Abstract
Blepharisma americanum, a member of the understudied ciliate class Heterotrichea, has a moniliform somatic macronucleus that resembles beads on a string. B. americanum is distinguishable by its pink coloration derived from the autofluorescent pigment blepharismin and tends to have a single somatic macronucleus with 3–6 nodes and multiple germline micronuclei. We used fluorescence confocal microscopy to explore the DNA content and amplification between the somatic and germline nuclei of B. americanum through its life cycle. We estimate that the DNA content of the macronucleus and micronucleus are 43±8 Gbp and 83±16 Mbp respectively. This correlates to an approximate DNA content difference of 500-fold from micronucleus to macronucleus and a macronuclear ploidy of ~1100N as compared to the presumably-diploid micronucleus. We also investigate a previously reported macronuclear inclusion, which is present sporadically across all life cycle stages; this inclusion looks as if it contains blepharismin based on its fluorescent properties, but its function remains unknown. We also provide additional detail to our understanding of life cycles changes in B. americanum by analyses of fluorescent images. Overall, the data analyzed here contribute to our understanding of the diversity of nuclear architecture in ciliates by providing details on the highly-polyploid somatic macronucleus of B. americanum.
Keywords: Ciliate, Confocal, Fluorescence, Genome Amplification, Life Cycle, Nucleus
CILIATES are unicellular organisms characterized by the presence of cilia and nuclear dualism (i.e. they have both a somatic macronucleus and a germline micronucleus within each cell/organism). The macronucleus is responsible for most cellular activity, while the micronucleus is quiescent throughout most of the life cycle. During conjugation, meiotic products of micronuclei are exchanged and fused, forming a new zygotic micronucleus that then divides mitotically (McGrath et al. 2007, Prescott 1994). One of these nuclei then undergoes extensive processing (including DNA elimination, chromosome fragmentation and amplification) resulting in a newly developed macronucleus (McGrath et al. 2007, Prescott 1994).
Most ciliates have polyploid macronuclei, but the level of ploidy varies greatly among the classes studied to date. The macronuclei of some ciliates such as Tetrahymena thermophila (Cl: Oligohymenophorea) have a roughly equal ploidy of 45N (Raikov 1996, Turkewitz et al. 2002) while ciliates in the classes Armophorea, Phyllopharyngea and Spirotrichea have ‘gene-sized’ chromosomes that are independently amplified and can be present in thousands of copies (Huang and Katz 2014, Xu et al. 2012). The class Heterotrichea, the focus of this study, has been the subject of numerous morphological studies, yet few focus on the structure of their nuclei (e.g. Guttes and Guttes 1960, Ovchinnikova et al. 1965, Kovaleva et al. 1997a, Kovaleva et al. 1997b).
We analyze the macronuclear and micronuclear genome content of Blepharisma americanum (Cl: Heterotrichea), and then describe changes in macronuclear morphology throughout the life cycle. Blepharisma is marked by pink coloration due to the presence of the photosensitizing pigment blepharismin (Giese 1973). Additional unusual features of the genus Blepharisma include cannibalism, a phenomenon that allows the organism to grow giant (McLoughlin 1957, Young 1938) and multi-conjugation, by which sets of three, or, more rarely up to five organisms mate (Weisz 1950, Giese 1973). The macronucleus of Blepharisma americanum is described as moniliform, resembling “beads-on-a-string” (Giese 1973, Suzuki 1954). This species is also reported to have a ‘nuclear inclusion’ in its macronucleus, though the chemical identity and function of this inclusion is unknown (Kennedy 1965, Young 1938).
Previous investigations of the nuclear morphology of Blepharisma species were done in an era before modern microscopy techniques (Giese 1973, Kennedy 1965, McLoughlin 1957, Suzuki 1954, Young 1938). With the exception of the examination of Blepharisma undulans Stein using TEM (Kennedy 1965), observations have been collected using nuclear stains with light microscopy and results reported with drawings, demonstrating the variable nature of Blepharisma americanum in both cell size and changes in macronuclear morphology throughout its life cycle (Giese 1973, Kennedy 1965, McLoughlin 1957, Suzuki 1954, Young 1938). Fluorescent microscopy, the approach used here, allows for detailed observations of nuclear morphology throughout ciliate life cycles (Maurer-Alcalá and Katz 2016, Postberg et al. 2005), as well as estimates of ploidy levels.
