after paclitaxel administration is unknown. This issue is especially intriguing, because paclitaxel is known to induce cell apoptosis(11) and apoptotic cells are potent activators of complement(12, 13).
In addition, most of the experiments relating complement to neuropathic pain were performed in mice(9), on the other hand, rats are physiologically closer than mice to humans in many aspects, including their complement system (14). In addition, modeling of human disease, especially in physiological monitoring and behavioral studies, is superior in rats(15). Using our newly developed C3 KO rats and rat neuronal cells, we found that complement is important in the development of peripheral neuropathy associated with paclitaxel administration, and that MAC-mediated neuronal damage could be an underlying mechanism.
Methods
Animals
Wild-type (WT) and C3 knockout (KO) rats were maintained under pathogen-free conditions in the animal facility of Lerner Research Institute, Cleveland Clinic. All procedures involving the rats were approved by the Institutional Animal Care and Use Committee of both the University of Michigan and Cleveland Clinic, and all were done in accordance with the U.S. Department of Health and Human Services Guide for the Care and Use of Laboratory Animals and institutional guidelines.
Development of C3 KO mosaic rats by CRISPR/Cas9 technology
CRISPR/Cas9 technology(16) was used to generate a genetically modified rat strain with a complement C3 gene knockout. The insertion of a premature termination codon in exon 2 is predicted to result in loss of protein expression due to nonsense-mediated decay of mRNA(17). Two single guide RNA (sgRNA) targets and protospacer adjacent motifs (PAM) were identified in rat complement C3 exon 2 (ENSRNOE00000506722) with the algorithm described by Doench et al.(18). The sgRNA targets were cloned into plasmid pX330 (Addgene.org plasmid #42230, a kind gift of Feng Zhang) as described(19). The guide targets were C40G1: 5’ TCATTACTCCCAATGTCCTG 3’ PAM: CGG with an algorithm score of 0.87 and C40G2: 5’ CTAGAGGCCCATGATGCTCA 3’ PAM: GGG with an algorithm score of 0.38. Predicted cut sites were in codons 35 and 51, respectively. Circular pX330 plasmids constructs were co-electroporated into rat embryonic fibroblasts with a PGKpuro plasmid(20). Genomic DNA was prepared from the cells after transient selection with puromycin. A 341 bp DNA fragment spanning the expected Cas9 cut sites was PCR amplified with forward primer 5’ GTCTGCTCTGGCAGGTGTC 3’ and reverse primer 5’ GAAGGGGCTATGGCTGACTA 3’. Amplicons were subjected to T7 endonuclease digestion essentially as described(21). Briefly, DNA fragments were amplified, melted and annealed, then subjected to T7EI digestion. The resulting digests are separated by agarose gel electrophoresis. The gel was stained with Sybr Gold (Invitrogen S11494) instead of ethidium bromide to achieve greater sensitivity in the detection of insertion/deletion mutations (indels) in the amplicons. The presence of indels produced by non-homologous end joining repair of Cas9 induced double-strand breaks resulted in the presence of lower molecular weight DNA fragments following T7 endonuclease I digestion for both sgRNA targets. Rat zygote microinjection was carried out at described(22).
Characterization of C3 KO rats at both DNA and protein levels
C3 mosaic rats were bred to WT rats and germline transmission was identified by PCR followed by sequencing to identify the Indel events. The identified heterozygous C3 KO rats (F344/Sprague-Dawley mixed background) were bred to each other, and the pups were genotyped as described above to identify the WT and C3 KO littermates. To confirm the deficiency of the C3 protein in the C3 KO rats, C3 levels were measured in sera collected from both WT and KO rats using a commercially available rat C3 ELISA kit (Innovative Research, Novi, MI) following the manufacturer’s protocol. In addition, C3 deposition assays were performed by incubating sera from WT and C3 KO rats with Zymosan, followed by flow cytometric analysis of deposited C3 on the surface of Zymosan following a previously established protocol (23) using a mouse anti-human/mouse C3 mAb that cross-reacts with rat C3 (Cedarlane, clone 10C7) (Suppl. Fig. 1).
Paclitaxel-induced peripheral neuropathy model
CIPN was induced in rats by daily intraperitoneal (IP) injection of 1.0 mg/kg paclitaxel (Tocris, Bristol, UK) dissolved in dimethyl sulfoxide (DMSO, Sigma) (as suggested by the manufacturer) for 4 consecutive days for a final cumulative dose of 4 mg/kg; the injection volume was 1 mL/kg (24, 25). As cremophor EL, the commonly-used solvent for paclitaxel has been reported to activate complement in vitro after incubating with serum(10), we used DMSO as the vehicle for our complement studies. Previous studies by others demonstrated that DMSO administration does not interfere with mechanical allodynia studies (26–28). Baseline responses to mechanical stimulation of the hind paw were established one day (day 0) before the first paclitaxel injection (day 1).
Pain behavioral testing
Rats were placed in a compartment with a wire mesh bottom and allowed to acclimate for 30 minutes before testing. Sensory thresholds for the development of allodynia to mechanical stimuli were assessed. Mechanical sensitivity was assessed by using a series of von Frey filaments with logarithmic incremental stiffness (Stoelting Co., Wood Dale, IL), as previously described(29) and 50% probability paw withdrawal thresholds were calculated by the up-down method. In brief, filaments were applied to the plantar surface of a hind paw for approximately 6 seconds in an ascending or descending order after a negative or positive withdrawal response, respectively. Six consecutive responses after the first change in response were used to calculate the paw withdrawal threshold (in grams). When response thresholds occurred outside the range of detection, the paw withdrawal threshold was assigned at 15.00 g for continuous negative responses and at 0.25 g for continuous positive responses. The behavioral testing was performed blindly with respect to drug administration.
In vivo complement activation assay by C3 western blotting
WT and C3 KO rats were intraperitoneally (i.p.) administered with paclitaxel or DMSO vehicle for 4 consecutive days (day 1 to 4), and serum samples were collected at day 5 when the behavioral tests confirmed the development of mechanical allodynia. Animals were deeply anesthetized with 2–4% isoflurane in 100% oxygen. Blood (10 ml) was collected from each rat by cardiac puncture. Serum was diluted 1:1000 with NP40 buffer and then mixed with an equal volume of 2× sample buffer (125 mM Tris-HCl pH 6.8; 20% glycerol; 4% SDS; 5% β-mercaptoethanol; 0.02% bromophenol blue). The samples were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred onto a PVDF membrane and probed with a polyclonal goat anti-mouse C3 antibody, which cross-reacts with rat C3 (1:500 dilution; MP Biomedicals, Solon, Ohio) (Suppl. Fig. 1).
