Abstract
The mature HIV-1 reverse transcriptase is a heterodimer that comprises 66 kDa (p66) and 51 kDa (p51) subunits. The latter is formed by HIV-1 protease-catalyzed removal of a C-terminal ribonuclease H domain from a p66 subunit. This proteolytic processing is a critical step in virus maturation and essential for viral infectivity. Here, we report that tRNA significantly enhances in vitro processing even at a substoichiometric tRNA:p66/p66 ratio. Other double stranded RNAs have considerably less pronounced effect. Our data support a model where interaction of p66/p66 with tRNA introduces conformational asymmetry in the two subunits, permitting specific proteolytic processing of one p66 to provide the mature RT p66/p51 heterodimer.
Keywords: HIV-1, proteolysis, maturation, tRNA, reverse transcriptase, RNase H
Graphical Abstract
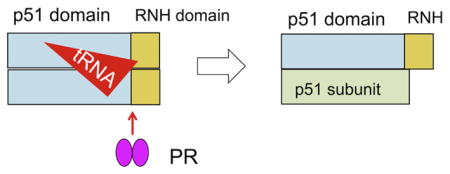
Introduction
HIV-1 reverse transcriptase (RT) is a multifunctional enzyme with both DNA polymerase and ribonuclease H (RNH) activities and is essential for HIV-1 replication 1. While the gene for RT encodes a 66 kDa protein, RT is initially translated as part of the 160 kDa Gag-Pol precursor polyprotein, which is proteolytically processed by HIV-1 protease (PR) during virion assembly and maturation. The mature RT is a heterodimer comprising 66 kDa (p66) and 51 kDa (p51) subunits; the p51 subunit is generated upon proteolytic cleavage of the p66 subunit between residues 440 and 441, thereby removing most of the RNH domain from this p66 subunit 1–5. This cleavage event is essential for viral infectivity 6; 7.
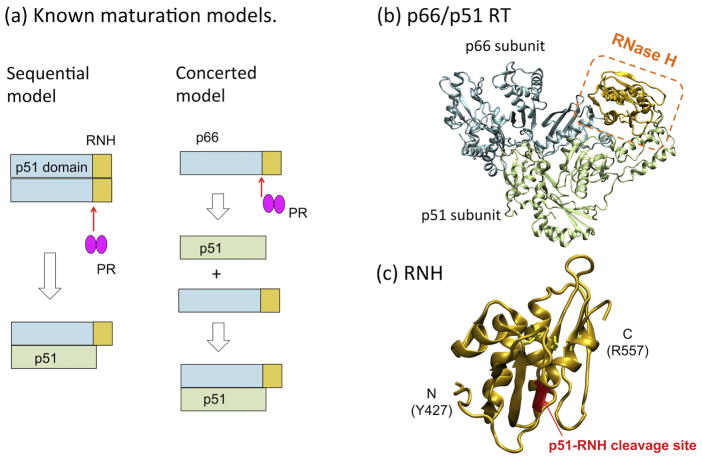
Formation of the mature p66/p51 heterodimer is generally thought to proceed via a p66/p66 homodimer intermediate (Sequential model in Fig. 1a), rather than generation of p51 followed by p66/p51 heterodimer formation (Concerted model in Fig. 1a) 8–10. However, data in support of the Sequential model are primarily based on studies that introduce mutations leading to dissociation of subunits in p66/p66 in vitro or at a viral level 8–10. Although lack of p66/p51 production using the mutants may support the requirement of the homodimer formation for the RT maturation (i.e., Sequential model), the mutants were originally known to affect p66/p51 heterodimer formation 11–14. Therefore, such experiments do not consider whether the mutations themselves diminish maturation of p66/p66 or whether they cause p66/p51 dissociation which in turn results in decreased RT detection. Further, structures of both p66/p51 (Fig. 1b) and the isolated RNH domain indicate that the p51-RNH cleavage site is buried within the core β-sheet of the folded RNH domain (Fig. 1c) and likely inaccessible to the protease 15–20. Despite efforts to determine the structure of the RNH domain in the immature RT p66/p66 homodimer, a detailed structure of p66/p66 is not available 21–24.
Figure 1.
(a) Concerted and Sequential Models for RT maturation 5; 8–10; (b) location of the RNH domain in the known p66/p51 RT structure, and (c) an expanded view of the RNH domain. In (a)–(c), the p51 and the RNH domains in the p66 subunit are shown in cyan and orange, respectively, while the p51 subunit is shown in lime color. In (b) and (c) the p51-RNH cleavage site (F440-Y441) is shown as a red ribbon. The images were generated using PDB code 1DLO52.
Our previous NMR studies suggested that the p51-RNH cleavage sites in both subunits of p66/p66 were buried and thus poorly accessible to protease 24. We therefore postulated that some virion-associated factor may play a role in promoting the RT maturation process. HIV-1 virions are known to contain substantial amounts of cellular tRNA 25–29, and RT can bind such tRNAs with nanomolar affinity 30. Here, we describe biochemical experiments to assess the impact of tRNA on the in vitro HIV-1 PR-catalyzed cleavage of p66/p66 to mature RT p66/p51. We show that processing of p66/p66 is slow and results in numerous aberrant products in the absence of tRNA. Surprisingly, in the presence of even a sub-stoichiometric tRNA:p66/p66 ratio, processing of p66/p66 by HIV-1 PR is greatly accelerated and with few or no aberrant cuts. Our data, obtained using two different p66 concentrations, show that the mature p66/p51 arises from proteolytic processing of the p66/p66, and not from processing of p66 monomers, in agreement with previously published data 8; 9; 31; 32. Although a certain processing enhancement was observed in the presence of ds-RNA, based on the fact that HIV-1 virions are known to contain substantial amounts of cellular tRNA 25–29, we propose a model where virion-encapsidated tRNA facilitates RT maturation to p66/p51 that is essential for HIV-1 replication.
