Abstract
Background
To determine the feasibility of radiomic analysis for predicting the therapeutic response of gastric carcinoma (GC) with abdominal cavity metastasis (GCACM) to pulsed low dose rate radiotherapy (PLDRT) using contrast-enhanced computed tomography (CECT) images.
Methods
Pretreatment CECT images of 43 GCACM patients were analyzed. Patients with complete response (CR) and partial response (PR) were considered responders, while stable disease (SD) and progressive disease (PD) as non-responders. A total of 1,117 image features were quantified from tumor region that segmented from arterial phase CT images. Intra-class correlation coefficient (ICC) and absolute correlation coefficient (ACC) were calculated for selecting influential feature subset. The capability of each influential feature on treatment response classification was assessed using Kruskal-Wallis test and receiver operating characteristic (ROC) analysis. Moreover, artificial neural network (ANN) and k-nearest neighbor (KNN) predictive models were constructed based on the training set (18 responders, 14 non-responders) and the testing set (6 responders, 5 non-responders) validated the reliability of the models. Comparison between the performances of the models was performed by using McNemar’s test.
Results
The analyses showed that 6 features (1 first order-based, 1 texture-based, 1 LoG-based, and 3 wavelet-based) were significantly different between responders and non-responders (AUCs range from 0.686 to 0.728). Both two prediction models based on features extracted from CECT showed potential in predicting the treatment response with higher accuracies (ANN: 0.714, KNN: 0.749 for the training set; ANN: 0.816, KNN: 0.816 for the testing set). No statistical difference was observed between the performance of ANN and KNN (P=0.999).
Conclusions
Pretreatment radiomic analysis using CECT can potentially provide important information regarding the therapeutic response to PLDRT for GCACM, improving risk stratification.
Keywords: Gastric carcinoma (GC), abdominal cavity metastasis, pulsed low dose rate radiotherapy (PLDRT), radiomic analysis, predictor, treatment response
Introduction
Gastric carcinoma (GC) is the fifth most commonly occurring types of malignancy, including more than one million new cancer diagnoses yearly, and also the third leading cause of cancer-related mortality with approximately 723,000 deaths every year (1). Patients with advanced GC always develop abdominal cavity metastasis (GCACM) with symptoms of pain, bleeding, and intestinal obstruction, severely affecting the quality of life. Generally, these patients are intolerant of surgery or further aggressive treatment. Radiotherapy is an option for such patients. However, the peritoneal radiation dose was restricted due to the low tolerable dose of the organ at risk near the target, for which patients with GCACM are not suitable for conventional radiotherapy.
Pulsed low-dose-rate radiotherapy (PLDRT) has been proved as an effective treatment strategy across a range of cancer type (2-8) by taking advantage of low-dose hyper-radiosensitivity (LDHRS) (7,9), with maximizing tumor control and without producing severe adverse normal tissue complication. Nevertheless, the response to PLDRT is highly individual, with many patients being insensitive to standard treatment regimens. On the other hand, patients who are unresponsive to PLDRT may be harmed by unnecessary radiation toxicity, resulting in a delay of the modification of treatment plan and, consequently, it implies a high risk of tumor progression and poor prognosis. Therefore, early identification of patients who are at higher risk of poor response before PLDRT would lead to the optimization of the therapeutic management and improvement of outcomes.
Recently, investigators have explored and validated a series of clinical biomarkers with the potential to be used in prediction of therapeutic response (10-14). For example, Nagashima et al. showed that biomarkers of immunohistochemical examination could be useful in predicting the clinical outcomes of unresectable GC patients with chemotherapy (10). De Cobelli et al. implied that apparent diffusion coefficient (ADC) of diffusion-weighted MRI (DW-MRI) can be used to assess tumor response to neoadjuvant chemotherapy for patients with gastro-oesophageal (12) and GC (13). More recently, a review suggested that molecular and proteomics analyses showed promising response prediction in gastric cancer (14). Although these clinical indicators were demonstrated promising results over the past years, little attention has been paid to the predictive capability of CT, which is a common pretreatment examination for patients with GC. As such, new tools based on the pretreatment CT images are expected of the prediction of tumor response, especially for the patients with GCACM before PLDRT.
Radiomics is a non-invasive approach that extracts quantitative features from medical images and allows for comprehensive visualization and characterization of the tumor region and corresponding microenvironment, which has been found to show significant predictive power for gene expression, pathological classification, treatment response, and clinical outcome (15-19). This novel method focused on the improvement of image analysis by converting imaging data into a high-dimensional mineable feature set using a series of the data-characterization algorithm. Recent studies on gastric cancer have shown that radiomic features based on CT images are associated with pathological types, response to neoadjuvant therapy, and survival (20-24). To our knowledge, no published research has determined whether the treatment response to PLDRT in patients with GCACM could be early predicted by radiomic analysis using pretreatment contrast-enhanced CT (CECT). Thus, the purpose of this study is to use radiomic features derived from baseline CECT combined with supervised machine learning algorithms to predict treatment response to PLDRT in GCACM patients.
Methods
Patient database
Forty-three patients who were treated with PLDRT for GCACM at Nanjing Drum Tower Hospital Cancer Centre from February 2014 to November 2017 were enrolled in this study under institutional ethics committee approval. Of the 43 patients, 15 were in stage III, and 28 were in stage IV. Patients in stage III underwent curative D2 gastrectomy after a multidisciplinary meeting and received 5-fluorouracil and oxaliplatin based chemotherapy for least four cycles after pathological diagnosis. Patients in stage IV received 5-fluorouracil-based palliative chemotherapy after diagnosis. The other inclusion criteria were met: (I) developed with metastases/peritoneal carcinosis after surgery or chemotherapy; (II) pre-PLDRT CECT was available; (III) with poor performance status unsuitable for further aggressive treatment (reoperation or conventional radiotherapy) after evaluation by the multi-disciplinary team; (IV) without received radiotherapy before PLDRT. Finally, all 43 patients completed PLDRT and enrolled in this study. Table 1 summarizes baseline patient characteristics.
