Abstract
Exchange between conformational states is required for biomolecular catalysis, allostery, and folding. A variety of NMR experiments have been developed to quantify motional regimes ranging from nanoseconds to seconds. In this work, we describe an approach to speed up the acquisition of chemical exchange saturation transfer (CEST) experiments that are commonly used to probe millisecond to second conformational exchange in proteins and nucleic acids. The standard approach is to obtain CEST datasets through the acquisition of a series of 2D correlation spectra where each experiment utilizes a single saturation frequency to 1H, 15N or 13C. These pseudo 3D datasets are time consuming to collect and are further lengthened by reduced signal to noise stemming from the long saturation pulse. In this article, we show how usage of a multiple frequency saturation pulse (i.e., MF-CEST) changes the nature of data collection from series to parallel, and thus decreases the total acquisition time by an integer factor corresponding to the number of frequencies in the pulse. We demonstrate the applicability of MF-CEST on a Src homology 2 (SH2) domain from phospholipase Cγ and the secondary active transport protein EmrE as model systems by collecting 13C methyl and 15N backbone datasets. MF-CEST can also be extended to additional sites within proteins and nucleic acids. The only notable drawback of MF-CEST as applied to backbone 15N experiments occurs when a large chemical shift difference between the major and minor populations is present (typically greater than ~8 ppm). In these cases, ambiguity may arise between the chemical shift of the minor population and the multiple frequency saturation pulse. Nevertheless, this drawback does not occur for methyl group MF-CEST experiments or in cases where somewhat smaller chemical shift differences are present.
Keywords: solution NMR, CEST, sensitivity enhancement, fast data acquisition, proteins
INTRODUCTION
NMR experiments over the last two decades have unveiled the importance of slow conformational motions to the discovery of molecular mechanisms underlying allostery, enzyme catalysis, and protein folding1–3. This progress has been fostered by methodological improvements to probe biomolecular dynamics using solution NMR on a range of timescales. Faster timescales are analyzed with rotating frame and free precession relaxation dispersion experiments that under ideal conditions are able to quantify the rate of conformational exchange, the populations between the interconverting states, and the chemical shifts of higher energy conformers within the ensemble of states. Developments for probing μsec to msec timescale motion include the in-phase/anti-phase CPMG relaxation dispersion experiment4,5 and the use of adiabatic pulses as the element to induce relaxation dispersion6,7. Slower motions on the msec to sec timescale necessitate the usage of exchange spectroscopy (EXSY)8–12 or chemical exchange saturation transfer (CEST) techniques13–15. The former method can be employed when the populations between states are comparable, while the latter methods are useful when a minor population is in exchange with the major conformational state14 or where the molecule is in exchange with a large molecular weight species that is unobservable by solution NMR16. Biological processes occurring on this timescale include conformational changes needed for molecular transport17–19, protein folding20–22, enzyme catalysis23, and molecular recognition mechanisms24,25.
The common experimental feature of relaxation dispersion spectroscopy and CEST experiments applied to biomolecules is that the variable exchange period represents an effective third dimension14. In other words, these datasets require a series of 2D correlation spectroscopies such as 1H/15N HSQC or 1H/13C HSQC to resolve the resonances in order to acquire site-specific information on the exchange process. Thus, the total time for these acquisitions is substantial and often requires more experimental time than a standard 3D experiment due to the signal to noise needed to derive quantitative conclusions from relaxation rates or saturation transfer stemming from chemical exchange. The best available solution for reducing the experimental duration is to employ non-linear sampling that reduces the number of indirect increments in the 2D correlation plane to resolve the signals26,27.
In this article, we propose an alternative method to reduce the acquisition times in CEST experiments applied to biomolecules by making use of multiple frequency saturation pulses. CEST was developed to improve contrast in magnetic resonance imaging13,28 and the use of multiple frequency pulses has been a valuable addition for distinguishing magnetization transfer from true CEST mechanisms29. Our experiments utilize multiple frequency saturation pulses to reduce the total time required to obtain pseudo 3D acquisitions by an integer factor corresponding to the number of frequencies incorporated within the pulse. This multiple frequency CEST method we refer to as MF-CEST is demonstrated with two protein systems on backbone amide 15N and methyl group 13C sites.
MATERIALS AND METHODS
Preparation of the C-Terminal SH2 Domain from Phospholipase Cγ
The Src Homology 2 (SH2) domain from PLCγ corresponding to residues N662 to L774 was expressed in E. coli using a modified M9 media where the sole nitrogen source was 15N ammonium chloride. In addition to 15N incorporation, MILVAT methyl labeling30 was incorporated into the SH2 domain by adding the following precursors or amino acids 1 hr prior to induction: L-methionine (13Cε) (250 mg/L); 2-ketobutyric acid-4-13C,3,3-2H2 (50 mg/L); 2-keto-3-methyl-13C-butyric-4-13C,3-2H acid (80 mg/L); L-alanine (3-13C,2-2H) (100 mg/L); L-threonine (4-13C;2,3-2H2) (50 mg/L); glycine (2H5) (100 mg/L). Protein expression was induced overnight at 20 °C using 0.1 mM IPTG. The cell pellet was resuspended in 20 mM sodium phosphate buffer containing 600 μM phenylmethylsulfonyl fluoride (PMSF), 1.25 mM EDTA, 2.3 μM leupeptin, 1.45 μM pepstatin A, 5% glycerol, 8.8 μM lysozyme, and 400 μM DTT, pH 6.5, and lysed using a French press. Cell debris were removed by centrifugation, and the supernatant was concentrated using an Amicon centrifuge spin concentrator (3 kDa cutoff). Concentrated lysate was injected onto a cation exchange column (Source S, GE Healthcare, Piscataway, NJ) and eluted using a gradient of 0.025 M to 1.0 M NaCl. Purity was assessed using SDS-PAGE. Elution from the cation exchange column was buffer exchanged into 25 mM HEPES (pH=7.5), 150 mM NaCl, 0.1% sodium azide, and 5% 2H2O, and concentrated to make the NMR sample consisting of 1.0 mM protein.