Results and Discussion
p66/p66 homodimer formation is essential for efficient RT proteolytic processing
We evaluated the kinetics of in vitro proteolytic processing by HIV-1 PR at different concentrations of RT p66 in the absence and in the presence of tRNA (Fig. 2).
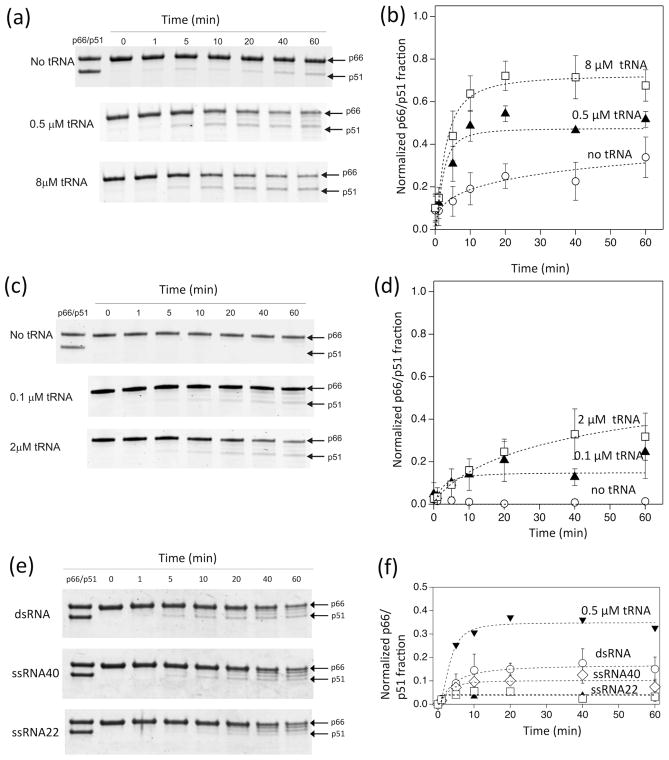
Figure 2.
Time dependence of p66 processing by HIV-1 PR in 20 mM sodium acetate buffer at pH 5.2 and 37 °C, monitored by SDS-PAGE (a and b) at a high concentration of p66 (8 μM as a p66 monomer, 4 μM as a dimer) proteolytically processed by 1 μM HIV-1 PR in the presence of 0, 0.5, 8 μM tRNA concentrations, (c and d) at a low concentration of p66 (1 μM p66 monomer concentration, 0.5 μM as a dimer) processed by 0.25 μM HIV-1 PR in the presence of 0, 0.1, and 2 μM tRNA concentrations, and (e and f) at a high concentration of p66 (8 μM as a p66 monomer, 4 μM as a dimer) proteolytically processed by 1 μM HIV-1 PR in the presence of 0.5 μM ds-RNA (40 nt/22 nt), ss-RNA (40 nt), and ss-RNA (22 nt). In (b, d, and f), p66/p51 fractions, determined from the gel images in (a, c, and e), are shown, respectively. Average data points were used to fit a curve with one standard deviation as an uncertainty of each data point. Since p66/p51 production occurs in parallel with p66 degradation by PR, the build-up curves do not necessarily reach a plateau. Because of these multiple factors, normalized χ2, χn2, values for the curve fits were between 0.2 and 10.7.
With 8 μM p66, over 50% of RT is predicted to be the p66/p66 homodimer, based on a p66/p66 dissociation constant of ~4 μM 33–35. In the absence of tRNA, minimal formation (<10%) of p51 is noted after 1 hour (Fig. 2a and 2b). In contrast, with stoichiometric amounts of tRNA, essentially complete processing to equivalent amounts of p66 and p51 (the RT p66/p51 heterodimer) is seen within 30 – 40 minutes. Under the same conditions, enhancement of processing is noted even with sub-stoichiometric levels of tRNA (0.5 μM). Surprisingly, the extent of processing under these sub-stoichiometric conditions, after 20 minutes, was more than 50% of that observed for the 8 μM tRNA samples. This suggests that the tRNA may act as a catalytic factor in the processing event.
In contrast, at low p66 concentrations (1 μM) where p66/p66 homodimer formation is minimal (<40%) 33–35, very little p51 was formed even after 1 h incubation, either in the absence of tRNA or in the presence of excess tRNA (2 μM) (Fig. 2c and 2d). Significant aberrant processing was also evident with low p66 and excess tRNA (Fig. S1). Because of such p66 degradation, p66/p51 production did not necessarily saturate as a function of time (Fig. 2). These results suggest that p66/p66 formation is important for proper maturation of RT to the p66/p51 heterodimer.
To determine whether maturation enhancement occurs in the presence of other RNAs, a similar set of experiments was performed using some other small RNA molecules: ds-RNA (40/22 nt), ss-RNA (40 nt) and ss-RNA (22 nt) (Fig. 2e and 2f). We chose the sub-stoichiometric nucleic acid concentration, 0.5 μM, to see the effect of RNA on the RT maturation but not the effect of stabilizing the matured RT. The positive control, 0.5 μM tRNA, showed similar, although slightly lower, activity compared to that obtained in our first set of experiments (see Fig. 2a & 2b), presumably due to slightly lower activity of PR used in the experiments in Fig. 2e. The efficiency of RT maturation in the presence of ds-RNA was about half, ~47%, of that obtained in the presence of tRNA (Fig. 2e & 2f). In the presence of the ss-RNAs, the efficiency was even lower, 29%. The results demonstrate that ds-RNA, in addition to tRNA, also enhances RT maturation, although to a lesser degree for those tested in our study.
