Abstract
Rotavirus A (RVA) G9 genotype is recognized as an emerging genotype which is spreading worldwide, however, our knowledge on pathogenicity of this virus is limited. In this study, porcine RVA strain HN03 was successfully isolated on MA-104 cells, and the isolate was propagated continuously for 7 passages after a virus cloning at passage 3. The virus titers from 4 to 10 passages ranged from 107.1 to 108.1 TCID50/ml. The growth curve of HN03 strain in cell culture was determined, and the virus production dynamics was confirmed by immunoperoxidase monolayer assay (IPMA). Sequence and phylogenetic analyses based on full-length VP7 and partial VP4 genes indicated that HN03 strain belongs to genotype G9P[23]. In addition, the sixth passage of strain HN03 in cell culture was subjected to 3-day-old piglets. All infected piglets developed severe watery diarrhea within 24 hr post-inoculation (hpi), but recovered from disease after 72 hpi. RVA antigen could be detected by IHC in the cytoplasm of villous enterocytes as early as 2 hr after appearance of clinical symptoms and virus antigen load kept increasing in the next 30 hr. The dynamics of RVA HN03 strain proliferation on cells and in pigs extended our understanding of rotavirus pathogenicity.
Keywords: G9 genotype, G9P[23], pathogenicity, porcine rotavirus
Rotaviruses (RVs) belong to the family Reoviridae, and they have a genome of 11 segments of double-stranded RNA encoding 6 structural proteins (VP1−VP4, VP6, and VP7) and five non-structural proteins (NSP1−NSP5/6) [7, 11]. RVs are classified into 8 major groups (RVA−RVH) based on the antigenic and genetic characteristics of the inner capsid protein VP6 [16]. RVA, RVB, RVC, RVE, and RVH have been detected in pigs [7, 31]. RVA is considered the main cause of severe diarrhea in young children and piglets worldwide [15, 18].
Based on molecular characterization of VP7 and VP4, RVA strains are further classified into different G and P types [17]. To date, at least 27 G genotypes and 38 P genotypes have been identified, several of them being detected from both humans and animals [6, 8]. Among porcine rotaviruses, the main G genotypes detected in pigs are G3, G4, G5, G9 and G11, whereas genotypes G1, G2, G6, G8, G10, G12 and G26 have been identified occasionally [5, 33]. Recently, RAV G9 genotype is recognized as an emerging genotype which is spreading worldwide, including mainland China, Taiwan, Korea, Japan, Thailand, Belgium, Italy, U.S.A., and Brazil [1, 13, 19, 24, 26, 27, 29, 32]. The rapidly increasing prevalence of this virus raises questions regarding its origin, epidemiological and pathogenicity importance. Until recently, there are only few studies on pathogenicity of porcine G9 rotavirus [12, 25, 34].
In the present study, we obtained a porcine RVA strain, named as HN03, which was isolated from one porcine rotavirus positive intestinal sample collected from a piglet with diarrhea in Henan Province, China. The isolation, molecular characterization, and an evaluation of pathogenicity in piglets of this virus were described in this study. Immunochemistry straining was applied to different parts of small intestine at different time points after infection to understand the progression of virus infection.
MATERIALS AND METHODS
Samples
In December 2015, 3 intestinal samples were obtained from diarrheic 5-day-old piglets, which were collected from a farm in Henan Province, China. The samples were examined by RT-PCR for porcine epidemic diarrhea virus (PEDV), transmissible gastroenteritis virus (TGEV), and RVA at National Research Center for Veterinary Medicine. As the results of all samples positive for RVA and negative for PEDV and TGEV, and one of positive sample was selected for virus isolation attempt. Small intestine tissue was homogenated in sterile phosphate-buffered saline (PBS), vortexed briefly, and centrifuged at 8,000 × g for 10 min at 4°C. The supernatant was passed through 0.22 µm Millipore filters, and was used as inoculum for virus isolation.
Virus isolation and propagation
Virus isolation of RVA was conducted on MA-104 cells (ATCC CRL-2378.1) as previously described [2] with some modifications. Briefly, MA-104 cells were grown in T25 flasks with α-Minimum Essential Medium (α-MEM, Gibco, Tulsa, OK, U.S.A.) supplemented with 5% fetal bovine serum (Hyclone, Logan, UT, U.S.A.) and antibiotics (100 units/ml of penicillin and 100 µg/ml of streptomycin). Confluent MA-104 cell monolayers were used for virus inoculation. The inoculum was pretreated with trypsin (type IX, Sigma, St. Louis, MO, U.S.A.) at a final concentration of 10 µg/ml at 37°C for 60 min. Growth medium was removed, and the monolayers of cells were washed with PBS twice before inoculation. Then 0.5 ml of treated inoculum and 0.5 ml maintenance medium was added. Maintenance medium was consisted of α-MEM supplemented with antibiotics (100 units/ml of penicillin and 100 µg/ml of streptomycin) and 0.5 µg/ml trypsin (type IX, Sigma). Following virus adsorption for 60 min at 37°C, inoculum was removed and the cell monolayers were washed with PBS once. Finally, 5 ml maintenance medium were added to each flask. Cultures were incubated at 37°C with 5% CO2, and examined daily for cytopathic effect (CPE). When CPE appeared in more than 80% of cells (−5 days after inoculation), the flasks were subjected to two freeze-thaw cycles. The cells and supernatants were harvested and stored at −70°C. These samples were used as seed stocks for the next passage. Serial four blind passages were performed if no CPE appeared within 5 days.