In vitro rat neuronal cell C3b/iC3b deposition assay
Rat neuronal PC-12 cells(30, 31) (~ 0.5×106) were first cultured in the presence of 50 nM of paclitaxel or DMSO for 12, 24 and 48 hr. Then the cells were harvested, washed and incubated at 37°C with 20% of WT or C3 KO Rat serum in 100 µL GVB++ buffer (0.1 % gelatin, 5 mM Veronal, 0.15 mM CaCl2 and 0.5 mM MgCl2, 145 mM NaCl, pH 7.3). After 30 min, cells were washed again with PBS plus 0.5% BSA, then stained with 5 µg/mL of FITC-labeled mouse anti-human/mouse C3 IgG that cross-reacts with Rat C3 (clone 10C7, Suppl. Fig. 1) to detect cell surface C3b/iC3b deposition using a flow cytometer.
In vitro complement-mediated rat neuronal cell cytotoxicity assay
An established fluorescence dye BCECF leakage-based cell cytotoxicity assay (32–34) was used to assess the extent of the complement-mediated rat neuronal cell injury after paclitaxel treatment. In brief, 2×105 PC-12 cells were first cultured in the presence of 50 nM of paclitaxel or DMSO for 12, 24 and 48 hr, then labeled with 5µM of BCECF-AM (Thermo Fisher Scientific, CA) for 30 minutes at 37°C. After washing (2×), the labeled cells were incubated with 20% WT or C3 KO Rat serum in 100 µL of GVB++ buffer for another 30 minutes at 37°C before supernatants were harvested, and the levels of leaked BCECF were measured by a fluorescence microtiter plate reader (PerkinElmer, MA) with excitation and emission wavelengths of 485 nm and 530 nm. To calculate the percentage of BCECF release (complement-mediated injury), the following formula was used: The percentage of BCECF release = [(A−B)/(C−B)] ×100%; where A represents the mean experimental BCECF release, B represents the mean spontaneous BCECF release, and C represents the mean maximum BCECF released that was induced by incubating cells with 0.1% Triton X-100.
Immunofluorescence staining for intra-epidermal nerve fibers
Immunostaining of the intra-epidermal nerve fibers was performed to assess their density as previously described (35, 36). In brief, after 4-day i.p. administration of paclitaxel and development of mechanical allodynia, animals were deeply anesthetized with 60 mg/kg sodium pentobarbital i.p. and perfused transcardially with 200 ml heparinized normal saline followed by 200 ml of 4% formaldehyde solubilized in 0.1 M phosphate buffered saline (PBS). The glabrous skin of the hind paw was excised and post-fixed in 4% formaldehyde overnight, then cryopreserved in 30% sucrose in PBS at 4°C for 24h. The whole-thickness of the paw skin was then cut to 16 µm thickness and collected free-floating in 0.01 M PBS. Tissue sections were washed with 0.1% Triton-X 100 in PBS and blocked with donkey serum followed by washing with PBS. A rabbit polyclonal anti-protein gene product 9.5 primary antibody (RB-9202, 1:200, ThermoScientific, Rockford, IL, USA) was used as the primary antibody and incubated for 2 hours at room temperature and then overnight at 4°C. Sections were then washed with PBS and incubated with Cy3 (goat anti-rabbit, 111-165-144, 1:200) conjugated secondary antibodies (Jackson Immuno Laboratories, West Grove, PA, USA) for 2 h at room temperature. Sections were then rinsed with wash buffer and mounted with Slow Fade anti-fade reagents (Invitrogen, Carlsbad, CA, USA). The omission of primary or secondary antibodies resulted in no immunostaining. Five sections per animal were chosen randomly to quantify the intra-epidermal nerve fiber density using a confocal microscope by an investigator, who was blind to the origin of the tissue being examined. All nerve fibers crossing into the epidermis were counted. Nerve fibers that branched within the epidermis were counted as one fiber. The number of intra-epidermal nerve fibers per viewing field was counted.
Statistical analyses
Data are presented as group mean ± standard error of the means (SEM) and analyzed using GraphPad Prism 6 software (GraphPad Inc., La Jolla, CA). Behavioral data were analyzed with two-way ANOVA (treatment × time) followed by Bonferroni post hoc multiple comparison tests. Immunofluorescence data were analyzed with one-way ANOVA (treatment) followed by post hoc Student-Newman-Keuls tests. p<0.05 was considered statistically significant.
Results
Design and validation of the rat C3 targeted CRISPR/CAS9
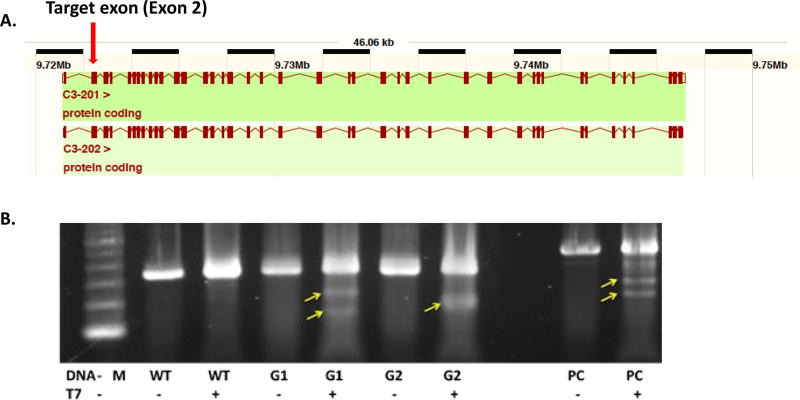
Based on the available rat genome sequence information, we designed two CRISPR/Cas9 constructs that targeted rat C3 exon 2 (C40) so that a premature termination codon was introduced into the C3 mRNA transcribed from exon 2, leading to nonsense-mediated mRNA decay and a null allele (Fig. 1A). Construct C40G1 targets the sequence 5'-TCA TTA CTC CCA ATG TCC TG - 3' followed by the protospacer adjacent motif (PAM) CGG, while construct C40G2 targets the sequence 5'- CTA GAG GCC CAT GAT GCT CA - 3' followed by the PAM GGG.
Figure. 1.