Preparation of EmrE Sample
The expression and purification of EmrE was carried out as previously described17,31,32. EmrE was expressed as a fusion construct with maltose-binding protein and purified using affinity and size exclusion chromatography in n-dodecyl-β-D-maltopyranoside (DDM, Anatrace). [Ile-13CH3, Leu-13CH3, Val-13CH3 U-15N, 2H]-labeled EmrE was expressed with addition of 50 mg/L 2-ketobutyric acid-4-13C, 3,3-2H2 sodium salt hydrate, and 80 mg/L of 2-keto-3-(methyl-2H3)-butyric acid-4-13C, 3-2H sodium salt 1 hr prior to induction. EmrE in DDM was reconstituted into dimyristoyl-sn-glycero-3-phosphocholine (DMPC) and dihexanoyl-sn-glycero-3-phosphocholine (DHPC) bicelles at a molar ratio of 1:3. The acyl chains of DMPC and DHPC were perdeuterated (14:0 PC 2H54 and 6:0 PC 2H22; Avanti Polar Lipids) to reduce the lipid signals in 1H/13C heteronuclear correlation experiments. The final NMR samples contained 0.8 mM EmrE bound to tetraphenylphosphonium with a lipid to protein ratio 150:1 (DMPC:EmrE, mol:mol). The buffer was 100 mM Na2HPO4 (pH=5.8) and 20 mM NaCl.
MF-CEST Acquisitions
All NMR datasets were acquired using a Bruker Avance III spectrometer at a 1H frequency of 600 MHz equipped with a TCI cryogenic probe. The pulse sequences used to incorporate multiple frequency saturation pulses were the same as those described by Kay and co-workers14,33. Shaped pulses were generated with one-, two-, or three-frequencies with a pulse length of 0.4 sec (cSH2) or 0.5 sec (EmrE) using a total of 40000 points. The Bruker python script used to create the multiple frequency pulses is available for download at https://github.com/jaeseung16/NMR_Bruker/blob/master/python/multifreq.py. During the saturation period, 1H decoupling was applied as a composite block of 90x240y90x14,34 at rf field strengths (ω1/2π) of 3.1 kHz and 5 kHz for 15N and 13C experiments, respectively. Note that the presence of small decoupling dips has been reported as a result of 1H composite pulse decoupling14. Although we did not observe these dips in our experiments, it is possible these artifacts will appear in very concentrated samples, which will be positioned ±1/(2pw) from the primary resonance, where pw is 1.17/(1H rf field) as reported by Vallurupalli et al14. If these decoupling dips are observed, one can increase the decoupling field strength to avoid interference with a true exchange dip or ignore them in subsequent data analysis.
One-frequency CEST experiments on 15N labeled cSH2 were acquired using a total of 59 × 2D acquisitions (1 reference and 58 CEST experiments) with saturation frequencies ranging from 103.5 ppm to 132 ppm in steps of 0.5 ppm at an applied rf field strength of 25 Hz. Two-frequency MF-CEST experiments were acquired using 29 × 2D experiments (1 reference and 28 MF-CEST experiments) with saturation frequencies ranging from 104 ppm to 131.5 ppm. Due to the two-frequency nature of the saturation pulse, the first experiment utilized saturation frequencies of 104 and 118 ppm. In each subsequent experiment, the saturation offset was increased by 0.5 ppm for both frequencies. The three-frequency MF-CEST experiments were collected using 20 × 2D experiments (1 reference and 19 MF-CEST experiments) with saturation frequencies ranging from 103.5 ppm to 131.5 ppm. The three-frequency experiments utilized offsets of 103.5 ppm, 113 ppm, and 122.5 ppm in the first experiment. In each subsequent experiment the offsets were shifted by 0.5 ppm. The spectral widths and sampling times were 1,703 Hz and 23.5 msec for 15N and 10000 Hz and 82.9 msec for 1H, respectively. The recycle delay was 1.5 sec.
Methyl MF-CEST experiments applied to MILVAT labeled cSH2 were acquired in a similar fashion as the backbone 15N experiments at a temperature of 25 °C. One-frequency CEST was acquired using 100 × 2D experiments (1 reference, 99 CEST) with the 13C saturation frequency ranging from 7.24 ppm to 31.74 ppm in steps of 0.25 ppm. Three-frequency MF-CEST consisted of 34 × 2D experiments (1 reference, 33 MF-CEST) where the 13C saturation frequencies were spaced by 8.25 ppm. The initial experiment employed saturation frequencies at 7.24 ppm, 15.49 ppm, and 23.74 ppm. Subsequent 2D experiments were frequency shifted by 0.25 ppm. The field strength for the one-frequency saturation pulse was 15 Hz (ω1/2π). The three-frequency MF-CEST pulse was increased in power by 9.54 dB. The spectral widths and sampling times were 3,773 Hz and 13.3 msec for 13C and 10,000 Hz and 59.9 msec for 1H, respectively. The recycle delay was 1.5 sec and each experiment was collected with 4 scans at a temperature of 25 °C.
Methyl one-frequency CEST and two-frequency MF-CEST experiments were acquired on EmrE at a temperature of 37 °C. The one-frequency experiment was acquired with 24 × 2D experiments (1 reference, 23 CEST) with saturation frequencies ranging from 9.6 ppm to 16.2 ppm in steps of 0.3 ppm. Two-frequency MF-CEST was acquired with the same number of experiments, but also utilized a second frequency excitation shifted by 10 ppm (i.e., start at 19.6 ppm). The spectral widths and sampling times were 4,000 Hz and 9.75 msec for 13C and 10000 Hz and 59.9 msec 1H, respectively. The recycle delay was 1.5 sec and each experiment was acquired with 32 scans. The experiment was repeated in duplicate and the average of these datasets is displayed. All pseudo 3D acquisitions on cSH2 or EmrE were acquired in an interleaved fashion.
Determination of Exchange Rate from CEST Curves
CEST curves were fit using chemex (https://github.com/gbouvignies/chemex) in a similar manner as previously described14. Namely, the relaxation rate R1 was fixed to be the same for the two populations in exchange and we modeled the B1 homogeneity in a similar manner, although changes to the homogeneity did not influence the fitted parameters. Due to the unique features of the EmrE asymmetric dimer17,19,31, the populations of the two states were each fixed to 0.5 and the 13C chemical shifts of the two exchanging species were also fixed to their values measured directly from the 1H/13C HSQC spectrum17,31. We also chose to fix the relaxation rate R2 to the same value for two exchanging populations, because the linewidths for the two populations were quite similar. Global and individual fits to the exchange rate (kex) were carried out for CEST and MF-CEST curves acquired on EmrE. A total of six curves were fit for CEST or MF-CEST profiles that corresponded to three residues (I54, I58, and I68). The global fits yielded a single value for each dataset, while individual fits yielded six values for the six sites. The average and standard deviation among individual fitted residues were also calculated and reported in the main text.