Virus titration was performed in 96-well plates with 10-fold serial dilutions performed in eight replicates per dilution. Virus titers were determined after five days of inoculation according to the method of Reed and Muench [22] and endpoints were expressed as the 50% tissue culture infective dose (TCID50)/ml.
Determination of virus growth curve
The 6th passage of porcine RVA HN03 was inoculated onto cell monolayers in 35 mm dishes at a multiplicity of infection (MOI) of 1.0 to determine virus growth curve. Following adsorption for 60 min at 37°C, virus inoculum was removed and cell monolayers were washed once by PBS, and then cells were fed with maintenance medium, and incubated at 37°C with 5% CO2. At 6, 9, 12, 15, 18, 21, 24, 27 and 30 hr post inoculation (hpi), cell culture supernatants and cells were collected. After freezing and thawing twice, the mixtures of cell culture supernatants and cell lysates were used for virus titration. Virus titration at various time points was carried out in triplicates.
Immunoperoxidase monolayer assay (IPMA)
MA-104 cells in 35 mm dishes that mock infected or infected with porcine rotavirus were fixed with 80% acetone for 30 min at 4°C, and then washed with PBS and air-dried. Dishes were incubated for 30 min at 37°C with a 1:500 dilution mouse monoclonal antibody (in-house made) specifically against rotavirus VP6 in a humidity chamber, and then 4°C overnight. Next, dishes were washes with PBS and incubated for 60 min at 37°C with HRP-labeled goat anti-mouse IgG (Biomedical Technologies Inc., Stoughton, MA, U.S.A.) diluted 1:100 in PBS. Finally, dishes were washed 3 times with PBS, followed by incubation for 5−10 min at room temperature in diaminobenzidine solution (ZSGB-BIO, Beijing, China). The sections were lightly counterstained with Mayer’s haematoxylin, dehydrated through graded concentrations of ethanol and xylene, and mounted. Cell staining was examined under an inverted light microscope.
VP7, VP4 and VP6 gene sequencing and phylogenetic analysis
Viral RNA extraction was performed with 200 µl sixth passage of the HN03 strain by using a viral nucleic acid extraction kit (Geneaid Biotech Ltd., Taiwan, China) following the instructions of the manufacturers. cDNA was generated by TransScript II First-Strand cDNA Synthesis SuperMix (TransGen Biotech, Beijing, China) using random primer. The full-length VP7 and partial VP4 genes were amplified using TransStart Taq DNA Polymerase (TransGen Biotech, Beijing, China). The primers and program used (Beg9/End9 for VP7, Con3/Con2 for VP4 and GEN_VP6F/GEN_VP6R for VP6) were previously described by Gouvea et al. [10], Gentsch et al. [9] and Okitsu et al. [21], respectively. Purified PCR products were cloned using pEASY-T1 Cloning Kit (TransGen Biotech, Beijing, China) according to the manufacturer’s instructions. Recombinant DNA clones were sequenced by GENEWIZ (Beijing, China). For each gene, three to four clones were sequenced, and the consensus sequence were used. Sequences obtained were submitted to the automated online rotavirus genotyping tool RotaC2.0 [14]. The sequences were also aligned with reference sequences from GenBank using ClustalX v1.81. Neighbor-joining trees based on the VP7 and VP4 genes were generated using PHYLIP v3.67 [23]. The reliability of branches was assessed by bootstrap analysis using 1,000 replicates. The degree of similarity among sequences was determined using MegAlign v7.0 (a tool in the software DNAStar). The full-length VP7, VP6, and partial VP4 gene nucleotide sequences of strain HN03 in this study were deposited in GenBank database under accession numbers KY649279, MH021179 and KY649280.
Experimental infection of 3-day-old piglets
Animal experiments were conducted and approved by the Animal Care and Ethics Committee of China National Research Center for Veterinary Medicine. Colostrum-deprived piglets used in two experiments were delivered by natural delivery from sows free from both antibodies and gene detection for porcine RVA, porcine epidemic diarrhea virus, porcine transmissible gastroenteritis virus, porcine circovirus 2, porcine reproductive and respiratory syndrome virus, and pseudorabies virus. Piglets were housed in pens (5−7 pigs per pen) in temperature-controlled rooms, and control groups were transferred to a different room prior to inoculation. The room temperature was maintained at 28−32°C. Pigs were fed every 2−3 hr with a commercial milk (Dumex PRECINUTRICARETM, Shanghai, China).
In experiment 1, eight 3-day-old piglets were assigned randomly to two groups: porcine RVA-infected group (n=5) and negative control group (n=3) in order to determine the virulence of strain HN03. Animals in infected group were inoculated orally 2 ml (107.6TCID50/ml) HN03 of passage 6. Piglets in negative control group were mock inoculated with 2 ml virus-free cell culture media. All 5 infected piglets developed diarrhea within 3 days post-inoculation (dpi), and recovered after 3 dpi.