Table 1. Baseline characteristics of patients.
| Characteristic | Value (n=43) |
|---|---|
| Gender, n [%] | |
| Male | 30 [70] |
| Female | 13 [30] |
| Age, median [range] (years) | 63 [28–79] |
| AJCC staging, n [%] | |
| III | 15 [35] |
| IV | 28 [65] |
| Irradiation site, n [%] | |
| Metastasis lesions | 6 [14] |
| Retroperitoneal and/or abdominal lymph nodes | 29 [68] |
| Hilar lymph nodes | 7 [16] |
| Stomach and abdominal lymph nodes | 1 [2] |
| PLDRT total dose, median [range] (Gy) | 50 [50–54] |
PLDRT
All patients were irradiated with a series of 0.4 Gy pulses separated by 5-min interval for 3-dimensional conformal radiotherapy, it created a time-averaged dose-rate of 0.0667 Gy/min. The dose prescriptions were designed to cover at least 96% of planning target volume (PTV).
Response assessment
One month after treatment, therapeutic responses were assessed by dedicated radiologist and radiation oncologist (Y Yang), according to the Response Evaluation Criteria in Solid Tumors (RECIST v1.1) (25). The follow-up was updated every two months. Patients were subsequently grouped according to RECIST using pre/post-PLDRT CT images with contrast or PET scans if applicable; specifically, patients with complete response (CR) or partial response (PR) were considered as responders, while those with stable disease (SD) or progressive disease (PD) were classified as non-responders.
CECT image acquisition and tumor segmentation
CECT examinations were performed 3–5 days before radiation treatment. All patients were scanned using 3 mm CT slice thickness, (Philips Brilliance 64; Philips Healthcare, Best, the Netherlands), with a resolution of approximately 1mm in the axial plane, according to the following acquisition parameters: tube voltage, 120 kVp; tube current, 200–250 mAs; rotation time, 0.75 s; pitch, 0.9; matrix, 512×512; convolution kernel, standard. Following a non-contrast scan, 100–120 mL iodinated contrast agent (Omnipaque, GE Healthcare, Shanghai, China) was injected intravenously at a rate of 3.0 mL/s with a pump injector (Medrad Stellant CT Injector System; Medrad, Indianola, PA, USA), yielding contrast-enhanced CT (CECT) (CECT was performed 30 s (arterial phase), 70 s (portal phase) after an infusion of contrast material). To obtain volume of interest (VOIs) for further radiomic analysis, a semi-automatically segmentation was performed by two senior board-certified radiation oncologists (Y Yang for VOI-1 and S Li for VOI-2) from arterial phase CT images using open-source available software (3D Slicer software, version 4.9.0, http://www.slicer.org) and then reviewed by an experienced radiation oncologist (J Yan). The contours for each VOI were drawn slightly within the border of the tumor lesion. The contours were consistent with gross tumor volume (GTV) delineated by radiation oncologists for radiotherapy treatment planning design and were manually modified to avoid adjacent air, fat, blood vessels and surrounding organs. Voxels related to air and adipose tissues [below 0 Hounsfield units (HU)] were automatically excluded from the analysis. All contoured volumes with a voxel size of 1×1×3 mm3 were resampled to an isotropic voxel size of 1×1×1 mm3 using cubic interpolation to unify the voxel size across the cohort.
Radiomic feature extraction
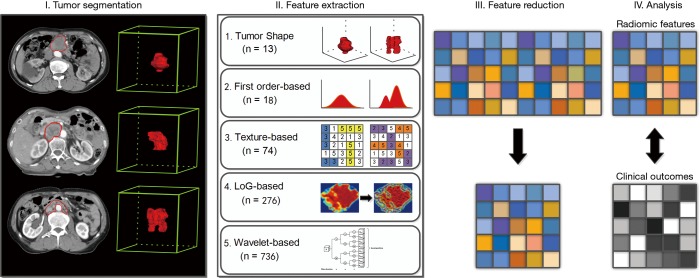
Pyradiomics V1.3.0 (26) was used to extract radiomic features from delineated three-dimensional (3D) VOIs (Figure 1). Pyradiomics is an open-source python package for the procession and extraction of radiomic features from medical image data using a large panel of automatically extracted data-characterization algorithms. Using this package, several categories features were extracted from original images, including shape and size (morphological feature), intensity histogram (IH, first order feature), gray-level co-occurrence matrix [GLCM; directions: 13 angles in 3D (26-connectivity); distance: 1 pixel], gray-level size-zone matrix (GLSZM; directions: 13 angles in 3D), gray-level run-length matrix (GLRLM, directions: 13 angles in 3D), neighboring gray-tone difference matrix (NGTDM; neighborhood size: 3×3×3) and gray-level dependence matrix (GLDM, distance: 1 pixel). As for GLCM, GLSZM, and GLRLM, the value of a feature was calculated on each angle separately, after which the mean of these values was obtained. Additionally, the aforementioned texture features (GLCM, GLRLM, NGTDM, and GLDM-based) were also extracted from the images preprocessed using Laplacian of Gaussian (LoG) band pass filter and wavelet filter. In particular, LoG band pass filter was applied to the input original image for fine to coarse texture (filter width: fine, σ=0.5; medium, σ=1.5; coarse, σ=2.5) (24) and wavelet filter was applied for focusing features on different frequency ranges within the tumor volume respectively.
Figure 1.
Workflow of radiomic analysis for gastric carcinoma with abdominal cavity metastasis.
Overall, as for each lesion, the radiomic features were extracted from both filtered and unfiltered image, using three principal methods: shape-based (shape and size), first-order based (IH), texture-based (GLCM, GLSZM, GLRLM, GLDM), LoG-based (LoGσ =0.5/1.5/2.5_GLCM, LoGσ =0.5/1.5/2.5_GLSZM, LoGσ =0.5/1.5/2.5_GLRLM, LoGσ =0.5/1.5/2.5_GLDM), and wavelet_based (Waveletlevel_GLCM, Waveletlevel_GLSZM, Waveletlevel_GLRLM, Waveletlevel_GLDM). A detailed list of the features is provided in Table S1.
Statistical analysis
Statistical analyses were performed using R software version 3.3.2 (R Foundation for Statistical Computing, Vienna, Austria).
