Abstract
The intestinal microbiota was revealed with the recent advances in molecular techniques, such as high-throughput sequencing analysis. As a result, the microbial changes are thought to influence the health of humans and animals and such changes are affected by several factors including diet, genetics, age, sex, and diseases. Similar studies are being conducted in dogs, and the knowledge of intestinal microbiota in dogs is expanding. Nonetheless, basic information on intestinal microbiota in dogs is less than that of humans. Our aim was to study toy poodles (n=21), a popular companion dog, in terms of basic characteristics of the faecal microbiota by 16S rRNA gene barcoding analysis. In the faecal microbiota, Firmicutes, Bacteroidetes, Proteobacteria, and Fusobacteria were the dominant phyla (over 93.4% of faecal microbiota) regardless of the attributes of the dogs. In family level, Enterobacteriaceae, Bacteroidaceae, and Lachnospiraceae were most prevalent. In case of a dog with protein-losing enteropathy, the diversity of faecal microbiota was different between before and after treatment. This study provides basic information for studying on faecal microbiota in toy poodles.
Keywords: faecal microbiota, Toy Poodle, 16S rRNA gene barcoding
The intestinal microbiota has great diversity and is known to affect human health and diseases. The recent advances in molecular techniques including 16S rRNA barcoding analysis on a high-throughput sequencer have resulted in a dramatic improvement in researching intestinal microbiota. These techniques revealed that changes in the intestinal microbiota of humans are affected by several factors including diet, genetics, age, sex, and diseases [1, 6, 15].
The influence of host health and physiology on microbial changes was also investigated in dogs. Gastrointestinal diseases are associated with alterations of the intestinal microbiota. The faecal microbial communities in dogs with diarrhoea were significantly different from those in healthy dogs, and the faecal dysbiosis in acute diarrhoea could be associated with altered systemic metabolic states [7, 21]. Other gastrointestinal diseases such as inflammatory bowel diseases (IBD) could be associated with alterations in the microbiota of the small intestine and faeces [9]. Dogs with calcium oxalate stones also have different diversity of intestinal microbiota in comparison with healthy dogs [5].
Feeding also affects intestinal microbiota in dogs. A prolonged period of fasting leads to significant changes in the small-intestinal microbiota formation and could have a significant impact on the metabolic function of the microbiota [12]. Feeding of dietary fibre also affects community formation of the microbiota in dogs, and Firmicutes appear to play an important role in by-diet clustering [13].
Pharmacotherapy has an effect on the gastrointestinal microbiota in dogs. The bacterial diversity of healthy dogs is significantly decreased by metronidazole administration [11]. Additionally, the intestinal microbiota could influence neuronal signalling to the brain. Park and colleagues showed that the diversity of the intestinal microbiota is less in obese dogs than in lean dogs, fed a restricted amount of the diet to maintain optimal body weight, according to 16S rRNA gene pyrosequencing analysis, and the level of serotonin, which is related to energy consumption, in the central nervous system is lower in obese dogs than in lean dogs [17].
The intestinal microbiota of dogs has been investigated as described above. In these studies, however, the dog breed was either not specified, or mainly laboratory Beagles, English Setters, and Labrador Retrievers were used, and the impact of genetic diversity of dogs on intestinal microbiota was not considered [18]. Additionally, little information on the relations among intestinal microbiota, sex, age, and breed of dogs is available. In this study, we focus on one of the most popular breed of companion dogs, toy poodles. The toy poodle is the most popular breed of all the registered purebred dogs in Japan according to the Japan Kennel Club (http://www.jkc.or.jp/). The aim of this study was to describe the basic information of microbiota in faecal samples of toy poodles by 16S rRNA gene barcoding analysis and to evaluate the effects of age, sex, and disease.