RESULTS AND DISCUSSION
Multiple Frequency CEST (MF-CEST)
CEST experiments on proteins and nucleic acids are collected through the acquisition of a series of HSQC-type spectra where each 2D experiment employs a single saturation frequency to longitudinal magnetization14,33. For 15N labeled proteins at the backbone, this involves sweeping over ~30 ppm in steps of 0.25 ppm to 0.5 ppm, which requires 60–120 × 2D correlation experiments. In addition to the pseudo 3D nature of the experiment, the relatively long saturation pulse needed to observe conformational exchange further lowers the signal to noise and increases the experimental acquisition time. With the goal of decreasing the total time required for CEST data collection, we envisioned an approach that uses saturation pulses consisting of multiple frequencies to enable data acquisition in parallel rather than the standard manner in series. We will refer to this method as multiple frequency CEST or MF-CEST (see scheme in Figure 1A). It should be noted that the major limitation to MF-CEST is that application of multiple frequency pulses can concurrently saturate both the major and minor populations, which would mask the observed conformational exchange. This will occur if the chemical shift difference between the exchanging sites were coincident with the difference in frequencies used for MF-CEST. In our applications detailed below, the difference in frequencies for MF-CEST is ~10 ppm or more, and therefore would lead to masking when the two populations have relatively large chemical shift differences. To the contrary, if the chemical shift differences are less than ~5–7 ppm (i.e., the typical case), then MF-CEST will have no ambiguity and allow for substantial reductions in data collection times. A second limitation related to the ambiguity created by the multiple frequency saturation pulse may occur if the minor site has a broad-band spectral signature, such as when an aggregate is studied16,35 or in the presence of paramagnetic interactions36. Under these circumstances the MF-CEST experiment may not provide a sufficiently unambiguous assignment of the exchanging sites.
Figure 1.
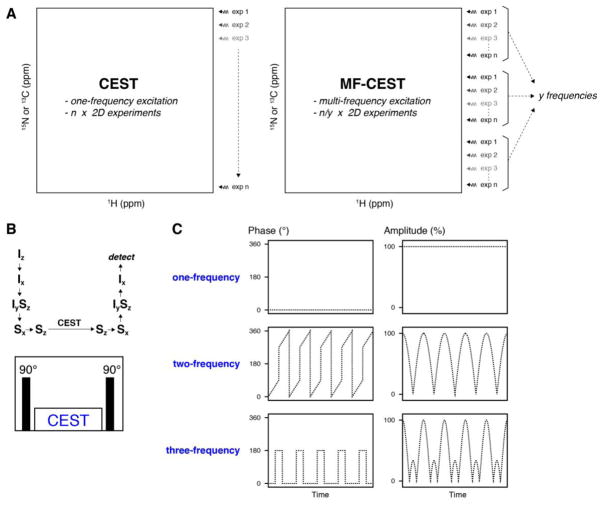
(A) Schematic of CEST and MF-CEST. Standard CEST acquisitions exhaustively sample all offset frequencies (i.e., in series), while the MF-CEST approach applies n frequencies at the same time and therefore can reduce the number of 2D acquisitions necessary (i.e., in parallel). (B) Magnetization flow for implementation of heteronuclear CEST experiments with saturation pulses on X nuclei (e.g., 15N or 13C). The CEST period in the pulse sequence schematic is shown for the X nuclei only. (C) Schematic of amplitude and phase modulation applied during the CEST period within panel B.
To implement MF-CEST, we designed shaped pulses incorporating one, two, or three frequencies with equally spaced offsets by combining the appropriately modulated components (see schematic example in Figure 1B). As an example of two-frequency MF-CEST applied to 15N, we set the two CEST frequencies in the first 2D experiment to 104 ppm and 118 ppm. The next experiment shifted the excitation frequencies by 0.5 ppm to produce the two-frequency saturations at 104.5 ppm and 118.5 ppm. In each subsequent experiment, the frequencies were shifted by 0.5 ppm until the final frequency set of 117.5 ppm and 131.5 ppm. All offsets used in the MF-CEST experiments are shown in Table I. The second difference from the standard CEST experiment is that the overall power with which the shaped saturation pulse is executed requires adjustment for the number of frequency components. Specifically, the number of applied frequencies (n) increases the power in dB proportional to 20 log n relative to a single frequency saturation. Hence, a two-frequency saturation requires 6.02 dB greater power, while a three-frequency pulse requires 9.45 dB more power relative to one-frequency CEST. In other words, the power in watts will be four-fold higher for two frequencies and nine-fold higher for three frequencies (i.e., power scales with n2). As a result, each individual saturation component would be executed at the same nominal nutation frequency as a single-frequency saturation pulse. The increased power is not a limitation for two- or three-frequency MF-CEST since the rf field strength (ω1/2π) is proportional to the peak linewidth and thus typical nutation frequencies range from 10 Hz to 40 Hz. This relatively small field strength means that the increased power for MF-CEST is not a limiting factor on the probe or spectrometer hardware and does not lead to any adverse effects.
Table I.
Saturation frequencies for 15N MF-CEST experiments on cSH2.