Inter-observer variability of radiomic features extraction was evaluated by calculating the intra-class correlation coefficient (ICC) using “irr” package (vers. 0.84) in R software (27). It was performed to assess feature reproducibility in repeated delineation (ICC <0.40, poor; 0.40≤ ICC <0.60, moderate; 0.60≤ ICC <0.80, good; ICC ≥0.80, excellent). Radiomic features with ICC value greater than 0.8 were selected.
Furthermore, pair-wise correlations among the above-selected features (ICC ≥0.80) were also considered. For these highly reproducible features, we constructed a correlation matrix and then calculated the absolute correlation coefficient (ACC) using “caret” package (ver.6.0-77). The ACC values close to 1 represented the features are strongly correlated. In our study, for example, if two variables have a high correlation (ACC ≥0.8), the function looks at the mean absolute correlation of each variable (with the remaining features) and removes the variable with the largest mean absolute correlation.
The capability of each influential feature (ICC ≥0.8 and ACC <0.8) to classify patients concerning therapeutic response were investigated by using the Kruskal-Wallis test (P<0.05) (28). Receiver operating characteristic (ROC) curves analysis was used to assess the diagnostic efficacy of each studied features for discrimination different treatment responses by measuring associated areas under the ROC curves (AUC). 95% confidence intervals (CIs), specificity and sensitivity, were also obtained.
Feature selection and radiomic model construction
Influential features (ICC ≥0.8 and ACC <0.8) extracted from CECT were subsequently modeled by supervised machine-learning algorithms using Weka software (University of Waikato, Hamilton, New Zealand): ANN (the number of hidden layers was 1) and KNN [5 neighbors (k=5) were chosen in this study]. To build and validate the predictive models, the patients were randomized into two groups: 32 for training (18 responders, 14 non-responders) and 11 for testing (6 responders, 5 non-responders).
To avoid model over-fitting, the feature vector dimensionality should be reduced firstly. Based on the training group (18 responders, 14 non-responders), wrapper-based feature selection method (29) was used to select optimal feature subset for the specific model (ANN or KNN). This method performed on the previously calculated feature subset (features with ICC ≥0.8 and ACC <0.8) by recursively removing features and then testing the predictive ability of the remaining features without missing any critical ones. After that, feature vector dimensionality was further reduced.
Once the optimal feature subsets had been obtained, ANN and KNN were trained and models generated for prediction. To assess predictive performance, 10-fold cross-validation (CV) was served as the internal validation in the training group. The associated metrics including specificity, precision, and accuracy of the predictions were calculated from true negatives (TN), false negatives (FN), true positives (TP), and false positives (FP). Additionally, Matthews correlation coefficient (MCC) was also calculated for providing additional reassurance on the model’s reliability.
Statistical comparison between ANN and KNN models
Significant differences between the performance of different models were evaluated with McNemar’s test (30). This test was conducted based on the results obtained from the 10-fold CV.
Predictive validation of radiomic models
We applied the established models to the validation group (6 responders, 5 non-responders), and the performance was assessed with specificity, precision, and accuracy.
Results
Treatment response after PLDRT
The treatment response was assessed one month after the PLDRT treatment. Two patients with no evidence of disease after treatments were considered as CRs. PLDRT treatment led to PR in 22 patients, whereas 17 and 2 were SD and PD respectively under treatment according to RECIST.
Overall, 24 patients were classified as responders (2 CR, 22 PR), while 19 patients were classified as non-responders (17 SD, 2 PD).
Predictive capabilities
A total of 1,117 radiomic features, preprocessed with or without LoGσ =0.5/1.5/2.5 filter and wavelet filter, were calculated on each of the 43 cases: 13 shape features, 18 first order features, 74 texture features, 276 LoG features, and 736 wavelet features. ICC and ACC were performed on the feature metrics and yielded 41 influential features. More details on the influential features are summarized in Table 2.
Table 2. The features obtained from preprocessing.
| Feature types | Features |
|---|---|
| Shape-based | None |
| First order-based | Skewness |
| Texture-based | gldm_DNUN, gldm_DV, glcm_Imc1, glcm_CS |
| LoG-based | LoG0.5_glcm_MP, LoG0.5_F_Skewness, LoG0.5_glszm_ SZNUN, LoG0.5_ glszm_ SALGLE, LoG1.5_glcm_Imc1, LoG2.5_glcm_Imc1, LoG2.5_ glszm_ LALGLE, LoG2.5_ glszm_ SALGLE |
| Wavelet-based | WHHH_F_Skewness, WHHH_F_Median, WHHH_F_Mean, WHHH_glszm_SAE, WHHH_ngtdm_Strength, WHLL_glcm_CS, WHLL_F_Skewness, WHLL_F_Median, WHLL_F_Kurtosis, WHLL_glszm_ SZNUN, WLHL_F_Skewness, WLHL_glszm_ SZNUN, WLHH_F_Mean, WLHH_glszm_SAE, WLHH_ngtdm_Strength, WLLH_F_Skewness, WLLH_F_Mean, WLLH_glszm_ SZNUN, WLLH_glszm_GLNU, WLLL_glcm_CS, WLLL_glcm_IV, WLLL_F_Mean, WLLL_F_Skewness, WHHL_F_Skewness, WHHL_glszm_ SZNUN, WHLH_F_Mean, WHLH_glcm_IDM, WHLH_glszm_SAE |
gldm, gray-level dependence matrix; DNUN, dependence non-uniformity normalized; DV, dependence variance; glcm, gray-level co-occurrence matrix; Imc1, informal measure of correlation 1; CS, cluster shade; LoG0.5/1.5/2.5, volume preprocessed using Laplacian of Gaussian band pass filter with 0.5/1.5/2.5 filter width; MP, maximum probability; F, first order; glszm, gray-level size-zone matrix; SZNUN, size zone non-uniformity normalized; SALGLE, small area low gray-level emphasis; LALGLE, large area low gray-level emphasis; WHLL, volume with a wavelet high-pass filter along x-direction, a low-pass filter along y-direction and a low-pass filter along z-direction; SAE, small area emphasis; ngtdm, neighboring gray-tone difference matrix; GLNU, gray-level non-uniformity; IV, inverse variance; IDM, inverse difference moment.