In experiment 2, to understand the progression of porcine rotavirus infection, eleven 3-day-old piglets were randomly allocated into infected group (pig number 1−4, 6−10, n=9) and mock-infected group (pig number 5 and 11, n=2), and treated by the same dose and route as in experiment 1. Clinical signs were monitored continuously. In infected group, pig 7 was euthanized at 12 hpi before the appearance of clinical signs. The remaining animals in infected group showed clinical symptoms between 15 and 22.5 hpi, and were euthanized at 2 (pig 6), 10 (pig 4), 20 (pig 2), 30 (pig 10), 40 (pig 1), 50 (pig 3) and 60 (pig 9) hr post-appearance of clinical symptoms (hpacs) of each individual piglets. Pig 5 and 11 in negative control group were euthanized at 24 and 75 hr after mock inoculation, respectively. Rectal swabs were collected daily for detection of virus shedding by RT-PCR. Formalin-fixed small intestine sections including duodenum, proximal jejunum, mid-jejunum, distal jejunum, and ileum were subjected to immunohistochemistry for antigen detection.
Quantification of virus shedding
Ten-fold serial dilutions of each rectal swab were prepared in PBS, from 100 to 10−4. RT-PCR was performed as previously described [35]. The highest dilution that yielded PCR amplicon of expected size (333 bp) was considered as the endpoint, and PCR titers of virus shedding were calculated based on the endpoint dilutions.
Immunohistochemistry (IHC)
Tissue sections were de-waxed in xylene, rehydrated through a graded series of alcohols, and air dried. Endogenous peroxidase activity was quenched with 3% hydrogen peroxide for 20 min at room temperature, and then the slides were washed twice with PBS, pH 7.2 (3 min each). Antigen retrieval was accomplished with 0.25% trypsin for 20 min at 37°C, followed by three rinses with PBS. All slides were incubated with 1:20 dilution normal horse serum (ZSGB-BIO) for 20 min at room temperature to saturate nonspecific protein binding sites. The slides were next treated at 37°C for 30 min, and then 4°C overnight in a humidity box using in-house monoclonal antibody against RVA VP6, diluted 1:500 with PBS. After three rinses with PBS, sections were flooded and incubated for 1 hr at 37°C with HRP-goat anti-mouse IgG (Biomedical Technologies Inc., Stoughton, MA, U.S.A.) diluted 1:100 with PBS. The slides were washed 3 times in PBS, and incubated in 3-Amino-9-ethylcarbazole solution (ZSGB-BIO) for 6−7 min at room temperature. Finally, the sections were lightly counterstained with Mayer’s hematoxylin. The slides were naturally dried and sealed with water-soluble tablets seal (GVA). Antigen detection was scored semi-quantitatively according to the following criteria: − =no signal, + =1–25% of villous enterocytes in twenty microscopic fields (magnification 100×) within the section showing a positive signal, ++ =26–50% of villous enterocytes showing a positive signal, +++ =51–75% of villous enterocytes showing a positive signal, ++++ =greater than 75% of villous enterocytes showing a positive signal.
RESULTS
Virus isolation and propagation
Virus isolation from one porcine RVA PCR-positive intestine homogenate was performed on MA-104 cells. Porcine RVA HN03 was successfully isolated. At passage 3, typical cytopathic effect (CPE) characterized by cell shrinking round, cell layer splitting, lysis, detachment, and shedding was observed at 3 dpi. The virus isolate was plaque-purified once at passage 3. In passages 4−10, CPE was visible within 30 hpi. No distinctive CPE was found in mock-infected control cultures. VP7, VP4 and VP6 genes of purified virus were sequenced at passages 3, 6, 9 and they shared 100% nucleotide similarity among each passage which implied that the virus stock contains only a single virus strain. During passages 4−10, the virus titers of HN03 ranged from 107.1 to 108.1 TCID50/ml, as summarized in Table 1.
Table 1. Summary of virus titers of porcine RVA HN03 during serially passages on MA-104 cells.
| Passage no. | Infectious titer (log10 TCID50/ml) |
|---|---|
| P1 | ND |
| P2 | ND |
| P3 | 6.3 |
| P4 | 7.1 |
| P5 | 7.3 |
| P6 | 7.6 |
| P7 | 8.0 |
| P8 | 7.9 |
| P9 | 8.0 |
| P10 | 8.1 |
Plaque purification was performed at P3. ND, not determined.
