Abstract
Drought stress triggers remarkable physiological changes and growth impediments, which significantly diminish plant biomass and crop yield. However, certain plant species show notable resilience, maintaining nearly normal yields under severe water deficits. For example, sorghum is a naturally drought-tolerant crop, which is ideal for studying plant adaptive responses to drought. Here we used sorbitol treatments to simulate drought-induced osmotic stress in sorghum cell suspension cultures and analysed fractions enriched for extracellular matrix proteins using isobaric tags for relative and absolute quantification technology. Sorbitol induced an overall increase in protein secretion, with putative redox proteins, proteases, and glycosyl hydrolases featuring prominently among the responsive proteins. Gene expression analysis of selected candidates revealed regulation at the transcriptional level. There was a notable differential gene expression between drought-tolerant and drought-sensitive sorghum varieties for some of the candidates. This study shows that protein secretion is a major component of the sorghum response to osmotic stress. Additionally, our data provide candidate genes, which may have putative functions in sorghum drought tolerance, and offer a pool of genes that could be developed as potential biomarkers for rapid identification of drought tolerant lines in plant breeding programs.
Introduction
Water is an essential solvent for cell biochemical reactions and is indispensable for life. Extreme dehydration reduces cell turgor and adversely affects cellular metabolic processes. Prolonged water deficits, such as imposed by severe droughts, result in leaf wilting and ultimately ends in plant death. While the majority of plants are very sensitive to water loss and capitulate under drought stress, several plant species have genetic adaptations ensuring their survival in marginal lands and extreme environments with limited water. There is intense research interest in understanding the molecular responses of plants to drought stress.
Upon sensing soil water deficits, plants activate transcriptional changes enabling them to deploy mechanisms for conserving water, metabolic reprogramming for adaptation to drought stress, and redirection of growth patterns to follow moisture gradients. The signalling events underpinning the adaptive responses to drought are complex and involve abscisic acid (ABA)-dependent and ABA-independent pathways. Dehydration triggers the biosynthesis of ABA1, which regulates plant water balance and osmotic stress tolerance via control of stomatal aperture2 and activation of stress tolerance genes3. ABA binds to its soluble receptor complex, pyrabactin resistance1/PYR1-Like/regulatory component of PYR1/PYRL/RCAR ABA receptors4,5. Receptor binding inhibits protein phosphatase 2C activity4–11, triggering autophosphorylation of SnRK2 kinases12–14, which in turn phosphorylate numerous substrates and activate multiple pathways including guard cell closure and drought stress-adaptive gene expression15.
A conserved ABA-responsive element in the gene promoter is an essential cis-acting element for regulating ABA-inducible gene expression16. MYB and MYC recognition sites are additional cis-acting elements identified in the promoters of some ABA-regulated genes17. Activation of ABA-dependent pathways in transgenic Arabidopsis by constitutive overexpression of the transcription factors ABF2, MYC2, or MYB2, leads to improved tolerance to drought/osmotic stress18,19. ABA-independent signalling pathways also operate in activation of stress-responsive genes during drought. Neither the primary receptors involved nor the signalling components that lead to drought-induced gene expression via ABA-independent pathways are known. However, the responsive genes possess a conserved cis-acting element in the promoter sequence known as the dehydration-responsive element (DRE)20. DRE-binding Protein 2A (DREB2A) specifically binds the DRE sequence21 to activate Arabidopsis gene expression in response to drought, high salinity, and heat-shock stress21,22. Constitutive activation of the ABA-independent pathways by overexpression of DREB2A confers increased drought tolerance in Arabidopsis21.
Transcriptomic changes driven by drought-induced signalling reprogram the proteome and cellular metabolism. The functional significance of most of the proteins is not fully understood. However, some of these have a role in signal transduction and activation of further gene expression, while others clearly support the adaptive response strategy to re-establish cellular homeostasis and survival under drought stress. The classes of proteins deployed during plant adaptation to drought were reviewed by Shinozaki and Yamaguchi-Shinozaki3. They include aquaporins for water movement across membranes and enzymes for the biosynthesis of osmolyte sugars, proline, and glycine-betaine, which are important for osmotic rebalancing. Cellular detoxification enzymes, such as ascorbate peroxidase, glutathione-S-transferase, catalase, and superoxide dismutase prevent oxidative damage, while protection of membranes and macromolecules is maintained by chaperones, messenger RNA-binding proteins, late embryogenesis abundant proteins, and similar proteins. The adaptive reprogramming of the transcriptome and proteome is supported by increased protein turnover facilitated by enzymes and proteins, such as ubiquitin, Clp protease, and thiol proteases. Transgenic plants overexpressing some of these genes acquire drought tolerance23, indicating that the gene products really function in stress tolerance.
Most of the research into plant molecular responses to drought has been conducted using drought-sensitive model species, such as Arabidopsis thaliana. Sorghum (Sorghum bicolor L. Moench), a naturally drought tolerant cereal24 with high genetic diversity, is a good model system for studying drought stress-adaptive responses25, especially with a view to identify novel genes that could be used to generate drought tolerant crops. The sorghum genome has been sequenced26 and some transcriptomic27 and proteomic28 analysis of leaf responses to osmotic stress and drought have been reported. We have a longstanding interest in understanding how the extracellular matrix proteome changes during stress-adaptive responses29,30. Our hypothesis is that the extracellular matrix is a repository of signal molecules used for cell-cell communications during stress adaptation, and analysis of this compartment may lead to identification of signal-regulatory proteins with a pivotal role in drought tolerance. Here, we used a sorghum cell suspension culture system to identify differentially expressed proteins in the extracellular matrix during osmotic stress and show that selected targets are differentially expressed in drought-tolerant and sensitive sorghum lines during drought stress.
Results
Identification of sorghum cell suspension culture ECM proteins
We designed experiments to isolate fractions enriched for secreted proteins in the soluble phase of the sorghum extracellular matrix (ECM). Our goal was to identify these proteins and analyse their response to osmotic stress. We used sorghum cell suspension cultures as a source of easily extractable soluble ECM proteins from the culture growth medium. Basing on preliminary data obtained from the growth curve, we used exponential phase 8-day-old cultures for stress treatments. Sorghum cell cultures were treated with 400 mM sorbitol31 and cells harvested every 24 h until 72 h for RNA extraction. We analysed expression profiles of sorghum homologues of Arabidopsis drought marker genes, ERD1 and DREB2A, to monitor the osmotic stress response and establish the optimal time for harvesting cells for protein extraction. We identified sorghum homologues of Arabidopsis ERD1 and DREB2A, which we named ERD1-1 (SORBI_3004G162400), ERD1-2 (SORBI_3006G065100), DREB2A-1 (SORBI_3009G101400), and DREB2A-2 (SORBI_3003G058200). With the exception of DREB2A-2, all the genes were activated by sorbitol treatment, with expression peaking at 48 h (Fig. 1). Therefore, in subsequent experiments, 48 h was selected as the time after sorbitol addition to harvest cell cultures for protein extraction. Use of 4 biological replicates for both sorbitol treatments and controls ensured that proteins with highly reproducible responses were identified.
Figure 1.