| Experiment | Two-frequency (ppm) | Three-frequency (ppm) | |||
|---|---|---|---|---|---|
| 1 | 104.0 | 118.0 | 103.5 | 113.0 | 122.5 |
| 2 | 104.5 | 118.5 | 104.0 | 113.5 | 123.0 |
| 3 | 105.0 | 119.0 | 104.5 | 114.0 | 123.5 |
| 4 | 105.5 | 119.5 | 105.0 | 114.5 | 124.0 |
| 5 | 106.0 | 120.0 | 105.5 | 115.0 | 124.5 |
| 6 | 106.5 | 120.5 | 106.0 | 115.5 | 125.0 |
| 7 | 107.0 | 121.0 | 106.5 | 116.0 | 125.5 |
| 8 | 107.5 | 121.5 | 107.0 | 116.5 | 126.0 |
| 9 | 108.0 | 122.0 | 107.5 | 117.0 | 126.5 |
| 10 | 108.5 | 122.5 | 108.0 | 117.5 | 127.0 |
| 11 | 109.0 | 123.0 | 108.5 | 118.0 | 127.5 |
| 12 | 109.5 | 123.5 | 109.0 | 118.5 | 128.0 |
| 13 | 110.0 | 124.0 | 109.5 | 119.0 | 128.5 |
| 14 | 110.5 | 124.5 | 110.0 | 119.5 | 129.0 |
| 15 | 111.0 | 125.0 | 110.5 | 120.0 | 129.5 |
| 16 | 111.5 | 125.5 | 111.0 | 120.5 | 130.0 |
| 17 | 112.0 | 126.0 | 111.5 | 121.0 | 130.5 |
| 18 | 112.5 | 126.5 | 112.0 | 121.5 | 131.0 |
| 19 | 113.0 | 127.0 | 112.5 | 122.0 | 131.5 |
| 20 | 113.5 | 127.5 | |||
| 21 | 114.0 | 128.0 | |||
| 22 | 114.5 | 128.5 | |||
| 23 | 115.0 | 129.0 | |||
| 24 | 115.5 | 129.5 | |||
| 25 | 116.0 | 130.0 | |||
| 26 | 116.5 | 130.5 | |||
| 27 | 117.0 | 131.0 | |||
| 28 | 117.5 | 131.5 | |||
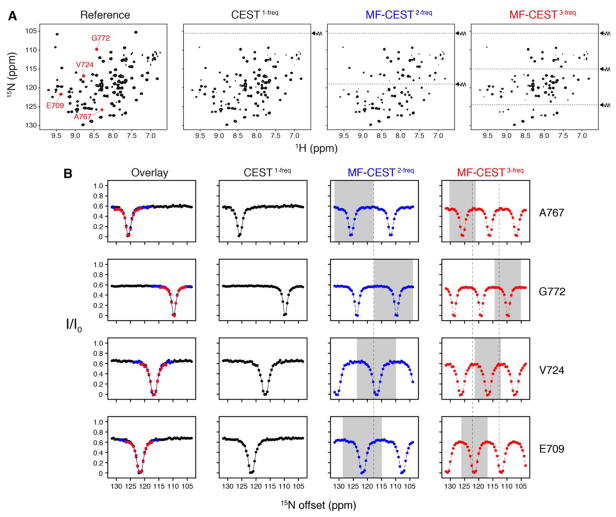
We incorporated our MF-CEST pulses into the 1H/15N HSQC-CEST pulse sequence as previously described14 and applied this to the C-terminal SH2 domain (cSH2) from phospholipase Cγ (PLCγ). This enzyme is activated upon recruitment and subsequent phosphorylation by receptor tyrosine kinases and was recently shown to be involved in a 1:2 complex between PLCγ and the fibroblast growth factor receptor37. The cSH2 protein was labeled with 15N and 13C at methyl group residues corresponding to Ile, Leu, Val, Met, Thr, and Ala. For 15N CEST experiments, we acquired the traditional one-frequency CEST experiment as well as two- and three-frequency MF-CEST experiments through the application of a 0.4 sec saturation pulse in the presence of 1H decoupling as previously described14,33. A reference spectrum with no saturation delay was also acquired in order to calculate reference peak intensities (I0). The total acquisition times for the one-, two-, and three-frequency experiments were 20 hr, 10 hr, and 7 hr, respectively. Example 2D spectra from the reference, one-frequency CEST, and two- and three-frequency MF-CEST are shown in Figure 2A. As observed in these spectra, the primary difference from the standard one-frequency CEST experiment is that the MF-CEST datasets have two or three horizontal holes in the spectra as indicated by dotted lines. CEST profiles (I/I0 vs. offset) from our pseudo 3D experiments are shown for several residues within the cSH2 domain in Figure 2B. In one-frequency CEST curves, we did not observe additional dips in the CEST profiles at 25 °C that would support the presence of slow chemical exchange between two or more conformations, and therefore this sample served as a control for comparing the standard CEST acquisition with MF-CEST. The overlay of CEST and MF-CEST results shows that our approach reproduces the shape of the standard one-frequency experiment and therefore can be used to reduce the time for acquisition by two to three-fold depending on whether two- or three-frequency MF-CEST is carried out. It is important to note that the rf field strength for saturation was set to be relatively high (ω1/2π = 25 Hz) in the experiments shown in Figure 2, which led to relatively broad CEST dips. For exchange processes involving small chemical shift differences, this saturation field should be reduced, which would narrow the width of the dip and enable better resolution of lowly populated exchange populations. Finally, we note that Gly residues are often located in a specific frequency location in the 15N spectrum, and therefore a minimalist use of 15N MF-CEST would be to employ a two-frequency shaped pulse where one frequency scans from ~100 to 115 ppm and a second that ranges from ~115 to 130 ppm. This would further reduce possible masking of lowly populated states undergoing slow conformational exchange with the major state and the ambiguity of the resulting dataset.
Figure 2.
15N CEST experiments applied to the cSH2 domain of PLCγ. (A) Example 2D spectra of the reference dataset, one-frequency CEST, and two- and three-frequency MF-CEST. The latter two spectra emphasize that two and three dips are created within the spectra (dotted lines indicate the application of rf saturation pulses). (B) CEST profiles for select residues highlighted in the reference spectrum shown in panel A. The overlay displayed on the left column illustrates the region of the MF-CEST curve that can be interpreted in an unambiguous manner to exchange (also highlighted in grey in individual MF-CEST profiles). The width of the saturation dip can be reduced by employing a reduced rf field (i.e., less than 25 Hz), which would enable better resolution of exchanging populations. However, no residues in cSH2 showed the presence of conformational exchange. Note that for the MF-CEST2-freq and MF-CEST3-freq columns in panel B, the white and grey regions are identical data points, which stems from the ambiguity of the multiple frequency excitations.