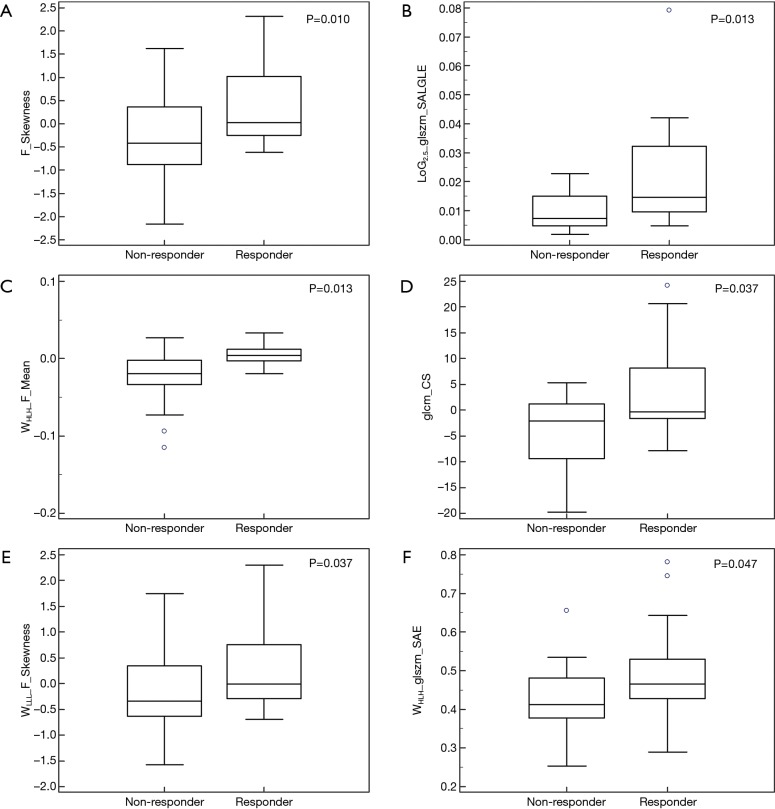
The result of Kruskal-Wallis test showed that 6 features (1 first-order feature, 1 texture feature, 1 LoG feature, 3 wavelet features; P value: 0.010–0.047) could differentiate between responders (CRs and PRs) and non-responders (SDs and PDs). Details of the result are summarized in Table 3 and the distribution of these statistically significant features was shown in Figure 2. As for differentiation between responders and non-responders, we analyzed the significant features with ROC curves and the corresponding AUC values were obtained (range from 0.686 to 0.728). For example, F_Skewness, with an AUC of 0.728 (sensitive =0.684, specificity =0.792), was capable of differentiating responders from non-responders, using a threshold of −0.271 (Table 3), indicating that tumor lesions whose F_Skewness was lower than −0.271 were most likely from non-responders. Similar results were obtained from the ROC analysis of other significant features. Figure 3 shows the ROC curves for the significant features.
Table 3. Features showing statistical difference between non-responders and responders.
| Feature | P value | Standard error | 95% CI | AUC | Sens | Spec | Cut-off |
|---|---|---|---|---|---|---|---|
| F_Skewness | 0.010 | 0.083 | 0.571–0.852 | 0.728 | 0.684 | 0.792 | ≤−0.271 |
| LoG2.5_glszm_SALGLE | 0.013 | 0.081 | 0.564–0.847 | 0.721 | 0.421 | 0.958 | ≤4.99×10−3 |
| WHLH_F_Mean | 0.013 | 0.081 | 0.564–0.847 | 0.721 | 0.684 | 0.791 | −3.42×10−3 |
| glcm_CS | 0.037 | 0.086 | 0.527–0.819 | 0.686 | 0.421 | 0.958 | ≤−4.547 |
| WLLL_F_Skewness | 0.037 | 0.087 | 0.527–0.819 | 0.686 | 0.736 | 0.667 | ≤−0.087 |
| WHLH_glszm_SAE | 0.047 | 0.084 | 0.518–0.812 | 0.678 | 0.526 | 0.833 | ≤0.401 |
F, first order; LoG2.5, volume preprocessed using Laplacian of Gaussian band pass filter with 2.5 filter width; GLSZM, gray-level size-zone matrix; SALGLE, small area low gray level emphasis; WHLH, volume with a wavelet high-pass filter along x-direction, a low-pass filter along y-direction and a high-pass filter along z-direction; GLCM, gray-level co-occurrence matrix; CI, confidence interval; AUC, area under the curve; Sens, Sensitivity; Spec, Specificity; Responders, patients with CR and PR; non-responders, patients with SD.
Figure 2.
Box plots of the amplitude features, successfully differentiating non-responders [stable disease (SD), progressive disease (PD)] from responders [complete response (CR), partial response (PR)]. (A) F_Skewness (P=0.010); (B) LoG2.5_glszm_SALGLE (P=0.013); (C) WHLH_F_Mean (P=0.013); (D) glcm_CS (P=0.037); (E) WLLL_F_Skewness (P=0.037); (F) WHLH_glszm_SAE (P=0.047).
Figure 3.
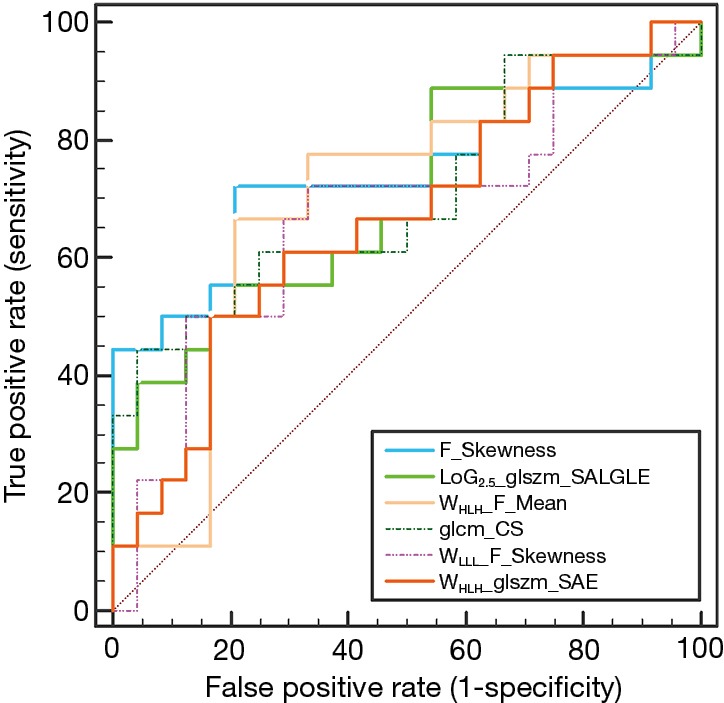
Receiver operating characteristic (ROC) curves on the basis of the significant features.