Virus growth characterization
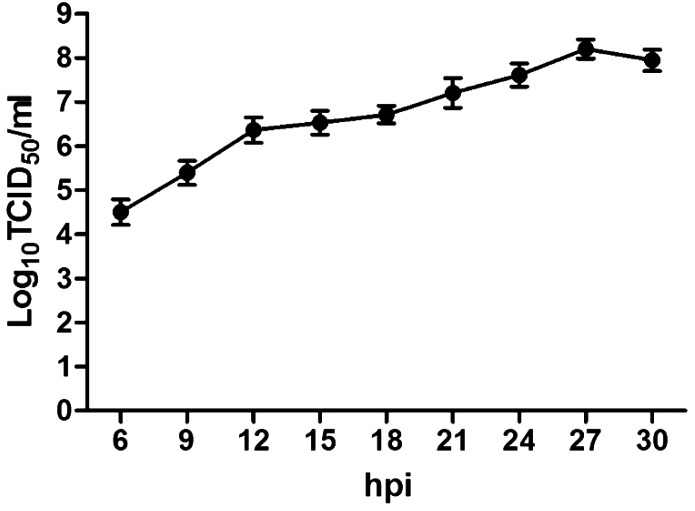
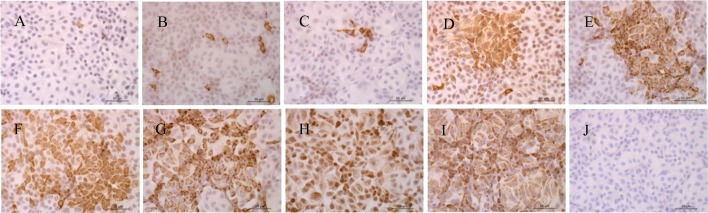
The cell-culture porcine RVA HN03 was adapted to propagation on MA-104 cells, and at the 6th passage, growth curve of the virus was determined (Fig. 1). CPE appeared at 15 hpi and spread rapidly to the entire cell monolayer. Virus titer reached the maximum (108.2 TCID50/ml) at 27 hpi. Virus growth was also confirmed by IPMA using anti-porcine RVA rotavirus monoclonal antibody (in-house made). As shown in Fig. 2, consistent with above virus titration results, the positive signal of staining could be detected as early as 6 hpi and reached the maximum at 30 hpi before infected cells detached from the well.
Fig. 1.
Growth curve of cell-adapted porcine RVA strain HN03 in MA-104 cell culture. Data was presented as mean ± SD by triplicates.
Fig. 2.
Detection of porcine RVA antigen from MA-104 cell cultures by IPMA at different time points. Panels A−I represent 6, 9, 12, 15, 18, 21, 24, 27 and 30 hpi, respectively, and panel J represents MA-104 cells without infection at 30 dpi. Magnification 200 ×. A–J: Bar=50 μm.
VP7, VP4 and VP6 gene sequencing and phylogenetic analysis
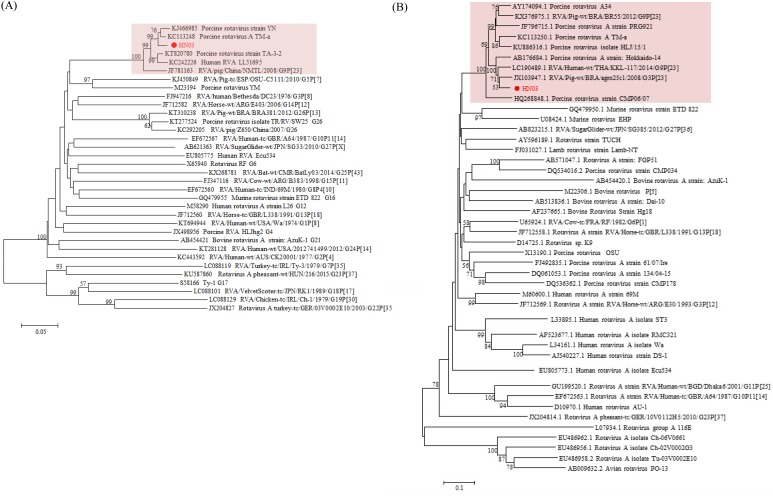
Porcine RVA strain HN03 was genotyped as G9P[23]. For VP7 gene, HN03 was most closely related to two Chinese strains YN and TM-a (Fig. 3A), with highest nucleotide sequence identity (97.6%). For partial VP4 gene, HN03 was most closely related to a Brazilian strain agro25c1 (Fig. 3B), with highest nucleotide sequence identity (92.3%), and shared only 88.2% to 88.6% nucleotide sequence identity with another two Chinese strains HLJ/15/1 and TM-a. For VP6 gene, HN03 was genotyped as I5, and exhibited highest nucleotide sequence identity (96%) to some porcine or human strains.
Fig. 3.
Phylogenetic relationship among porcine RVA HN03 in this study and reference sequences in GenBank, as inferred by neighbor-joining analyses of the full-length VP7 (A) and partial VP4 (B) nucleotide sequences based on distance calculated using the Kimura 2-parameter model. Bootstrap values (in percentage) above 50 from 1,000 pseudoreplicates are shown. The isolate in this study is marked by closed circle.
Pathogenicity on 3-day-old piglets
In experiment 1, all five piglets developed severe watery diarrhea, and displayed anorexia at 15−24 hpi. 60% animals (3/5) developed vomiting within 48 hpi. During 36−72 hpi, challenged animals exhibited mild to moderate diarrhea and anorexia. After 72 hpi, all challenged pigs had normal or pasty feces and normal appetite. The animals in mock-infected control group had normal feces and no clinical signs.