MATERIALS AND METHODS
Samples
A total of 22 faecal samples were analyzed in this study (Table 1). All samples were collected in Tokyo, Japan. Of those, 20 faecal samples were collected from 20 (Sample ID: 1 to 20) toy poodles without diarrhoea and medication, including 12 males and 8 females. As an attribute of the dog, only age and sex were collected. The other 2 samples (Sample ID: 21-1 and 21-2) were collected twice from 1 female toy poodle with protein-losing enteropathy (PLE). The second sampling was performed in 20 days after the first sampling. The steroid administration was started 2 months before initial sampling and was continuing to the second sampling of faecal swabs. This dog had been on medication during the sample collection period, and the levels of serum albumin was measured with Bromocresol Green Albumin Assay at each sampling time in this dog. Collected faecal samples were stored at −80°C until DNA extraction.
Table 1. Summary of collecting samples.
| Sample ID | Age (Year) | Sex | Sampling date |
|---|---|---|---|
| 1 | 0 | C | 2016.4.8 |
| 2 | 0 | M | 2016.5.7 |
| 3 | 1 | M | 2016.5.14 |
| 4 | 2 | S | 2016.4.8 |
| 5 | 2 | S | 2016.4.15 |
| 6 | 2 | S | 2016.5.7 |
| 7 | 3 | S | 2016.4.16 |
| 8 | 3 | F | 2016.5.2 |
| 9 | 4 | C | 2016.4.13 |
| 10 | 4 | C | 2016.5.9 |
| 11 | 5 | S | 2016.4.25 |
| 12 | 5 | C | 2016.4.30 |
| 13 | 5 | S | 2016.5.13 |
| 14 | 6 | C | 2016.4.21 |
| 15 | 7 | M | 2016.4.8 |
| 16 | 7 | C | 2016.4.23 |
| 17 | 9 | M | 2016.5.11 |
| 18 | 10 | S | 2016.5.8 |
| 19 | 11 | C | 2016.4.19 |
| 20 | 11 | C | 2016.4.22 |
| 21-1 | 12 | S | 2016.4.9 |
| 21-2 | 12 | S | 2016.5.14 |
M: male, F: female, C: castrotion male, S: spayed female.
DNA extraction and 16S rRNA gene sequencing
Before extraction, each faecal swab was resuspended in 2 ml of sterile PBS and centrifuged at max speed for 3 min. DNA was extracted from pellet samples with the QIAamp Fast DNA Stool Mini Kit (Qiagen Hilden, Germany).
The V3-V4 region of 16S rRNA genes was amplified using universal primers F341 and R805 according to the high-throughput sequencing protocol (Illumina, San Diego, CA, U.S.A.) as previously described [8]. The barcoded amplicons were processed with Premix EX Taq (TaKaRa-Bio, Kusatsu, Japan). The PCR conditions were as follows: 94°C for 3 min; 25 cycles of 94°C for 30 sec, 55°C for 30 sec, 72°C for 30 sec; and a final extension at 72°C for 10 min. The PCR products were purified using Agencourt AMPure XP (Beckman Coulter, Brea, CA, U.S.A.) and served as templates in the second round of PCR to attach dual indices and Illumina sequencing adapters. In the second round of PCR, thermal conditions were as follows: 98°C for 30 sec; 8 cycles of 98°C for 30 sec, 60°C for 30 sec, 72°C for 30 sec; and a final extension at 72°C for 5 min. The second round of PCR products was purified as described above. The amplicons were sequenced on the MiSeq platform (Illumina) using the MiSeq Reagent kit v3 (600 cycles) with 300 paired-end reads (Illumina).
Diversity analysis
A total of 50,000 raw sequence reads per sample were extracted from all the raw reads obtained on MiSeq. The extracted reads were quality trimmed, filtered, combined with fastq-join script, and chimera depleted using QIIME 1.9.1. Raw reads were combined using fastq-join script with the following settings: N-percent maximum difference=8, N-minimum overlap=6. Then, the joined reads were trimmed based on a quality score (<33). Chimeric sequences were depleted with USEARCH v6.1. Clustering into the operational taxonomic units (OTUs) was carried out at 97% similarity against the Greengenes database 13.8, and taxonomy was assigned to OTUs in QIIME. Sequence data obtained from 22 samples were subjected to the diversity analysis.