Supervised classification and statistical comparison
Wrapper-based feature selection method was performed on the influential feature set (features with ICC ≥0.8 and ACC <0.8, Table 2) before ANN and KNN modeling for generating different optimal feature sets for each model, with the results showing that 7 features were selected for ANN and 4 features were selected for KNN. Specifically, 3 features (LoG2.5_glszm_LALGLE, WLHL_glszm_SZNUN, WHHL_F_SKewness) were both selected for these two models. Table 4 displays details of the optimal feature sets for each model.
Table 4. Optimal feature set obtained from Wrapper-based feature selection.
| Feature type | ANN | KNN |
|---|---|---|
| Shape-based | None | None |
| First order-based | None | None |
| Texture-based | None | glcm_CS |
| LoG-based | LoG2.5_glszm_LALGLE | LoG2.5_glszm_LALGLE |
| LoG0.5_glcm_MP | ||
| Wavelet-based | WLHL_glszm_ SZNUN | WLHL_glszm_SZNUN |
| WHHL_F_SKewness | WHHL_F_SKewness | |
| WHLL_F_Median | ||
| WHLL_glcm_CS | ||
| WLLL_glcm_CS |
glcm, gray-level co-occurrence matrix; CS, Cluster Shade; LoG2.5, volume preprocessed using Laplacian of Gaussian band pass filter with 2.5 filter width; glszm, gray-level size-zone matrix; LALGLE, large area low gray level emphasis; MP, Maximum Probability; SZNUN, size zone non-uniformity normalized; F, first order; WLHL, volume with a wavelet low-pass filter along x-direction, a high-pass filter along y-direction and a low-pass filter along z-direction; ANN, artificial neural network; KNN, k-nearest neighbor.
The classification results based on training group from repeating 10-fold CV is presented in Table 5, including parameters of specificity, precision, weighted accuracy, and MCC for the two models. Both ANN and KNN models achieved higher prediction, with the accuracies of 0.714 and 0.749 respectively.
Table 5. Summary of classification results obtained from training set using 10-fold CV.
| Algorithm | TP rate | FP rate | Precision | F-measure | MCC | Accuracy |
|---|---|---|---|---|---|---|
| ANN | 0.719 | 0.298 | 0.717 | 0.717 | 0.425 | 0.714 |
| KNN | 0.750 | 0.242 | 0.758 | 0.751 | 0.504 | 0.749 |
ANN, artificial neural network; KNN, k-nearest neighbor; FP, false-positive; TP, true-positive; MCC, Matthews correlation coefficient.
The results from pairwise comparisons in McNemar’s test revealed no statistically significant difference between the performance of ANN and KNN models, indicating that the choice of the models was not of substantial importance (P=0.999).
Validation result
Table 6 summarizes the detail results of model testing based on the testing group. The two predictive models achieved same accuracies (ANN: 0.816, KNN: 0.816).
Table 6. Classification results obtained from testing set.
| Algorithm | TP rate | FP rate | Precision | F-measure | MCC | Accuracy |
|---|---|---|---|---|---|---|
| KNN | 0.818 | 0.185 | 0.818 | 0.818 | 0.663 | 0.816 |
| ANN | 0.818 | 0.185 | 0.818 | 0.818 | 0.663 | 0.816 |
ANN, artificial neural network; KNN, k-nearest neighbor; FP, false-positive; TP, true-positive; MCC, Matthews correlation coefficient.
Discussion
Prior works have documented the value of radiomic analysis in GC. Liu et al. (20), for example, correlated CT texture features with differentiation degree, Lau classification and vascular invasion status of GC. An Austria group reported that CT textural information might aid radiologists in the classification of gastric adenocarcinoma, lymphoma, and gastrointestinal stromal tumors (21). Similar findings were observed by Ma et al. that found radiomic analysis has the potential to accurately differentiate Borrmann type IV gastric cancer from primary gastric lymphoma (22). Based on pretreatment CECT, Giganti et al. suggested that radiomic analysis can provide biomarkers regarding the response rate to neoadjuvant therapy for GC (23). This same group subsequently demonstrated a correlation between CT-based radiomic features and overall survival in patients with GC (24). To date, there have been few studies to investigate the application of radiomic analysis for predicting therapeutic response in gastric cancer. Promising results have been reported for GC treated with neoadjuvant therapy (23). However, studies have yet to investigate the application of radiomic analysis to predict the response of patients with GCACM to PLDRT.
In present work, we analyzed 1,117 radiomic features quantifying tumor phenotypic differences and found that six parameters were able to discriminate responders from non-responders before PLDRT initiation with AUC values range from 0.686 to 0.728. This suggested that there is a significant difference in the tumor tissue between patients who responded to PLDRT versus those who do not, and this underlying difference can be characterized by using of radiomic features calculated from pretreatment CECT. The radiomic features were mathematical measurements concerned with the quantitative description of pixel arrangement within the tumor region (31), and spatial distribution of pixel within homogeneous tumors appeared more regular than those heterogeneous ones (32). In other words, radiomic features evaluated in this study highlight tumor heterogeneity at a regional and local level, depending on types of feature matrix, may be associated with hypoxia, proliferation, cellularity, vascularization, and necrosis (33). Therefore, radiomic features could be correlated to physiologic process and consequently response to PLDRT could be predicted, as it is closely related to intratumoral heterogeneity. Our results provide compelling evidence for using imaging biomarkers as potential predictors of tumor response to PLDRT for GCACM patients.