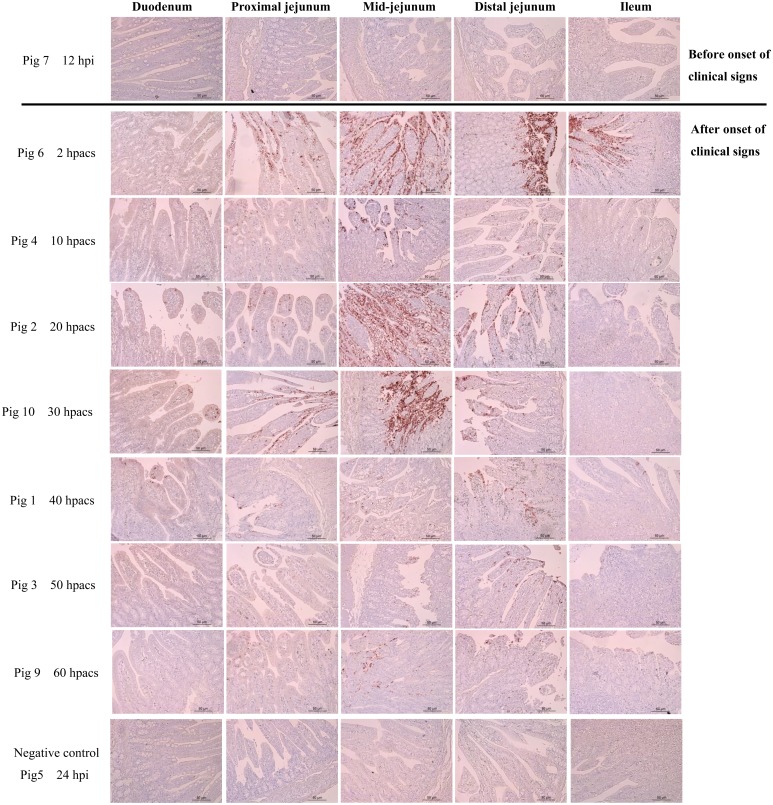
In experiment 2, onset of clinical signs was similar to that in experiment 1. The piglets in infected group developed severe diarrhea at 15−22.5 hpi. Detection of fecal viral shedding coincided with onset of clinical signs. High-level viral RNA PCR titer (10−3 to 10−4 dilution) was detected from rectal swabs within 9 hr after onset of clinical signs, and decreased subsequently (10−3 to 10−1) (Table 2). RVA antigen was initially detected by IHC in the cytoplasm of villous enterocytes of infected pig 6 at 18 hpi (2 hr after onset of clinical signs), and apparent positive signal was visible in entire jejunum and ileum (Fig. 4). The highest quantities of RV antigen was detected in the jejunum (especially in mid-jejunum) of infected piglets within 30 hpacs (hour post appearance of clinical symptoms), and declined at latter time points (Fig. 4).
Table 2. Fecal shedding of virus, clinical signs and immunohistochemistry findings after inoculation of pigs with porcine RVA strain HN03.
| Pig status, pig no. | Fecal shedding, PCR titer a), by hpi |
Onset of clinical signs, hpi | Hpi at euthanasia (after onset) | Antigen detection by IHC |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 12 | 24 | 36 | 48 | 60 | 72 | Duodenum | Proximal jejunum | Mid-jejunum | Distal jejunum | Ileum | |||
| HN03-inoculated | ||||||||||||||
| 7 | − | − | ND | ND | ND | ND | ND | None | 12.0 | − | − | − | − | − |
| 6 | − | 0 | ND | ND | ND | ND | ND | 16.0 | 18.0 (2) | − | +++ | ++++ | ++++ | +++ |
| 4 | − | 0 | −3 | ND | ND | ND | ND | 17.5 | 27.5 (10) | + | + | +++ | + | − |
| 2 | − | 0 | −3 | −3 | ND | ND | ND | 22.5 | 42.5 (20) | + | ++ | ++++ | ++ | + |
| 10 | − | 0 | −4 | −3 | ND | ND | ND | 15.0 | 45.0 (30) | + | +++ | ++++ | ++ | + |
| 1 | − | 0 | −3 | −2 | −2 | −2 | ND | 22.5 | 62.5 (40) | ++ | ++ | ++ | ++ | + |
| 3 | − | 0 | −3 | −3 | −2 | −1 | ND | 17.5 | 67.5 (50) | − | + | + | + | + |
| 9 | − | 0 | −4 | −3 | −2 | −2 | −2 | 15.0 | 75.0 (60) | − | + | +++ | + | + |
| Negative control | ||||||||||||||
| 5 | − | − | − | ND | ND | ND | ND | None | NA | − | − | − | − | − |
| 11 | − | − | − | − | − | − | − | None | NA | − | − | − | − | − |
−, negative; a) dilution 10n; hpi, hour post-inoculation; ND, not determined; NA, not applicable.
Fig. 4.
Immunohistochemistry staining of small-intestine sections from infected and control pigs. Porcine RVA antigen (brown reaction product) was detected in the villous enterocytes. Magnification 200 ×. Bar=50 μm.