Ploidy and genome content have been estimated in diverse eukaryotes with a variety of techniques (Cousin et al. 2009, Prescott 1994, Xu et al. 2012). Quantification of fluorescence from DAPI is one method that has been used to estimate genome sizes in diverse protists (LaJeunesse et al. 2005, Parfrey and Katz 2010, Whittaker et al. 2012, Mukherjee et al. 2009) as well as in plants where genome sizes between genera can span several orders of magnitude (e.g. 125 Mbp in Arabidopsis thaliana to 22 Gbp in Pinus taeda; Suda and Trávníček 2006, Kaul et al. 2000, Zimin et al. 2014). There are limitations when using fluorescence to estimate genomic features, including preferential binding by DAPI for A-T rich regions of chromosomes, which can lead to inaccurate estimations of the genome size (Doležel et al. 1992, Noirot et al. 2002). Despite this potential bias, estimates with DAPI still provide useful approximations of genome size, especially in organisms where few genomic/molecular data currently exist.
In this study, we use DAPI staining and confocal microscopy to investigate several features of B. americanum including: 1) its macronuclear morphology throughout life cycle stages 2) the relative DNA amplification during macronuclear development and 3) a nuclear inclusion in the somatic macronucleus. These data provide insights into the nuclear dynamics of Blepharisma americanum and highlight the need for further work in this understudied ciliate lineage.
MATERIALS AND METHODS
Culturing
Blepharisma americanum were obtained from Carolina Biological (131430) and cultured in either filtered pond water or Blepharisma culturing solution (Giese 1973). Cells were cultured in 6-well plates at room temperature with a rice grain to support bacterial growth. Cultures were renewed bi-weekly by moving a small number of B. americanum to a new well with fresh filtered pond water and a rice grain.
Fixation and DAPI staining
To prepare samples for fixation, concentrated cells were collected via pipet and placed in 1.5 ml-centrifuge tubes and then filled to 1 ml with pond water or culturing solution depending on culturing conditions. Cells were centrifuged at 1000 g for 5 min and excess water was removed. Allium cepa root tips, Saccharomyces cerevisiae, and epithelial cells from human oral mucosa were also prepared for fixation. Cells were fixed with either 8% PFA/0.2% Triton-x100 in a 1:5 ratio, or 20% PFA/50% RNALater/5% Trizol in a 1:6 ratio, on Superfrost (Fisher) microscope slides for 15 minutes, and then were washed twice for 2 min with 1x PBS. Slides were incubated with DAPI (0.1 mg/ml; Fisher) in the dark for five min. Cells were then rinsed three times with 1x PBS. A drop of SlowFade Gold (Invitrogen) was added and slides were sealed with nail polish.
Imaging
Images were collected on a Leica TCS SP5 laser scanning confocal microscope (63× oil immersion objective). The UV laser with an excitation wavelength of 405 nm was used to visualize DAPI, the argon laser with an excitation wavelength of 488 nm was used to collect DIC images, and the 514 nm argon laser was used to collect red light from blepharismin. Images of the nuclear inclusion were captured with a resolution of 1024×1024, with an acquisition speed of 200 Hz, and line average 7. Z-stacks of A. cepa, S. cerevisiae, H. sapiens, and B. americanum nuclei for determination of DNA content were collected at 1024×1024, 400 Hz, with a line average of 5, and a z-step of 0.29 µm. For the macronuclear inclusion, Z-stacks were imaged with a z-step of 0.5 µm. All cells and their nuclei were selected for imaging so as to reduce artifacts introduced from their preparation (i.e. we focused on cells that retained their morphology).
Image Quantification
Genome amplification
The fluorescence intensities and volumes for a total of 50 S. cerevisiae nuclei, 53 H. sapiens cheek nuclei, 38 A. cepa root tip nuclei, 24 B. americanum macronuclei, and 65 B. americanum micronuclei were used for analysis. Z-stack profiles were analyzed using ImageJ (Rasband, W.S. ImageJ. U. S. National Institutes of Health, Bethesda, Maryland, USA, http://imagej.nih.gov/ij/, 1997–2014; Table S1). The macro 3D Objects Counter, was used to quantify the total fluorescence of the nuclei integrated across volume (Cordelieres, F. P. 3D Objects Counter; ImageJ). Fluorescence data from A. cepa, S. cerevisiae, H. sapiens were used as standards as they have well-established genome sizes. These standards were used to assess the variability in the intensity of DAPI staining. Estimates of the relationship between each unit of fluorescence intensity (FI) and known genome size are 356 bp/FI for S. cerevisiae, 410 bp/FI for H. sapiens, and 522 bp/FI for A. cepa nuclei. To account for the variability in the relationship between fluorescence signal and genome size, we made three estimates of the DNA content for B. americanum: 1) an ‘average’ estimate based on our three standards (429 bp/FI); 2) a ‘maximum’ calculated using A. cepa value of 522 bp/FI; and 3) ‘minimum’based on S. cerevisiae value of 356 bp/FI.