Other factors potentially impacting tRNA-mediated p66 processing
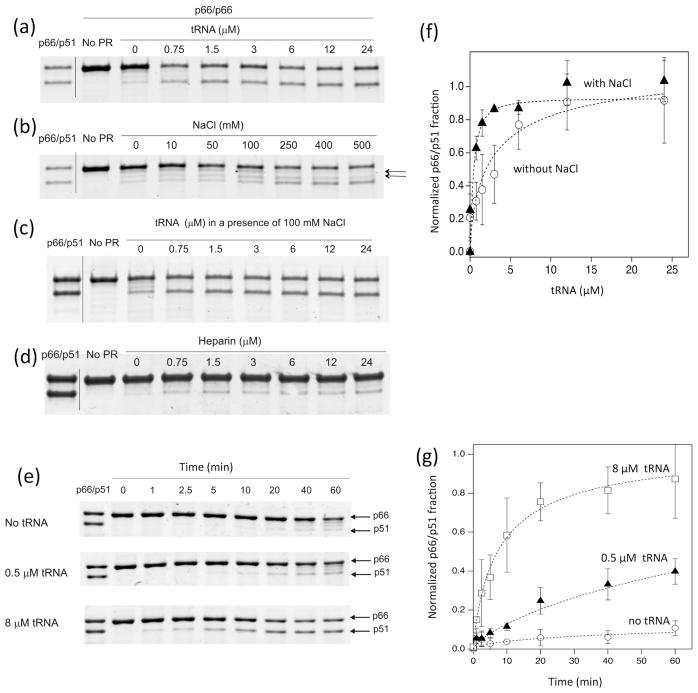
The data presented in Fig. 2 show that tRNA facilitates the formation of RT p66/p51 heterodimers. Confirmation was obtained using dose-dependency experiments, in which p66/p51 production by PR was assessed at different tRNA concentrations (Fig. 3a). Since both p66/p66 dimer formation and PR activity may be influenced by ionic strength 36; 37, and tRNA may act as a polyanion 38–40, we investigated the impact of different salt concentrations on p66 processing. In the absence of tRNA, at physiological NaCl concentrations (50–150 mM), aberrant non-specific cleavage products (Fig. 3b, arrows), with molecular sizes between those of p51 and p66, were seen; significant p66/p51 formation became evident only above 400 mM NaCl (Fig. 3b). In contrast, even at low levels of tRNA, substantial RT p66/p51 formation was noted in the presence of 100 mM NaCl (Fig. 3c). Heparin did not enhance p66/p66 maturation (Fig. 3d), and PR activity in the assay with fluorescent HIV protease substrate 1 (Sigma) was actually less in the presence of 0.5, 1.0 or 5.0 μM tRNA (concentrations at which we observed enhanced p66 processing efficiency) compared to its activity without tRNA or at 10 μM tRNA (Fig. S2), indicating that a simple PR activity enhancement by polyanions does not explain our results 40. These data show that while higher ionic strength may enhance proteolytic activity, as well as dimer formation, it does not impact the total production of p66/p51 (Fig. 3f).
Figure 3.
Effects of varying amounts of (a) tRNA, (b) NaCl, (c) tRNA in the presence of 100 mM NaCl, and (d) heparin on p66 processing by HIV-1 PR for 20 min at 37 ºC, and (e) the time dependence of p66 processing by HIV-1 PR in the presence of 100 mM NaCl at 20 ºC. In all the experiments, reactions were initiated by the addition of 1 μM HIV-1 PR to p66 at a high concentration of p66 (8 μM as a monomer, 4 μM as a dimer) and monitored by SDS-PAGE, and the buffer contains 20 mM sodium acetate at pH 5.2. In (e), experiments were carried out in the presence of 0, 0.5, and 8 μM tRNA. (f) Plots of intensity changes and the fit curves, χn2 values 2.7 and 4.5, are shown for (a) and (c), respectively. (g) Plots of intensity changes and the fit curves, χn2 values from 0.88 to 2.2, are shown for (f). In (b), aberrant non-specific cleavage products are shown by arrows.
Kinetic experiments of p66 processing in a buffer that contained 100 mM NaCl (Fig. 3e and 3g and Fig. S3) also showed a pattern of the p66/p51 production similar to that observed in a buffer lacking 100 mM NaCl (Fig. 2a), i.e., tRNA impacts p66/p51 production. Of note, the reaction was performed at 20 ºC instead of 37 ºC because the PR catalytic rate increases in the presence of 100 mM NaCl, compared to the absence of NaCl (37 ºC, in Fig. S3). Overall, our data, obtained with different NaCl conditions (Fig. 2 and 3), consistently show that tRNA influences the selectivity of cleavage at the processing site. Although there are reports that tRNA may act as a polyanion 38–40, tRNA clearly increases selectivity of the processing site for p66/p51 production in the presented RT maturation experiments.
p66/p66 interaction with tRNA
Our biochemical in vitro proteolytic processing studies strongly suggest that RT p66/p66 homodimer is the substrate for HIV-1 PR processing to mature RT p66/p51 heterodimer. To further evaluate this, we carried out biophysical analysis of the RT species using size-exclusion chromatography (Fig. 4).