DISCUSSION
RAV HN03 strain was plaque-purified once before testing its pathogenicity on pigs. Since VP7, VP4 and VP6 genes of purified virus were sequenced at passages 3, 6, 9 and they shared 100% nucleotide similarity among each passage, the possibility of contamination with other rotavirus or enteric viruses could be low based on the nucleotide identities among the clones for each gene segment. RAV G9 genotype is recognized as an emerging genotype in pigs and humans worldwide, and porcine G9 strains usually are found in combination with P[7], P[13], P[19] and P[23] [32]. Two recent reports showed that two G9 genotype porcine rotaviruses including G9P[7] and G9P[23] are circulating in pig herds in mainland China [26, 34]. In this study, porcine RVA strain HN03 obtained was identified as genotype G9P[23]. Porcine RVA G9P[23] genotype has been reported in several other Asian countries or regions, such as Taiwan [32], Thailand [20, 21, 33], Japan [28], and South Korea [13], as well as in Italy [19], Belgium [30], and Brazil [27].
Until recently, there are limited studies on pathogenicity of porcine G9 rotavirus (G9P[7], G9P[13] and G9P[23]) [12, 25, 34]. Kim et al. [12] reported that porcine G9P[23] strain PRG942 is able to induce diarrhea at DPI 1−8 in 3-day-old piglets, and inoculated piglets did not show any other clinical signs in addition to diarrhea. Zhang et al. [34] reported that all three 4-day-old piglets showed clinical signs (diarrhea with yellow and watery feces) at 16−24 hr after oral inoculation with rotavirus G9P[7] strain JS-01-2014; one infected piglet died, and the other two became moribund at 50 hr after inoculation. The report by Shao et al. [25] showed that all the inoculated one-week-old piglets with G9P[13] rotavirus developed diarrhea. In this study, all infected piglets developed severe watery diarrhea at 15−24 hpi and recovered from disease with normal or pasty feces and normal appetite at 72 hpi. Interestingly, a recent report by Wu et al. [32] showed that all 29 porcine G9 RVA strains were collected from healthy pigs of various ages including suckling piglets. Thus, more studies are needed to be carried out to improve our knowledge on pathogenicity of porcine RVA G9 rotavirus.
Rotavirus infects mature enterocytes in the mid and upper villous epithelium of the host’s small intestine which ultimately leads to cell death, villous atrophy, and diarrhea [4]. In this study, significantly larger amounts of antigen were present in the jejunum than in duodenum and ileum, especially in the mid-jejunum within 30 hpacs; hereafter, the amounts of viral antigen in the entire small intestines decreased, and that in the duodenum disappeared after 50 hpacs. The result differed from previous report by Kim et al. [12] that following G9P[7] and G9P[23] infection, RV antigen was almost equally distributed among duodenum, jejunum and ileum during the period (DPI 1−7) with diarrhea. The cause of the difference in colonized area may derive from different virulence of strains, and more virulent strain can spread more effectively, which was similar to previous report by Bridger et al. [3] in study on bovine rotaviruses.
In conclusion, porcine rotavirus G9P[23] strain HN03 was successfully isolated, serially propagated in cell culture and characterized. Phylogenetic analysis based on VP7 and VP4 genes indicates that HN03 strain belongs to genotype G9P[23]. The results of animal experiments reveal that this strain could cause watery diarrhea but led to no death in 3-day-old piglets. The information presented in this study expands our understanding the prevalence, genetic characterization and pathogenesis of porcine G9 rotavirus.
Acknowledgments
This work was supported by grant from National Key Research and Development Program (Grant No.2016YFD0500703), Major science and technology projects in Henan Province (Grant No.171100110200), and Luoyang HeLuo Talent Plan (Dr. Kegong Tian).
REFERENCES
- 1.Amimo J. O., Vlasova A. N., Saif L. J.2013. Detection and genetic diversity of porcine group A rotaviruses in historic (2004) and recent (2011 and 2012) swine fecal samples in Ohio: predominance of the G9P[13] genotype in nursing piglets. J. Clin. Microbiol. 51: 1142–1151. doi: 10.1128/JCM.03193-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bohl E. H., Theil K. W., Saif L. J.1984. Isolation and serotyping of porcine rotaviruses and antigenic comparison with other rotaviruses. J. Clin. Microbiol. 19: 105–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bridger J. C., Hall G. A., Parsons K. R.1992. A study of the basis of virulence variation of bovine rotaviruses. Vet. Microbiol. 33: 169–174. doi: 10.1016/0378-1135(92)90044-T [DOI] [PubMed] [Google Scholar]