Design and validation of the CRISPR/Cas9 construct to introduce Indel into the exon 2 of the rat C3 gene. A. Map of the rat C3 gene; B. Validation of the C40G1 (G1) and C40G2 (G2) constructs in vitro. Activity of sgRNA C40G1 and C40G2 in cultured rat fibroblasts. A DNA fragment spanning the expected Cas9 cut site was PCR amplified and treated with T7 endonuclease I. Both C40G1 and C40G2 produced indel mutations in fibroblasts as shown by the presence of low molecular weight product in Lanes 5 and 7 (arrows). Lane 1: 100 bp ladder. Lanes 2 and 3: wild-type rat fibroblast PCR amplicon untreated (−) and treated with T7EI (+). Lanes 3 and 4: C40G1 sgRNA treated rat fibroblast PCR amplicon untreated (−) and treated with T7EI (+). Lanes 6 and 7: C40G2 sgRNA treated rat fibroblast PCR amplicon untreated (−) and treated with T7EI (+). Lanes 8 and 9: PC (positive control) PCR amplicon untreated (−) and treated with T7EI (+) shows the expected pattern.
To validate the designed constructs and their ability to induce Indels in exon 2 of the rat C3 gene, we electroporated constructs C40G1 or C40G2 into rat embryonic fibroblast cells and prepared DNA from the transfected cells. Following an established protocol, we PCR-amplified the DNA fragments spanning the expected Cas9-cut sites and digested them with T7 endonuclease. The presence of Indel mutations produced by non-homologous end joining repair of Cas9-induced double-strand breaks result in the presence of lower molecular weight DNA fragments. We found that both constructs introduced Indels (arrows) in rat cells, with construct C40G2 appearing to be more effective than construct C40G1 (Fig. 1B).
Development and characterization of the C3 KO rats
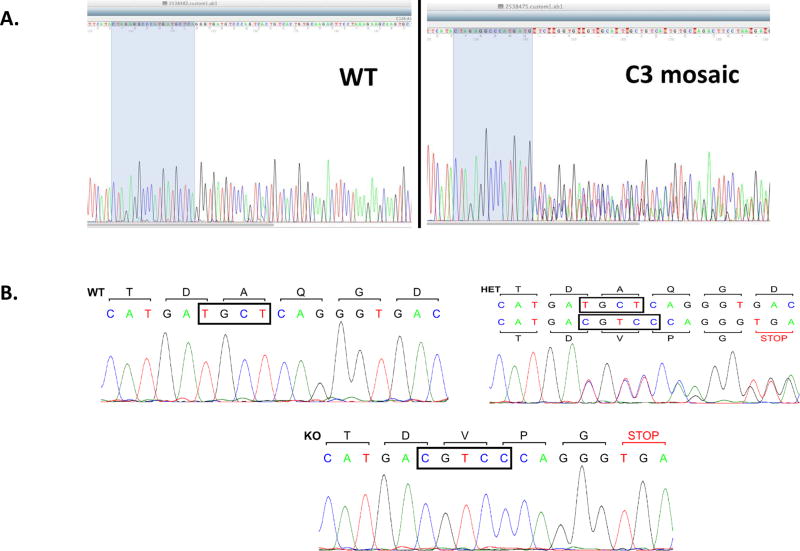
We microinjected fertilized rat eggs with the validated CRISPR/Cas 9 construct C40G2 according to previously published protocols, and genotyped the resultant pups by PCR amplification of the targeted region followed by sequencing. We identified 3 founder rats that carried Indel in the targeted region, in which TGCT was changed to CGTCC, resulting in a frame-shift mutation in the C3 transcript (Fig. 2). This mutation should lead to its degradation and, therefore, deficiency of C3 protein. After breeding the mosaic rats carrying the C3 Indels with WT rats, we identified germline-transmitted rats by sequencing. We then bred the C3 heterologous rats with each other and genotyped the resultant pups as described above. As shown in Fig. 2B, while the WT rats had the normal TGCT sequence, the heterozygous rats showed both the WT TGCT sequence and the KO CGTCC sequence, and the homozygous KO rats only had the KO CGTCC sequences, demonstrating successful C3 exon 2 editing.
Figure. 2.
Identification of the founder mosaic and C3 KO rats by sequencing. The edited regions of exon 2 of the rat C3 gene were PCR amplified and sequenced; the data showed an in-frame reading in the WT rats (A and B, left panels) but not in the mosaic founder rat (A, right panel). The heterozygous (HET) rats (B, right panel) had both the WT allele (TGCT) and the KO allele (CGTCC), while the homozygous Rats (B, bottom panel) had only the KO allele (CGTCC).
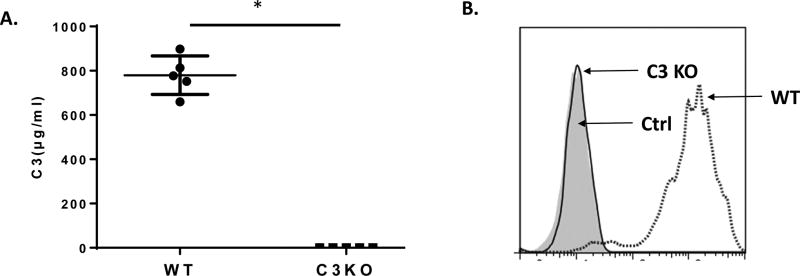
To confirm that the resultant frame-shift mutation caused by our CRISPR/Cas9 leads to C3 deficiency in the C3 KO rats, we measured C3 protein levels in the rat sera by ELISA. We detected approximately 0.8–0.9 mg/ml C3 protein in the WT rats, and no detectable C3 protein in the C3 KO rats (Fig. 3A). In addition, we incubated zymosan with 20% WT or C3 KO serum and then measured the level of C3b deposition on the cell/particle surface by flow cytometric analysis using a polyclonal anti-mouse C3 IgG that cross-reacts with rat C3. We found that while C3b/iC3b were readily detected on zymosan after incubation with WT rat serum, no C3/iC3b deposits were detectable after incubation with C3 KO serum (Fig. 3B). These data confirmed that C3 KO rats are deficient in C3 protein as the result of CRISPR/Cas9 editing of exon 2 of the rat gene.
Figure. 3.
Characterization of the C3 KO rats. A. Determination of serum C3 concentration. C3 levels in the sera from WT and C3 KO rats (n=5 in each group) were measured by ELISA following the manufacturer provided protocol; approximately 0.8 mg/ml C3 protein was detected in WT rats with no detectable C3 protein in the C3 KO rats. *p<0.05 B. C3 deposition assay. Serum (20%) from WT (dotted line) or C3 KO rats (solid line) were incubated with zymosan for complement activation. Then the deposited C3b/iC3b was assessed by staining the zymosan with a polyclonal anti-mouse C3 IgG (cross-react with rat C3) followed by flow cytometric analysis (shaded area, isotype control, ctrl).