MF-CEST Applied to 13C Methyl Groups
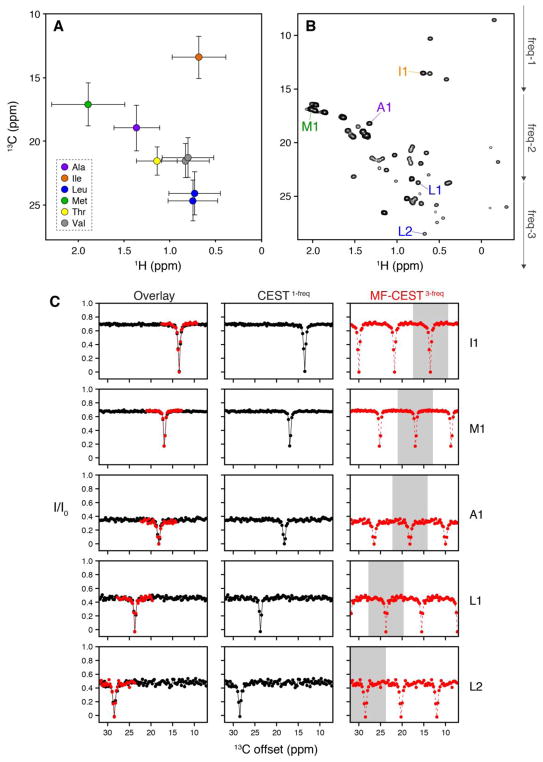
Methyl group labeling of proteins with –13CH3 in a perdeuterated background has substantial benefits that include increased sensitivity, resolution, and the ability to study macromolecular systems not accessible with detection of the polypeptide backbone38. Initial labeling procedures focused on Ile, Leu, and Val (ILV)39 and recent efforts have further extended labeling to all methyl sites in proteins (MILVAT)30. The broad usage of this labeling strategy in the protein NMR field motivated us to investigate the use of MF-CEST to methyl labeled proteins. 13C chemical shifts of methyl groups span ~25 ppm and due to the narrower linewidths of these experiments, the required step size for CEST experiments is ~0.25 ppm or less. This requires at least 100 × 2D HSQC-type experiments in order to saturate each possible methyl chemical shift frequency. A major benefit of MF-CEST applied to methyl groups is that 13C chemical shifts cluster in specific regions, which is emphasized in the plot in Figure 3A where the values were obtained from the BMRB database on diamagnetic proteins40. As can be surmised from this plot, it is uncommon to observe 13C methyl chemical shift changes by more than ~5 ppm. Therefore, the benefit for using 13C MF-CEST to probe slow conformational exchange in methyl labeled samples is even greater than the 15N based MF-CEST experiments (i.e., the masking effect is essentially negligible).
Figure 3.
Application of MF-CEST to 13C MILVAT labeled cSH2. (A) Average and standard deviation for methyl group chemical shifts from diamagnetic proteins as obtained from the BMRB database40. (B) HMQC spectrum shown for MILVAT labeled cSH2. (C) CEST profiles for select unassigned residues highlighted in the spectrum shown in panel (B). The overlay shows the part of the MF-CEST curve that is interpretable to exchanging populations on a slow timescale (also highlighted in grey in individual MF-CEST profiles). Note that for the MF-CEST3-freq column in panel C, the white and grey regions correspond to identical data points, which stems from the ambiguity of the multiple frequency excitations. Nevertheless, due to the frequency separation of specific methyl 13C chemical shifts shown in panel A, it is unlikely that methyl-based MF-CEST experiments will lead to ambiguous data interpretation.
To carry out 13C CEST experiments, we used the 1H/13C HSQC-CEST pulse sequence described by Bouvignies and Kay33 and acquired a standard one-frequency CEST dataset along with a three-frequency MF-CEST experiment on MILVAT labeled cSH2 domain (reference 2D correlation spectrum shown in Figure 3B). The one-frequency CEST dataset consisted of 100 × 2D experiments (1 reference, 99 CEST) where the 13C saturation frequency was changed from 7.24 ppm to 31.74 ppm in steps of 0.25 ppm. The three-frequency CEST consisted of 34 × 2D experiments (1 reference, 33 CEST) where the 13C saturation frequencies were spaced by 8.25 ppm. The first MF-CEST experiment used a pulse consisting of saturation frequencies at 7.24 ppm, 15.49 ppm, and 23.74 ppm. Subsequent 2D experiments employed a frequency shift by 0.25 ppm in the same way as the one-frequency CEST dataset (all frequencies are listed in Table II). A comparison of CEST profiles is shown in Figure 3C. Similar to 15N experimental results, the CEST I/I0 intensities as a function of offset overlay closely with those from MF-CEST, which validates the use of MF-CEST experiments to reduce the total acquisition time by a factor of three-fold when using a three-frequency saturation pulse.
Table II.
Saturation frequencies for 13C MF-CEST experiments on cSH2.
| Experiment | Three-frequency (ppm) | ||
|---|---|---|---|
| 1 | 7.24 | 15.49 | 23.74 |
| 2 | 7.49 | 15.74 | 23.99 |
| 3 | 7.74 | 15.99 | 24.24 |
| 4 | 7.99 | 16.24 | 24.49 |
| 5 | 8.24 | 16.49 | 24.74 |
| 6 | 8.49 | 16.74 | 24.99 |
| 7 | 8.74 | 16.99 | 25.24 |
| 8 | 8.99 | 17.24 | 25.49 |
| 9 | 9.24 | 17.49 | 25.74 |
| 10 | 9.49 | 17.74 | 25.99 |
| 11 | 9.74 | 17.99 | 26.24 |
| 12 | 9.99 | 18.24 | 26.49 |
| 13 | 10.24 | 18.49 | 26.74 |
| 14 | 10.49 | 18.74 | 26.99 |
| 15 | 10.74 | 18.99 | 27.24 |
| 16 | 10.99 | 19.24 | 27.49 |
| 17 | 11.24 | 19.49 | 27.74 |
| 18 | 11.49 | 19.74 | 27.99 |
| 19 | 11.74 | 19.99 | 28.24 |
| 20 | 11.99 | 20.24 | 28.49 |
| 21 | 12.24 | 20.49 | 28.74 |
| 22 | 12.49 | 20.74 | 28.99 |
| 23 | 12.74 | 20.99 | 29.24 |
| 24 | 12.99 | 21.24 | 29.49 |
| 25 | 13.24 | 21.49 | 29.74 |
| 26 | 13.49 | 21.74 | 29.99 |
| 27 | 13.74 | 21.99 | 30.24 |
| 28 | 13.99 | 22.24 | 30.49 |
| 29 | 14.24 | 22.49 | 30.74 |
| 30 | 14.49 | 22.74 | 30.99 |
| 31 | 14.74 | 22.99 | 31.24 |
| 32 | 14.99 | 23.24 | 31.49 |
| 33 | 15.24 | 23.49 | 31.74 |
Note that MF-CEST can also be carried out with –CHD2 labeling as has recently been shown to give sensitivity enhancement factors of two- to five-fold over the standard –CH3 labeling experiments41. Additional speedup for methyl groups is conceivable by implementing ultrafast CEST methodology whereby the saturation frequencies would be spatially encoded42,43. The use of this approach would likely come at a sensitivity cost, and may not be universally applicable, although advantages from one-shot operation could be exploited especially when polarization would vary, such as in hyperpolarization experiments, or when real-time conformational changes are tracked.