Moreover, we further incorporated ANN and KNN machine-learning algorithm into radiomic features to build predictive models. For modeling, in present work, ANN and KNN were constructed on the training group and subsequently tested on the validation group. Additionally, to obtain more robust prediction model and minimize the risk of modeling over-fitting, we followed a series of preprocessing procedures: assessment for feature reproducibility and correlation, wrapper-based method for feature selection as well as 10-fold CV approach for internal validation. After that, both two models achieved higher predictive accuracies in the training set (ANN, 0.714; KNN, 0.749) and testing set (ANN, 0.816; KNN, 0.816). These observations might be attributed to the ability of radiomic analysis to indirectly capture the information of intratumoral heterogeneity, including parameters not easily visible by a simple visual inspection, which they might be associated with tumor response to PLDRT. These findings extend those of Giganti et al. (23), confirming that radiomic analysis is not only used to predict preoperative response to neoadjuvant therapy in patients with gastric cancer but also to predict the response to treatment in GCACM patients treated with PLDRT. This study therefore indicates that the benefits gained from multivariate radiomic analysis could improve patient risk stratification. Most notably, this is the first study to our knowledge to explore the potential of radiomic analysis for the assessment of phenotypic properties of GCACM tumors which could be related to different treatment response, and the first study that explores the potential of joint supervised machine-learning algorithms and CECT radiomic features for the prediction of treatment response in GCACM patients treated with PLDRT. Our study provides a good enlightenment to the coming studies to prospectively establish patient cohorts.
However, several limitations are worth noting. First, due to relatively small sample cohort of patients in a single center and the retrospective nature of this study, our radiomic models remain to be externally validated in multiple centers with a larger and prospective patient cohort in the future. Second, although the predictive power of radiomic analysis has been demonstrated in our study, the radiomic-biology correlations have not yet to be identified in published literature and clinical experience. Therefore, future studies are necessary to explore the potential mechanism further. Third, due to a comprehensive consideration of clinical factors and economic conditions of patients, not all patients had performed PET scans before and after treatment. In this case, we are still rely on CECT for response evaluation, which could potentially affect assessment result. Finally, in this work, we only assessed early responses (e.g., one month after treatment) without assessment for long-term effectiveness. Further studies are on-going to follow up with the patients to ensure the accuracy of response evaluation.
In conclusion, radiomic features derived from CECT in combination with supervised machine learning algorithm could serve as a clinical tool to facilitate early prediction of GCACM patients’ response to PLDRT, with the advantage of low cost, using existing CECT images, without subjecting patients to unnecessary radiation exposure or imaging.
Table S1. Feature type and associated features.
| Feature type | Methods | Feature name |
|---|---|---|
| Morphology -based | Shape | Maximum 3D diameter (M3D) |
| Maximum 2D diameter slice (M2DS) | ||
| Sphericity | ||
| Minor axis (MA) | ||
| Elongation | ||
| Surface volume ratio (SVR) | ||
| Volume | ||
| Major axis (MA1) | ||
| Surface area (SA) | ||
| Flatness | ||
| Least axis (LA) | ||
| Maximum 2D diameter column (M2DC) | ||
| Maximum 2D diameter row (M2DR) | ||
| First order-based | Histogram | Interquartile range (IQR) |
| Skewness | ||
| Uniformity | ||
| Median | ||
| Energy | ||
| Robust mean absolute deviation (RMAD) | ||
| Mean absolute deviation (MAD) | ||
| Total energy (TE) | ||
| Maximum | ||
| Root mean squared (RMS) | ||
| 90 percentile | ||
| Minimum | ||
| Entropy | ||
| Range | ||
| Variance | ||
| 10 percentile | ||
| Kurtosis | ||
| Mean | ||
| Texture-based | GLCM | Joint average (JA) |
| Sum average (SA) | ||
| Joint entropy (JE) | ||
| Cluster shade (CS) | ||
| Maximum probability (MP) | ||
| Idmn | ||
| Joint energy (JE) | ||
| Contrast | ||
| Difference entropy (DE) | ||
| Inverse variance (IV) | ||
| Difference variance (DV) | ||
| Idn | ||
| Idm | ||
| Correlation | ||
| Autocorrelation | ||
| Sum entropy (SE) | ||
| Sum squares (SS) | ||
| Cluster prominence (CP) | ||
| Imc2 | ||
| Imc1 | ||
| Difference average (DA) | ||
| Id | ||
| Cluster tendency (CT) | ||
| GLSZM | Gray level variance (GLV) | |
| Zone variance (ZV) | ||
| Gray level non uniformity normalized (GLNUN) | ||
| Size zone non uniformity normalized (SZNUN) | ||
| Size zone non uniformity (SZNU) | ||
| Gray level non uniformity (GLNU) | ||
| Large area emphasis (LAE) | ||
| Small Area high gray level emphasis (SAHGLE) | ||
| Zone percentage (ZP) | ||
| Large area low gray level emphasis (LALGLE) | ||
| Large area high gray level emphasis (LAHGLE) | ||
| High gray level zone emphasis (HGLZE) | ||
| Small area emphasis (SAE) | ||
| Low gray level zone emphasis (LGLZE) | ||
| Zone entropy (ZE) | ||
| Small area low gray level emphasis (SALGLE) | ||
| GLRLM | Short run low gray level emphasis (SRLGLE) | |
| Gray level variance (GLV) | ||
| Low gray level run emphasis (LGLRE) | ||
| Gray level non uniformity normalized (GLNUN) | ||
| Run variance (RV) | ||
| Gray level non uniformity (GLNU) | ||
| Long run emphasis (LRE) | ||
| Short Run high gray level emphasis (SRHGLE) | ||
| Run length non uniformity (RLNU) | ||
| Short run emphasis (SRE) | ||
| Long run high gray level emphasis (LRHGLE) | ||
| Run percentage (RP) | ||
| Long run low gray level emphasis (LRLGLE) | ||
| Run entropy (RE) | ||
| High gray level run emphasis (HGLRE) | ||
| Run length non uniformity normalized (RLNUN) | ||
| NGTDM | Coarseness | |
| Complexity | ||
| Strength | ||
| Contrast | ||
| Busyness | ||
| GLDM | Gray level variance (GLV) | |
| High gray level emphasis (HGLE) | ||
| Dependence entropy (DE) | ||
| Dependence non uniformity (DNU) | ||
| Gray level non uniformity (GLNU) | ||
| Small dependence emphasis (SDE) | ||
| Small dependence high gray level emphasis (SDHGLE) | ||
| Dependence non uniformity normalized (DNUN) | ||
| Large dependence emphasis (LDE) | ||
| Large dependence low gray level emphasis (LDLGLE) | ||
| Dependence variance (DV) | ||
| Large dependence high gray level emphasis (LDHGLE) | ||
| Small dependence low gray level emphasis (SDLGLE) | ||
| Low gray level emphasis (LGLE) | ||
| LoG-based | First-order statistic and texture of Laplacian of Gaussian (LoG). Filter width: fine, σ=0.5; medium, σ=1.5; coarse, σ=2.5 | First-order features |
| GLCM features | ||
| GLSZM features | ||
| GLRLM features | ||
| NGTDM features | ||
| GLDM features | ||
| Wavelet-based | First-order statistic and texture of wavelet decomposition. Decomposition levels: LLL, LLH, LHL, LHH, HLL, HLH, HHL, HHH. | First-order features |
| GLCM features | ||
| GLSZM features | ||
| GLRLM features | ||
| NGTDM features | ||
| GLDM features |
GLCM, gray-level co-occurrence matrix; GLSZM, gray-level size-zone matrix; GLRLM, gray-level run-Length matrix; NGTDM, neighboring gray-tone difference matrix; GLDM, gray-level dependence matrix; Decomposition levels, i.e., LLH interpreted as the high-pass sub band, resulting from directional filtering of the volume with a low-pass filter along x-direction, a low pas filter along y-direction and a high-pass filter along z-direction.