- 4.Burke B., Desselberger U.1996. Rotavirus pathogenicity. Virology 218: 299–305. doi: 10.1006/viro.1996.0198 [DOI] [PubMed] [Google Scholar]
- 5.Collins P. J., Martella V., Sleator R. D., Fanning S., O’Shea H.2010. Detection and characterisation of group A rotavirus in asymptomatic piglets in southern Ireland. Arch. Virol. 155: 1247–1259. doi: 10.1007/s00705-010-0713-1 [DOI] [PubMed] [Google Scholar]
- 6.De Grazia S., Giammanco G. M., Potgieter C. A., Matthijnssens J., Banyai K., Platia M. A., Colomba C., Martella V.2010. Unusual assortment of segments in 2 rare human rotavirus genomes. Emerg. Infect. Dis. 16: 859–862. doi: 10.3201/eid1605.091826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Estes M. K., Kapikian A. Z.2007. Rotaviruses. pp. 1917–1974. In: Fields Virology, 5th ed. (Knipe, D. M., Griffin, D. E., Lamb, R. A., Straus, S. E., Howley, P. M., Martin, M. A. and Roizman, B. eds), Lippincott Williams & Wilkins, Philadelphia. [Google Scholar]
- 8.Fujii Y., Mitake H., Yamada D., Nagai M., Okadera K., Ito N., Okada K., Nakagawa K., Mizutani T., Sugiyama M.2016. Genome Sequences of Rotavirus A Strains Ty-1 and Ty-3, Isolated from Turkeys in Ireland in 1979. Genome Announc. 4: 4. doi: 10.1128/genomeA.01565-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gentsch J. R., Glass R. I., Woods P., Gouvea V., Gorziglia M., Flores J., Das B. K., Bhan M. K.1992. Identification of group A rotavirus gene 4 types by polymerase chain reaction. J. Clin. Microbiol. 30: 1365–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gouvea V., Glass R. I., Woods P., Taniguchi K., Clark H. F., Forrester B., Fang Z. Y.1990. Polymerase chain reaction amplification and typing of rotavirus nucleic acid from stool specimens. J. Clin. Microbiol. 28: 276–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.International Committee on Taxonomy of Viruses.2012. Virus taxonomy: classification and nomenclature of viruses. In: Ninth report of the International Committee on Taxonomy of Viruses. (King, A. M. Q., Adams, M. J., Carstens, E. B. and Lefkowitz, E. J. eds). Elsevier Academic Press, San Diego. [Google Scholar]
- 12.Kim H. H., Park J. G., Matthijnssens J., Kim H. J., Kwon H. J., Son K. Y., Ryu E. H., Kim D. S., Lee W. S., Kang M. I., Yang D. K., Lee J. H., Park S. J., Cho K. O.2013. Pathogenicity of porcine G9P[23] and G9P[7] rotaviruses in piglets. Vet. Microbiol. 166: 123–137. doi: 10.1016/j.vetmic.2013.05.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim H. H., Matthijnssens J., Kim H. J., Kwon H. J., Park J. G., Son K. Y., Ryu E. H., Kim D. S., Lee W. S., Kang M. I., Yang D. K., Hyun B. H., Park S. I., Park S. J., Cho K. O.2012. Full-length genomic analysis of porcine G9P[23] and G9P[7] rotavirus strains isolated from pigs with diarrhea in South Korea. Infect. Genet. Evol. 12: 1427–1435. doi: 10.1016/j.meegid.2012.04.028 [DOI] [PubMed] [Google Scholar]
- 14.Maes P., Matthijnssens J., Rahman M., Van Ranst M.2009. RotaC: a web-based tool for the complete genome classification of group A rotaviruses. BMC Microbiol. 9: 238. doi: 10.1186/1471-2180-9-238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marthaler D., Homwong N., Rossow K., Culhane M., Goyal S., Collins J., Matthijnssens J., Ciarlet M.2014. Rapid detection and high occurrence of porcine rotavirus A, B, and C by RT-qPCR in diagnostic samples. J. Virol. Methods 209: 30–34. doi: 10.1016/j.jviromet.2014.08.018 [DOI] [PubMed] [Google Scholar]
- 16.Matthijnssens J., Otto P. H., Ciarlet M., Desselberger U., Van Ranst M., Johne R.2012. VP6-sequence-based cutoff values as a criterion for rotavirus species demarcation. Arch. Virol. 157: 1177–1182. doi: 10.1007/s00705-012-1273-3 [DOI] [PubMed] [Google Scholar]
- 17.Matthijnssens J., Ciarlet M., Rahman M., Attoui H., Bányai K., Estes M. K., Gentsch J. R., Iturriza-Gómara M., Kirkwood C. D., Martella V., Mertens P. P., Nakagomi O., Patton J. T., Ruggeri F. M., Saif L. J., Santos N., Steyer A., Taniguchi K., Desselberger U., Van Ranst M.2008. Recommendations for the classification of group A rotaviruses using all 11 genomic RNA segments. Arch. Virol. 153: 1621–1629. doi: 10.1007/s00705-008-0155-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Medici M. C., Abelli L. A., Martinelli M., Corradi D., Dodi I., Tummolo F., Albonetti V., Martella V., Dettori G., Chezzi C.2011. Clinical and molecular observations of two fatal cases of rotavirus-associated enteritis in children in Italy. J. Clin. Microbiol. 49: 2733–2739. doi: 10.1128/JCM.01358-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Monini M., Zaccaria G., Ianiro G., Lavazza A., Vaccari G., Ruggeri F. M.2014. Full-length genomic analysis of porcine rotavirus strains isolated from pigs with diarrhea in Northern Italy. Infect. Genet. Evol. 25: 4–13. doi: 10.1016/j.meegid.2014.03.024 [DOI] [PubMed] [Google Scholar]