Application of MF-CEST to a Membrane Protein with Millisecond Timescale Dynamics
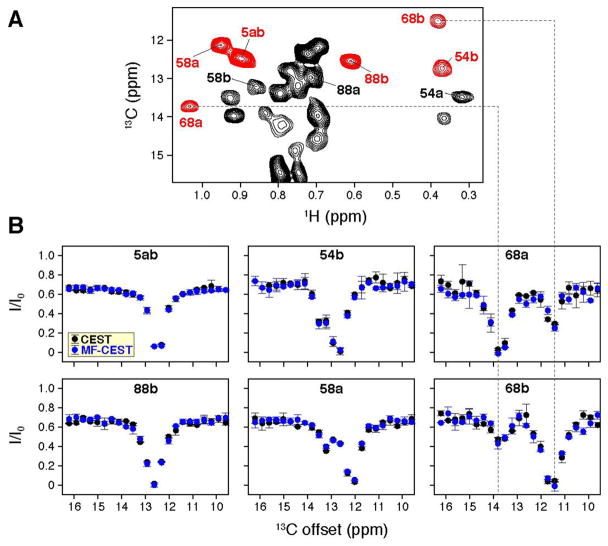
MF-CEST applied to the cSH2 domain from PLCγ demonstrated the feasibility of our method for reducing the total acquisition time for CEST acquisitions. To demonstrate this approach can also be used in cases of conformational exchange, we used the multidrug transporter EmrE bound to tetraphenylphosphonium that has previously been shown to undergo slow conformational exchange19,31. EmrE forms an anti-parallel and asymmetric dimer44–46 important for bacterial detoxification47 such that each monomer within the dimer gives rise to separate chemical shift frequencies. It has been established that the two populations for each residue within EmrE exchange in both the drug-free and drug-bound forms17,19,31. Recently, we demonstrated that the conformational exchange rate can be modulated by the presence of a substrate, either proton or drug17,31,48. To carry out MF-CEST, we labeled EmrE at 13C methyl sites using the ILV labeling strategy in a perdeuterated background (see 2D 1H/13C spectrum in Figure 4A). Previously, we published the chemical shift assignment of Ile residues and therefore focused on these sites17. While the use of CEST is not required to quantify conformational exchange in EmrE since it consists of a 50/50 population split between the two monomers, it provides a control for CEST experiments since the exchange has been validated using exchange spectroscopy17,19. We acquired one-frequency CEST and two-frequency MF-CEST over the Ile region of the spectrum. The second frequency in MF-CEST was applied over the Leu/Val region of the 13C spectrum. Similar to the experimental results obtained for the cSH2 domain of PLCγ, we observed that CEST profiles were superimposable between one-frequency CEST and two-frequency MF-CEST. However, in the case of EmrE, we observed two dips in I/I0 versus offset profiles for several residues, which stemmed from exchange between the asymmetric monomers within the dimer. Specifically, Figure 4B shows CEST curves for residues that have different chemical shifts for the two populations (see I54, I58, and I68). As expected, the populations for the two I68 methyl resonances each show the dip that matches the frequency of the other in the respective CEST curves (see dotted lines in Figure 4). Residues where the 13C chemical shifts for the two populations are the same (I5) or very similar (I88) do not show two discrete dips in their respective CEST profiles. In order to determine the exchange rate for EmrE using these datasets, we carried out global fits of CEST curves for residues displaying two distinguishable dips (i.e., I54, I58, I68) for the CEST and MF-CEST experiments and obtained kex values of 2.4 ± 0.2 sec−1 and 2.1 ± 0.2 sec−1, respectively. Globally fitted curves are shown in the supporting information. We also carried out individual fits to residues in CEST and MF-CEST, and these yielded average kex values of 2.7 ± 0.9 sec−1 and 2.3 ± 0.5, respectively, where the error is the standard deviation among the six fitted residues. These fitted values suggest CEST and MF-CEST yield indistinguishable exchange rates given the error in the measurements, and therefore validate the use of MF-CEST to probe systems that undergo slow conformational exchange.
Figure 4.
CEST and MF-CEST applied to a system with conformational exchange. (A) Reference HMQC spectrum for ILV labeled EmrE in a perdeuterated background and bound to tetraphenylphosphonium. (B) Overlay of CEST (black) and MF-CEST (blue) curves for the Ile region of the spectrum. I5 and I88 show minimal chemical shift differences in the 13C dimension, while I54, I58, and I68 show the presence of two CEST dips that support the presence of exchange. The CEST dips for 68a and 68b match the respective curves (see dotted lines), which further emphasizes the ability to use MF-CEST for reducing the time needed for data collection.
CONCLUSION
In this work, multiple frequency saturation pulses for CEST acquisitions were implemented and are referred to as MF-CEST. Since the standard way to collect CEST datasets utilizes a series of 2D experiments that each employ a single saturation frequency, the use of MF-CEST can reduce the total experimental time by a factor of two or three-fold depending on the number of saturation frequencies incorporated into the pulse. Alternatively, one can employ MF-CEST by maintaining the same overall experimental acquisition time and increase the number of points in the profile by two to three-fold, which would enable one to more clearly identify the exchange dip. A limitation of MF-CEST arises when the frequency difference between the multiple saturation pulses is similar to the frequency difference between peaks undergoing conformational exchange. In this circumstance, MF-CEST can potentially mask the minor population dip within the CEST profile. This would occur for relatively large chemical shift differences between the states in exchange (i.e., greater than ~8 ppm). Nevertheless, for systems with smaller chemical shift differences, this problem does not appear. In fact, application of this method to methyl groups in macromolecules offers one of the greatest advantages due to the relatively large 13C spectral range and narrow linewidths for these sites. Use of MF-CEST in these cases is unlikely to lead to ambiguity due to the fact that methyl-containing residues tend to cluster into specific frequency ranges. Finally, MF-CEST can be extended to other sites within proteins and nucleic acids, such as Cα or Cβ for protein based experiments, which have excellent frequency separation yet cover a large range of 13C chemical shifts.
Supplementary Material
Acknowledgments
The NMR methodology was supported by NSF award MCB 1506420 (to N.J.T.) and NIH grant R01 EB016045 (to A.J.). Applications to EmrE and the SH2 domain were funded by NIH grants R01 AI108889 and GM 117118, respectively. ML was supported from a Dean’s Dissertation Fellowship from New York University and W.M.M. acknowledges support from an NIH career transition award (F99 CA212474). All NMR data were collected with a cryoprobe at NYU that was supported by an NIH S10 grant (OD016343). We thank Professor Moosa Mohammadi for the SH2 domain plasmid and Dr. Jae-Seung Lee for scientific discussions. We also thank Dr. Jae-Seung Lee for sharing a Python script for generating multiple frequency saturation pulses. This script is freely available from the link: https://github.com/jaeseung16/NMR_Bruker/blob/master/python/multifreq.py.