Complement C3 is activated in rats treated with paclitaxel
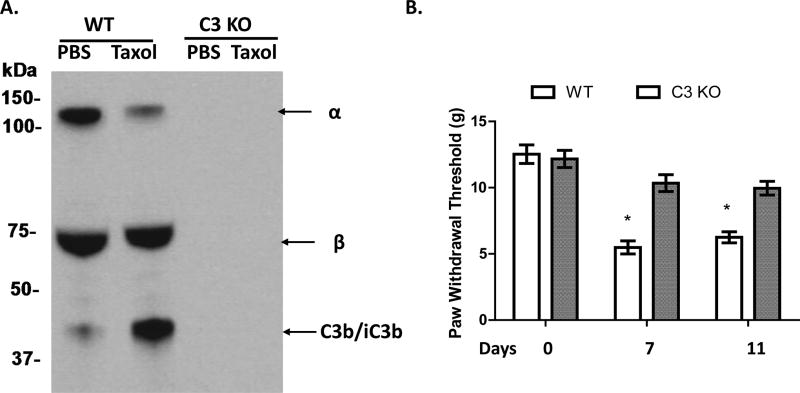
Whether complement is activated in vivo after paclitaxel administration is unknown. To investigate this, we collected blood from WT and C3 KO rats following treatment with either paclitaxel or DMSO (vehicle) for 4 days, and analyzed the sera for complement activation products of iC3b by western blot. These assays showed that while sera from DMSO-treated rats had typically strong α and β bands of the C3 protein and a weak band for α2 chain of iC3b that was probably a result of baseline complement activation, sera from paclitaxel-treated rats displayed a significantly reduced α band of C3 and a markedly increased amount of the α2 chain of iC3b (Fig. 4), demonstrating that complement was strongly activated in the paclitaxel-treated rats. Not surprisingly, no C3 band was detected in the sera from DMSO or paclitaxel-injected C3 KO rats (Fig. 4A).
Figure. 4.
Complement is important in paclitaxel-induced mechanical allodynia in CIPN. A. Complement is activated after paclitaxel administration. WT or C3 KO rats were i.p. injected with DMSO (Vehicle) or 1 mg/kg paclitaxel (in DMSO according to the instructions of the manufacturer, Tocris, Bristol, UK) for 4 days (day 1 to 4), and then sera were collected on day 5 when the behavioral tests confirmed the development of mechanical allodynia and probed with an anti-C3 IgG to assess complement (C3) activation. There was a significant reduction of the α band (~130KDa) of C3 and an increased α2 chain of iC3b (~40 KDa) in the paclitaxel-treated WT rats. No C3 protein was detectable in sera from the C3 KO rats. B. C3 KO rats showed increased paw withdrawal threshold (PWT). WT and C3 KO rats (n=10/group) were injected with paclitaxel for 4 days (day 1 to 4), PWT was assessed on days 0, 7 and 11.; * p<0.05.
C3 KO rats demonstrate reduced mechanical allodynia
Mechanical allodynia is commonly used to measure pain in CIPN and can be quantitated by paw withdrawal threshold (PWT) assays. To explore the potential role of complement in CIPN, we first measured the PWT in naïve WT and C3 KO rats and then in the same WT and C3 KO rats on days 3 and 7 following administration of paclitaxel by i.p. injection. These assays showed that while WT rats had PWTs of 5.2±0.4 grams and 5.4±0.5 grams on day 3 and day 7, C3 KO rats had significantly higher PWTs (11.0±0.4 grams and 10.5±0.5 grams, respectively), indicating less mechanical allodynia development in C3 KO rats in CIPN (Fig 4B).
C3 KO rats have reduced intradermal nerve fibers loss after paclitaxel treatment
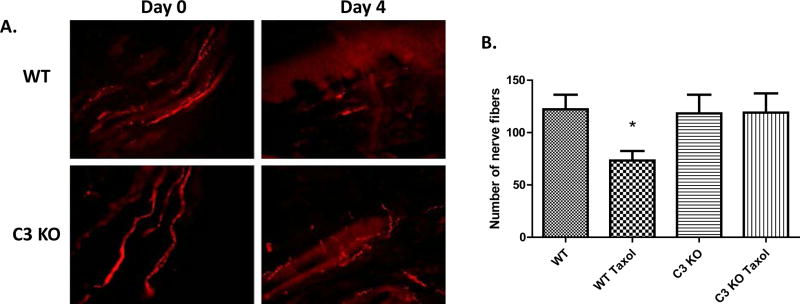
The loss of the intra-epidermal nerve fibers has been implicated in a host of chronic painful neuropathic conditions(37), including CIPN(38). To explore this as a possible mechanism by which C3 KO rats are protected from pain in CIPN, we investigated whether loss of the intra-epidermal nerve fibers was alleviated in C3 KO rats 4 days after paclitaxel treatment by quantitating the nerve fibers by immunofluorescence staining for PGP9.5, a pan-neuronal marker(39). We found that in WT and C3 KO rats without paclitaxel injection, intra-epidermal nerve fibers had normal morphology, originating from the cutaneous layer and extending into the intra-epidermis as long nerve fibers (Fig. 5). However, 4 days after paclitaxel administration, we found a significant decrease in PGP9.5 staining in the WT rats, but not in the C3 KO rats, indicating that while intra-epidermal nerve fiber loss was largely preserved in C3 KO rats in CIPN.
Figure. 5.
C3 KO rats have reduced paclitaxel-induced loss of intradermal nerve fibers. WT and C3 KO rats were administered with i.p. paclitaxel for 4 consecutive days (day 1 to 4). Animals were perfused and glabrous skin of hind paws collected at day 5 when the behavioral tests confirmed the development of mechanical allodynia. A. intradermal nerve fibers in the glabrous hind paws of WT (upper panels) and C3 KO (lower panels) rats were identified by PGP 9.5 staining (red) before (left panels) and after (right panels) 4-day i.p. paclitaxel administration. DAPI (4',6-diamidino-2-phenylindole) staining (Blue) was used to for the nuclear counterstains. Pictures were taken at 20 × objectives with a fluorescent microscope. B. Quantification of surviving nerve fibers from 10 random slides taken from 3 animals/group. *p<0.05
Paclitaxel-treated Rat neuronal cells activate complement that leads to cell damage
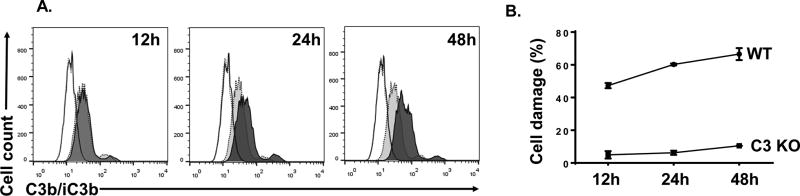
To demonstrate that rat neuronal cells activate complement after paclitaxel treatment, and are directly injured by the activated complement, we exposed rat neuronal PC12 cells with 50 nM of paclitaxel or DMSO for 12, 24, and 48 hours first, then incubated them with 20% WT or C3 KO rat serum. We then assessed complement activation by detecting the deposition of complement activation products C3b/iC3b on the PC12 cells using flow cytometric analysis. As shown in Fig. 6A, we found modest C3b/iC3b deposition on PC12 cells treated with paclitaxel or DMSO for 12h after their incubation with WT but not C3 KO rat serum. However, at 24 and 48 hr time points, there was significantly higher deposition of C3b/iC3b on the cells treated with paclitaxel than those treated with DMSO, clearly demonstrating that the rat neuronal cells activate complement after paclitaxel treatment. In addition, we loaded some of the paclitaxel-treated cells with BCECF, incubated them with WT or C3KO rat serum, then assessed MAC-mediated cellular damage by quantitating levels of BCECF leaked into the culture supernatants. These experiments showed that these paclitaxel-treated rat neuronal cells had significantly higher levels of leaked BCECF after incubating with WT rat serum than those with C3 KO serum (Fig. 6B), suggesting that the rat neuronal cells are directly damaged by the activated complement MAC after paclitaxel treatment.