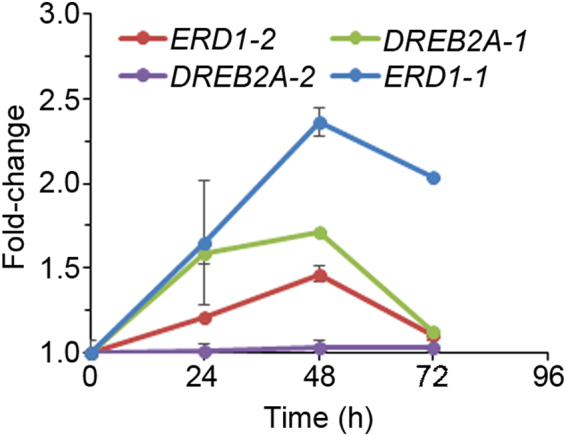
Activation of sorghum ERD1 and DREB2A expression in response to sorbitol. Sorghum cell suspension cultures were treated with sorbitol and cells harvested at the indicated time-points. Gene expression was analysed using qRT-PCR. Error bars represent means ± S.D. (n = 3).
Cell cultures were treated with sorbitol and secreted proteins were isolated from the culture medium by simple filtration of the cell culture and acetone precipitation of the filtrate. ECM protein samples from control and osmotic stressed cultures were then digested with trypsin, labelled with iTRAQ, fractionated by liquid chromatography, and analysed using tandem mass spectrometry. Only proteins with at least 2 sequenced peptides, each with a statistical confidence threshold ≥ 95%, were considered positively identified. A total of 179 different proteins were positively identified in the ECM fractions of sorghum cell cultures. The full mass spectrometry data of these proteins is provided in Supplementary Dataset (Table S2). This dataset represents a snapshot of the sorghum cell culture secretome at 10 days post-subculturing. Some of the 179 proteins have functional annotations in the protein database derived from sequence identity, which include peroxidases, alpha-galactosidases, alpha-mannosidase, endoglucanases, purple acid phosphatase, malate dehydrogenase and xyloglucan endotransglucosylase. Other proteins are annotated as uncharacterized proteins since database annotation is still incomplete. All the functionally annotated and uncharacterized proteins identified here will require experimental validation of protein function. Apart from the sorghum specific proteins, we also identified trypsin and human keratin proteins, which are known contaminants in proteomic analysis. These contaminants serve as defacto positive controls and their identification in interrogating extensive protein databases indicates that protein identification was specific.
Differentially expressed ECM proteins in response to osmotic stress
For quantitative analysis of osmotic stress-related protein expression, a minimum threshold of 2-fold change in protein abundance at a significance level of p ≤ 0.05 was applied to filter the dataset. This resulted in a total of 92 proteins that were differentially expressed in response to sorbitol-induced osmotic stress (Table 1). With the exception of one down-regulated protein, the rest were up-regulated, indicating that sorbitol triggered an overall increase in protein secretion. Next we used the SignalP tool to analyse the protein sequences for the presence of a signal peptide, which targets proteins to the secretory pathway. A predicted N-terminal signal peptide was identified in 54 of these proteins (Table 1), indicating that they are secreted via the classical secretory pathway requiring a leader sequence. The remaining proteins were predicted not to have an N-terminal signal peptide (Table 1). Bioinformatic analysis of the primary sequences was used to detect putative functional domains in the differentially expressed proteins, which were then assigned to specific protein families (Table 1). There were 18 proteins assigned to glycosyl-hydrolases/glycosidases, 5 to cell wall modifying enzymes, 12 to proteases, 27 to redox proteins, and 30 proteins were left unclassified.
Table 1.
List of sorghum secreted proteins that are responsive to sorbitol-induced osmotic stress.
| Prot. #a | Accessionb | Protein Name | Ratioc | SDd | p valuee | Signal Peptidef | Family nameg |
|---|---|---|---|---|---|---|---|
| Glycosyl-hydrolases/Glycosidases | |||||||
| 6 | A0A1B6QHZ6 | Uncharacterized protein OS = Sorghum bicolor GN = SORBI_001G089000 | 2.93 | 0.12 | 4.85E-06 | − | Glycoside hydrolase superfamily |
| 8 | C5X532 | Alpha-galactosidase OS = Sorghum bicolor GN = SORBI_002G123100 | 2.05 | 0.05 | 7.96E-04 | + | Glycoside hydrolase superfamily |
| 22 | A0A1B6QI05 | Uncharacterized protein OS = Sorghum bicolor GN = SORBI_001G089100 | 2.19 | 0.08 | 1.79E-04 | + | Glycoside hydrolase superfamily |
| 27 | C5XKE9 | Endoglucanase OS = Sorghum bicolor GN = SORBI_003G015700 | 2.84 | 0.35 | 2.25E-05 | − | Glycoside hydrolase family 9 |
| 28 | C5Y397 | Alpha-mannosidase OS = Sorghum bicolor GN = SORBI_005G132400 | 4.24 | 0.28 | 1.10E-06 | + | Glycosyl hydrolase family 38 |
| 29 | C5X8J4 | Xyloglucan endotransglucosylase/hydrolase OS = Sorghum bicolor GN = SORBI_002G302000 | 3.58 | 0.24 | 1.42E-06 | + | Xyloglucan endotransglucosylase/hydrolase |
| 36 | C5XB38 | Uncharacterized protein OS = Sorghum bicolor GN = SORBI_002G055600 | 2.33 | 0.16 | 7.22E-05 | + | Glycoside hydrolase family 18 |
| 72 | C5X022 | Uncharacterized protein OS = Sorghum bicolor GN = SORBI_001G525000 | 2.00 | 0.11 | 1.51E-04 | + | Glycoside hydrolase family 28 |
| 82 | A0A1B6QC86 | Uncharacterized protein OS = Sorghum bicolor GN = SORBI_002G189100 | 2.07 | 0.16 | 6.77E-04 | − | Glycoside hydrolase family 81 |
| 84 | C5XFX7 | Uncharacterized protein OS = Sorghum bicolor GN = SORBI_003G247000 | 2.29 | 0.20 | 5.08E-05 | + | Glycoside hydrolase family 5 |
| 86 | A0A1B6PTQ9 | Uncharacterized protein OS = Sorghum bicolor GN = SORBI_005G204700 | 2.07 | 0.48 | 5.04E-03 | + | Glycoside hydrolase family 28 |
| 88 | C5XB39 | Uncharacterized protein OS = Sorghum bicolor GN = SORBI_002G055700 | 2.35 | 0.11 | 1.15E-05 | + | Glycoside hydrolase family 18 |
| 95 | C5X5L7 | Alpha-galactosidase OS = Sorghum bicolor GN = SORBI_002G417800 | 4.06 | 0.21 | 1.34E-06 | + | Glycoside hydrolase family 27 |
| 133 | C5WP48 | Alpha-mannosidase OS = Sorghum bicolor GN = SORBI_001G268700 | 3.05 | 0.31 | 3.92E-04 | + | Glycoside hydrolase family 38 |