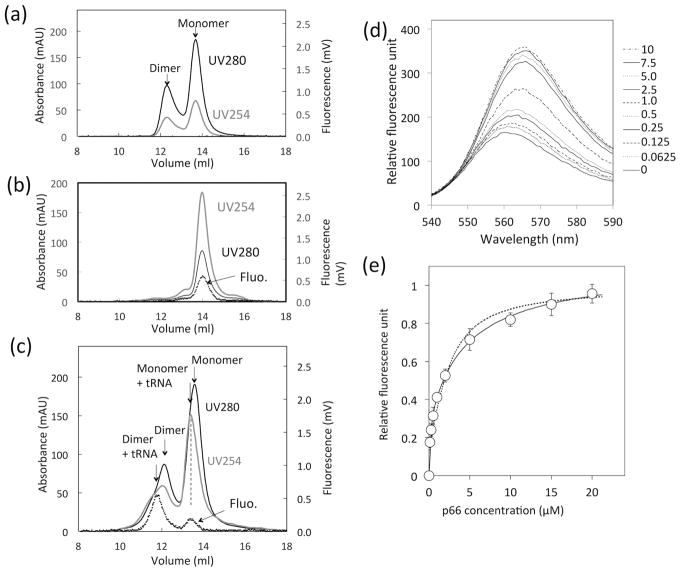
Figure 4.
tRNA binding to p66 protein, monitored by (a – c) SEC elution profiles of (a) p66 protein solution, (b) tRNA solution, and (c) that mixed with tRNA, and (d, e) change of fluorescence emission of Cy3-labeled tRNA at varying p66 concentrations obtained by spectrofluorometry. In (a – c), UV absorbance at 254 nm (gray line) and 280 nm (black line), and fluorescence emission at 560 nm (dashed line) are shown. In (d), changes in fluorescence of total 1 μM tRNA, containing 40 nM Cy3-labeled tRNA, as a function of p66 concentration were recorded. In (e), fluorescence intensity changes at 560 nm (d) are plotted. The dash line is a fit-curve calculated with a single binding mode model (χn2 = 6.5), and the solid line indicates a fit-curve calculated with a two-binding mode model (χn2 = 0.8). The null hypothesis was rejected with p = 0.00036.
In the absence of tRNA, both monomer and dimer peaks of p66 were observed, consistent with previous data (Fig. 4a) 24. tRNA alone eluted at a slightly greater volume than p66 monomer alone, based on both UV and the fluorescence detection (Fig. 4b). In the mixture of p66 and tRNA, two additional peak signals, compared to p66 alone, were noted (Fig. 4c); these stem from p66/p66-tRNA and p66-tRNA complex species, as confirmed by the fluorescence emission of Cy3-labeled tRNA. The loaded p66 and tRNA concentrations, 40 μM and 5 μM, respectively, are empirically estimated to be 4 μM p66 and 0.5 μM tRNA on a column, which are similar to the conditions used in the processing experiments (Fig. 2a).
The tRNA interaction with p66 protein was quantified by recording changes in fluorescence emission of Cy3-labeled tRNA at varying p66 concentration in the fluorescence spectroscopic analysis (Fig. 4d). The emission changes at 560 nm could not be described by a single-site binding model (Fig. 4e, dashed line) and were better explained with a two-binding mode model (Fig. 4e, solid line) that gave two Kd values: 65.8 ± 26.9 nM and 2.38 ± 0.69 μM. When considered in the context of the known concentrations of p66, we conclude that these dissociation constants reflect tRNA dissociation from p66/p66 (dimer) and from p66 (monomer), respectively. Competitive gel mobility shift assay of a solution containing tRNA, p66, and PR suggests that tRNA more strongly interacts with p66 than PR (Fig. S4). Taken together, we confirmed the p66/p66-tRNA interaction. In addition, observation of p66-tRNA species explains why p66 processing at low p66 concentration showed tRNA dependence (Fig. 2c and 2d).
A proposed model for RT maturation
Here, we demonstrate that tRNA interacts with p66/p66 homodimer and facilitates selective cleavage at the p51-RNH site by HIV-1 PR (Fig. 2). We also demonstrate that the effect of tRNA on the selectivity of p66 processing is distinct from the effect of ionic strength (Fig. 3 and S2). Consistent with the processing experiments, both tRNA-p66/p66 and tRNA-p66 forms were observed. Although tRNA can bind p66/p51 tightly (KD = 3–50 nM 5; 30; 41), tRNA interaction with p66/p66 is the same as, or weaker than, the p66/p51 binding (Fig. 4e). Because of this moderate binding and observation of the significant tRNA effect at a sub-stoichiometric concentration, it is possible that transient interaction of tRNA with p66/p66 mediates the selective processing at the p51-RNH site, with tRNA likely being released before heterodimer formation is complete 5; 35; 37. Such gain of RT maturation at the sub-stoichiometric concentration suggests that the tRNA may serve as a catalytic molecule in RT maturation, rather than just stabilizing the matured RT in solution.
Our in vitro data suggests that ds-RNA, in addition to tRNA, also enhances RT maturation, although to a lesser degree for those tested in our study. Considering this in vitro data, we can not conclude which RNA mediates the RT maturation in virio. However, these data are consistent with previously observed changes in enzymatic activities of p66 upon tRNA interaction 42; 43. The proposed model also explains why p66/p51 formation in cells occurs efficiently within one hour 44; 45, whereas in vitro, in the absence of nucleic acid, RT heterodimer formation takes significantly longer and with lower yield. HIV-1 is known to contain numerous copies of multiple tRNA species in addition to the essential primer tRNA 25–29, thus, these other tRNA species may play a role in directing appropriate proteolytic maturation of HIV polyproteins, especially the conversion of the RT p66/p66 homodimer to the mature RT p66/p51 heterodimer.
Conclusion
Our data show that the RT p66/p66 can interact with tRNA and that this interaction facilitates selective cleavage at the p51-RNH site by HIV PR. Importantly, we also show that this selective cleavage is independent of ionic strength, but dependent on the concentration of RT p66, a factor directly related to the RT p66 subunit dissociation strength. Facilitation of the selective cleavage at the p51-RNH site by HIV-1 PR is significant even at a sub-stoichiometric tRNA concentration. We propose a model in which interaction of the p66/p66 homodimer with tRNA introduces conformational asymmetry in the two subunits, permitting specific proteolytic processing of one of the p66 subunits leading to formation of the mature p66/p51 RT.