Figure 6.
Rat neuronal cells activate complement that leads to cell damage after paclitaxel treatment. A. PC12 rat neuronal cells were cultured in the presence of 50 nM of paclitaxel or DMSO for 12, 24 and 48 hr, then incubated with 20% WT or C3 KO rat serum. Complement activation was assessed by detecting C3b/iC3 deposition on the cells using flow cytometry. Dotted lines, isotype controls; Solid lines, cells treated with paclitaxel then incubated with C3 KO serum; shaded solid lines, cells treated with paclitaxel and incubated with WT serum; shaded dot lines: cells treated with DMSO then incubated with WT serum. B. PC12 cells were cultured in the presence of 50 nM paclitaxel for 12, 24 and 48 hr, then loaded with BCECF-AM and incubated with 20% WT (WT) or C3 KO (C3 KO) serum. MAC-mediated cell damage was assessed by measuring levels of leaked BCECF in the culture supernatants.
Discussion
In this study, we developed a C3 KO rat using CRISPR/Cas9 technology (Figs.1,2&3) and confirmed the deficiency of C3 protein (Fig 3) in the resultant C3 KO rats. Using this novel C3 KO rat model, we demonstrated that paclitaxel-induced mechanical allodynia was much less severe in C3 KO than in WT rats (Fig 4B). In mechanistic studies, we found that complement was activated in the WT rats after paclitaxel treatment (Fig 4A) and that paclitaxel-induced loss of intradermal nerve fibers was less severe in C3 KO than in WT rats (Fig 5). In addition, in vitro experiments using PC12 cells showed that rat neuronal cells activate complement after paclitaxel treatment, and suffered direct MAC-mediated cell damage (Fig. 6). These findings, to be best of our knowledge, are the first to indicate a mechanistic link between complement and the development of paclitaxel-induced CIPN.
Initially, rats were the first choice as an experimental animal for biomedical research because they are physiologically closer than mice to humans in many aspects. More importantly, it has long been recognized in the field that mouse complement is significantly different from human complement(40). For example, (i) >80% of C4-deficient and >95% of C1q-deficient patients develop severe lupus(41), but very few C4 or C1q KO mice spontaneously develop lupus(42, 43); and (ii) while human complement lyses antibody-sensitized sheep erythrocytes in in vitro assays even at greater than 100-fold dilutions, mouse complement is very poor at lysing any cells. In contrast, both human and rat complement lyse antibody-sensitized sheep erythrocytes with great efficiency, and native human complement regulators such as CD59 efficiently regulate rat complement and vice versa(14). Aside from these similarities between the human and rat complement systems, the larger size of rats and their more consistent responses in behavioral studies make them better models than mice in these studies. Thus, complement KO rats should be superior to complement KO mice for studying the role of complement in human pathological conditions, especially studies involving behavior experiments. However, until recently, the technology required to precisely edit the rat genome and generate gene-engineered rats was complicated, and hitherto only one strain of rat with spontaneous deficiency of a complement component, C6, is known(44). Despite its apparent limitations, the mouse became the primary animal model for studying human diseases, mainly because of the availability of various gene-engineered strains and relevant reagents. However, with the development of CRISPR technology(45), the rat genome can now be manipulated without much difficulty, and gene KO rats can be generated at a reasonable cost and with excellent efficiency. The development of the C3 KO rat model and its use in the CIPN studies provide an example of using non-mouse complement KO animals to study the precise role of complement in human pathological conditions.
CIPN is a common devastating neuropathic pain syndrome related to cancer chemotherapy(46), which can be the dose-limiting factor in cancer therapy(1). The current clinical therapies for CIPN are not effective(46). Therefore, there is an urgent need to understand the mechanisms underlying CIPN to guide identification of novel therapies. Studies have suggested a critical role of inflammation and neuroimmune activation in CIPN, especially in paclitaxel-induced CIPN(47, 48). Paclitaxel recruits neutrophils(49), macrophages(50), monocytes(49), and CD8+ T cells(2) into dorsal root ganglion (DRG), induces expression of pro-inflammatory cytokines (TNF-α, IL-1β, and IL-6) and suppresses the expression of anti-inflammatory cytokines (IL-10 and IL-4) in the lumbar DRG and spinal cord(3, 4). These pro-inflammatory cytokines may subsequently sensitize primary nociceptive nerves(51) to cause pain. Consequently, administration of exogenous anti-inflammatory cytokines, such as IL-10, has been found effective in attenuating paclitaxel-induced allodynia(2, 3). However, whether paclitaxel administration could activate complement in vivo or not was not clear. Even though paclitaxel itself does not directly activate complement(10), we found that complement is significantly activated in paclitaxel-treated rats (Fig. 4A) and rat neuronal cells (Fig. 6A). These results suggest that complement is activated indirectly through apoptotic and dead cells resulting from paclitaxel administration.
Although complement has been implicated in other pain models, the potential role of complement in CIPN was not clear. It has been reported that depletion of complement by Cobra venom factor (CVF) alleviates neuropathic pain in the spinal nerve ligation (SNL) rat(52), and activation of complement in spinal microglia leads to C5a-mediated pain hypersensitivity in peripheral nerve injury models of neuropathic pain(53). Indeed, C5aR antagonist suppressed heat hyperalgesia and mechanical allodynia induced by intra-plantar C5a injection or hind paw incision(7, 8, 54). Intrathecal infusion of C5aR antagonist significantly reduced cold allodynia in the SNL rats(53). We herein demonstrated that paclitaxel-induced mechanical allodynia was decreased in C3 KO compared to WT rats (Fig 4), and that MAC is integrally involved in the damage of neuronal cells (Fig. 6B). These data suggest that complement may be a novel target for the treatment of CIPN, however, since the deficiency of C3 will almost abolish the production of C5a in addition to MAC, a potential role of C5a-mediated signaling in this CIPN model warrants further studies.