| 140 | C5YCY4 | Uncharacterized protein OS = Sorghum bicolor GN = SORBI_006G160700 | 2.42 | 0.42 | 2.17E-04 | − | Glycosyl hydrolase family 32 |
| 141 | A0A1B6Q8G8 | Uncharacterized protein (Fragment) OS = Sorghum bicolor GN = SORBI_003G440900 | 2.92 | 0.11 | 4.18E-05 | − | Glycosyl hydrolase family 32 |
| 145 | C5YBF1 | Uncharacterized protein OS = Sorghum bicolor GN = SORBI_006G132700 | 2.79 | 0.29 | 2.59E-05 | + | Glycoside hydrolase family 19 |
| 150 | C5X3W3 | Uncharacterized protein OS = Sorghum bicolor GN = SORBI_002G246400 | 2.30 | 0.42 | 1.16E-03 | + | Glycoside hydrolase, family 28 |
| Cell wall modifying enzymes | |||||||
| 2 | C5WSF9 | Uncharacterized protein OS = Sorghum bicolor GN = SORBI_001G301500 | 3.18 | 0.19 | 3.58E-06 | + | Expansin/Lol pI |
| 17 | C5WSF0 | Uncharacterized protein OS = Sorghum bicolor GN = SORBI_001G300800 | 3.23 | 0.41 | 3.53E-05 | + | Expansin/Lol pI family |
| 33 | C5Z0P5 | Uncharacterized protein OS = Sorghum bicolor GN = SORBI_009G055900 |
2.95 | 0,23 | 3.77E-06 | − | Fasciclin-like arabinogalactan protein |
| 59 | C5WSE5 | Uncharacterized protein OS = Sorghum bicolor GN = SORBI_001G300400 | 3.14 | 0.23 | 7.20E-06 | + | Expansin/Lol pI |
| 87 | C5YVJ7 | Uncharacterized protein OS = Sorghum bicolor GN = SORBI_009G232100 | 2.36 | 0.11 | 1.38E-06 | + | Fasciclin 1 domain |
| Proteases | |||||||
| 14 | A0A1B6PLA9 | Uncharacterized protein OS = Sorghum bicolor GN = SORBI_006G104300 | 2.25 | 0.09 | 2.28E-05 | + | Gamma-glutamyl-transpeptidase |
| 20 | A0A1B6QMT3 | Uncharacterized protein OS = Sorghum bicolor GN = SORBI_001G348900 | 3.09 | 0.24 | 6.02E-06 | + | Peptidase S10, serine carboxypeptidase |
| 26 | C5XQ74 | Uncharacterized protein OS = Sorghum bicolor GN = SORBI_003G208800 | 2.05 | 0.10 | 4.70E-04 | − | Aspartic peptidase A1 family |
| 48 | A0A1B6PNM7 | Uncharacterized protein OS = Sorghum bicolor GN = SORBI_006G242000 | 3.27 | 0.13 | 7.96E-07 | + | Peptidase C1A |
| 85 | C5WT64 | Uncharacterized protein OS = Sorghum bicolor GN = SORBI_001G170700 | 2.05 | 0.18 | 4.15E-04 | + | Peptidase S8 subtilisin-related |
| 94 | C5WXN2 | Carboxypeptidase OS = Sorghum bicolor GN = SORBI_001G348800 | 2.10 | 0.14 | 2.50E-04 | + | Peptidase S10, serine carboxypeptidase |
| 98 | C5YNA1 | Uncharacterized protein OS = Sorghum bicolor GN = SORBI_007G172100 | 3.73 | 0.25 | 5.00E-06 | + | Peptidase C1A |
| 122 | A0A1B6PHE0 | Uncharacterized protein OS = Sorghum bicolor GN = SORBI_007G120800 | 3.13 | 0.41 | 8.07E-05 | − | Peptidase M1 family |
| 136 | C5WQK1 | Uncharacterized protein OS = Sorghum bicolor GN = SORBI_001G280000 | 2.77 | 0.28 | 3.61E-05 | + | Peptidase S10, serine carboxypeptidase |
| 138 | A0A1B6QEG2 | Uncharacterized protein OS = Sorghum bicolor GN = SORBI_002G315800 | 4.42 | 0.25 | 3.79E-07 | + | Peptidase C1A |
| 173 | C5XDR4 | Uncharacterized protein OS = Sorghum bicolor GN = SORBI_002G217200 | 2.87 | 0.25 | 8.52E-06 | + | Peptidase C1A |
| 178 | C5Y171 | Uncharacterized protein OS = Sorghum bicolor GN = SORBI_004G142800 | 5.62 | 0.52 | 1.25E-06 | + | Peptidase C1A domain and family |
| Redox proteins | |||||||
| 7 | A0A1B6QG95 | Uncharacterized protein OS = Sorghum bicolor GN = SORBI_002G416600 | 2.08 | 0.03 | 6.75E-05 | − | Plant peroxidase |
| 13 | C5Y360 | Peroxidase OS = Sorghum bicolor GN = SORBI_005G011300 | 2.73 | 0.31 | 4.80E-05 | + | Plant peroxidase |
| 23 | C5Z240 | Uncharacterized protein OS = Sorghum bicolor GN = SORBI_010G003100 | 2.40 | 0.19 | 3.69E-05 | + | Cupredoxin |
| 30 | C5WNY4 | Uncharacterized protein OS = Sorghum bicolor GN = SORBI_001G129700 | 2.07 | 0.18 | 1.57E-04 | + | Germin |
| 31 | C5YC92 | Uncharacterized protein OS = Sorghum bicolor GN = SORBI_006G018100 | 2.18 | 0.30 | 3.17E-04 | + | Germin |
| 35 | C5XIY1 | Peroxidase OS = Sorghum bicolor GN = SORBI_003G152100 | 2.98 | 0.14 | 5.62E-06 | + | Plant peroxidase |
| 38 | A0A1B6QN00 | Uncharacterized protein OS = Sorghum bicolor GN = SORBI_001G360500 | 2.16 | 0.41 | 1.46E-03 | + | Plant peroxidase |
| 41 | C6JSB7 | Peroxidase OS = Sorghum bicolor GN = Sb0246s002010 | 7.79 | 1.84 | 4.99E-05 | + | Plant peroxidase |
| 51 | A0A1B6QGB6 | Uncharacterized protein OS = Sorghum bicolor GN = SORBI_002G416800 | 2.21 | 0.07 | 1.86E-05 | + | Plant peroxidase |
| 69 | A0A1B6Q9F4 | Uncharacterized protein OS = Sorghum bicolor GN = SORBI_002G057900 | 5.63 | 0.26 | 6.39E-08 | − | Thioredoxin |
| 92 | C5XL59 | Uncharacterized protein OS = Sorghum bicolor GN = SORBI_003G024700 | −2.40 | 0.04 | 7.37E-04 | − | Plant peroxidase |
| 97 | C5XIY0 | Peroxidase OS = Sorghum bicolor GN = SORBI_003G152000 | 2.58 | 0.11 | 9.88E-06 | − | Plant peroxidase |
| 104 | A0A194YU12 | Uncharacterized protein OS = Sorghum bicolor GN = SORBI_004G341200 | 6.45 | 0.29 | 2.34E-08 | − | Glutathione-disulphide reductase |
| 110 | A0A1B6QN96 | Uncharacterized protein OS = Sorghum bicolor GN = SORBI_001G371900 | 13.59 | 1.99 | 1.14E-05 | − | Cu-Zn superoxide dismutase-like |
| 111 | A0A1B6Q818 | Uncharacterized protein OS = Sorghum bicolor GN = SORBI_003G416300 | 5.84 | 0.45 | 3.19E-07 | − | GST C-terminal domain-like |
| 129 | C5X6P7 | Uncharacterized protein OS = Sorghum bicolor GN = SORBI_002G140400 | 2.34 | 0.14 | 1.53E-05 | + | Cupredoxin |
| 131 | C5WWQ2 | Uncharacterized protein OS = Sorghum bicolor GN = SORBI_001G342600 | 8.47 | 2.43 | 6.43E-05 | − | Thioredoxin |
| 134 | C5YQ75 | Peroxidase OS = Sorghum bicolor GN = SORBI_008G010500 | 2.85 | 0.13 | 2.98E-06 | + | Plant peroxidase |
| 137 | C5X780 | Uncharacterized protein OS = Sorghum bicolor GN = SORBI_002G007200 | 2.70 | 0.12 | 9.70E-06 | + | Cupredoxin |