Nuclear inclusion
ImageJ transects were taken spanning the greatest diameter of the nuclear inclusion or the brightest area of fluorescence for blepharismin across 10 nm sections from 580–640 nm. Each set of measurements was averaged to get one value for each image at each point of emission and divided by the largest point to give comparable ratios.
RESULTS
Genomic amplification in Blepharisma americanum
Using fluorescence microscopy, we investigated the relative size and genome content of the macronucleus and micronucleus of Blepharisma americanum. Estimates for the average volume of B. americanum’s micronuclei and macronuclei are 11 µm3 and 5521 µm3 respectively (Tables 1 and S1; estimates were taken as the volume of fluorescence for each nucleus of the Z-stacks for each cell). Nuclear volume and fluorescent signal are strongly correlated (R = 0.975, P << 0.05; Fig. 1), allowing us to estimate DNA content from B. americanum.
Table 1.
DNA content and genome size estimates from fluorescence emission of five types of nuclei. We estimated the genome sizes of Blepharisma americanum macronucleus and micronucleus using Allium cepa, Homo sapiens, and Saccharomyces cerevisiae nuclei as standards.
| Nuclei | Avg. Fluorescence | DNA Content (Mbp) |
|---|---|---|
| Blepharisma americanum macronucleus | 99,187,747 | 42,560 (32,270/51,789) |
| Blepharisma americanum micronucleus | 193,231 | 82.9 (68.7/100.9) |
| Allium cepa | 30,412,972 | 15,876 |
| Homo sapiens | 8,053,061 | 3,300 |
| Saccharomyces cerevisiae | 35,145 | 12.5 |
Note: numbers in parentheses represent minimum and maximum estimates as discussed in methods.
Fig. 1.
Left: This scatterplot shows the linear relationship of DNA content to the volume of nuclei. DNA content of Blepharisma americanum macronuclei is estimated to be 43 ± 8 Gbp and micronuclei is estimated to be 83 ± 16 Mbp. Right: Boxplot representing the total DNA content for the nuclei studied in this work: Saccharomyces cerevisiae (Yeast - Brown), Blepharisma americanum micronucleus (Orange), Homo sapiens (Human - Green), Allium cepa (Onion - Purple), and B. americanum macronucleus (Blue). Shaded regions represent the interquartile range, with the upper limit representing the third quartile and the lower limit representing the first quartile. Black lines through the middle of the shaded region represent median values. Lines drawn through the boxes extend to maximum and minimum values with unfilled circles representing suspected outliers.
We estimated the DNA content for B. americanum’s macronucleus and micronuclei using the average fluorescence of the three ‘standards’ – yeast (Saccharomyces cerevisiae), human (Homo sapiens), and onion (Allium cepa) – that were chosen based on their range of genome sizes (Tables 1 and S1) to determine the approximate relationship between fluorescence intensity and DNA content. Here, we report the minimum and maximum estimates of Blepharisma’s nuclear DNA content based on these standards. Using this approach, the somatic macronuclear DNA content is 43 ± 8 Gbp (min: 32 Gbp; max: 52 Gbp) whereas the germline micronuclear DNA content is 84 ± 16 Mbp (min: 69 Mbp; max: 101 Mbp; Table 1). We then estimated the relative ploidy difference between macronucleus and micronucleus based on the assumption that the micronucleus is likely diploid (Prescott 1994). This approach yields an approximate macronuclear ploidy of 1027N. This value represents a rough approximation, as it does not account for processing of DNA during macronuclear development, which includes elimination of non-coding DNA in other ciliates lineages (Allen and Nowacki 2017, McGrath et al. 2007, Liu et al. 2005, Prescott 1994).