Loss of intra-epidermal nerve fibers in plantar hind paw skin has been implicated as one of the mechanisms of mechanical allodynia induced by the chemotherapy agents such as vincristine and paclitaxel(38, 55). We previously reported that loss of intra-epidermal nerve fibers also occurred in a hind limb ischemia-induced neuropathic pain model(36). A loss of intra-epidermal nerve fibers seems to be a common feature of nearly every chronic neuropathic pain syndrome(56). In the current study, we found that paclitaxel-induced loss of intra-epidermal nerve fibers was less severe in C3 KO than in WT rats (Fig 5). These results suggest that activated complement contributes significantly to the loss of intra-epidermal nerve fibers in this model. As a key component of the innate immune system, complement is well-known to form MACs to damage cells and tissues after activation. Paclitaxel treatment induces cell apoptosis and death(11), apoptotic/dead cells activate complement through the classical pathway of complement activation by binding to C1q, or directly activate complement through the alternative pathway of complement activation(12, 13). Once complement is activated, MACs are assembled that could directly damage the epidermal nerve. Using flow cytometric assays and the BCECF leakage studies, we demonstrated C3b/iC3b deposition on rat neuronal PC12 cells after paclitaxel treatment and these neuronal cells were directly damaged by activated complement (MACs) as evidenced by the increased BCECF leakage (Fig 6). We also detected some complement activation on the PC12 cells treated with DMSO, which could be background complement activation as a result of the presence of serum antibodies against rat allogenic antigens and/or xenogenic antigens on the PC12 cells that were cultured in media containing FBS.
In summary, we have generated the first complement gene engineered rat. We found that paclitaxel administration activates complement in vivo and that paclitaxel-induced mechanical allodynia is less severe in C3KO rats, in association of reduced loss of intra-epidermal nerve fibers. We also showed that rat neuronal cells activate complement and suffer direct complement-mediated cellular damage. These findings demonstrate a critical role of complement in the development of CIPN, and suggest that targeting complement may be useful in the prevention and treatment of complications in patients with CIPN.
Supplementary Material
Acknowledgments
We thank Elizabeth Hughes for the preparation of genome editing reagents, Wanda Filipiak for rat zygote microinjection, Michael Zeidler for genotyping and identification of C3 mutations, and the Transgenic Animal Model Core of the University of Michigan’s Biomedical Research Core Facilities.
Funding
This work was supported in parts by the Chronic Pain Research grant from the American Society of Regional Anesthesia and Pain Medicine (J.X) and a grant from the National Institute of Health NIH R01DK10358 (F.L.).
Abbreviations
- CIPN
Chemotherapy-induced peripheral neuropathy
- KO
Knockout
- WT
Wild-type
- C3aR
Complement 3a receptor
- C5aR
Complement 5a receptor
- CRISPR
Clustered Regularly Interspaced Short Palindromic Repeats
- ELISA
Enzyme-linked immunosorbent assay
- i.p.
Intraperitoneal
- DMSO
Dimethyl sulfoxide
- SDS
Sodium dodecyl sulfate
- PVDF
Polyvinylidene difluoride
- PBS
Phosphate buffered saline
- ANOVA
Analysis of variance
- PWT
Paw withdrawal threshold
- DRG
Dorsal root ganglion
- SNL
Spinal nerve ligation
- CVF
Cobra venom factor
- MAC
Membrane attack complex
- TNF
Tumor necrosis factor
- IL
Interleukins
Footnotes
The authors declare no potential conflicts of interest.
Competing interest
The authors declare that they have no competing interests.
Authors’ contribution
JX, LZ, MX, YL carried out the inflammation experiments, pain behavior experiments, PH did the fluorescent imaging experiments. TLS and DF helped with the generation of C3 KO rats, analyzed data, and contributed to writing the manuscript. JX, RR, and FL designed all experiments, helped analyze and interpret the resulting data, and contributed to writing the manuscript. All authors read and approved the final manuscript.
References
- 1.Quasthoff S, Hartung HP. Chemotherapy-induced peripheral neuropathy. J Neurol. 2002;249:9–17. doi: 10.1007/pl00007853. [DOI] [PubMed] [Google Scholar]
- 2.Krukowski K, Eijkelkamp N, Laumet G, Hack CE, Li Y, Dougherty PM, Heijnen CJ, Kavelaars A. CD8+ T Cells and Endogenous IL-10 Are Required for Resolution of Chemotherapy-Induced Neuropathic Pain. J Neurosci. 2016;36:11074–11083. doi: 10.1523/JNEUROSCI.3708-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ledeboer A, Jekich BM, Sloane EM, Mahoney JH, Langer SJ, Milligan ED, Martin D, Maier SF, Johnson KW, Leinwand LA, Chavez RA, Watkins LR. Intrathecal interleukin-10 gene therapy attenuates paclitaxel-induced mechanical allodynia and proinflammatory cytokine expression in dorsal root ganglia in rats. Brain Behav Immun. 2007;21:686–698. doi: 10.1016/j.bbi.2006.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Janes K, Wahlman C, Little JW, Doyle T, Tosh DK, Jacobson KA, Salvemini D. Spinal neuroimmune activation is independent of T-cell infiltration and attenuated by A3 adenosine receptor agonists in a model of oxaliplatin-induced peripheral neuropathy. Brain Behav Immun. 2015;44:91–99. doi: 10.1016/j.bbi.2014.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee JJ, Swain SM. Peripheral neuropathy induced by microtubule-stabilizing agents. J Clin Oncol. 2006;24:1633–1642. doi: 10.1200/JCO.2005.04.0543. [DOI] [PubMed] [Google Scholar]
- 6.Song WC, Sarrias MR, Lambris JD. Complement and innate immunity. Immunopharmacology. 2000;49:187–198. doi: 10.1016/s0162-3109(00)80303-3. [DOI] [PubMed] [Google Scholar]
- 7.Clark JD, Qiao Y, Li X, Shi X, Angst MS, Yeomans DC. Blockade of the complement C5a receptor reduces incisional allodynia, edema, and cytokine expression. Anesthesiology. 2006;104:1274–1282. doi: 10.1097/00000542-200606000-00024. [DOI] [PubMed] [Google Scholar]