| 151 | C5XC95 | Uncharacterized protein OS = Sorghum bicolor GN = SORBI_002G345800 | 2.76 | 0.18 | 1.45E-05 | + | Cupredoxin |
| 155 | A0A1B6QFT7 | Uncharacterized protein OS = Sorghum bicolor GN = SORBI_002G392300 | 38.70 | 5.94 | 6.01E-06 | + | Plant peroxidase |
| 159 | A0A1B6P9F6 | Uncharacterized protein OS = Sorghum bicolor GN = SORBI_009G190800 | 4.28 | 0.35 | 1.95E-06 | − | Thioredoxin |
| 161 | C5Z0N9 | Peroxidase OS = Sorghum bicolor GN = SORBI_009G055300 | 2.76 | 0.11 | 9.30E-06 | + | Plant peroxidase |
| 167 | C5XRU7 | Uncharacterized protein OS = Sorghum bicolor GN = SORBI_004G148100 | 3.01 | 0.42 | 8.74E-05 | + | Germin |
| 169 | A0A1B6QJR7 | Uncharacterized protein OS = Sorghum bicolor GN = SORBI_001G189000 | 2.10 | 0.39 | 2.17E-03 | − | Plant peroxidase |
| 174 | C5YN91 | Uncharacterized protein OS = Sorghum bicolor GN = SORBI_007G171000 | 3.42 | 2.19 | 8.12E-03 | − | FAD/NAD linked reductases, dimerization (C-terminal) domain |
| 179 | A0A1B6QB11 | Uncharacterized protein OS = Sorghum bicolor GN = SORBI_002G133800 | 5.62 | 0.92 | 1.42E-05 | − | FAD/NAD linked reductases, dimerization (C-terminal) domain |
| Unclassified | |||||||
| 19 | A0A194YMM6 | Uncharacterized protein OS = Sorghum bicolor GN = SORBI_010G262500 | 7.94 | 0.27 | 1.04E-08 | − | Glyceraldehyde-3-phosphate dehydrogenase, type I |
| 21 | C5Z6U2 | Uncharacterized protein OS = Sorghum bicolor GN = SORBI_010G210000 | 2.82 | 0.36 | 5.01E-05 | − | Ubiquitin |
| 40 | C5XWE5 | Uncharacterized protein OS = Sorghum bicolor GN = SORBI_004G197600 | 2.52 | 0.16 | 3.59E-05 | + | Glycerophosphoryl diester phosphodiesterase family |
| 43 | A0A1B6PD28 | Uncharacterized protein OS = Sorghum bicolor GN = SORBI_008G113000 | 3.08 | 0.18 | 7.65E-06 | + | Purple acid phosphatase, N-terminal domain family |
| 47 | C5XPK9 | Uncharacterized protein OS = Sorghum bicolor GN = SORBI_003G205600 | 2.69 | 0.09 | 8.53E-07 | + | Leucine-rich repeat domain family |
| 49 | A0A194YGY2 | Uncharacterized protein OS = Sorghum bicolor GN = SORBI_010G027000 | 5.90 | 0.35 | 1.21E-07 | − | Enolase-like |
| 53 | C5Z6U1 | Uncharacterized protein OS = Sorghum bicolor GN = SORBI_010G209900 | 2.96 | 0.16 | 5.80E-06 | + | Not predicted |
| 67 | C5Y587 | Uncharacterized protein OS = Sorghum bicolor GN = SORBI_005G049800 | 5.06 | 0.47 | 1.20E-06 | − | Alginate lyase |
| 68 | C5YBH7 | Uncharacterized protein OS = Sorghum bicolor GN = SORBI_006G135500 | 2.07 | 0.07 | 1.38E-04 | + | Galactose oxidase central domain |
| 70 | C5XX52 | Glyceraldehyde-3-phosphate dehydrogenase OS = Sorghum bicolor GN = SORBI_004G205100 | 4.49 | 0.29 | 5.19E-07 | − | Glyceraldehyde 3-phosphate dehydrogenase |
| 76 | C5WXD7 | Uncharacterized protein OS = Sorghum bicolor GN = SORBI_001G209300 | 2.67 | 0.13 | 2.62E-05 | + | Uncharacterised protein family, basic secretory protein |
| 79 | C5X502 | Dirigent protein OS = Sorghum bicolor GN = SORBI_002G119900 | 3.01 | 0.31 | 2.02E-05 | + | Allene oxide cyclase/Dirigent protein |
| 90 | A0A1B6QEI0 | Uncharacterized protein OS = Sorghum bicolor GN = SORBI_002G317600 | 4.45 | 0.12 | 9.13E-08 | − | YjgF/YER057c/UK114 family |
| 103 | C5YW21 | Malate dehydrogenase OS = Sorghum bicolor GN = SORBI_009G240700 | 5.43 | 0.89 | 2.77E-05 | − | L-Lactate/malate dehydrogenase |
| 106 | C5WT90 | Uncharacterized protein OS = Sorghum bicolor GN = SORBI_001G173300 | 2.48 | 0.28 | 2.43E-04 | − | Reversibly glycosylated polypeptide family |
| 113 | C5XYB4 | Uncharacterized protein OS = Sorghum bicolor GN = SORBI_004G229300 | 2.28 | 0.14 | 3.92E-05 | − | Phosphate-induced protein 1 |
| 115 | C5XQW7 | Uncharacterized protein OS = Sorghum bicolor GN = SORBI_003G087300 | 2.29 | 0.12 | 8.67E-05 | + | S1/P1 nuclease family |
| 116 | C5WQH5 | Uncharacterized protein OS = Sorghum bicolor GN = SORBI_001G149500 | 2.79 | 0.48 | 2.33E-04 | − | None predicted |
| 117 | C5Y1P6 | Uncharacterized protein OS = Sorghum bicolor GN = SORBI_005G099500 | 2.44 | 0.15 | 3.59E-05 | + | Nucleoside phosphatase GDA1/CD39 family |
| 120 | C5YSB1 | Uncharacterized protein OS = Sorghum bicolor GN = SORBI_008G048400 | 2.55 | 0.21 | 2.60E-05 | + | Alginate lyase |
| 121 | A0A1B6QAK5 | Uncharacterized protein OS = Sorghum bicolor GN = SORBI_002G113800 | 2.97 | 0.29 | 3.91E-05 | − | Spermidine/spermine synthases |
| 123 | C5XFH6 | Fructose-bisphosphate aldolase OS = Sorghum bicolor GN = SORBI_003G393900 |
4.01 | 0.53 | 2.75E-05 | − | Fructose-bisphosphate aldolase, class-I |
| 130 | C5XTG0 | Uncharacterized protein OS = Sorghum bicolor GN = SORBI_004G166500 | 6.10 | 0.45 | 5.54E-07 | − | N-carbamoylputrescine amidase |
| 132 | C5X9N2 | Uncharacterized protein OS = Sorghum bicolor GN = SORBI_002G039000 | 4.15 | 0.38 | 4.03E-06 | + | ML domain |
| 139 | C5YRS3 | Purple acid phosphatase OS = Sorghum bicolor GN = SORBI_008G037000 | 3.69 | 0.34 | 5.42E-06 | − | Purple acid phosphatase-like, N-terminal domain family |
| 148 | C5WT45 | Uncharacterized protein OS = Sorghum bicolor GN = SORBI_001G168500 | 3.55 | 0.33 | 6.16E-06 | − | Serpin family |
| 152 | C5XQ07 | Uncharacterized protein OS = Sorghum bicolor GN = SORBI_003G072300 | 5.32 | 1.29 | 1.52E-04 | − | Triosephosphate isomerase |
| 165 | A0A1B6PLT5 | Uncharacterized protein OS = Sorghum bicolor GN = SORBI_006G133000 | 2.14 | 0.38 | 5.89E-04 | + | Galactose-binding domain-like |
| 168 | C5XG88 | Small ubiquitin-related modifier | 6.26 | 1.94 | 6.02E-04 | − | Ubiquitin-related |
| 181 | A0A1B6PJF1 | Uncharacterized protein OS = Sorghum bicolor GN = SORBI_006G014400 | 2.20 | 0.32 | 8.41E-04 | − | AmbAllergen |
aProtein number assigned in ProteinPilot.