Materials and Methods
Protein expression and purification
p66/p51 HIV-1 RT was prepared using the p6HRT-PROT plasmid 46 as previously described 47; 48. The p66 sequence from p6HRT-PROT used for p66/p51 expression 46 was cloned into the pPSG-IBA3 vector using the StarGate cloning system (IBA Solutions for Life Sciences, Göttingen, Germany). p66 protein was expressed in BL21 (DE3) E. coli cells and purified using a Strep-Tactin gravity flow column (IBA Solutions). Protein concentration (calculated as p66 monomer) was determined by measuring absorbance at 280 nm with an extinction coefficient of 137405 M−1 cm−1. Purified p66 and p66/p51 were stored in 25 mM sodium phosphate, pH 7.0, 250 mM NaCl and 50% v/v glycerol at −80 °C. HIV-1 PR was expressed and purified as previously described 49–51.
HIV-1 PR-catalyzed processing of p66/p66
Proteolytic processing of p66 protein was evaluated using kinetic (time-course) experiments that determined the rate of processing, and fixed-time experiments that assessed the impact of differences in tRNA:protein ratio on extent of processing. Since a monomer:dimer ratio of p66 depends on protein concentration and tRNA concentration, all p66 concentrations are reported as monomer protein concentration for the Materials and Methods purpose. Processing experiments were carried out in 20 mM sodium acetate buffer, pH 5.2, at 37 ºC, unless otherwise noted.
Kinetic experiments were conducted at two p66 concentrations: “high p66 concentration” (8 μM), where RT is predominantly in the p66/p66 homodimer form and processed by 1 μM PR in the presence of 0, 0.5, or 8 μM synthetized tRNALys3 of human sequence (TriLink BioTechnologies LLC, San Diego, CA), and “low p66 concentration” (1 μM), where RT is predominantly in the p66 monomer form and processed using 0.25 μM PR in the presence of 0, 0.1, or 2 μM tRNA. Additional kinetic experiments at high p66 (8 μM) were conducted in the sodium acetate buffer containing 100 mM NaCl at 20 ºC. Fixed-time experiments were conducted using high p66 (8 μM) conditions only, at different concentrations of tRNA or NaCl or heparin (Sigma-Aldrich. St Louis, MO), which were added to p66 protein prior to starting the reaction, incubated at room temperature for 5 min, and allowed to react at 37 ºC for 10 – 20 min. The fixed-time experiments at different tRNA concentrations were performed in sodium acetate buffer with/without 100 mM NaCl.
The kinetic (time-course) modes of p66 proteolytic processing was evaluated at 8 μM p66 and 1 μM PR, in the presence of 0.5μM ds-RNA (40/22 nt), ss-RNA (40 nt) and ss-RNA (20 nt), of which 40 nt and 20 nt sequences are 5′-AGGUGAGUGAGAUGAUAACAAAUUUGCGAGCCCCAGAUG and 5′-GCAUCUGGGGCUCGCAAAUUUG, respectively (TriLink BioTechnologies LLC, San Diego, CA).
In all the processing experiments, reactions were stopped by addition of Tricine sample loading buffer (Bio-Rad Laboratories, Berkeley, CA) and denaturation at 95 ºC for 5 min. Samples were loaded onto precast 4–15% Tris-glycine gels (Bio-Rad) and stained with Bio-safe Coomassie stain (Bio-Rad). Band intensities were quantified using an Odyssey CLX gel imaging system by Image Studio software (Li-Cor Biosciences, Lincoln, NE) or an Amersham Imager 600 (GE Healthcare Life Sciences). For quantification, these gel experiments were repeated at least three times. Production of RT p66/p51 heterodimer was determined based on the ratio of p66 to p51 band intensities in the following way: the ratio of p66 to p51 band intensities of a reference heterodimer was first quantified in the same gel, and, production of the heterodimer against the initial p66 intensity was determined using the p51 band intensity normalized by the reference p66/p51 intensity ratio. An average of the three quantified p66/p51 production was plotted with the standard deviation as an error bar. Trends of intensity changes were shown by fit curves using Igor (Wavemetrics, Inc., Lake Oswego, OR).
Analytical Size Exclusion Chromatography to monitor p66/p66-tRNA interaction
All Size Exclusion Chromatography (SEC) experiments used a 24-ml analytical Superdex 200 Increase 10/300 GL column (GE Healthcare), mobile phase of 25 mM Bis-tris buffer, pH 7.0, containing 100 mM NaCl with 0.02% sodium azide at a flow rate of 0.5 ml/min. Injection volume was 50 μl, and protein elution was monitored by UV absorbance at 254 and 280 nm. Elution profiles of 40 μM p66 RT were evaluated in the absence of tRNA, or following preincubation with 5 μM tRNA containing tracer tRNA 3′-end labeled with pCp-Cy3 (Jena Bioscience, Jena, Germany). As a control, the labeled tRNA was injected without mixing with p66. With the SEC experiments that contain labeled tRNA, in addition to UV, the fluorescence emission at 560 nm (excitation 485 at nm) was also measured using an in-line Shimadzu RF-10AXL Fluorescence Detector.
Fluorescence spectroscopic analysis of RT p66/p66 - tRNA interaction
The interaction of tRNA with RT p66 was evaluated in 25 mM Bis-Tris, pH 7.0, 100 mM NaCl. RT p66 protein in various concentrations was incubated for 30 min with 1 μM total tRNA containing tracer Cy3-labeled tRNA. Emission spectra were collected using a FluoroMax-4 (Horiba Scientific, Edison, NJ) with excitation at 485 nm. All experiments were carried out at least three separate times to determine an average and a standard deviation of the data. The change in fluorescence at 560 nm was plotted at different protein concentrations 0 – 20 μM with error bars representing one standard deviation. The tRNA dissociation constant, KD, was determined assuming two models, one with a single KD and the other with two independent KDs, using χ2-minimization routine in Matlab (MathWorks, Natick, MA), and evaluated using F-test.