- 8.Jang JH, Clark JD, Li X, Yorek MS, Usachev YM, Brennan TJ. Nociceptive sensitization by complement C5a and C3a in mouse. Pain. 2010;148:343–352. doi: 10.1016/j.pain.2009.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fritzinger DC, Benjamin DE. The Complement System in Neuropathic and Postoperative Pain. Open Pain J. 2016;9:26–37. doi: 10.2174/1876386301609010026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Szebeni J, Muggia FM, Alving CR. Complement activation by Cremophor EL as a possible contributor to hypersensitivity to paclitaxel: an in vitro study. J Natl Cancer Inst. 1998;90:300–306. doi: 10.1093/jnci/90.4.300. [DOI] [PubMed] [Google Scholar]
- 11.Wang TH, Wang HS, Soong YK. Paclitaxel-induced cell death: where the cell cycle and apoptosis come together. Cancer. 2000;88:2619–2628. doi: 10.1002/1097-0142(20000601)88:11<2619::aid-cncr26>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 12.Fishelson Z, Attali G, Mevorach D. Complement and apoptosis. Mol Immunol. 2001;38:207–219. doi: 10.1016/s0161-5890(01)00055-4. [DOI] [PubMed] [Google Scholar]
- 13.Hochreiter-Hufford A, Ravichandran KS. Clearing the dead: apoptotic cell sensing, recognition, engulfment, and digestion. Cold Spring Harbor perspectives in biology. 2013;5:a008748. doi: 10.1101/cshperspect.a008748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lehto T, Morgan BP, Meri S. Binding of human and rat CD59 to the terminal complement complexes. Immunology. 1997;90:121–128. doi: 10.1046/j.1365-2567.1997.00120.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iannaccone PM, Jacob HJ. Rats! Dis Model Mech. 2009;2:206–210. doi: 10.1242/dmm.002733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Popp MW, Maquat LE. Leveraging Rules of Nonsense-Mediated mRNA Decay for Genome Engineering and Personalized Medicine. Cell. 2016;165:1319–1322. doi: 10.1016/j.cell.2016.05.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doench JG, Hartenian E, Graham DB, Tothova Z, Hegde M, Smith I, Sullender M, Ebert BL, Xavier RJ, Root DE. Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nature biotechnology. 2014;32:1262–1267. doi: 10.1038/nbt.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McBurney MW, Fournier S, Jardine K, Sutherland L. Intragenic regions of the murine Pgk-1 locus enhance integration of transfected DNAs into genomes of embryonal carcinoma cells. Somat Cell Mol Genet. 1994;20:515–528. doi: 10.1007/BF02255842. [DOI] [PubMed] [Google Scholar]
- 21.Sakurai T, Watanabe S, Kamiyoshi A, Sato M, Shindo T. A single blastocyst assay optimized for detecting CRISPR/Cas9 system-induced indel mutations in mice. BMC Biotechnol. 2014;14:69. doi: 10.1186/1472-6750-14-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Filipiak WE, Saunders TL. Advances in transgenic rat production. Transgenic Res. 2006;15:673–686. doi: 10.1007/s11248-006-9002-x. [DOI] [PubMed] [Google Scholar]
- 23.Tu Z, Bu H, Dennis JE, Lin F. Efficient osteoclast differentiation requires local complement activation. Blood. 2010;116:4456–4463. doi: 10.1182/blood-2010-01-263590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Polomano RC, Mannes AJ, Clark US, Bennett GJ. A painful peripheral neuropathy in the rat produced by the chemotherapeutic drug, paclitaxel. Pain. 2001;94:293–304. doi: 10.1016/S0304-3959(01)00363-3. [DOI] [PubMed] [Google Scholar]
- 25.Xu JJ, Diaz P, Bie B, Astruc-Diaz F, Wu J, Yang H, Brown DL, Naguib M. Spinal gene expression profiling and pathways analysis of a CB2 agonist (MDA7)-targeted prevention of paclitaxel-induced neuropathy. Neuroscience. 2014;260:185–194. doi: 10.1016/j.neuroscience.2013.12.028. [DOI] [PubMed] [Google Scholar]
- 26.Colucci M, Maione F, Bonito MC, Piscopo A, Di Giannuario A, Pieretti S. New insights of dimethyl sulphoxide effects (DMSO) on experimental in vivo models of nociception and inflammation. Pharmacol Res. 2008;57:419–425. doi: 10.1016/j.phrs.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 27.Le AM, Lee M, Su C, Zou A, Wang J. AMPAkines have novel analgesic properties in rat models of persistent neuropathic and inflammatory pain. Anesthesiology. 2014;121:1080–1090. doi: 10.1097/ALN.0000000000000351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suter MR, Kirschmann G, Laedermann CJ, Abriel H, Decosterd I. Rufinamide attenuates mechanical allodynia in a model of neuropathic pain in the mouse and stabilizes voltage-gated sodium channel inactivated state. Anesthesiology. 2013;118:160–172. doi: 10.1097/ALN.0b013e318278cade. [DOI] [PubMed] [Google Scholar]
- 29.Chaplan SR, Bach FW, Pogrel JW, Chung JM, Yaksh TL. Quantitative assessment of tactile allodynia in the rat paw. Journal of neuroscience methods. 1994;53:55–63. doi: 10.1016/0165-0270(94)90144-9. [DOI] [PubMed] [Google Scholar]
- 30.Greene LA, Tischler AS. Establishment of a noradrenergic clonal line of rat adrenal pheochromocytoma cells which respond to nerve growth factor. Proc Natl Acad Sci U S A. 1976;73:2424–2428. doi: 10.1073/pnas.73.7.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Levi A, Eldridge JD, Paterson BM. Molecular cloning of a gene sequence regulated by nerve growth factor. Science. 1985;229:393–395. doi: 10.1126/science.3839317. [DOI] [PubMed] [Google Scholar]