bProtein accession numbers obtained from the UniProt database searches against sequences of S. bicolor only.
cRatio represents the average fold-change (n = 4) in response to sorbitol-induced osmotic stress relative to the control. A negative value indicates down-regulation.
dStandard deviation of the fold-changes (n = 4).
eProbability value obtained from a Student’s t-test comparing the fold changes between the sorbitol-induced osmotic stress treatments and the control (n = 4).
fSignal peptide prediction using SignalP 4.1 (http://www.cbs.dtu.dk/services/SignalP). A positive sign denotes the presence of a predicted signal peptide; a negative sign denotes the absence of a signal peptide.
gFamily name as predicted using the InterPro (http://www.ebi.ac.uk/interpro/) and Superfamily (www.supfam.org) database.
Analysis of sorbitol-induced gene expression
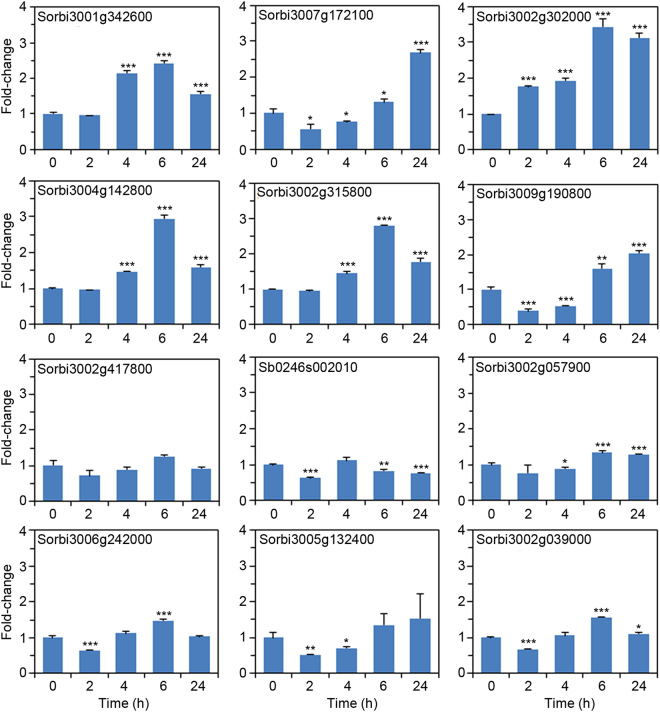
The observed increase in the amount of secreted proteins may be a result of increased expression of the genes encoding these proteins or increased translation of the corresponding mRNA. To investigate if osmotic stress transcriptionally regulated some of these candidates, we used qRT-PCR analysis on randomly selected 12 genes from the top 30 proteins of differentially expressed proteins that had been ranked in descending order of the fold-change magnitude (Supplementary Dataset - Table S3). Sorghum cell cultures were treated with sorbitol and samples for RNA extraction harvested 0, 2, 4, 6 and 24 h later. We focused on early transcriptional responses, which precede changes at the protein level analysed 48 h after sorbitol addition. With the exception of SORBI_3002G417800, whose expression did not respond to osmotic stress at any time-point, all the other 11 genes investigated responded significantly to sorbitol at least at one time-point (Fig. 2). However, for Sb0246s002010 and SORBI_3005G132400 the significant response within the first 24 h was transcriptional repression. For the other genes, there was either an initial suppression of gene expression at the early time-points followed by activation (e.g., SORBI_3007G172100), or gene activation without any suppression (e.g., SORBI_3002G302000) (Fig. 2). Taken together, these results show that increased protein secretion into the ECM observed in this study could be driven by transcriptional regulation, post-transcriptional regulation, or regulated at both transcription and translation levels, depending on the specific proteins. Moreover, the different expression profiles across the sampled 12 genes suggest that there is complex coordination of the gene network governing the proteome response to osmotic stress.
Figure 2.
Sorbitol-induced gene expression. Sorghum cell suspension cultures were treated with sorbitol and cells harvested at the indicated time-points for qRT-PCR analysis. Error bars represent means ± S.D. (n = 3). One, two and three asterisks indicate statistically significant differences between control and sorbitol treatment means at each time-point, p ≤ 0.05, 0.01, and 0.001, respectively.
Analysis of drought-induced gene expression in sorghum plants
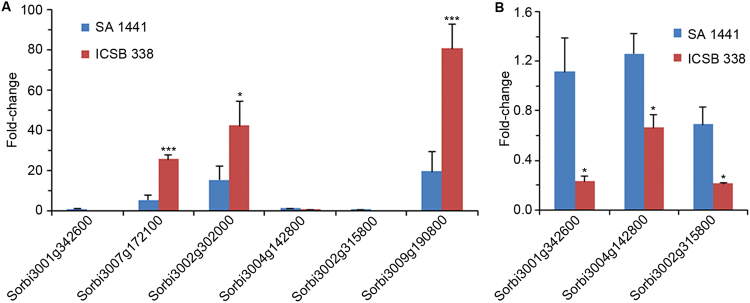
Six of the 12 genes analysed by qRT-PCR were activated ≥2-fold in response to sorbitol treatment of sorghum cell suspension cultures (Fig. 2). We then investigated if activation of these 6 genes (S0RBI_3001G342600, SORBI_3007G172100, SORBI_3002G302000, SORBI_3004G142800, SORBI_3002G315800 and SORBI_3009G190800) in the in vitro cell culture system is recapitulated in sorghum plants exposed to drought stress. We selected two sorghum varieties with contrasting drought response phenotypes; the drought-tolerant SA 1441 and “drought-sensitive” ICSB 338. After a period of growth with optimal soil water content, the plants were exposed to drought stress by withholding water for 11 days. Across all the 6 genes, there was a significant difference in drought-induced expression in root tissues of the two sorghum varieties (Fig. 3A,B). Expression of SORBI_3007G172100, SORBI_3002G302000 and SORBI_3009G190800 increased in response to drought, with up-regulation in the drought-sensitive ICSB 338 variety being significantly greater than the tolerant SA 1441 variety (Fig. 3A). Conversely, SORBI_3001G342600, SORBI_3004G142800 and SORBI_3002G315800 were significantly suppressed in the drought-sensitive ICSB 338 while remaining largely unchanged in the drought tolerant variety SA 1441 (Fig. 3B).
Figure 3.
Drought stress-induced gene expression in sorghum roots. Drought-tolerant SA 1441 and drought-sensitive ICSB 338 sorghum plants were exposed to drought for 11 days and gene expression analysed by qRT-PCR. The control plants were not exposed to drought and had a gene expression value set at 1-fold. Error bars represent means ± S.D. (n = 5). One and three asterisks indicate statistically significant differences between the SA 1441 and ICSB 338 means, p ≤ 0.05 and 0.001, respectively.
