Abstract
In Haemophilus parasuis, the rfa cluster has been identified as a virulence-associated factor that is involved in lipooligosaccharide (LOS) biosynthesis. In this study, we assessed the roles of rfaD and rfaF genes in H. parasuis SC096 on LOS-induced pro-inflammatory factors and the related signaling pathways in porcine alveolar macrophages (PAMs) by real-time PCR and western blotting. The results showed that the LOSs of both rfaD and rfaF mutants (ΔrfaD-LOS and ΔrfaF-LOS) significantly decreased the mRNA expression of pro-inflammatory factors (IL-1α, IL-1β, IL-6, IL-8 and TNF-α) in PAMs compared with H. parasuis SC096 LOS (WT-LOS). Furthermore, in ΔrfaD-LOS- and ΔrfaF-LOS-treated cells, IκBα degradation was significantly inhibited and levels of phospho-p65 and phospho-p38 were significantly reduced in PAMs. These findings suggested that the rfaD and rfaF genes mediated LOS induction of pro-inflammatory cytokines in PAMs by regulating the NF-κB and MAPKs signaling pathways during H. parasuis infection.
Keywords: Haemophilus parasuis, pro-inflammatory factor, rfaD, rfaF, signaling pathway
Haemophilus parasuis is an important respiratory-tract pathogen of Glässer’s disease in swine, which is characterized by pleuritis, pericarditis, peritonitis, pneumonia, arthritis, and meningitis [6]. Recent studies have demonstrated that H. parasuis infections in hosts could stimulate inflammatory cytokines released through the regulation of nuclear factor-κB (NF-κB) and mitogen-activated protein kinase (MAPK) signaling pathways [1, 2].
Lipopolysaccharides (LOSs) are a major component of the outer membranes of gram-negative bacteria. LOSs can be identified by cell-surface molecules such as TLR4 and MD-2 when cells were stimulated by LOS, then by the activation of a variety of transcription factors through regulation of the NF-κB and MAPK signaling pathways and secretion of inflammatory cytokines [4]. Haemophilus parasuis LOS could induce pro-inflammatory responses and upregulated the expression of interleukin-1α (IL-1α), IL-1β, IL-6 and IL-8 in host cells [11]. Nevertheless, it is uncertain which LOS residues participate in the process of pro-inflammatory responses. As virulence-associated factors, both rfaD and rfaF genes are important parts of the rfa cluster involved in LOS biosynthesis of H. parasuis [8, 10]. The ΔrfaD mutant showed impaired abilty to adhere to and invade host cells [10]. Loss of the rfaF gene resulted in a severely truncated LOS structure and decreased abilities of serum resistance, adhesion, and invasion [8], which suggested that the full LOS structure influenced the ability of the bacteria to interact with the host cells. However, the roles of rfaD and rfaF genes in pro-inflammatory responses are still unknown on H. parasuis LOSs. In this study, we purified the LOS from ΔrfaD and ΔrfaF mutants to demonstrate the expression of pro-inflammatory factors and their related signaling pathways in porcine alveolar macrophages (PAMs).
H. parasuis SC096 and its rfaD and rfaF mutant strains were cultivated in liquid medium [8, 10]. LOSs were extracted using the hot-phenol method [5] and quantified using the anthrone-sulfuric acid method [11]. Compared to LOS from the H. parasuis SC096 strain (WT-LOS), the LOS from the rfaD mutant strain (ΔrfaD-LOS) migrated faster (Fig. S1) and exhibited the truncated LOS structure. PAMs (3D4/2 cell line from the American Type Culture Collection) were plated in 12-well microplates in RPMI 1640 medium (Invitrogen, Carlsbad, CA, U.S.A.) supplemented with 10% (V/V) heat-inactivated fetal bovine serum and cultured with 5% CO2 at 37°C for 24 hr, and stimulated with WT-LOS, ΔrfaD-LOS, and ΔrfaF-LOS at concentrations of 5 or 10 µg/ml. Cell pellets and supernatants were collected at 6, 12 and 24 hr after incubation. Total RNA was extracted with TRIzol (Invitrogen) and cDNA was synthesized with PrimeScriptTM RT reagent Kit (TaKaRa, Dalian, China). Real-time PCR was performed with the primer pairs (Table 1) using an Applied Biosystems 7300 Real-time PCR System (ABI, Foster City, CA, U.S.A.). The data were analyzed using the 2–∆∆CT method in triplicates for three independent experiments. A LPS preparation from Echerichia coli O111: B4 (Sigma Aldrich, St. Louis, MO, U.S.A.) was used as a positive control, and the unstimulated PAMs cells were used as a mock-stimulus. The ribosomal protein L4 (RPL4), stably expressed in PAMs, was used as a reference gene to normalize the results of gene expression detected by the real-time PCR assay [3].