Statistical analysis
Bacterial diversity of each sample (α-diversity) was calculated, and rarefaction curves were plotted using QIIME. Differences in intestinal microbiota in dogs (β-diversity) were investigated using the phylogeny-based unweighted and weighted UniFrac distance matric, and principal coordinates analysis (PCoA) plots were plotted using QIIME. Differences in microbial composition between samples were analyzed using the one-way analysis of similarity (ANOSIM) in QIIME, and statistical analysis was conducted in software package EzR. In these statistical analyses, data from 20 samples, excluding data from the dog with PLE, were analyzed.
RESULTS
Animals and sequence analysis
At first, 22 faecal swab samples from toy poodles were analyzed. In toy poodles without diarrhoea, the numbers of dogs in age ranges 0–3, 4–8 and 9–12 were 8 (5 females and 3 males), 8 (6 females and 2 males), and 4 (3 females and 1 male), respectively. Two samples, Sample ID 21-1 and 21-2, were from the same 12-year-old female toy poodle with a diagnosis of protein-losing enteropathy (PLE) without any clinical symptoms for 6 months before initial sampling. The serum albumin level of the dog was 1.4 g/dl at first sampling and 3.8 g/dl at the second sampling.
A total of 18,734,955 reads of 16S rRNA genes were obtained from 22 samples, with a range between 654,396 and 1,034,920 reads per sample. The resulting sequences were deposited in the National Center for Biotechnology Information Sequence Read Archive under BioProject PRJDB5398 (Accession ## experiment: DRX076298–DRX076319, run: DRR082462–DRR082483).
Characterization of the faecal microbiota from toy poodles without symptoms and with PLE
To reveal the characteristics of the faecal microbiota composition in dogs, the obtained sequence data of 22 faecal samples from 21 toy poodles was analyzed. At 97% similarity, a total of 106 OTUs, with a mean of 30.5 ± 9.6, were observed at the genus level in these faecal samples from the dogs (Table 2).
Table 2. The number of OTUs at each biological classification.
| Phylum | Class | Order | Family | Genus | |
|---|---|---|---|---|---|
| Total | 9 | 21 | 32 | 66 | 106 |
| Min | 3 | 5 | 6 | 11 | 15 |
| Max | 9 | 18 | 24 | 40 | 58 |
| Mean ± S.D. | 4.9 ± 1.2 | 9.1 ± 2.8 | 11.5 ± 4.2 | 19.1 ± 7.0 | 30.5 ± 9.6 |
min: minimum, max: maximum, S.D.: standard deviation.
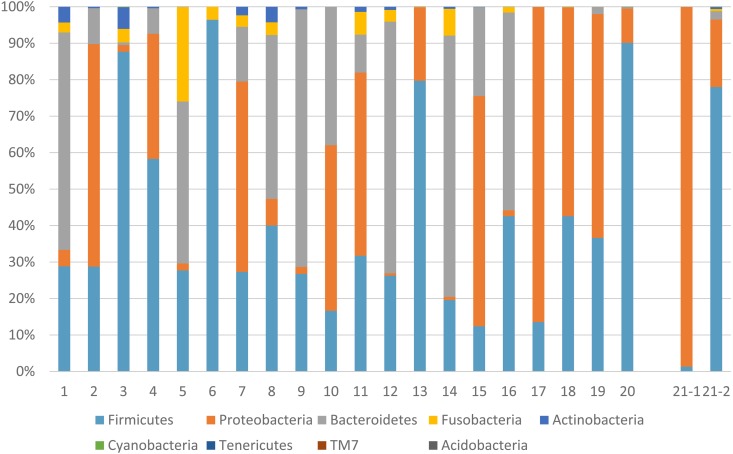
In the dogs without any symptoms (Sample ID 1 to 20), the maximum and minimum number of OTUs in phylum level was 9 and 3 per sample, respectively. In phylum level, the intestinal microbiota were classified into a total of 8 phyla (Fig. 1, ID 1 to 20). Firmicutes (mean, 30.3%; range, 12.4–90.1%), Proteobacteria (mean, 14.8%; range, 0.03–98.6%), Bacteroidetes (mean, 12.7%; range, 0.0–71.6%), or Fusobacteria (mean, 0.82%; range, 0.0–25.9%) were predominant in all samples; these four phyla were responsible for over 93.4% of the intestinal microbiota. Nevertheless, the ratio of these four phyla was different among samples. In family level, the maximum and minimum number of OTUs was 40 and 11 per sample, respectively. Although the dominant families were varied among samples, Clostridiaceae (0.5–71.1%), Lachnospiraceae (3.1–30.8%), and Enterobaacteriaceae (0.01–47.5%) were detected in all samples (Supplemental Table 1). In healthy dog group, Enterobacteriaceae (24.2 ± 0.29%), Bacteroidaceae (19.0 ± 0.25%), and Lachnospiraceae (13.4 ± 0.15%) were most prevalent at family level.