HIV-1 protease-catalyzed processing of premature reverse transcriptase p66 to yield p66/p51 heterodimer was investigated at various conditions in vitro.
p66/p66 homodimer is the substrate for HIV-1 protease processing to become a mature p66/p51 heterodimer.
tRNA interacts with p66/p66 homodimer and facilitates selective cleavage at the p51-RNH site by HIV-1 protease.
Acknowledgments
This study was supported by grants from the National Institutes of Health (R01GM105401 to RI and RLS, R01GM109767 to RI, R01AI00890 to TI, SGS, and MAP, P50GM103368 to JHE, SGS, and MAP). We thank Michel Guerrero for his technical assistance, Michael Tsang at the University of Pittsburgh for use of Odyssey CLX gel imaging system and Teresa Brosenitsch for reading the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Coffin JM, Hughes SH, Varmus HE. Retroviruses. Cold Spring Harbor Laboratory Press; Plainview, NY: 1997. [PubMed] [Google Scholar]
- 2.Chattopadhyay D, Evans DB, Deibel MR, Jr, Vosters AF, Eckenrode FM, Einspahr HM, Hui JO, Tomasselli AG, Zurcher-Neely HA, Heinrikson RL, et al. Purification and characterization of heterodimeric human immunodeficiency virus type 1 (HIV-1) reverse transcriptase produced by in vitro processing of p66 with recombinant HIV-1 protease. J Biol Chem. 1992;267:14227–32. [PubMed] [Google Scholar]
- 3.Sharma SK, Fan N, Evans DB. Human immunodeficiency virus type 1 (HIV-1) recombinant reverse transcriptase. Asymmetry in p66 subunits of the p66/p66 homodimer. FEBS Lett. 1994;343:125–30. doi: 10.1016/0014-5793(94)80303-x. [DOI] [PubMed] [Google Scholar]
- 4.Katz RA, Skalka AM. The retroviral enzymes. Annu Rev Biochem. 1994;63:133–73. doi: 10.1146/annurev.bi.63.070194.001025. [DOI] [PubMed] [Google Scholar]
- 5.Divita G, Rittinger K, Geourjon C, Deleage G, Goody RS. Dimerization kinetics of HIV-1 and HIV-2 reverse transcriptase: a two step process. J Mol Biol. 1995;245:508–21. doi: 10.1006/jmbi.1994.0042. [DOI] [PubMed] [Google Scholar]
- 6.Abram ME, Parniak MA. Virion instability of human immunodeficiency virus type 1 reverse transcriptase (RT) mutated in the protease cleavage site between RT p51 and the RT RNase H domain. Journal of Virology. 2005;79:11952–11961. doi: 10.1128/JVI.79.18.11952-11961.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abram ME, Sarafianos SG, Parniak MA. The mutation T477A in HIV-1 reverse transcriptase (RT) restores normal proteolytic processing of RT in virus with Gag-Pol mutated in the p51-RNH cleavage site. Retrovirology. 2010;7:6. doi: 10.1186/1742-4690-7-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sluis-Cremer N, Arion D, Abram ME, Parniak MA. Proteolytic processing of an HIV-1 pol polyprotein precursor: insights into the mechanism of reverse transcriptase p66/p51 heterodimer formation. International Journal of Biochemistry & Cell Biology. 2004;36:1836–1847. doi: 10.1016/j.biocel.2004.02.020. [DOI] [PubMed] [Google Scholar]
- 9.Wapling J, Moore KL, Sonza S, Mak J, Tachedjian G. Mutations that abrogate human immunodeficiency virus type 1 reverse transcriptase dimerization affect maturation of the reverse transcriptase heterodimer. Journal of Virology. 2005;79:10247–10257. doi: 10.1128/JVI.79.16.10247-10257.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Figueiredo A, Moore KL, Mak J, Sluis-Cremer N, de Bethune MP, Tachedjian G. Potent nonnucleoside reverse transcriptase inhibitors target HIV-1 Gag-Pol. PLoS Pathog. 2006;2:e119. doi: 10.1371/journal.ppat.0020119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ghosh M, Jacques PS, Rodgers DW, Ottman M, Darlix JL, leGrice SFJ. Alterations to the primer grip of p66 HIV-1 reverse transcriptase and their consequences for template-primer utilization. Biochemistry. 1996;35:8553–8562. doi: 10.1021/bi952773j. [DOI] [PubMed] [Google Scholar]
- 12.Tachedjian G, Aronson HEG, Goff SP. Analysis of mutations and suppressors affecting interactions between the subunits of the HIV type 1 reverse transcriptase. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:6334–6339. doi: 10.1073/pnas.97.12.6334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tachedjian G, Aronson HE, de los Santos M, Seehra J, McCoy JM, Goff SP. Role of residues in the tryptophan repeat motif for HIV-1 reverse transcriptase dimerization. J Mol Biol. 2003;326:381–96. doi: 10.1016/s0022-2836(02)01433-x. [DOI] [PubMed] [Google Scholar]
- 14.Tachedjian G, Moore KL, Goff SP, Sluis-Cremer N. Efavirenz enhances the proteolytic processing of an HIV-1 pol polyprotein precursor and reverse transcriptase homodimer formation. FEBS Lett. 2005;579:379–84. doi: 10.1016/j.febslet.2004.11.099. [DOI] [PubMed] [Google Scholar]
- 15.Davies JF, 2nd, Hostomska Z, Hostomsky Z, Jordan SR, Matthews DA. Crystal structure of the ribonuclease H domain of HIV-1 reverse transcriptase. Science. 1991;252:88–95. doi: 10.1126/science.1707186. [DOI] [PubMed] [Google Scholar]