Life cycle
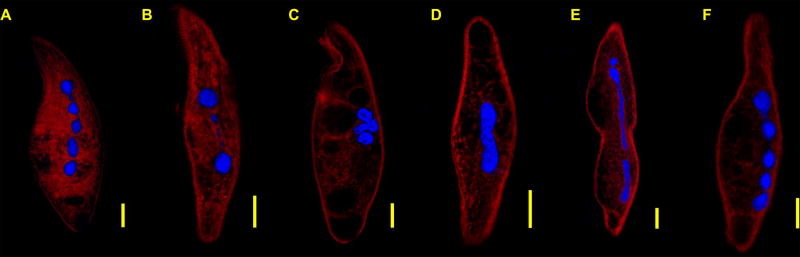
We assessed macronuclear morphology and arrangement in the various life cycle stages of Blepharisma americanum. Due to the inability to make synchronous cultures of B. americanum, we scanned mixed populations for cells in various stages (e.g. conjugation, actively dividing and log-growth) and described patterns for each stage that we observed at least 5 times (Table 2). A total of 93 B. americanum imaged in this study produced images of suitable quality for further analyses. We excluded B. americanum cells that had clear irregularities in their membranes and/or disarrayed macronuclei (likely fixation artifacts). We observed three major forms of B. americanum macronuclei during cell division: 1) moniliform arrangement of macronuclear nodes (Fig. 2A, F), 2) tight clustering of separated macronuclear nodes (Fig. 2C) and 3) the elongation of the macronucleus associated with cell division (Fig. 2E). The largest proportion of somatic nuclei (53/93; 56.9%) appeared as the typical “beads-on-a-string” (moniliform) arrangement of macronuclear nodes with interspersed micronuclei present in highly variable numbers, often in close proximity to the macronucleus (Fig. 2A, F). For vegetative cells (e.g. cells that are neither dividing nor conjugating), there are on average four macronuclear nodes, most often ranging from three to six, with larger terminal nodes than interior nodes (Fig. 2A, F), although the number and size of these nodes is highly variable with some terminal nodes not being distinctly larger than any internal node.
Table 2.
Nuclear inclusions present in nearly all life cycle stages in Blepharisma americanum.
| Cell Life Stage | # Cells | Cells with Nuclear Inclusion |
|---|---|---|
| Vegetative | 53 | 22 |
| Amitosis | 30 | 17 |
| Conjugation | 10 | 9 |
Fig. 2.
Morphological changes in the Blepharisma americanum macronucleus during amitosis. A. Macronucleus with 5 distinct nodes in asexual growth. B. Dissolution of connecting strands and migration of middle nodes. C. Condensing macronuclear nodes. D. Macronuclei extending into rod-like form. E. Rod-like form pinching across middle and enlargement of terminal nodes. F. Return to the non-dividing macronuclear morphology. DNA (blue) and Blepharismin (red). Scale bar = 20µm.
During the onset of division (and presumably amitosis), the thin nuclear envelope connections between macronuclear nodes disappear and the nodes migrate together (12/93; 12.9%, Fig. 2B, C), eventually forming a large mass. Following this condensation of somatic nodes, the large macronuclear mass elongates into either very thick or thin strands, where the macronuclear nodes are underdeveloped, which we propose is towards the final step of amitosis and prior to cell division (18/93; 19.4%, Fig. 2D, E). We did fix a small number of cells (10/93; 10.8%) during conjugation, as determined by the connection of two B. americanum across their oral apparatuses. However, we were unable to identify micronuclei undergoing mitosis for two potential reasons: 1) it is a brief event that occurs and the number of cells imaged was insufficient to observe their mitosis and 2) their close proximity to the macronucleus makes it very difficult to distinguish changes in micronuclear morphology due to their relative small size.
Nuclear Inclusion
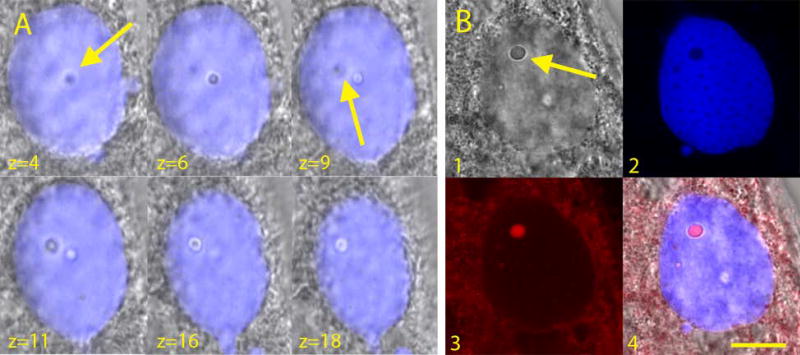
Examination of the nuclear morphology of Blepharisma americanum led to several insights into the presence of a DNA-poor inclusion found in the macronuclei. Of the 93 life cycle images captured of B. americanum, 52 showed a small nuclear inclusion in one or more macronuclear nodes across all stages of life (52/93; 55.9%). Of the 18 non-dividing macronuclei in the “beads on a string” formation showing the presence of nuclear inclusions (18/24; Table 2), 18 terminal nodes showed inclusions versus 10 non-terminal nodes, indicating a predominance in their presence in terminal nodes. These inclusions are roughly spherical, as determined by Z-series scans (Fig. 3), and have an average diameter of 1.9 µm based on 60 inclusions in 32 individuals. About 79% of the macronuclear inclusions measured, when excited with a UV laser, emitted red light with spectra highly matching that of blepharismin (Fig. 4).
Fig. 3.