References
- 1.Palmer AG, 3rd, Massi F. Characterization of the dynamics of biomacromolecules using rotating-frame spin relaxation NMR spectroscopy. Chemical reviews. 2006;106:1700–1719. doi: 10.1021/cr0404287. [DOI] [PubMed] [Google Scholar]
- 2.Massi F, Grey MJ, Palmer AG., 3rd Microsecond timescale backbone conformational dynamics in ubiquitin studied with NMR R1rho relaxation experiments. Protein science : a publication of the Protein Society. 2005;14:735–742. doi: 10.1110/ps.041139505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mittermaier AK, Kay LE. Observing biological dynamics at atomic resolution using NMR. Trends Biochem Sci. 2009;34:601–11. doi: 10.1016/j.tibs.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 4.Loria JP, Rance M, Palmer AG., III A Relaxation-Compensated Carr-Purcell-Meiboom-Gill Sequence for Characterizing Chemical Exchange by NMR Spectroscopy. J Am Chem Soc. 1999;121:2331–2. [Google Scholar]
- 5.Korzhnev DM, Kloiber K, Kanelis V, Tugarinov V, Kay LE. Probing slow dynamics in high molecular weight proteins by methyl-TROSY NMR spectroscopy: application to a 723-residue enzyme. J Am Chem Soc. 2004;126:3964–73. doi: 10.1021/ja039587i. [DOI] [PubMed] [Google Scholar]
- 6.Traaseth NJ, et al. Heteronuclear Adiabatic Relaxation Dispersion (HARD) for quantitative analysis of conformational dynamics in proteins. J Magn Reson. 2012;219:75–82. doi: 10.1016/j.jmr.2012.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mangia S, Traaseth NJ, Veglia G, Garwood M, Michaeli S. Probing slow protein dynamics by adiabatic R(1rho) and R(2rho) NMR experiments. J Am Chem Soc. 2010;132:9979–81. doi: 10.1021/ja1038787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gutowsky HS, Saika A. Dissociation, Chemical Exchange, and the Proton Magnetic Resonance in Some Aqueous Electrolytes. Journal of Chemical Physics. 1953;21:1688–1694. [Google Scholar]
- 9.Li Y, Palmer AG., 3rd TROSY-selected ZZ-exchange experiment for characterizing slow chemical exchange in large proteins. J Biomol NMR. 2009;45:357–60. doi: 10.1007/s10858-009-9385-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sahu D, Clore GM, Iwahara J. TROSY-based z-exchange spectroscopy: application to the determination of the activation energy for intermolecular protein translocation between specific sites on different DNA molecules. J Am Chem Soc. 2007;129:13232–7. doi: 10.1021/ja074604f. [DOI] [PubMed] [Google Scholar]
- 11.Montelione GT, Wagner G. 2d Chemical-Exchange Nmr-Spectroscopy by Proton-Detected Heteronuclear Correlation. Journal of the American Chemical Society. 1989;111:3096–3098. [Google Scholar]
- 12.Farrow NA, Zhang O, Forman-Kay JD, Kay LE. A heteronuclear correlation experiment for simultaneous determination of 15N longitudinal decay and chemical exchange rates of systems in slow equilibrium. J Biomol NMR. 1994;4:727–34. doi: 10.1007/BF00404280. [DOI] [PubMed] [Google Scholar]
- 13.Ward KM, Aletras AH, Balaban RS. A new class of contrast agents for MRI based on proton chemical exchange dependent saturation transfer (CEST) J Magn Reson. 2000;143:79–87. doi: 10.1006/jmre.1999.1956. [DOI] [PubMed] [Google Scholar]
- 14.Vallurupalli P, Bouvignies G, Kay LE. Studying “invisible” excited protein states in slow exchange with a major state conformation. J Am Chem Soc. 2012;134:8148–61. doi: 10.1021/ja3001419. [DOI] [PubMed] [Google Scholar]
- 15.Tang C, Louis JM, Aniana A, Suh JY, Clore GM. Visualizing transient events in amino-terminal autoprocessing of HIV-1 protease. Nature. 2008;455:693–U92. doi: 10.1038/nature07342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fawzi NL, Ying J, Ghirlando R, Torchia DA, Clore GM. Atomic-resolution dynamics on the surface of amyloid-beta protofibrils probed by solution NMR. Nature. 2011;480:268–272. doi: 10.1038/nature10577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gayen A, Leninger M, Traaseth NJ. Protonation of a glutamate residue modulates the dynamics of the drug transporter EmrE. Nat Chem Biol. 2016;12:141–5. doi: 10.1038/nchembio.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Akyuz N, Altman RB, Blanchard SC, Boudker O. Transport dynamics in a glutamate transporter homologue. Nature. 2013 doi: 10.1038/nature12265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morrison EA, et al. Antiparallel EmrE exports drugs by exchanging between asymmetric structures. Nature. 2012;481:45–50. doi: 10.1038/nature10703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sekhar A, et al. Thermal fluctuations of immature SOD1 lead to separate folding and misfolding pathways. Elife. 2015;4:e07296. doi: 10.7554/eLife.07296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma RS, et al. Determination of pseudocontact shifts of low-populated excited states by NMR chemical exchange saturation transfer. Phys Chem Chem Phys. 2016;18:13794–8. doi: 10.1039/c6cp01127f. [DOI] [PubMed] [Google Scholar]
- 22.Fizil A, Gaspari Z, Barna T, Marx F, Batta G. “Invisible” conformers of an antifungal disulfide protein revealed by constrained cold and heat unfolding, CEST-NMR experiments, and molecular dynamics calculations. Chemistry. 2015;21:5136–44. doi: 10.1002/chem.201404879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gladkova C, et al. An invisible ubiquitin conformation is required for efficient phosphorylation by PINK1. EMBO J. 2017;36:3555–3572. doi: 10.15252/embj.201797876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao B, Guffy SL, Williams B, Zhang Q. An excited state underlies gene regulation of a transcriptional riboswitch. Nat Chem Biol. 2017;13:968–974. doi: 10.1038/nchembio.2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Delaforge E, et al. Large-Scale Conformational Dynamics Control H5N1 Influenza Polymerase PB2 Binding to Importin alpha. J Am Chem Soc. 2015;137:15122–34. doi: 10.1021/jacs.5b07765. [DOI] [PubMed] [Google Scholar]