Fig. 1.
Population representation of the faecal microbiome at the phylum level in each sample. The OTUs obtained using QIIME was classified into the level of the phylum, 9 phyla, for each faecal swab sample from 20 dogs without diarrhoea and 1 dog with PLE.
In the dog with PLE, The microbiota was dramatically changed before and after treatment (Fig. 1, ID 21-1 and 21-2). At 97% similarity, 3 and 9 OTUs at phylum level and 15 and 58 OTUs at genus level were observed before and after treatment, respectively. At the phylum level, the predominant taxon of microbiota was Proteobacteria (98.5%) at the first sampling. By contrast, at the second sampling, the predominant taxon of microbiota was Firmicutes (78.0%) followed by Proteobacteria (18.5%). At the family level, Enterobacteriaceae was dominant (98.6%) at the first sampling. Nonetheless, the ratio decreased to 17.7% at the second sampling (Supplemental Table 1). The level of serum albumin increased (1.4 to 3.8 g/dl) with an increase in diversity of the faecal microbiota.
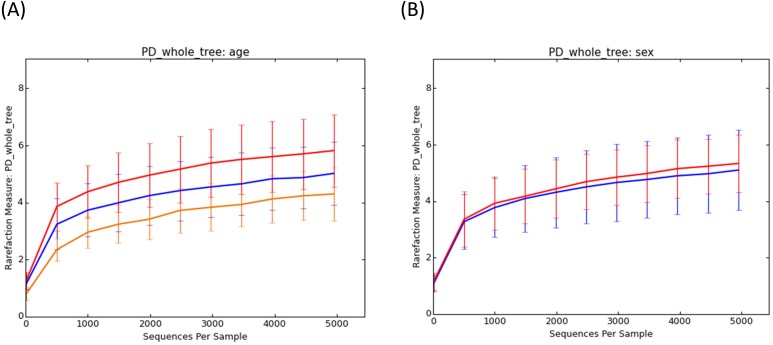
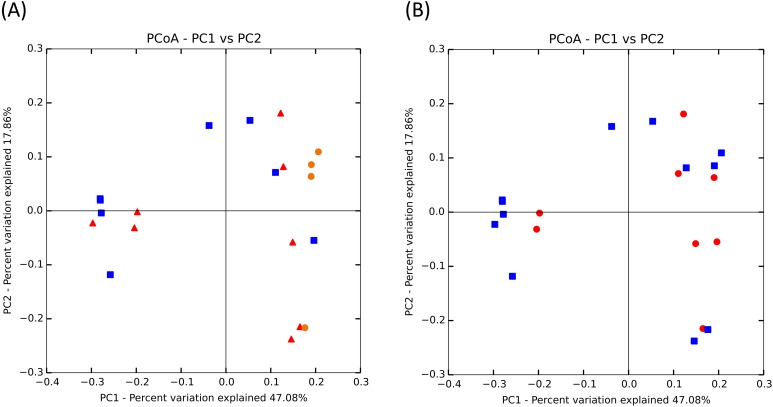
In faecal samples from healthy dogs, the relationships between microbiota and the age or the sex were evaluated. The rarefaction curves on Phylogenetic Diversity (PD) whole tree for each of the 20 healthy dogs, age group, and sex are shown in Fig. 2. Statistical analysis was performed to a depth of 4,950 reads. Rarefaction of the observed species was significantly different among age groups (Kruskal-Wallis; P<0.001). In contrast, there was no significant difference between females and males (Mann-Whitney U test: P=0.19). Additionally, there was a statistically significant difference between the young and middle-aged group, young and elderly group, and middle-aged and elderly group (Bonferroni-corrected Mann-Whitney U test: P<0.01, P<0.001, P<0.05, respectively). The PCoA plots based on the weighted UniFrac distance metric of the faecal microbiome by age group and sex are shown in Fig. 3. No clear-cut separation was observed (sex; P=0.45, age; P=0.18).