Macronuclear inclusion of Blepharisma americanum. A. Z-series through the macronuclear inclusion, where each slice corresponds to 0.5µm. B. 1. DIC image of a macronuclear node. 2. DAPI staining. 3. Red emission. 4. Overlay of DIC, DAPI staining and red emission. Arrows indicate the location of the nuclear inclusion. Scale bar = 10µm.
Fig. 4.
Comparative emission spectra for the macronuclear inclusion and blepharismin (n = 15 for blepharismin and n = 14 for macronuclear inclusion). Both spectra show a maximum emission at 600–610 nm and a similar shape.
DISCUSSION
In this study, we estimate the DNA content of Blepharisma americanum macronuclei and micronuclei, propose three macronuclear configurations that vary across major life cycle stages, and present data on the macronuclear inclusion as analyzed by fluorescence confocal microscopy. We also demonstrate the utility of DAPI staining as a means of estimating DNA content and genome size in poorly studied lineages of eukaryotes.
Genome Amplification
Based on the extreme size difference between Blepharisma americanum’s macronuclei and micronuclei, we predicted that macronuclei would contain far more DNA than micronuclei. The linear relationship of genome size to DAPI fluorescence presented in this study suggests that fluorescent microscopy is accurate enough to make preliminary estimates of genome sizes (Fig. 1). Using the correlation between fluorescence and known DNA content for our three standard eukaryotes (yeast, onion and human), we estimate DNA content for the macronucleus (43 ± 8 Gbp; Table 1) and micronucleus of Blepharisma americanum (84 ± 16 Mbp; Table 1) and calculate that the macronuclear genome is approximately 1,000 times the amplification of the germline DNA. The level of amplification is probably an underestimate as some amount of the germline genome is likely to be eliminated during the development of the somatic genome of Blepharisma americanum. Among other ciliates, Tetrahymena thermophila eliminates ~30% of its germline DNA sequences from the developing macronucleus (Liu et al. 2005, Yao and Gorovsky 1974), while Chilodonella uncinata, Oxytricha trifallax and Stylonychia lemnae eliminate from 35–95% of their germline sequences (Prescott 1994).
The relationship of nuclear size to fluorescence supports the insights by Cavalier-Smith (2005) that nuclear size is directly related to DNA content. Under this model, the large macronuclear volume of Blepharisma americanum may have evolved to support the relatively large cell size (~250–350 µm). This relationship holds across taxa as the yeast S. cerevisiae had the smallest nuclei by volume and much lower total fluorescence as compared to the larger nuclei of A. cepa and B. americanum (Fig. 1).
Life Cycle
Changes in the morphology of the macronucleus throughout the life cycle of B. americanum proposed here are comparable to previously observed macronuclear behaviors (Giese 1973, Kennedy 1965, McLoughlin 1957, Suzuki 1954, Young 1938). During the dominant non-dividing life cycle stage, B. americanum’s macronucleus is present as a set of nodes interconnected with a thin string of macronuclear DNA (Fig. 2A, F). The individuals in our study were found to have three to six macronuclear nodes, consistent with Giese’s findings (1973), although other studies have indicated that the number of macronuclear nodes in B. americanum can range from two to ten (Giese 1973, Kennedy 1965, McLoughlin 1957). Suzuki (1954) observed that the terminal nodes of the macronucleus are always larger in size than the interior nodes although these differences in size are highly variable, which is again consistent with our observations.
Our analysis of cells in division is consistent with published work that describes amitosis in Blepharisma americanum. Initially, the thin strands between inner nodes disappear, and the inner nodes begin to congregate (Fig. 2B; Suzuki 1954, Young 1938, Weisz 1949). This leads to a relatively large mass of non-fused macronuclear nodes, generally localized towards the center of the organism (Giese 1973, Brasier et al. 2006, Suzuki 1954, Young 1938), visible in our observation of clustering of macronuclear nodes (Fig. 2C). As no cells visualized in this study formed a uniform single mass, despite tight clustering, we imagine the condensation into a uniform mass prior to elongation is short-lived. After this congregation, the nodes fuse to form a single rod-like macronucleus, which begins stretching throughout much of the cell (Fig. 2D & E; McLoughlin 1957, Suzuki 1954, Young 1938). The elongation of this stage is paralleled with an increase in cell length. The rod-like form is split into distinct macronuclei with the onset of cytokinesis (Fig 2E; Suzuki 1954, Young 1938). After cytokinesis, the macronucleus reforms its typical ‘beads-on-a-string’ arrangement (Fig. 2A & F).