Acknowledgements
The authors would like to thank the reviewers for their insightful suggestions, which helped improve the manuscript.
Ethical Statement: This study was approved by the Nanjing Drum Tower Hospital’s ethics committee (No. 2014-052-06) and informed consent from the patients was provided.
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015;136:E359-E386. 10.1002/ijc.29210 [DOI] [PubMed] [Google Scholar]
- 2.Wobb J, Krueger SA, Kane JL, Galoforo S, Grills IS, Wilson GD, Marples B. The effects of pulsed radiation therapy on tumor oxygenation in 2 murine models of head and neck squamous cell carcinoma. Int J Radiat Oncol Biol Phys 2015;92:820-8. 10.1016/j.ijrobp.2015.04.002 [DOI] [PubMed] [Google Scholar]
- 3.Zhang P, Wang B, Chen X, Cvetkovic D, Chen L, Lang J, Ma CM. Local Tumor Control and Normal Tissue Toxicity of Pulsed Low-Dose Rate Radiotherapy for Recurrent Lung Cancer: An In Vivo Animal Study. Dose Response 2015;13:1559325815588507. 10.1177/1559325815588507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meyer K, Krueger SA, Kane JL, Wilson TG, Hanna A, Dabjan M, Hege KM, Wilson GD, Grills I, Marples B. Pulsed radiation therapy with concurrent cisplatin results in superior tumor growth delay in a head and neck squamous cell carcinoma murine model. Int J Radiat Oncol Biol Phys 2016;96:161-9. 10.1016/j.ijrobp.2016.04.031 [DOI] [PubMed] [Google Scholar]
- 5.Richards GM, Tomé WA, Robins HI, Stewart JA, Welsh JS, Mahler PA, Howard SP. Pulsed reduced dose-rate radiotherapy: a novel locoregional retreatment strategy for breast cancer recurrence in the previously irradiated chest wall, axilla, or supraclavicular region. Breast Cancer Res Treat 2009;114:307-13. 10.1007/s10549-008-9995-3 [DOI] [PubMed] [Google Scholar]
- 6.Cannon GM, Tomé WA, Robins HI, Howard SP. Pulsed reduced dose-rate radiotherapy: case report. J Neurooncol 2007;83:307-11. 10.1007/s11060-007-9329-z [DOI] [PubMed] [Google Scholar]
- 7.Lee DY, Chunta JL, Park SS, Huang J, Martinez AA, Grills IS, Krueger SA, Wilson GD, Marples B. Pulsed versus conventional radiation therapy in combination with temozolomide in a murine orthotopic model of glioblastoma multiforme. Int J Radiat Oncol Biol Phys 2013;86:978-85. 10.1016/j.ijrobp.2013.04.034 [DOI] [PubMed] [Google Scholar]
- 8.Adkison JB, Tomé W, Seo S, Richards GM, Robins HI, Rassmussen K, Welsh JS, Mahler PA, Howard SP. Reirradiation of large-volume recurrent glioma with pulsed reduced-dose-rate radiotherapy. Int J Radiat Oncol Biol Phys 2011;79:835-41. 10.1016/j.ijrobp.2009.11.058 [DOI] [PubMed] [Google Scholar]
- 9.Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature 2003;421:499-506. 10.1038/nature01368 [DOI] [PubMed] [Google Scholar]
- 10.Nagashima F, Boku N, Ohtsu A, Yoshida S, Hasebe T, Ochiai A, Sakata Y, Saito H, Miyata Y, Hyodo I. Biological markers as a predictor for response and prognosis of unresectable gastric cancer patients treated with irinotecan and cisplatin. Jpn J Clin Oncol 2005;35:714-9. 10.1093/jjco/hyi194 [DOI] [PubMed] [Google Scholar]
- 11.Blackham AU, Greenleaf E, Yamamoto M, Hollenbeak C, Gusani N, Coppola D, Pimiento JM, Wong J. Tumor regression grade in gastric cancer: Predictors and impact on outcome. J Surg Oncol 2016;114:434-9. 10.1002/jso.24307 [DOI] [PubMed] [Google Scholar]
- 12.De Cobelli F, Giganti F, Orsenigo E, Cellina M, Esposito A, Agostini G, Albarello L, Mazza E, Ambrosi A, Socci C. Apparent diffusion coefficient modifications in assessing gastro-oesophageal cancer response to neoadjuvant treatment: comparison with tumour regression grade at histology. Eur Radiol 2013;23:2165-74. 10.1007/s00330-013-2807-0 [DOI] [PubMed] [Google Scholar]
- 13.Giganti F, De Cobelli F, Canevari C, Orsenigo E, Gallivanone F, Esposito A, Castiglioni I, Ambrosi A, Albarello L, Mazza E. Response to chemotherapy in gastric adenocarcinoma with diffusion - weighted MRI and 18F - FDG - PET/CT: Correlation of apparent diffusion coefficient and partial volume corrected standardized uptake value with histological tumor regression grade. J Magn Reson Imaging 2014;40:1147-57. 10.1002/jmri.24464 [DOI] [PubMed] [Google Scholar]
- 14.Aichler M, Luber B, Lordick F, Walch A. Proteomic and metabolic prediction of response to therapy in gastric cancer. World J Gastroenterol 2014;20:13648-57. 10.3748/wjg.v20.i38.13648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aerts HJ, Velazquez ER, Leijenaar RT, Parmar C, Grossmann P, Carvalho S, Bussink J, Monshouwer R, Haibe-Kains B, Rietveld D. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun 2014;5:4006. 10.1038/ncomms5006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Limkin EJ, Sun R, Dercle L, Zacharaki EI, Robert C, Reuzé S, Schernberg A, Paragios N, Deutsch E, Ferté C. Promises and challenges for the implementation of computational medical imaging (radiomics) in oncology. Ann Oncol 2017;28:1191-206. 10.1093/annonc/mdx034 [DOI] [PubMed] [Google Scholar]