Fig. 2.
Rarefaction analysis of the 16S rRNA gene obtained from faecal samples of dogs without diarrhoea. Lines represent the average of each group, and error bars represent the standard deviations. The analysis was classified by age (A); red line: 0–3 years old, blue line: 4–8 years old, and yellow line: 9–12 years old, and by sex (B); red line: females, and blue line: males.
Fig. 3.
PCoA plots of the 16S rRNA gene from faecal samples. PCoA was calculated using weighted UniFrac distance. The analysis was classified by age (A) and sex (B). There was no significant difference among age groups (ANOSIM, P=0.18, R=0.069) and between sex groups (ANOSIM, P=0.45, R=–0.013).
DISCUSSION
In this study, we demonstrated the characteristics of the faecal swab microbiota in toy poodles without intestinal dysbiosis, and the sequential change of the microbiota in a toy poodle with PLE. The high-throughput sequencing analysis targeting 16S rRNA gene revealed the intestinal microbiota. In the 16S rRNA gene analysison healthy dogs, the predominant phyla were Fusobacteria, Firmicutes, Bacteroidetes, Actinobacteria, and Proteobacteria, and in the shotgun sequencing analysis of the faecal samples, the predominant phyla were Bacteroidetes, Firmicutes, Proteobacteria, Fusobacteria, and Actinobacteri [2, 10]. We detected that Firmicutes, Bacteroidetes, Proteobacteria, and Fusobacteria were the dominant phyla, composing over 95% of the total microbes, even though the dogs were under various conditions, such as breeding environment, age, diet, genetic background and health conditions. Additionally, Firmicutes, Bacteroidetes, and Proteobacteria composed 90% or more of the total microbes in the faecal samples from all but one animal. And in family level of microbiota in toy poodles, Enterobacteriaceae in Proteobacteria, Bacteroidaceae in Bacteroidetes, and Lachnospiraceae in Firmicutes were predominant in healthy toy poodles. In beagle dog analysis, Firmicutes, followed by Actinobacteria, Proteobacteria, and Bacteroidetes in lean dogs were predominant, and Proteobacteria, followed by Firmicutes in obese dogs was predominant, and Lactobacillus was predominant family in Firmicutes in lean and obese dogs and Unclassified Proteobacteria and Psychobacter was predominant family in Proteobacteria in lean and obese dogs, respectively [17]. In the analysis of faecal samples from healthy dogs with diverse home environments, diets, and living area, Firmicutes was dominant phylum and Clostridiaceae was dominant family [21]. In adult dogs with hound bloodlines, Firmicutes was the predominant phylum, followed by Fusobacteria and Tenericutes, and Clostridiaceae, Lachnospiraceae, Ruminococcaceae, and Fusobacteriaceae were predominant families [16]. These results indicates that four bacterial phyla, Firmicutes, Bacteroidetes, Proteobacteria, and Fusobacteria, may be necessary for the analysis of intestinal microbiota, but predominant phylum and family might be different among the dog breeds. Therefore, further research of microbiota focused on the differences in dog breeds might be necessary.
The diversity of intestinal microbiota in humans changes with age [4, 14]. In companion animals, such as dogs and cats, there are only a few reports on the correlation of diversity of their intestinal microbiota to age. In cats, the faecal microbiota is predominantly determined by age [3]. In dogs, however, the diversity of the faecal microbiota was not associated with age or sex [19]. Although this study could not use diet, home environment, and health condition for statistical adjustments, it indicates that the age might affect to the diversity of the faecal microbiota in toy poodles. On the other hands, the influence of sex was not observed in the diversity. To reveal more clearly the effect of age and sex to microbiota, large-scale analysis should be performed including various metadata collection including diets.