- 20.Okitsu S., Khamrin P., Thongprachum A., Maneekarn N., Mizuguchi M., Ushijima H.2011. Predominance of porcine P[23] genotype rotaviruses in piglets with diarrhea in northern Thailand. J. Clin. Microbiol. 49: 442–445. doi: 10.1128/JCM.02263-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Okitsu S., Khamrin P., Thongprachum A., Kongkaew A., Maneekarn N., Mizuguchi M., Hayakawa S., Ushijima H.2013. Whole-genomic analysis of G3P[23], G9P[23] and G3P[13] rotavirus strains isolated from piglets with diarrhea in Thailand, 2006−2008. Infect. Genet. Evol. 18: 74–86. doi: 10.1016/j.meegid.2013.05.005 [DOI] [PubMed] [Google Scholar]
- 22.Reed L. J., Muench H.1938. A simple method of estimating fifty percent endpoints. Am. J. Hyg. 27: 493–497. [Google Scholar]
- 23.Retief J. D.2000. Phylogenetic analysis using PHYLIP. Methods Mol. Biol. 132: 243–258. [DOI] [PubMed] [Google Scholar]
- 24.Saikruang W., Khamrin P., Chaimongkol N., Suantai B., Kongkaew A., Kongkaew S., Ushijima H., Maneekarn N.2013. Genetic diversity and novel combinations of G4P[19] and G9P[19] porcine rotavirus strains in Thailand. Vet. Microbiol. 161: 255–262. doi: 10.1016/j.vetmic.2012.07.036 [DOI] [PubMed] [Google Scholar]
- 25.Shao L., Fischer D. D., Kandasamy S., Rauf A., Langel S. N., Wentworth D. E., Stucker K. M., Halpin R. A., Lam H. C., Marthaler D., Saif L. J., Vlasova A. N.2015. Comparative in vitro and in vivo studies of porcine rotavirus G9P[13] and human rotavirus Wa G1P[8]. J. Virol. 90: 142–151. doi: 10.1128/JVI.02401-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shi H., Chen J., Li H., Sun D., Wang C., Feng L.2012. Molecular characterization of a rare G9P[23] porcine rotavirus isolate from China. Arch. Virol. 157: 1897–1903. doi: 10.1007/s00705-012-1363-2 [DOI] [PubMed] [Google Scholar]
- 27.Silva F. D., Espinoza L. R., Tonietti P. O., Barbosa B. R., Gregori F.2015. Whole-genomic analysis of 12 porcine group A rotaviruses isolated from symptomatic piglets in Brazil during the years of 2012−2013. Infect. Genet. Evol. 32: 239–254. doi: 10.1016/j.meegid.2015.03.016 [DOI] [PubMed] [Google Scholar]
- 28.Teodoroff T. A., Tsunemitsu H., Okamoto K., Katsuda K., Kohmoto M., Kawashima K., Nakagomi T., Nakagomi O.2005. Predominance of porcine rotavirus G9 in Japanese piglets with diarrhea: close relationship of their VP7 genes with those of recent human G9 strains. J. Clin. Microbiol. 43: 1377–1384. doi: 10.1128/JCM.43.3.1377-1384.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Theuns S., Heylen E., Zeller M., Roukaerts I. D., Desmarets L. M., Van Ranst M., Nauwynck H. J., Matthijnssens J.2015. Complete genome characterization of recent and ancient Belgian pig group A rotaviruses and assessment of their evolutionary relationship with human rotaviruses. J. Virol. 89: 1043–1057. doi: 10.1128/JVI.02513-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Theuns S., Vyt P., Desmarets L. M. B., Roukaerts I. D. M., Heylen E., Zeller M., Matthijnssens J., Nauwynck H. J.2016. Presence and characterization of pig group A and C rotaviruses in feces of Belgian diarrheic suckling piglets. Virus Res. 213: 172–183. doi: 10.1016/j.virusres.2015.12.004 [DOI] [PubMed] [Google Scholar]
- 31.Wakuda M., Ide T., Sasaki J., Komoto S., Ishii J., Sanekata T., Taniguchi K.2011. Porcine rotavirus closely related to novel group of human rotaviruses. Emerg. Infect. Dis. 17: 1491–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu F. T., Bányai K., Jiang B., Liu L. T., Marton S., Huang Y. C., Huang L. M., Liao M. H., Hsiung C. A.2017. Novel G9 rotavirus strains co-circulate in children and pigs, Taiwan. Sci. Rep. 7: 40731. doi: 10.1038/srep40731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yodmeeklin A., Khamrin P., Chuchaona W., Saikruang W., Kongkaew A., Vachirachewin R., Kumthip K., Okitsu S., Ushijima H., Maneekarn N.2016. Great genetic diversity of rotaviruses detected in piglets with diarrhea in Thailand. Arch. Virol. 161: 2843–2849. doi: 10.1007/s00705-016-2976-7 [DOI] [PubMed] [Google Scholar]
- 34.Zhang H., Zhang Z., Wang Y., Wang X., Xia M., Wu H.2015. Isolation, molecular characterization and evaluation of the pathogenicity of a porcine rotavirus isolated from Jiangsu Province, China. Arch. Virol. 160: 1333–1338. doi: 10.1007/s00705-015-2347-9 [DOI] [PubMed] [Google Scholar]
- 35.Zhang K., He Q.2010. Establishment and clinical application of a multiplex reverse transcription PCR for detection of porcine epidemic diarrhea virus, porcine transmissible gastroenteritis virus and porcine group A rotavirus. Acta Veterinaria et Zootechnica Sinica 41: 1001–1005 (In Chinese). [Google Scholar]