- 17.Lambin P, Rios-Velazquez E, Leijenaar R, Carvalho S, van Stiphout RG, Granton P, Zegers CM, Gillies R, Boellard R, Dekker A. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer 2012;48:441-6. 10.1016/j.ejca.2011.11.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gillies RJ, Kinahan PE, Hricak H. Radiomics: Images Are More than Pictures, They Are Data. Radiology 2016;278:563-77. 10.1148/radiol.2015151169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumar V, Gu Y, Basu S, Berglund A, Eschrich SA, Schabath MB, Forster K, Aerts HJ, Dekker A, Fenstermacher D. Radiomics: the process and the challenges. Magn Reson Imaging 2012;30:1234-48. 10.1016/j.mri.2012.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu S, Liu S, Ji C, Zheng H, Pan X, Zhang Y, Guan W, Chen L, Guan Y, Li W. Application of CT texture analysis in predicting histopathological characteristics of gastric cancers. Eur Radiol 2017;27:4951-9. 10.1007/s00330-017-4881-1 [DOI] [PubMed] [Google Scholar]
- 21.Ba-Ssalamah A, Muin D, Schernthaner R, Kulinna-Cosentini C, Bastati N, Stift J, Gore R, Mayerhoefer ME. Texture-based classification of different gastric tumors at contrast-enhanced CT. Eur J Radiol 2013;82:e537-e543. 10.1016/j.ejrad.2013.06.024 [DOI] [PubMed] [Google Scholar]
- 22.Ma Z, Fang M, Huang Y, He L, Chen X, Liang C, Huang X, Cheng Z, Dong D, Liang C. CT-based radiomics signature for differentiating Borrmann type IV gastric cancer from primary gastric lymphoma. Eur J Radiol 2017;91:142-7. 10.1016/j.ejrad.2017.04.007 [DOI] [PubMed] [Google Scholar]
- 23.Giganti F, Marra P, Ambrosi A, Salerno A, Antunes S, Chiari D, Orsenigo E, Esposito A, Mazza E, Albarello L. Pre-treatment MDCT-based texture analysis for therapy response prediction in gastric cancer: Comparison with tumour regression grade at final histology. Eur J Radiol 2017;90:129-37. 10.1016/j.ejrad.2017.02.043 [DOI] [PubMed] [Google Scholar]
- 24.Giganti F, Antunes S, Salerno A, Ambrosi A, Marra P, Nicoletti R, Orsenigo E, Chiari D, Albarello L, Staudacher C. Gastric cancer: texture analysis from multidetector computed tomography as a potential preoperative prognostic biomarker. Eur Radiol 2017;27:1831-9. 10.1007/s00330-016-4540-y [DOI] [PubMed] [Google Scholar]
- 25.Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, Dancey J, Arbuck S, Gwyther S, Mooney M, Rubinstein L, Shankar L, Dodd L, Kaplan R, Lacombe D, Verweij J. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer 2009;45:228-47. 10.1016/j.ejca.2008.10.026 [DOI] [PubMed] [Google Scholar]
- 26.van Griethuysen JJ, Fedorov A, Parmar C, Hosny A, Aucoin N, Narayan V, Beets-Tan RG, Fillion-Robin J-C, Pieper S, Aerts HJ. Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Res 2017;77:e104-e107. 10.1158/0008-5472.CAN-17-0339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gamer M, Lemon J, Singh IFP. irr: Various Coefficients of Interrater Reliability and Agreement. R Package Version 2012.
- 28.Tixier F, Le RC, Hatt M, Albarghach N, Pradier O, Metges JP, Corcos L, Visvikis D. Intratumor heterogeneity characterized by textural features on baseline 18F-FDG PET images predicts response to concomitant radiochemotherapy in esophageal cancer. J Nucl Med 2011;52:369-78. 10.2967/jnumed.110.082404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kohavi R, John GH. Wrappers for feature subset selection. Artificial Intelligence 1997;97:273-324. 10.1016/S0004-3702(97)00043-X [DOI] [Google Scholar]
- 30.Salzberg SL. On Comparing Classifiers: Pitfalls toAvoid and a Recommended Approach. Kluwer Academic Publishers, 1997. [Google Scholar]
- 31.Castellano G, Bonilha L, Li L, Cendes F. Texture analysis of medical images. Clin Radiol 2004;59:1061-9. 10.1016/j.crad.2004.07.008 [DOI] [PubMed] [Google Scholar]
- 32.Ng F, Ganeshan B, Kozarski R, Miles KA, Goh V. Assessment of primary colorectal cancer heterogeneity by using whole-tumor texture analysis: contrast-enhanced CT texture as a biomarker of 5-year survival. Radiology 2013;266:177-84. 10.1148/radiol.12120254 [DOI] [PubMed] [Google Scholar]
- 33.Ganeshan B, Goh V, Mandeville HC, Ng QS, Hoskin PJ, Miles KA. Non-small cell lung cancer: histopathologic correlates for texture parameters at CT. Radiology 2013;266:326-36. 10.1148/radiol.12112428 [DOI] [PubMed] [Google Scholar]