In leaf tissues, expression of all 6 genes was up-regulated in the drought-tolerant variety SA 1441 (Fig. 4). Thus, at least within this 6 gene selection, SA 1441 recruited all genes in leaf tissues responding to drought, while only half of them responded in the roots. In contrast, ICSB 338 had very marginal or no response across all genes in leaves, while the roots had a very robust upregulation of 3 genes and suppression of the other 3 genes. Collectively, these results demonstrate that candidates selected from our protein dataset are differentially expressed in sorghum lines with contrasting drought responses.
Figure 4.
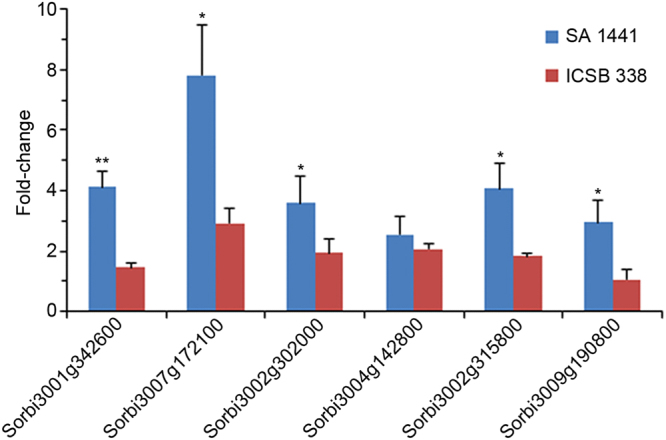
Drought stress-induced gene expression in sorghum leaves. Drought-tolerant SA 1441 and drought-sensitive ICSB 338 sorghum plants were exposed to drought for 11 days and gene expression analysed by qRT-PCR. The control plants were not exposed to drought and had a gene expression value set at 1-fold. Error bars represent means ± S.D. (n = 5). One and two asterisks indicate statistically significant differences between the SA 1441 and ICSB 338 means, p ≤ 0.05 and 0.01, respectively.
Discussion
Drought stress triggers remarkable physiological responses and growth perturbations, which significantly diminish plant biomass and seed yield. These responses are underpinned by changes in gene expression, which are governed by poorly understood signalling processes. As sorghum is a crop that thrives under drought, it is an attractive model crop for gene discovery and studying the mechanisms driving adaptation to drought. Here we used a sorghum cell suspension culture system to obtain fractions enriched for ECM proteins. The ECM is a functional space in which secreted proteins, carbohydrates and other metabolites play a pivotal role in cell growth, cell-cell communication, and responses to changes in environmental factors. A cell culture system is scalable for production of high amounts of secretory molecules for analysis. Moreover, cell cultures are a useful in vitro system, which has been instrumental in key plant science discoveries, such as discovery of the roles of oxidative cross-linking of the cell wall32 or of ROS and nitric oxide33,34 in plant pathogen interactions.
We made three key observations relating to the ECM and sorghum adaptive responses to drought stress. First, there was an overall increase in protein secretion when cells were exposed to osmotic stress. Secretion of over 50% of the soluble ECM proteins identified in this study was upregulated by ≥2-fold. Similarly, an increase in protein secretion was observed in chickpea cell cultures responding to polyethylene glycol treatment35. Previous studies have demonstrated that increased protein secretion is essential for mounting a defensive response to pathogen attack36,37. Because most pathogens invade the ECM space, secretion of a cocktail of antimicrobial proteins is essential in terminating the attack. The surge in protein secretion in response to osmotic stress appears to suggest a key role for the ECM in drought adaptive responses. This might be important, particularly in switching metabolism from optimal growth to stress adaptation. Upon sensing soil water deficits, shoot growth is suppressed and resources are funnelled towards root growth in pursuit of the receding ground water. Programmed cell death may be invoked to kill off root meristems to break apical dominance38 as a strategy to redirect root growth away from water-depleted zones towards available water gradients. The changes in protein expression observed here constitute part of the gene network underpinning these physiological and morphological changes. Proteins are part of the molecular cargo exported into the plant ECM to build the cell wall infrastructure, decorate the external face of the plasma membrane with receptor complexes, and regulate cell division and differentiation30,39. The heightened protein secretion triggered by osmotic stress could play a crucial role in mediating the changes in growth and cellular physiology associated with drought.
The second key finding relates to identification of specific differentially expressed ECM proteins. These fell into four broad functional categories, namely glycosyl-hydrolases/glycosidases, cell wall modifying enzymes, proteases, and redox proteins. Glycosyl-hydrolases/glycosidases are known carbohydrate metabolising enzymes and have diverse substrate specificity40,41. In this study, we identified 18 hydrolases from different families, indicating the wide spectrum of substrate specificity and mechanisms of action. Although the precise role of these enzymes in osmotic stress response is not clear, carbohydrates are important biomolecules, which have structural42 and signalling40 functions. Interestingly, none of these glycosyl hydrolases/glucosidases identified in the present secretome study were reported in a sorghum drought study, which focused on the leaf proteome28. However, glycosyl-hydrolases/glycosidases have also been identified in secretome studies of Arabidopsis responding to both pathogen attack43 and nutritional phosphate deficiency44. A computational functional annotation study attempted to assign putative functions to the 238 uncharacterised sorghum glycoside hydrolases, with stress response functions being ascribed to these enzymes45.
There were 5 cell wall modifying proteins that responded to osmotic stress, which included putative expansin-like and fascilin-like protein families (Table 1). Expansins are known extracellular proteins involved in remodelling cell walls by facilitating cell wall relaxation and extension46; while fascilin domain containing proteins may be involved in cell adhesion processes47. Expansins have been identified in rice secretome studies exposed to rice blast fungus and elicitor48, while a fascilin-like arabinogalactan protein was identified in Arabidopsis secretome following pathogen infection43. Our study indicates that the role for these proteins span several types of plant stress.
Of particular note was the increased secretion of proteases and redox proteins. The identified proteases are putative members of the peptidase, serine carboxypeptidase, aspartic peptidase, gamma-glutamyl-transpeptidase and peptidase subtilisin-related protein families. Proteolytic cleavage of proteins and peptides could be useful in regulating enzyme activity49 and post-translational activation of peptide signals via cleavage of inhibitory domains of pro-peptides50,51. Deployment of these signal regulatory proteins could play critical roles during stress adaptation. Proteolysis could also function in the control of protein turnover, which becomes critical during stress response52. These enzymes have also been identified in previous secretome studies31,44. Several redox proteins, including peroxidases and thioredoxin had increased secretion after imposition of osmotic stress. Peroxidases are important in cell wall lignification53, but are also part of a large protein network that controls the homeostasis of ROS. At low concentration, ROS serve a signalling role54,55, but function in cell death activation at high concentration55,56. Thioredoxin is a molecular switch used for regulating enzyme activity via reducing disulphide bridges linking cycteine residues57,58. Overall, our results indicate that ECM protein networks could play very wide-ranging functions in drought stress adaptive responses.