Table 1. Sequences of the PCR primers used in this study.
| Primers | Nucleotide sequence (5′–3′) | PCR product length (bp) |
|---|---|---|
| IL-1β-F | ACCTGGACCTTGGTTCTCTG | 83 |
| IL-1β-R | CATCTGCCTGATGCTCTTGT | |
| IL-1α-F | GAAGAAGAGACGGTTGAG | 109 |
| IL-1α-R | GCTGTATGTTGCTGATCT | |
| IL-6-F | AATCCAGACAAAGCCACCAC | 79 |
| IL-6-R | TCCACTCGTTCTGTGACTGC | |
| IL-8-F | TAGGACCAGAGCCAGGAAGA | 92 |
| IL-8-R | AGCAGGAAAACTGCCAAGAA | |
| TNF-α-F | CCACCAACGTTTTCCTCACT | 82 |
| TNF-α-R | TTGATGGCAGAGAGGAGGTT | |
| RPL4-F | GCTCTATGGCACTTGGCGT | 124 |
| RPL4-R | GCGGAGGGCTCTTTGGAT |
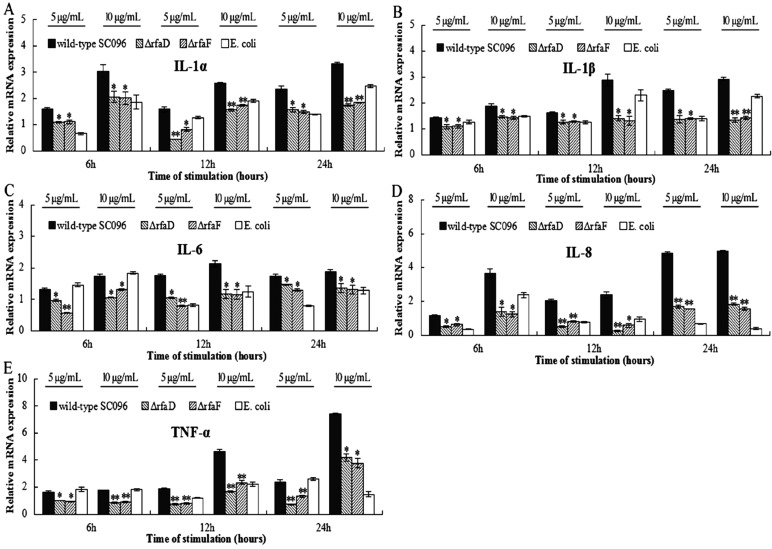
In Neisseria meningitidis, both the rfaD and rfaF mutant strains downregulated expression of pro-inflammatory factors in mouse serum [7, 9]. However, the roles of rfaD and rfaF in the pathogenesis of H. parasuis LOS were largely unknown. In this study, compared to the WT-LOS, LOSs from both mutants have significantly poorer abilities to induce mRNAs of inflammatory cytokine in treated PAMs at 6, 12 and 24 hr (P<0.05), including IL-1α, IL-1β, IL-6, IL-8 and TNF-α (Fig. 1). The results demonstrated that the loss of rfaD and rfaF genes in H. parasuis SC096 resulted in decreased LOS-mediated pro-inflammatory cytokine expressions in PAMs, which suggested that the truncated LOS structure might attenuate the inflammatory response during a H. parasuis infection.
Fig. 1.
The mRNA expression of pro-inflammatory cytokines in lipooligosaccharide (LOS)-stimulated porcine alveolar macrophages (PAMs). PAMs were stimulated with ΔrfaD-LOS, ΔrfaF-LOS and WT-LOS (5 and 10 µg/ml) for 6, 12 and 24 hr. The levels of IL-1α (A), IL-1β (B), IL-6 (C), IL-8 (D) and TNF-α (E) mRNAs were measured by qRT-PCR. The values presented are mean ± SD of three independent experiments, and data were analyzed using one-way ANOVA. *P<0.05; **P<0.001 compared to WT-LOS-treated PAMs.