- 26.Long D, Delaglio F, Sekhar A, Kay LE. Probing Invisible, Excited Protein States by Non-Uniformly Sampled Pseudo-4D CEST Spectroscopy. Angewandte Chemie-International Edition. 2015;54:10507–10511. doi: 10.1002/anie.201504070. [DOI] [PubMed] [Google Scholar]
- 27.Matsuki Y, Konuma T, Fujiwara T, Sugase K. Boosting Protein Dynamics Studies Using Quantitative Nonuniform Sampling NMR Spectroscopy. Journal of Physical Chemistry B. 2011;115:13740–13745. doi: 10.1021/jp2081116. [DOI] [PubMed] [Google Scholar]
- 28.Zhou JY, van Zijl PCM. Chemical exchange saturation transfer imaging and spectroscopy. Progress in Nuclear Magnetic Resonance Spectroscopy. 2006;48:109–136. [Google Scholar]
- 29.Lee JS, Regatte RR, Jerschow A. Isolating chemical exchange saturation transfer contrast from magnetization transfer asymmetry under two-frequency rf irradiation. J Magn Reson. 2012;215:56–63. doi: 10.1016/j.jmr.2011.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Proudfoot A, Frank AO, Ruggiu F, Mamo M, Lingel A. Facilitating unambiguous NMR assignments and enabling higher probe density through selective labeling of all methyl containing amino acids. J Biomol NMR. 2016;65:15–27. doi: 10.1007/s10858-016-0032-2. [DOI] [PubMed] [Google Scholar]
- 31.Cho MK, Gayen A, Banigan JR, Leninger M, Traaseth NJ. Intrinsic conformational plasticity of native EmrE provides a pathway for multidrug resistance. J Am Chem Soc. 2014;136:8072–80. doi: 10.1021/ja503145x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gayen A, Banigan JR, Traaseth NJ. Ligand-induced conformational changes of the multidrug resistance transporter EmrE probed by oriented solid-state NMR spectroscopy. Angew Chem Int Ed Engl. 2013;52:10321–4. doi: 10.1002/anie.201303091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bouvignies G, Kay LE. A 2D (1)(3)C-CEST experiment for studying slowly exchanging protein systems using methyl probes: an application to protein folding. J Biomol NMR. 2012;53:303–10. doi: 10.1007/s10858-012-9640-7. [DOI] [PubMed] [Google Scholar]
- 34.Levitt MH. Symmetrical Composite Pulse Sequences for Nmr Population-Inversion .2. Compensation of Resonance Offset. Journal of Magnetic Resonance. 1982;50:95–110. [Google Scholar]
- 35.Milojevic J, Esposito V, Das R, Melacini G. Understanding the molecular basis for the inhibition of the Alzheimer’s Abeta-peptide oligomerization by human serum albumin using saturation transfer difference and off-resonance relaxation NMR spectroscopy. J Am Chem Soc. 2007;129:4282–90. doi: 10.1021/ja067367+. [DOI] [PubMed] [Google Scholar]
- 36.Hansen DF, Led JJ. Determination of the geometric structure of the metal site in a blue copper protein by paramagnetic NMR. Proc Natl Acad Sci U S A. 2006;103:1738–43. doi: 10.1073/pnas.0507179103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang Z, et al. Two FGF Receptor Kinase Molecules Act in Concert to Recruit and Transphosphorylate Phospholipase Cgamma. Mol Cell. 2016;61:98–110. doi: 10.1016/j.molcel.2015.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tugarinov V, Kay LE. Methyl groups as probes of structure and dynamics in NMR studies of high-molecular-weight proteins. Chembiochem : a European journal of chemical biology. 2005;6:1567–1577. doi: 10.1002/cbic.200500110. [DOI] [PubMed] [Google Scholar]
- 39.Rosen MK, et al. Selective methyl group protonation of perdeuterated proteins. Journal of Molecular Biology. 1996;263:627–636. doi: 10.1006/jmbi.1996.0603. [DOI] [PubMed] [Google Scholar]
- 40.Ulrich EL, et al. BioMagResBank. Nucleic Acids Res. 2008;36:D402–8. doi: 10.1093/nar/gkm957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rennella E, Huang R, Velyvis A, Kay LE. (13)CHD2-CEST NMR spectroscopy provides an avenue for studies of conformational exchange in high molecular weight proteins. J Biomol NMR. 2015;63:187–99. doi: 10.1007/s10858-015-9974-z. [DOI] [PubMed] [Google Scholar]
- 42.Xu X, Lee JS, Jerschow A. Ultrafast scanning of exchangeable sites by NMR spectroscopy. Angew Chem Int Ed Engl. 2013;52:8281–4. doi: 10.1002/anie.201303255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Boutin C, Leonce E, Brotin T, Jerschow A, Berthault P. Ultrafast Z-Spectroscopy for (129)Xe NMR-Based Sensors. J Phys Chem Lett. 2013;4:4172–4176. doi: 10.1021/jz402261h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen YJ, et al. X-ray structure of EmrE supports dual topology model. Proc Natl Acad Sci U S A. 2007;104:18999–9004. doi: 10.1073/pnas.0709387104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tate CG, Ubarretxena-Belandia I, Baldwin JM. Conformational changes in the multidrug transporter EmrE associated with substrate binding. J Mol Biol. 2003;332:229–42. doi: 10.1016/s0022-2836(03)00895-7. [DOI] [PubMed] [Google Scholar]
- 46.Ubarretxena-Belandia I, Baldwin JM, Schuldiner S, Tate CG. Three-dimensional structure of the bacterial multidrug transporter EmrE shows it is an asymmetric homodimer. EMBO J. 2003;22:6175–81. doi: 10.1093/emboj/cdg611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schuldiner S. EmrE, a model for studying evolution and mechanism of ion-coupled transporters. Biochim Biophys Acta. 2009;1794:748–62. doi: 10.1016/j.bbapap.2008.12.018. [DOI] [PubMed] [Google Scholar]
- 48.Banigan JR, Leninger M, Her AS, Traaseth NJ. Assessing Interactions Between a Polytopic Membrane Protein and Lipid Bilayers Using Differential Scanning Calorimetry and Solid-State NMR. J Phys Chem B. 2018;122:2314–2322. doi: 10.1021/acs.jpcb.8b00479. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.