The third key observation we made was that genes encoding selected candidate proteins are differentially expressed between drought-tolerant and “drought-sensitive” sorghum varieties exposed to drought. We found that selected genes are transcriptionally regulated by sorbitol-induced osmotic stress in the in vitro cell suspension culture system. Analysis of these genes in sensitive versus drought-tolerant sorghum varieties exposed to drought revealed significant differences in expression profiles. Drought activation of gene expression in the sensitive sorghum line was limited to 3 genes mainly in root tissues, with very modest changes in leaves (Figs 3 and 4). Although activation of the same 3 genes in roots of the drought-tolerant SA 1441 was lower than in ICSB 338, the latter also activated all six genes in leaves (Fig. 4). This may indicate that successful drought tolerance requires adaptive gene expression in both subterranean and aerial plant organs. Future genetic experiments could provide functional data of single or multiple genes in adaptive responses to drought. Collectively, these expression profiles indicate two things. First, that the ECM could provide targets for use in enhancing drought tolerance in crops. Because the response of the sorghum varieties to drought differs from each other, then genes/proteins differing in their response to drought between the two varieties could be potential key regulators of drought adaptation. Second, that datasets of differentially expressed ECM proteins under osmotic stress may provide biomarkers that could be used in breeding programmes to rapidly identify drought-tolerant and sensitive varieties.
In conclusion, the ECM is replete with proteins involved in cell growth control, cell communication and cell signalling during responses to environmental stress. A wide range of plant species and experimental systems has been used to study the ECM proteins, including sorghum. This study extends the number of proteins identified in the sorghum ECM from 14 proteins29 to 179 proteins (Supplementary Dataset - Table S2). A large proportion of these (∼72%) possess a predicted signal peptide (Supplementary Dataset - Table S2), which targets them to the secretory pathway59, while the remainder do not possess a signal peptide. This raises the concern of whether the apparent increase in secretion of some of these proteins actually arises from sorbitol-induced cell death and release of intracellular proteins. However, we discount this possibility on the basis of three observations. First, all the proteins identified with increased abundance after sorbitol treatment were also identified in the stress-free control cell cultures. Their secretion in exponentially growing viable control cultures makes cell death an unlikely cause. Secondly, if cell death was responsible, we would have expected to identify many abundant cytosolic house-keeping proteins appearing in sorbitol samples only and absent from control samples. This was not the case. Finally, sorbitol at the concentrations used and time-scale of treatment causes cells to lose water and shrink, with no reduction in cell viability (data not shown). Proteins without a signal peptide identified in our ECM fractions add to the growing number of animal and plant proteins, which are secreted into the ECM via alternative mechanisms not requiring the signal peptide60,61. For example, a leaderless CaRRP1 protein has been confirmed to be a bona fide ECM protein using a YFP-tagged recombinant version of the protein35. Increased secretion of both signal peptide-containing and leaderless proteins is a strong indication that the ECM protein network is part of the molecular machinery deployed when sorghum encounters deficits in soil water content. Importantly, differential expression of some of the target proteins between drought-tolerant and drought-sensitive sorghum varieties implicates the candidates in mediating drought tolerance, though genetic experiments will be required to definitively confirm this.
Methods
Plant material and growth conditions
Seeds of white sorghum previously used for the generation of cell suspension cultures62 were obtained from Professor Bongani Ndimba (Agricultural Research Council (ARC), South Africa). In this study, white sorghum callus and cell suspension cultures were initiated and maintained on Murashige and Skoog Minimal Organics medium under dark conditions as described previously62. The cell cultures were sub-cultured every two weeks and used for sorbitol-induced osmotic stress treatments 8 days post sub-culture. Drought-tolerant (SA 1441) and drought-sensitive (ICSB 338) sorghum varieties were obtained from the ARC-Grain Crop Institute, Potchefstroom, South Africa. Sorghum seeds were sown in potted soil and grown at 25–30 °C under a 16 h-photoperiod. Plants were grown in square pots with a volume of 216 cm3 filled with Levington F2 + Sand compost (ICL Ltd., Ipswich, UK). All plants were well watered until they reached the V3 stage (3 fully expanded leaves with the fourth one emerging) before imposing drought stress by cessation of watering.
Osmotic and drought stress treatments
Eight days after subculturing, sorghum cell suspension cultures were exposed to osmotic stress by treating with 400 mM sorbitol. Control cell cultures were spiked with an equivalent volume of sterile distilled water for the same duration. A time-course sorbitol treatment experiment of sorghum cell suspension cultures was carried over a 72 h period, and expression analysis of drought marker genes ERD1 (early responsive to dehydration 1) and DREB2A was analysed at 0, 24, 48 and 72 h in order to establish the most appropriate time for proteome analysis. For protein analysis, 4 biological replicates of 30 mL each were treated with sorbitol and harvested 48 h later. For RNA analysis, 3 biological replicates of 10 mL each were treated and harvested 0, 2, 4, 6, and 24 h later. For drought stress treatments, well-watered plants at the V3 stage were divided into two groups; control and drought stressed plants. The control plants were watered throughout the experiment as necessary, while water was withheld for 11 days from the drought-stressed plants. Five biological replicates were generated for each group. For leaf samples, each biological replicate was a pool of 3 leaves, each coming from an independent plant. For root samples, a biological replicate consisted of roots pooled from 2 plants. The leaf or root material was snap-frozen in liquid nitrogen and stored at −80 °C for use in RNA extraction.
RNA extraction and analysis
Total RNA was extracted from cell cultures, root and leaf samples using RNeasy Plant Kits (Qiagen, Manchester, UK) according to the manufacturer’s instructions. First strand complementary DNA (cDNA) synthesis was performed using 1.5 µg total RNA template and oligo-(dT)15 using the GoScriptTM Reverse Transcription System (Promega, Southampton, UK) according to the manufacturer’s instructions. Quantitative real-time PCR (qRT-PCR) was performed on the Rotar-Gene 3000 (Corbett Research, Sydney, Australia) using the SensiFAST™ SYBR® No-ROX kit (Bioline, London, UK). The reaction consisted of 10 µl SensiFAST reagent, 0.4 µM each of the forward and reverse primers, and 5 µl of 8-fold dilution cDNA in a final volume of 20 µl. The thermal cycling conditions were as follows: denaturation at 95 °C for 3 min followed by 40 cycles of 95 °C for 10 sec, annealing at 56 °C for 15 sec and extension at 72 °C for 25 sec. All reactions were carried out on 3 technical replicates for each of the biological replicate. Data analysis was carried out using the REST2009 version 2.0.13 software (Qiagen) using Sb03g038910 as a constitutive reference control gene, whose expression does not alter in response to drought stress27. The primer sequences of all genes used are listed in supplementary Table S1.
Protein sample preparation and iTRAQ Labelling
Control and sorbitol-treated cell cultures were filtered through 2 layers of Miracloth to separate the cells from the growth medium. Secreted proteins were isolated from the growth medium by acetone precipitation as described previously63 and solubilised in a solution containing 9 M urea, 2 M thiourea and 4% (w/v) CHAPS. There were 4 biological replicates of controls and the same for sorbitol treatments. Labelling of protein samples with iTRAQ tags was performed as described previously63 with minor modifications. Briefly, for each sample, 50 μg of protein were reduced with tris(2-carboxyethylphosphine) (TCEP) and alkylated with methyl-methane-thiol-sulfonate (MMTS). Thereafter, protein samples were digested at 37 °C for ~16 h using a 1:10 (w/w) trypsin to protein sample ratio, vacuum-dried, re-suspended in triethylammonium bicarbonate buffer (pH 8.5), and labelled with an 8-plex iTRAQ reagent kit (Applied Biosystems Sciex, Foster City, USA) according to the manufacturer’s instructions.