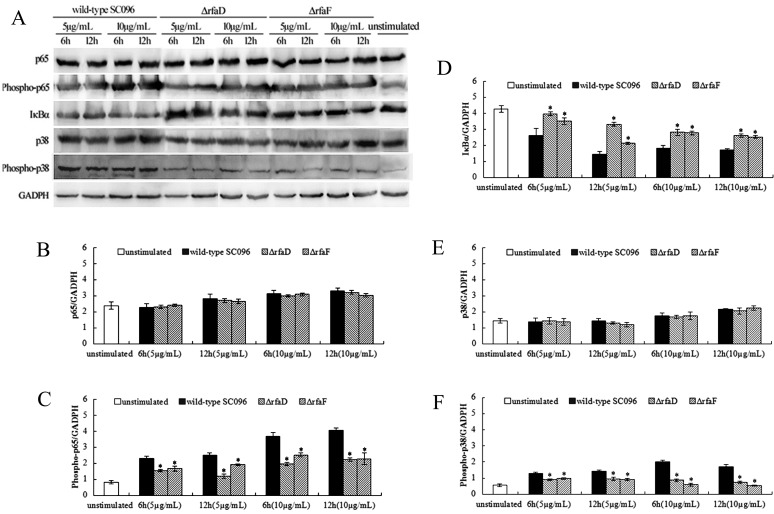
To further investigate the signaling mechanisms underlying the induction of the inflammatory response, we measured the expression of NF-κB and MAPK signaling molecules by western blotting, including NF-κB p65, phospho-NF-κB p65, IκBα, p38 and phospho-p38. PAMs were treated with WT-LOS, ΔrfaD-LOS, and ΔrfaF-LOS (5 and 10 µg/ml) for 6 and 12 hr. Western blot analysis was performed as described previously [2]. Anti-NF-κB p65, anti-phospho-NF-κB p65, anti-IκBα and anti-GADPH monoclonal antibodies as well as anti-p38 and anti-phospho-p38 polyclonal antibodies were obtained from Cell Signaling Technology (CST, Danvers, MA, U.S.A.). The HRP-conjugated goat anti-mouse or goat anti-rabbit IgG were obtained from Abbkine (Redlands, CA, U.S.A.). Densitometry values of immunoblot signals were obtained from three separate experiments using FusionCapt Advance software (Vilber Lourmat, Eberhardzell, Germany).
The phosphorylation of p65 and p38 were noticeably decreased in a dose-dependent manner in both ΔrfaD-LOS- and ΔrfaF-LOS-treated PAMs compared to the WT-LOS-treated group (Fig. 2). Both ΔrfaD-LOS and ΔrfaF-LOS resulted in a higher concentration of IκBα in a dose-dependent manner in stimulated PAMs compared to WT-LOS. Furthermore, an analysis of the densitometry values showed that the relative ratios of IκBα/GAPDH significantly increased (P<0.05), while the phosphorylation of p65/GAPDH and p38/GAPDH significantly decreased in both the ΔrfaD-LOS- and ΔrfaF-LOS-treated PAMs compared with the WT-LOS-treated group (P<0.05). Nevertheless, there was no obvious difference in the relative ratios of p65/GAPDH and p38/GAPDH. Therefore, the results suggested that the LOSs of H. parasuis participated in the activation of NF-κB and MAPK signaling pathways during infection. Both ΔrfaD-LOS and ΔrfaF-LOS resulted in a decrease in phosphorylation of p65 and p38 in the NF-κB and MAPK signaling pathways. Based on the above results, we postulated that the reduced ability to induce the inflammatory cytokines in ΔrfaD-LOS- and ΔrfaF-LOS-treated PAMs may be related to the reduction of p65 and p38 phosphorylation during NF-κB and MAPK signaling.
Fig. 2.
Both ΔrfaD-lipooligosaccharide (ΔrfaD-LOS) and ΔrfaF-LOS resulted in a decrease in phosphorylation of p65 and p38 during NF-κB and MAPK signaling in PAMs. PAMs were stimulated with ΔrfaD-LOS, ΔrfaF-LOS, and WT-LOS (5 and 10 µg/ml) for 6 and 12 hr. Western blot analyses of p65, phospho-p65, IκBα, p38 and phospho-p38 were performed (A). Western blot of the above-mentioned proteins and quantifications of p65 (B), phospho-p65 (C), IκBα (D), p38 (E) and phospho-p38 (F). GAPDH was used as a loading control. Bar graphs show the relative protein expression quantified from three separate experiments. The values presented are mean ± SD, and data were analyzed using one-way ANOVA. *P<0.05 compared to WT-LOS-treated PAMs.