Macronuclear Inclusion
We provide additional data on the nuclear inclusion of B. americanum. The macronuclear inclusion of B. americanum investigated in this study is approximately 1.9 µm in diameter and tends to be in a DAPI poor region. Young (1938) proposed that this macronuclear inclusion was a protein, perhaps a storage product, used in metabolic activities of macronuclear division. Kennedy (1965) conducted further tests on the inclusion, describing it as a carbohydrate-protein complex, but also suggesting it to be a storage product that the macronucleus could use during division. The macronuclear inclusion existed in the macronuclei in all life cycle stages, inconsistent with Young’s hypothesis that it is cyclic in nature and disappears as the cell approaches amitotic division (Young 1938).
Interestingly, the macronuclear inclusion appears to contain blepharismin, the photosensitive pigment found in Blepharisma (Fig. 3 and 4). The presence of blepharismin in the macronuclear inclusion may either be a natural phenomenon or a fixation artifact. It is possible that blepharismin is chemically attracted to the compound in the inclusion and that the use of Triton-x100 (a chemical detergent) in the fixation of cells could have an effect on its appearance. If, however, the inclusion harboring blepharismin is a natural phenomenon, then its purpose is unclear, especially considering that studies have found blepharismin to be a photosensitizing pigment involved in transducing perceived light to the photoreceptor system of B. americanum (Sobierajska et al. 2006).
SYNTHESIS
We have demonstrated that the heterotrich ciliate B. americanum has nearly 1,000 fold as much DNA in its somatic macronucleus as compared to its germline micronucleus (43±8 Gbp compared to 83±16 Mbp, respectively). Such high levels of amplification are also documented in the classes Spirotrichea and Phyllopharyngea, two lineages in which extensive fragmentation generates gene-sized chromosomes that are then amplified 1,000 fold or more (Huang and Katz 2014, Xu et al. 2012). In contrast, Karyorelictea – the class sister to the Heterotrichea – are described as paradiploid (e.g ~2–2.5N; Raikov and Karadzhan 1985) while other ciliates such as Tetrahymena and Paramecium amplify their somatic genomes ~45 times and ~800 times, respectively (Eisen et al. 2006, Duret et al. 2008). Hence, ciliates contain a tremendous diversity of nuclear architectures, particularly among somatic macronuclear genomes.
Supplementary Material
Acknowledgments
We would like to thank Judith Wopereis for her help and advice with fluorescence microscopy as well as Dr. Jean-David Grattepanche for providing us with the Trizol-based fixative used in this study. This work was supported by both a NIH award (1R15GM113177-01) and NSF award (DEB-1208741) to LAK.
LITERATURE CITED
- Allen SE, Nowacki M. Necessity Is the Mother of Invention: Ciliates, Transposons, and Transgenerational Inheritance. Trends in Genetics. 2017;33(3):197–207. doi: 10.1016/j.tig.2017.01.005. [DOI] [PubMed] [Google Scholar]
- Ammermann D. Giant chromosomes in ciliates. Results Probs Cell Differ. 1987;14:59–67. doi: 10.1007/978-3-540-47783-9_4. [DOI] [PubMed] [Google Scholar]
- Brasier M, McLoughlin N, Green O, Wacey D. A fresh look at the fossil evidence for early Archaean cellular life. Philosophical Transactions of the Royal Society B-Biological Sciences. 2006;361:887–902. doi: 10.1098/rstb.2006.1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier-Smith T. Economy, speed and size matter: evolutionary forces driving nuclear genome miniaturization and expansion. Annals of Botany. 2005;95:147–75. doi: 10.1093/aob/mci010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cousin A, Heel K, Cowling WA, Nelson MN. An efficient high-throughput flow cytometric method for estimating DNA ploidy level in plants. Cytometry Part A. 2009;75A:1015–1019. doi: 10.1002/cyto.a.20816. [DOI] [PubMed] [Google Scholar]
- Doležel J, Sgorbati S, Lucretti S. Comparison of three DNA fluorochromes for flow cytometric estimation of nuclear DNA content in plants. Physiologia plantarum. 1992;85:625–631. [Google Scholar]