- 16.Jacobo-Molina A, Arnold E. HIV reverse transcriptase structure-function relationships. Biochemistry. 1991;30:6351–6. doi: 10.1021/bi00240a001. [DOI] [PubMed] [Google Scholar]
- 17.Hostomska Z, Matthews DA, Davies JF, 2nd, Nodes BR, Hostomsky Z. Proteolytic release and crystallization of the RNase H domain of human immunodeficiency virus type 1 reverse transcriptase. J Biol Chem. 1991;266:14697–702. [PubMed] [Google Scholar]
- 18.Kohlstaedt LA, Wang J, Friedman JM, Rice PA, Steitz TA. Crystal structure at 3.5 A resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science. 1992;256:1783–1790. doi: 10.1126/science.1377403. [DOI] [PubMed] [Google Scholar]
- 19.Jacobo-Molina A, Ding J, Nanni RG, Clark ADJ, Lu X, Tantillo C, Williams RL, Kamer G, Ferris AL, Clark P, Hizi A, Hughes SH, Arnold E. Crystal structure of human immunodeficiency virus type 1 reverse transcriptase complexed with double-stranded DNA at 3.0 A resolution shows bent DNA. Proc Natl Acad Sci U S A. 1993;90:6320–6324. doi: 10.1073/pnas.90.13.6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pari K, Mueller GA, DeRose EF, Kirby TW, London RE. Solution structure of the RNase H domain of the HIV-1 reverse transcriptase in the presence of magnesium. Biochemistry. 2003;42:639–50. doi: 10.1021/bi0204894. [DOI] [PubMed] [Google Scholar]
- 21.Zheng XH, Pedersen LC, Gabel SA, Mueller GA, Cuneo MJ, DeRose EF, Krahn JM, London RE. Selective unfolding of one Ribonuclease H domain of HIV reverse transcriptase is linked to homodimer formation. Nucleic Acids Research. 2014;42:5361–5377. doi: 10.1093/nar/gku143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zheng X, Perera L, Mueller GA, DeRose EF, London RE. Asymmetric conformational maturation of HIV-1 reverse transcriptase. Elife. 2015;4:e06359. doi: 10.7554/eLife.06359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zheng XH, Pedersen LC, Gabel SA, Mueller GA, DeRose EF, London RE. Unfolding the HIV-1 reverse transcriptase RNase H domain - how to lose a molecular tug-of-war. Nucleic Acids Research. 2016;44:1776–1788. doi: 10.1093/nar/gkv1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sharaf NG, Poliner E, Slack RL, Christen MT, Byeon IJ, Parniak MA, Gronenborn AM, Ishima R. The p66 immature precursor of HIV-1 reverse transcriptase. Proteins. 2014;82:2343–52. doi: 10.1002/prot.24594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kleiman L, Caudry S, Boulerice F, Wainberg MA, Parniak MA. Incorporation of tRNA into normal and mutant HIV-1. Biochem Biophys Res Commun. 1991;174:1272–80. doi: 10.1016/0006-291x(91)91559-u. [DOI] [PubMed] [Google Scholar]
- 26.Jiang M, Mak J, Ladha A, Cohen E, Klein M, Rovinski B, Kleiman L. Identification of tRNAs incorporated into wild-type and mutant human immunodeficiency virus type 1. J Virol. 1993;67:3246–53. doi: 10.1128/jvi.67.6.3246-3253.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mak J, Jiang M, Wainberg MA, Hammarskjold ML, Rekosh D, Kleiman L. Role of Pr160gag-pol in mediating the selective incorporation of tRNALys into human immunodeficiency virus type 1 particles. J Virol. 1994;68:2065–72. doi: 10.1128/jvi.68.4.2065-2072.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kleiman L, Cen S. The tRNALys packaging complex in HIV-1. Int J Biochem Cell Biol. 2004;36:1776–86. doi: 10.1016/j.biocel.2004.02.022. [DOI] [PubMed] [Google Scholar]
- 29.Pavon-Eternod M, Wei M, Pan T, Kleiman L. Profiling non-lysyl tRNAs in HIV-1. RNA. 2010;16:267–73. doi: 10.1261/rna.1928110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arion D, Harada R, Li XG, Wainberg MA, Parniak MA. HIV-1 reverse transcriptase shows no specificity for the binding of primer tRNA(LyS3) Biochemical and Biophysical Research Communications. 1996;225:839–843. doi: 10.1006/bbrc.1996.1260. [DOI] [PubMed] [Google Scholar]
- 31.Chiang CC, Wang SM, Pan YY, Huang KJ, Wang CT. A single amino acid substitution in HIV-1 reverse transcriptase significantly reduces virion release. J Virol. 2010;84:976–82. doi: 10.1128/JVI.01532-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chiang CC, Tseng YT, Huang KJ, Pan YY, Wang CT. Mutations in the HIV-1 reverse transcriptase tryptophan repeat motif affect virion maturation and Gag-Pol packaging. Virology. 2012;422:278–87. doi: 10.1016/j.virol.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 33.Sluis-Cremer N, Dmitrienko GI, Balzarini J, Camarasa MJ, Parniak MA. Human immunodeficiency virus type 1 reverse transcriptase dimer destabilization by 1-{spiro[4′-amino-2′,2′-dioxo-1′,2′-oxathiole-5′,3′-[2′,5′-bis-O-(tert-butyldimethylsilyl)-beta-D-ribofuranosyl]]}-3-ethylthymine. Biochemistry. 2000;39:1427–1433. doi: 10.1021/bi991682+. [DOI] [PubMed] [Google Scholar]
- 34.Venezia CF, Howard KJ, Ignatov ME, Holladay LA, Barkley MD. Effects of efavirenz binding on the subunit equilibria of HIV-1 reverse transcriptase. Biochemistry. 2006;45:2779–89. doi: 10.1021/bi051915z. [DOI] [PubMed] [Google Scholar]