Acute and chronic gastrointestinal diseases, including IBD, are associated with alterations of the intestinal and faecal microbiota in humans, cats, and dogs. In case of dogs with IBD, prevalence of Firmicutes and Bacteroidetes decreased while that of Proteobacteria, especially Enterobacteriaceae increased [9, 20, 21]. In this study, we showed that microbiota composition in the dog with PLE clearly varied before and after treatment. Before treatment, Proteobacteria was the major phylum and the diversity of the microbiota was lower, OTUs was 15. After treatment, however, the serum albumin level recovered and the diversity of the microbiota increased (OTUs was 58), and the dominant microbe move to Firmicutes These data indicates that intestinal microbial condition might affect to serum protein level. More detailed studies are needed to reveal the relation between intestinal microbiota and serum albumin level in PLE.
In this study, four phyla, Firmicutes, Proteobacteria, Bacteroidetes, and Fusobacteria, were found to be dominant in the intestinal microbiota of toy poodles, and the microbiota diversity decreased with age. In the toy poodle with PLE, microbiota composition changed in relation to the serum albumin level. While these findings are elementary, they still represent useful information for microbiota research on dogs.
Supplementary
REFERENCES
- 1.Cong X., Xu W., Janton S., Henderson W. A., Matson A., McGrath J. M., Maas K., Graf J.2016. Gut Microbiome Developmental Patterns in Early Life of Preterm Infants: Impacts of Feeding and Gender. PLOS ONE 11: e0152751. doi: 10.1371/journal.pone.0152751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deng P., Swanson K. S.2015. Gut microbiota of humans, dogs and cats: current knowledge and future opportunities and challenges. Br. J. Nutr. 113 Suppl: S6–S17. doi: 10.1017/S0007114514002943 [DOI] [PubMed] [Google Scholar]
- 3.Deusch O., O’Flynn C., Colyer A., Swanson K. S., Allaway D., Morris P.2015. A Longitudinal Study of the Feline Faecal Microbiome Identifies Changes into Early Adulthood Irrespective of Sexual Development. PLOS ONE 10: e0144881. doi: 10.1371/journal.pone.0144881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dillon R. J., Webster G., Weightman A. J., Keith Charnley A.2010. Diversity of gut microbiota increases with aging and starvation in the desert locust. Antonie van Leeuwenhoek 97: 69–77. doi: 10.1007/s10482-009-9389-5 [DOI] [PubMed] [Google Scholar]
- 5.Gnanandarajah J. S., Johnson T. J., Kim H. B., Abrahante J. E., Lulich J. P., Murtaugh M. P.2012. Comparative faecal microbiota of dogs with and without calcium oxalate stones. J. Appl. Microbiol. 113: 745–756. doi: 10.1111/j.1365-2672.2012.05390.x [DOI] [PubMed] [Google Scholar]
- 6.Goodrich J. K., Davenport E. R., Waters J. L., Clark A. G., Ley R. E.2016. Cross-species comparisons of host genetic associations with the microbiome. Science 352: 532–535. doi: 10.1126/science.aad9379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guard B. C., Barr J. W., Reddivari L., Klemashevich C., Jayaraman A., Steiner J. M., Vanamala J., Suchodolski J. S.2015. Characterization of microbial dysbiosis and metabolomic changes in dogs with acute diarrhea. PLOS ONE 10: e0127259. doi: 10.1371/journal.pone.0127259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hatta Y., Omatsu T., Tsuchiaka S., Katayama Y., Taniguchi S., Masangkay J. S., Puentespina R., Jr., Eres E., Cosico E., Une Y., Yoshikawa Y., Maeda K., Kyuwa S., Mizutani T.2016. Detection of Campylobacter jejuni in rectal swab samples from Rousettus amplexicaudatus in the Philippines. J. Vet. Med. Sci. 78: 1347–1350. doi: 10.1292/jvms.15-0621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Honneffer J. B., Minamoto Y., Suchodolski J. S.2014. Microbiota alterations in acute and chronic gastrointestinal inflammation of cats and dogs. World J. Gastroenterol. 20: 16489–16497. doi: 10.3748/wjg.v20.i44.16489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hooda S., Minamoto Y., Suchodolski J. S., Swanson K. S.2012. Current state of knowledge: the canine gastrointestinal microbiome. Anim. Health Res. Rev. 13: 78–88. doi: 10.1017/S1466252312000059 [DOI] [PubMed] [Google Scholar]