- Duret L, Cohen J, Jubin C, Dessen P, Goût JF, Mousset S, Aury JM, Jaillon O, Noël B, Arnaiz O, Bétermier M, Wincker P, Meyer E, Sperling L. Analysis of sequence variability in the macronuclear DNA of Paramecium tetraurelia: a somatic view of the germline. Genome research. 2008;18(4):585–596. doi: 10.1101/gr.074534.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giese AC. Blepharisma: the biology of a light-sensitive protozoan. Stanford University Press; 1973. [Google Scholar]
- Guttes E, Guttes S. Incorporation of tritium-labeled thymidine into the macronucleus of Stentor coeruleus. Experimental cell research. 1960;19:626–628. doi: 10.1016/0014-4827(60)90071-9. [DOI] [PubMed] [Google Scholar]
- Huang J, Katz LA. Nanochromosome Copy Number Does not Correlate with RNA Levels Though Patterns are Conserved between Strains of the Ciliate Morphospecies Chilodonella uncinata. Protist. 2014;165:445–451. doi: 10.1016/j.protis.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaul S, Koo HL, Jenkins J, Rizzo M, Rooney T, Tallon LJ, Feldblyum T, Nierman W, Benito MI, Lin XY, Town CD, Venter JC, Fraser CM, Tabata S, Nakamura Y, Kaneko T, Sato S, Asamizu E, Kato T, Kotani H, Sasamoto S, Ecker JR, Theologis A, Federspiel NA, Palm CJ, Osborne BI, Shinn P, Conway AB, Vysotskaia VS, Dewar K, Conn L, Lenz CA, Kim CJ, Hansen NF, Liu SX, Buehler E, Altafi H, Sakano H, Dunn P, Lam B, Pham PK, Chao Q, Nguyen M, Yu GX, Chen HM, Southwick A, Lee JM, Miranda M, Toriumi MJ, Davis RW, Wambutt R, Murphy G, Dusterhoft A, Stiekema W, Pohl T, Entian KD, Terryn N, Volckaert G, Salanoubat M, Choisne N, Rieger M, Ansorge W, Unseld M, Fartmann B, Valle G, Artiguenave F, Weissenbach J, Quetier F, Wilson RK, de la Bastide M, Sekhon M, Huang E, Spiegel L, Gnoj L, Pepin K, Murray J, Johnson D, Habermann K, Dedhia N, Parnell L, Preston R, Hillier L, Chen E, Marra M, Martienssen R, McCombie WR, Mayer K, White O, Bevan M, Lemcke K, Creasy TH, Bielke C, Haas B, Haase D, Maiti R, Rudd S, Peterson J, Schoof H, Frishman D, Morgenstern B, Zaccaria P, Ermolaeva M, Pertea M, Quackenbush J, Volfovsky N, Wu DY, Lowe TM, Salzberg SL, Mewes HW, Rounsley S, Bush D, Subramaniam S, Levin I, Norris S, Schmidt R, Acarkan A, Bancroft I, Quetier F, Brennicke A, Eisen JA, Bureau T, Legault BA, Le QH, Agrawal N, Yu Z, Martienssen R, Copenhaver GP, Luo S, Pikaard CS, Preuss D, Paulsen IT, Sussman M, Britt AB, Selinger DA, Pandey R, Mount DW, Chandler VL, Jorgensen RA, Pikaard C, Juergens G, Meyerowitz EM, Theologis A, Dangl J, Jones JDG, Chen M, Chory J, Somerville MC, In AG. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Kennedy JR. The morphology of Blepharisma undulans Stein. The Journal of Protozoology. 1965;12:542–561. [Google Scholar]
- Kovaleva VG, Raikov IB, Miyake A. Fine Structure of Conjugation of the Ciliate Blepharisma japonicum 1. Changes of the Old Macronucleus. Archiv Fur Protistenkunde. 1997a;148:343–350. [Google Scholar]
- Kovaleva VG, Raikov IB, Miyake A. Fine Structure of Conjugation of the Ciliate Blepharisma japonicum 2. Changes of Meiotic and Ameiotic Micronuclei and Development of Meiotic and Ameiotic Macronuclear Anlagen. Archiv Fur Protistenkunde. 1997b;148:351–363. [Google Scholar]
- LaJeunesse TC, Lambert G, Andersen RA, Coffroth MA, Galbraith DW. Symbiodinium (Pyrrhophyta) genome sizes (DNA content) are smallest among dinoflagellates. Journal of Phycology. 2005;41:880–886. [Google Scholar]
- Liu Y, Song X, Gorovsky MA, Karrer KM. Elimination of foreign DNA during somatic differentiation in Tetrahymena thermophila shows position effect and is dosage dependent. Eukaryotic Cell. 2005;4(2):421–431. doi: 10.1128/EC.4.2.421-431.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer-Alcalá XX, Katz LA. Nuclear architecture and patterns of molecular evolution are correlated in the ciliate Chilodonella uncinata. Genome Biology and Evolution. 2016:evw099. doi: 10.1093/gbe/evw099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath CL, Zufall RA, Katz LA. Variation in macronuclear genome content of three ciliates with extensive chromosomal fragmentation: a preliminary analysis. J Eukaryot Microbiol. 2007;54:242–6. doi: 10.1111/j.1550-7408.2007.00257.x. [DOI] [PubMed] [Google Scholar]