In conclusion, both rfaD and rfaF mutants of the H. parasuis SC096 strain, with the truncated LOS structures, had a significant effect on LOS-induced pro-inflammatory factors in PAMs, such as IL-1α, IL-1β, IL-6, IL-8 and TNF-α. Also, LOSs of both mutants had decreased p65 and p38 phosphorylation. The above results indicated that both rfaD and rfaF genes mediated LOS induction of pro-inflammatory cytokines in PAMs by regulating the NF-κB and MAPKs signaling pathways during H. parasuis infection, which suggested that the full LOS structure had a significant role on the inflammatory response in H. parasuis. Overall, this study focused on the LOS-induced inflammatory mechanism that will provide a theoretical basis for the pathogenic mechanism of H. parasuis.
Supplementary
Acknowledgments
This work was supported by the National Natural Science Foundation of China (31772766), the Innovative Research Team Program of Department of Education of Sichuan Province (13TD0057), and the Fundamental Research Funds for the Central Universities of Southwest University for Nationalities (2018HQZZ15).
REFERENCES
- 1.Chen Y., Jin H., Chen P., Li Z., Meng X., Liu M., Li S., Shi D., Xiao Y., Wang X., Zhou Z., Bi D., Zhou R.2012. Haemophilus parasuis infection activates the NF-κB pathway in PK-15 cells through IκB degradation. Vet. Microbiol. 160: 259–263. doi: 10.1016/j.vetmic.2012.05.021 [DOI] [PubMed] [Google Scholar]
- 2.Chen Y., Liu T., Langford P., Hua K., Zhou S., Zhai Y., Xiao H., Luo R., Bi D., Jin H., Zhou R.2015. Haemophilus parasuis induces activation of NF-κB and MAP kinase signaling pathways mediated by toll-like receptors. Mol. Immunol. 65: 360–366. doi: 10.1016/j.molimm.2015.02.016 [DOI] [PubMed] [Google Scholar]
- 3.Cinar M. U., Islam M. A., Uddin M. J., Tholen E., Tesfaye D., Looft C., Schellander K.2012. Evaluation of suitable reference genes for gene expression studies in porcine alveolar macrophages in response to LPS and LTA. BMC Res. Notes 5: 107. doi: 10.1186/1756-0500-5-107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cohen J.2002. The immunopathogenesis of sepsis. Nature 420: 885–891. doi: 10.1038/nature01326 [DOI] [PubMed] [Google Scholar]
- 5.Hitchcock P. J., Brown T. M.1983. Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver-stained polyacrylamide gels. J. Bacteriol. 154: 269–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oliveira S., Pijoan C.2004. Haemophilus parasuis: new trends on diagnosis, epidemiology and control. Vet. Microbiol. 99: 1–12. doi: 10.1016/j.vetmic.2003.12.001 [DOI] [PubMed] [Google Scholar]
- 7.Plant L., Sundqvist J., Zughaier S., Lövkvist L., Stephens D. S., Jonsson A. B.2006. Lipooligosaccharide structure contributes to multiple steps in the virulence of Neisseria meningitidis. Infect. Immun. 74: 1360–1367. doi: 10.1128/IAI.74.2.1360-1367.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu C., Zhang L., Zhang B., Feng S., Zhou S., Li J., Zou Y., Liao M.2013. Involvement of lipooligosaccharide heptose residues of Haemophilus parasuis SC096 strain in serum resistance, adhesion and invasion. Vet. J. 195: 200–204. doi: 10.1016/j.tvjl.2012.06.017 [DOI] [PubMed] [Google Scholar]
- 9.Zarantonelli M. L., Huerre M., Taha M. K., Alonso J. M.2006. Differential role of lipooligosaccharide of Neisseria meningitidis in virulence and inflammatory response during respiratory infection in mice. Infect. Immun. 74: 5506–5512. doi: 10.1128/IAI.00655-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang B., Xu C., Zhang L., Zhou S., Feng S., He Y., Liao M.2013. Enhanced adherence to and invasion of PUVEC and PK-15 cells due to the overexpression of RfaD, ThyA and Mip in the ΔompP2 mutant of Haemophilus parasuis SC096 strain. Vet. Microbiol. 162: 713–723. doi: 10.1016/j.vetmic.2012.09.021 [DOI] [PubMed] [Google Scholar]
- 11.Zhou S., He X., Xu C., Zhang B., Feng S., Zou Y., Li J., Liao M.2014. The outer membrane protein P2 (OmpP2) of Haemophilus parasuis induces proinflammatory cytokine mRNA expression in porcine alveolar macrophages. Vet. J. 199: 461–464. doi: 10.1016/j.tvjl.2013.12.010 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.