- 32.Li Y, Smith D, Li Q, Sheibani N, Huang S, Kern T, Nagaraj RH, Lin F. Antibody-mediated retinal pericyte injury: implications for diabetic retinopathy. Invest Ophthalmol Vis Sci. 2012;53:5520–5526. doi: 10.1167/iovs.12-10010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Y, Qiu W, Zhang L, Fung J, Lin F. Painting factor H onto mesenchymal stem cells protects the cells from complement- and neutrophil-mediated damage. Biomaterials. 2016;102:209–219. doi: 10.1016/j.biomaterials.2016.05.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Y, Lin F. Mesenchymal stem cells are injured by complement after their contact with serum. Blood. 2012;120:3436–3443. doi: 10.1182/blood-2012-03-420612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu CC, Lu N, Cui Y, Yang T, Zhao ZQ, Xin WJ, Liu XG. Prevention of paclitaxel-induced allodynia by minocycline: Effect on loss of peripheral nerve fibers and infiltration of macrophages in rats. Mol Pain. 2010;6:76. doi: 10.1186/1744-8069-6-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu J, Tang Y, Xie M, Bie B, Wu J, Yang H, Foss JF, Yang B, Rosenquist RW, Naguib M. Activation of cannabinoid receptor 2 attenuates mechanical allodynia and neuroinflammatory responses in a chronic post-ischemic pain model of complex regional pain syndrome type I in rats. Eur J Neurosci. 2016;44:3046–3055. doi: 10.1111/ejn.13414. [DOI] [PubMed] [Google Scholar]
- 37.Ebenezer GJ, Hauer P, Gibbons C, McArthur JC, Polydefkis M. Assessment of epidermal nerve fibers: a new diagnostic and predictive tool for peripheral neuropathies. J Neuropathol Exp Neurol. 2007;66:1059–1073. doi: 10.1097/nen.0b013e31815c8989. [DOI] [PubMed] [Google Scholar]
- 38.Boyette-Davis J, Xin W, Zhang H, Dougherty PM. Intraepidermal nerve fiber loss corresponds to the development of taxol-induced hyperalgesia and can be prevented by treatment with minocycline. Pain. 2011;152:308–313. doi: 10.1016/j.pain.2010.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang AR, May D, Bourne P, Scott G. PGP9.5: a marker for cellular neurothekeoma. Am J Surg Pathol. 1999;23:1401–1407. doi: 10.1097/00000478-199911000-00011. [DOI] [PubMed] [Google Scholar]
- 40.Ong GL, Mattes MJ. Mouse strains with typical mammalian levels of complement activity. J Immunol Methods. 1989;125:147–158. doi: 10.1016/0022-1759(89)90088-4. [DOI] [PubMed] [Google Scholar]
- 41.Rynes RI. Inherited complement deficiency states and SLE. Clin Rheum Dis. 1982;8:29–47. [PubMed] [Google Scholar]
- 42.Einav S, Pozdnyakova OO, Ma M, Carroll MC. Complement C4 is protective for lupus disease independent of C3. J Immunol. 2002;168:1036–1041. doi: 10.4049/jimmunol.168.3.1036. [DOI] [PubMed] [Google Scholar]
- 43.Walport MJ, Davies KA, Botto M. C1q and systemic lupus erythematosus. Immunobiology. 1998;199:265–285. doi: 10.1016/S0171-2985(98)80032-6. [DOI] [PubMed] [Google Scholar]
- 44.van Dixhoorn MG, Timmerman JJ, Van Gijlswijk-Janssen DJ, Muizert Y, Verweij C, Discipio RG, Daha MR. Characterization of complement C6 deficiency in a PVG/c rat strain. Clin Exp Immunol. 1997;109:387–396. doi: 10.1046/j.1365-2249.1997.4551354.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mashimo T. Gene targeting technologies in rats: zinc finger nucleases, transcription activator-like effector nucleases, and clustered regularly interspaced short palindromic repeats. Dev Growth Differ. 2014;56:46–52. doi: 10.1111/dgd.12110. [DOI] [PubMed] [Google Scholar]
- 46.Hershman DL, Lacchetti C, Dworkin RH, Lavoie Smith EM, Bleeker J, Cavaletti G, Chauhan C, Gavin P, Lavino A, Lustberg MB, Paice J, Schneider B, Smith ML, Smith T, Terstriep S, Wagner-Johnston N, Bak K, Loprinzi CL O. American Society of Clinical. Prevention and management of chemotherapy-induced peripheral neuropathy in survivors of adult cancers: American Society of Clinical Oncology clinical practice guideline. J Clin Oncol. 2014;32:1941–1967. doi: 10.1200/JCO.2013.54.0914. [DOI] [PubMed] [Google Scholar]
- 47.Zitvogel L, Apetoh L, Ghiringhelli F, Kroemer G. Immunological aspects of cancer chemotherapy. Nat Rev Immunol. 2008;8:59–73. doi: 10.1038/nri2216. [DOI] [PubMed] [Google Scholar]
- 48.Makker PG, Duffy SS, Lees JG, Perera CJ, Tonkin RS, Butovsky O, Park SB, Goldstein D, Moalem-Taylor G. Characterisation of Immune and Neuroinflammatory Changes Associated with Chemotherapy-Induced Peripheral Neuropathy. PLoS One. 2017;12:e0170814. doi: 10.1371/journal.pone.0170814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu XJ, Zhang Y, Liu T, Xu ZZ, Park CK, Berta T, Jiang D, Ji RR. Nociceptive neurons regulate innate and adaptive immunity and neuropathic pain through MyD88 adapter. Cell Res. 2014;24:1374–1377. doi: 10.1038/cr.2014.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang H, Li Y, de Carvalho-Barbosa M, Kavelaars A, Heijnen CJ, Albrecht PJ, Dougherty PM. Dorsal Root Ganglion Infiltration by Macrophages Contributes to Paclitaxel Chemotherapy-Induced Peripheral Neuropathy. J Pain. 2016;17:775–786. doi: 10.1016/j.jpain.2016.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang XM, Lehky TJ, Brell JM, Dorsey SG. Discovering cytokines as targets for chemotherapy-induced painful peripheral neuropathy. Cytokine. 2012;59:3–9. doi: 10.1016/j.cyto.2012.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Levin ME, Jin JG, Ji RR, Tong J, Pomonis JD, Lavery DJ, Miller SW, Chiang LW. Complement activation in the peripheral nervous system following the spinal nerve ligation model of neuropathic pain. Pain. 2008;137:182–201. doi: 10.1016/j.pain.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 53.Griffin RS, Costigan M, Brenner GJ, Ma CH, Scholz J, Moss A, Allchorne AJ, Stahl GL, Woolf CJ. Complement induction in spinal cord microglia results in anaphylatoxin C5a-mediated pain hypersensitivity. J Neurosci. 2007;27:8699–8708. doi: 10.1523/JNEUROSCI.2018-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jang JH, Liang D, Kido K, Sun Y, Clark DJ, Brennan TJ. Increased local concentration of complement C5a contributes to incisional pain in mice. J Neuroinflammation. 2011;8:80. doi: 10.1186/1742-2094-8-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Siau C, Xiao W, Bennett GJ. Paclitaxel- and vincristine-evoked painful peripheral neuropathies: loss of epidermal innervation and activation of Langerhans cells. Exp Neurol. 2006;201:507–514. doi: 10.1016/j.expneurol.2006.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lauria G, Lombardi R. Skin biopsy: a new tool for diagnosing peripheral neuropathy. BMJ. 2007;334:1159–1162. doi: 10.1136/bmj.39192.488125.BE. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.