- McLoughlin DK. Macro nuclear Morphogenesis during Division of Blepharisma undulans. The Journal of Protozoology. 1957;4:150–153. [Google Scholar]
- Mukherjee C, Majumder S, Lohia A. Inter-cellular variation in DNA content of Entamoeba histolytica originates from temporal and spatial uncoupling of cytokinesis from the nuclear cycle. PLoS Negl Trop Dis. 2009;3:e409. doi: 10.1371/journal.pntd.0000409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noirot M, Barre P, Louarn J, Duperray C, Hamon S. Consequences of stoichiometric error on nuclear DNA content evaluation in Coffea liberica var dewevrei using DAPI and propidium iodide. Annals of Botany. 2002;89:385–389. doi: 10.1093/aob/mcf056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovchinnikova L, Cheissin E, Selivanova G. Photometric study of the DNA content in the nuclei of Spirostomum ambiguum (Ciliata, Heterotricha) Acta Protozool. 1965;3(7):69–78. [Google Scholar]
- Parfrey LW, Katz LA. Genome dynamics are influenced by food source in Allogromia laticollaris strain CSH (Foraminifera) Genome Biol Evol. 2010;2:678–85. doi: 10.1093/gbe/evq051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postberg J, Alexandrova O, Cremer T, Lipps HJ. Exploiting nuclear duality of ciliates to analyse topological requirements for DNA replication and transcription. Journal of Cell Science. 2005;118:3973–3983. doi: 10.1242/jcs.02497. [DOI] [PubMed] [Google Scholar]
- Prescott DM. The DNA of Ciliated Protozoa. Microbiological Reviews. 1994;58:233–267. doi: 10.1128/mr.58.2.233-267.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raikov IB, Karadzhan BP. Fine structure and cytochemistry of the nuclei of the primitive ciliate Tracheloraphis crassus (Karyorelictida) Protoplasma. 1985;126(1–2):114–129. [Google Scholar]
- Raikov IB. Nuclei of Ciliates. Ciliates: cells as organisms. 1996:221–242. [Google Scholar]
- Sobierajska K, Fabczak H, Fabczak S. Photosensory transduction in unicellular eukaryotes: a comparison between related ciliates Blepharisma japonicum and Stentor coeruleus and photoreceptor cells of higher organisms. Journal of Photochemistry and Photobiology B: Biology. 2006;83:163–171. doi: 10.1016/j.jphotobiol.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Suda J, Trávníček P. Reliable DNA ploidy determination in dehydrated tissues of vascular plants by DAPI flow cytometry—new prospects for plant research. Cytometry Part A. 2006;69:273–280. doi: 10.1002/cyto.a.20253. [DOI] [PubMed] [Google Scholar]
- Suzuki S. Taxonomic studies on Blepharisma undulans Stein with special reference to the macronuclear variation. J. Sci. Hiroshima Univ. 1954;15:205–20. [Google Scholar]
- Turkewitz AP, Orias E, Kapler G. Functional genomics: the coming of age for Tetrahymena thermophila. Trends in Genetics. 2002;18(1):35–40. doi: 10.1016/s0168-9525(01)02560-4. [DOI] [PubMed] [Google Scholar]
- Weisz PB. Multiconjugation in Blepharisma. The Biological Bulletin. 1950;98:242–246. doi: 10.2307/1538671. [DOI] [PubMed] [Google Scholar]
- Whittaker KA, Rignanese DR, Olson RJ, Rynearson TA. Molecular subdivision of the marine diatom Thalassiosira rotula in relation to geographic distribution, genome size, and physiology. Bmc Evolutionary Biology. 2012;12:209. doi: 10.1186/1471-2148-12-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K, Doak TG, Lipps HJ, Wang JM, Swart EC, Chang WJ. Copy number variations of 11 macronuclear chromosomes and their gene expression in Oxytricha trifallax. Gene. 2012;505:75–80. doi: 10.1016/j.gene.2012.05.045. [DOI] [PubMed] [Google Scholar]
- Yao M, Gorovsky MA. Comparison of Sequences of Macro-Nutrients and Micronuclear DNA of Tetrahymena-Pyriformis. Chromosoma. 1974;48(1):1–18. doi: 10.1007/BF00284863. [DOI] [PubMed] [Google Scholar]
- Young D. Macronuclear reorganization in Blepharisma undulans. Journal of Morphology. 1938;64:297–353. [Google Scholar]
- Zimin A, Stevens KA, Crepeau M, Holtz-Morris A, Koriabine M, Marcais G, Puiu D, Roberts M, Wegrzyn JL, de Jong PJ, Neale DB, Salzberg SL, Yorke JA, Langley CH. Sequencing and Assembly of the 22-Gb Loblolly Pine Genome. Genetics. 2014;196:875–890. doi: 10.1534/genetics.113.159715. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.