- 11.Igarashi H., Maeda S., Ohno K., Horigome A., Odamaki T., Tsujimoto H.2014. Effect of oral administration of metronidazole or prednisolone on fecal microbiota in dogs. PLOS ONE 9: e107909. doi: 10.1371/journal.pone.0107909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kasiraj A. C., Harmoinen J., Isaiah A., Westermarck E., Steiner J. M., Spillmann T., Suchodolski J. S.2016. The effects of feeding and withholding food on the canine small intestinal microbiota. FEMS Microbiol. Ecol. 92: fiw085. doi: 10.1093/femsec/fiw085 [DOI] [PubMed] [Google Scholar]
- 13.Middelbos I. S., Vester Boler B. M., Qu A., White B. A., Swanson K. S., Fahey G. C., Jr.2010. Phylogenetic characterization of fecal microbial communities of dogs fed diets with or without supplemental dietary fiber using 454 pyrosequencing. PLoS ONE 5: e9768. doi: 10.1371/journal.pone.0009768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mihajlovski A., Doré J., Levenez F., Alric M., Brugère J. F.2010. Molecular evaluation of the human gut methanogenic archaeal microbiota reveals an age-associated increase of the diversity. Environ. Microbiol. Rep. 2: 272–280. doi: 10.1111/j.1758-2229.2009.00116.x [DOI] [PubMed] [Google Scholar]
- 15.O’Toole P. W., Jeffery I. B.2015. Gut microbiota and aging. Science 350: 1214–1215. doi: 10.1126/science.aac8469 [DOI] [PubMed] [Google Scholar]
- 16.Panasevich M. R., Kerr K. R., Dilger R. N., Fahey G. C., Jr., Guérin-Deremaux L., Lynch G. L., Wils D., Suchodolski J. S., Steer J. M., Dowd S. E., Swanson K. S.2015. Modulation of the faecal microbiome of healthy adult dogs by inclusion of potato fibre in the diet. Br. J. Nutr. 113: 125–133. doi: 10.1017/S0007114514003274 [DOI] [PubMed] [Google Scholar]
- 17.Park H. J., Lee S. E., Kim H. B., Isaacson R. E., Seo K. W., Song K. H.2015. Association of obesity with serum leptin, adiponectin, and serotonin and gut microflora in beagle dogs. J. Vet. Intern. Med. 29: 43–50. doi: 10.1111/jvim.12455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sabbioni A., Ferrario C., Milani C., Mancabelli L., Riccardi E., Di Ianni F., Beretti V., Superchi P., Ossiprandi M. C.2016. Modulation of the Bifidobacterial Communities of the Dog Microbiota by Zeolite. Front. Microbiol. 7: 1491. doi: 10.3389/fmicb.2016.01491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Šlapeta J., Dowd S. E., Alanazi A. D., Westman M. E., Brown G. K.2015. Differences in the faecal microbiome of non-diarrhoeic clinically healthy dogs and cats associated with Giardia duodenalis infection: impact of hookworms and coccidia. Int. J. Parasitol. 45: 585–594. doi: 10.1016/j.ijpara.2015.04.001 [DOI] [PubMed] [Google Scholar]
- 20.Suchodolski J. S.2016. Diagnosis and interpretation of intestinal dysbiosis in dogs and cats. Vet. J. 215: 30–37. doi: 10.1016/j.tvjl.2016.04.011 [DOI] [PubMed] [Google Scholar]
- 21.Suchodolski J. S., Markel M. E., Garcia-Mazcorro J. F., Unterer S., Heilmann R. M., Dowd S. E., Kachroo P., Ivanov I., Minamoto Y., Dillman E. M., Steiner J. M., Cook A. K., Toresson L.2012. The fecal microbiome in dogs with acute diarrhea and idiopathic inflammatory bowel disease. PLoS ONE 7: e51907. doi: 10.1371/journal.pone.0051907 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.