- 35.Venezia CF, Meany BJ, Braz VA, Barkley MD. Kinetics of association and dissociation of HIV-1 reverse transcriptase subunits. Biochemistry. 2009;48:9084–93. doi: 10.1021/bi9010495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tropea JE, Nashed NT, Louis JM, Sayer JM, Jerina DM. Effect of salt on the kinetic parameters of retroviral and mammalian aspartic acid proteases. Bioorg Chem. 1991;20:67–76. [Google Scholar]
- 37.Cabodevilla JF, Odriozola L, Santiago E, Martinez-Irujo JJ. Factors affecting the dimerization of the p66 form of HIV-1 reverse transcriptase. Eur J Biochem. 2001;268:1163–72. doi: 10.1046/j.1432-1327.2001.01939.x. [DOI] [PubMed] [Google Scholar]
- 38.Porter DJT, Hanlon MH, Carter LH, Danger DP, Furfine ES. Effectors of HIV-1 Protease Peptidolytic Activity. Biochemistry. 2001;40:11131–11139. doi: 10.1021/bi010418p. [DOI] [PubMed] [Google Scholar]
- 39.Lipfert J, Doniach S, Das R, Herschlag D. Understanding nucleic acid-ion interactions. Annu Rev Biochem. 2014;83:813–41. doi: 10.1146/annurev-biochem-060409-092720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Potempa M, Nalivaika E, Ragland D, Lee SK, Schiffer CA, Swanstrom R. A Direct Interaction with RNA Dramatically Enhances the Catalytic Activity of the HIV-1 Protease In Vitro. J Mol Biol. 2015;427:2360–78. doi: 10.1016/j.jmb.2015.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lanchy JM, Ehresmann C, Le Grice SF, Ehresmann B, Marquet R. Binding and kinetic properties of HIV-1 reverse transcriptase markedly differ during initiation and elongation of reverse transcription. EMBO J. 1996;15:7178–87. [PMC free article] [PubMed] [Google Scholar]
- 42.Robert D, Sallafranque-Andreola ML, Bordier B, Sarih-Cottin L, Tarrago-Litvak L, Graves PV, Barr PJ, Fournier M, Litvak S. Interactions with tRNA(Lys) induce important structural changes in human immunodeficiency virus reverse transcriptase. FEBS Lett. 1990;277:239–42. doi: 10.1016/0014-5793(90)80855-d. [DOI] [PubMed] [Google Scholar]
- 43.Andreola ML, Nevinsky GA, Barr PJ, Sarih-Cottin L, Bordier B, Fournier M, Litvak S, Tarrago-Litvak L. Interaction of tRNALys with the p66/p66 form of HIV-1 reverse transcriptase stimulates DNA polymerase and ribonuclease H activities. J Biol Chem. 1992;267:19356–62. [PubMed] [Google Scholar]
- 44.Lindhofer H, von der Helm K, Nitschko H. In vivo processing of Pr160gag-pol from human immunodeficiency virus type 1 (HIV) in acutely infected, cultured human T-lymphocytes. Virology. 1995;214:624–7. doi: 10.1006/viro.1995.0074. [DOI] [PubMed] [Google Scholar]
- 45.Speck RR, Flexner C, Tian CJ, Fu XF. Comparison of human immunodeficiency virus type 1 Pr55(Gag) and Pr160(Gag-Pol) processing intermediates that accumulate in primary and transformed cells treated with peptidic and nonpeptidic protease inhibitors. Antimicrobial Agents and Chemotherapy. 2000;44:1397–1403. doi: 10.1128/aac.44.5.1397-1403.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Le Grice SF, Gruninger-Leitch F. Rapid purification of homodimer and heterodimer HIV-1 reverse transcriptase by metal chelate affinity chromatography. Eur J Biochem. 1990;187:307–14. doi: 10.1111/j.1432-1033.1990.tb15306.x. [DOI] [PubMed] [Google Scholar]
- 47.Fletcher RS, Holleschak G, Nagy E, Arion D, Borkow G, Gu Z, Wainberg MA, Parniak MA. Single-step purification of recombinant wild-type and mutant HIV-1 reverse transcriptase. Protein Expr Purif. 1996;7:27–32. doi: 10.1006/prep.1996.0004. [DOI] [PubMed] [Google Scholar]
- 48.Gong Q, Menon L, Ilina T, Miller LG, Ahn J, Parniak MA, Ishima R. Interaction of HIV-1 reverse transcriptase ribonuclease H with an acylhydrazone inhibitor. Chem Biol Drug Des. 2011;77:39–47. doi: 10.1111/j.1747-0285.2010.01052.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee T, Laco GS, Torbett BE, Fox HS, Lerner DL, Elder JH, Wong CH. Analysis of the S3 and S3′ subsite specificities of feline immunodeficiency virus (FIV) protease: development of a broad-based protease inhibitor efficacious against FIV, SIV, and HIV in vitro and ex vivo. Proc Natl Acad Sci U S A. 1998;95:939–44. doi: 10.1073/pnas.95.3.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee TY, Le VD, Lim DY, Lin YC, Morris GM, Wong AL, Olson AJ, Elder JH, Wong CH. Development of a new type of protease inhibitors, efficacious against FIV and HIV variants. Journal of the American Chemical Society. 1999;121:1145–1155. [Google Scholar]
- 51.Heaslet H, Kutilek V, Morris GM, Lin YC, Elder JH, Torbett BE, Stout CD. Structural insights into the mechanisms of drug resistance in HIV-1 protease NL4-3. J Mol Biol. 2006;356:967–81. doi: 10.1016/j.jmb.2005.11.094. [DOI] [PubMed] [Google Scholar]
- 52.Hsiou Y, Ding J, Das K, Clark ADJ, Hughes SH, Arnold E. Structure of unliganded HIV-1 reverse transcriptase at 2.7 A resolution: implications of conformational changes for polymerization and inhibition mechanisms. Structure. 1996;4:853–860. doi: 10.1016/s0969-2126(96)00091-3. [DOI] [PubMed] [Google Scholar]