Peptides of the 4 control replicates were labelled with 113, 114, 115, and 116 iTRAQ tags, while sorbitol-treated samples were labelled with 117, 118, 119, and 121 tags. All eight samples were pooled to make one composite sample, which was then vacuum-dried and re-suspended in 3.8 mL of buffer A (10 mM K2HPO4/25% acetonitrile, pH 3.0). Thereafter, the sample was separated into 50 fractions on a PolySULFOETHYL A strong cation exchange column (Poly LC Inc. 200 × 2.1 mm, 5 μm) at 300 nL/min on an Ettan LC (GE Healthcare, Pittsburgh, USA) HPLC system. Peptide separation was performed using a biphasic gradient of: 0–150 mM KCl over 11.25 column volumes and 150–500 mM KCl in buffer A over 3.25 column volumes. A total of 50 fractions were collected over the gradient, and reduced to 30 by pooling those with low peptide concentration. The 30 fractions were dried down and re-suspended in 90 μL of 2% acetonitrile/0.1% formic acid. Aliquots of 20 μL from each fraction were analysed by LC-MS/MS using a QStar Pulsar i mass spectrometer (MDS-Sciex/Applied Biosystems).
Mass spectra data analysis
Mass spectra data were analysed as described previously63, with minor modifications. Briefly, ProteinPilot software 4.5 (Beta) Revision 1656 Paragon algorithm build 1654 (ABSciex) was used for data analysis against the UniProt database sequences for S. bicolor (downloaded in October 2013, 58756 entries) plus 162 known contaminants from proteomic experiments. A minimum score threshold of 2.0 (99% confidence) was set for protein identification and all proteins identified on the basis of a single peptide were filtered out of the dataset, resulting in a total of 179 unique proteins.
For quantitative analysis of the differentially expressed proteins, the abundance of each protein in all samples was calculated as a ratio to the 113-tagged control sample. Averages of the ratios for each protein across the four replicates were calculated. The fold-change in protein expression was denoted by the ratio of control to sorbitol-treated samples. For the down-regulated proteins, the osmotic stressed average was the numerator and the control was the denominator, with a negative sign denoting down-regulation. A probability value for the comparison of the control average to sorbitol average was obtained from the Student’s t-test at 95% confidence.
Bioinformatic analysis
The presence of an N-terminal signal peptide on all identified proteins was predicted using SignalP 4.164. The InterPro65 and Superfamily66 databases were used for protein sequence analysis to identify protein functional domains used for assignments to relevant protein families.
Data availability statement
The datasets generated and/or analysed during the current study are available from the corresponding author on request.
Electronic supplementary material
Acknowledgements
This research was supported by the National Research Foundation-South Africa grant 93612 to the R.N. group, Biotechnology and Biological Sciences Research Council grants (BB/N012623/1 and BB/MO28429/1) to the S.C. group, and the Royal Society-Newton Advanced Fellowship grant NA160140 jointly awarded to the R.N. and S.C. groups. E.R. was supported by National Research Foundation and Agricultural Research Council student bursaries. We thank Colleen Turnbull for technical assistance in the experimental setup.
Author Contributions
R.N. and S.C. designed the experiments, analysed data and wrote the manuscript; S.C. and E.R. conducted experiments; M.M. analysed qRT-PCR data; N.G.S. conducted the initial sorghum drought field screening experiment and A.P.B. performed mass spectrometric analysis. All authors reviewed the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-27003-1.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Rudo Ngara, Email: NgaraR@ufs.ac.za.
Stephen Chivasa, Email: stephen.chivasa@durham.ac.uk.
References
- 1.Harris MJ, Outlaw WH, Mertens R, Weiler EW. Water-stress-induced changes in the abscisic acid content of guard cells and other cells of Vicia faba L. leaves as determined by enzyme-amplified immunoassay. P. Natl. Acad. Sci. USA. 1988;85:2584–2588. doi: 10.1073/pnas.85.8.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jones RJ, Mansfield TA. Suppression of stomatal opening in leaves treated with abscisic acid. J. Exp. Bot. 1970;21:714–719. doi: 10.1093/jxb/21.3.714. [DOI] [Google Scholar]
- 3.Shinozaki K, Yamaguchi-Shinozaki K. Gene expression and signal transduction in water-stress response. Plant Physiol. 1997;115:327–334. doi: 10.1104/pp.115.2.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Park SY, et al. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science. 2009;324:1068–1071. doi: 10.1126/science.1173041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ma Y, et al. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science. 2009;324:1064–1068. doi: 10.1126/science.1172408. [DOI] [PubMed] [Google Scholar]
- 6.Melcher K, et al. A gate-latch-lock mechanism for hormone signalling by abscisic acid receptors. Nature. 2009;462:602–608. doi: 10.1038/nature08613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miyazono K, et al. Structural basis of abscisic acid signalling. Nature. 2009;462:609–614. doi: 10.1038/nature08583. [DOI] [PubMed] [Google Scholar]
- 8.Nishimura, N. et al. Structural mechanism of abscisic acid binding and signaling by dimeric PYR1. Science326 (2009). [DOI] [PMC free article] [PubMed]
- 9.Santiago J, et al. The abscisic acid receptor PYR1 in complex with abscisic acid. Nature. 2009;462:665–668. doi: 10.1038/nature08591. [DOI] [PubMed] [Google Scholar]
- 10.Yin P, et al. Structural insights into the mechanism of abscisic acid signaling by PYL proteins. Nat. Struct. Mol. Biol. 2009;16:1230–1236. doi: 10.1038/nsmb.1730. [DOI] [PubMed] [Google Scholar]
- 11.Zhang X, et al. Complex structures of the abscisic acid receptor PYL3/RCAR13 reveal a unique regulatory mechanism. Structure. 2012;20:780–790. doi: 10.1016/j.str.2012.02.019. [DOI] [PubMed] [Google Scholar]
- 12.Fujii H, et al. In vitro reconstitution of an abscisic acid signalling pathway. Nature. 2009;462:660–664. doi: 10.1038/nature08599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Soon FF, et al. Molecular mimicry regulates ABA signaling by SnRK2 kinases and PP2C phosphatases. Science. 2012;335:85–88. doi: 10.1126/science.1215106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ng LM, et al. Structural basis for basal activity and autoactivation of abscisic acid (ABA) signaling SnRK2 kinases. P. Natl. Acad. Sci. USA. 2011;108:21259–21264. doi: 10.1073/pnas.1118651109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Okamoto M, et al. Activation of dimeric ABA receptors elicits guard cell closure, ABA-regulated gene expression, and drought tolerance. P. Natl. Acad. Sci. USA. 2013;110:12132–12137. doi: 10.1073/pnas.1305919110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Busk PK, Pages M. Regulation of abscisic acid-induced transcription. Plant Mol. Biol. 1998;37:425–435. doi: 10.1023/A:1006058700720. [DOI] [PubMed] [Google Scholar]
- 17.Abe H, et al. Role of arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. Plant Cell. 1997;9:1859–1868. doi: 10.1105/tpc.9.10.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fujita Y, et al. AREB1 is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance in Arabidopsis. Plant Cell. 2005;17:3470–3488. doi: 10.1105/tpc.105.035659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abe H, et al. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell. 2003;15:63–78. doi: 10.1105/tpc.006130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamaguchi-Shinozaki K, Shinozaki K. A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell. 1994;6:251–264. doi: 10.1105/tpc.6.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Q, et al. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell. 1998;10:1391–1406. doi: 10.1105/tpc.10.8.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sakuma Y, et al. Dual function of an Arabidopsis transcription factor DREB2A in water-stress-responsive and heat-stress-responsive gene expression. P. Natl. Acad. Sci. USA. 2006;103:18822–18827. doi: 10.1073/pnas.0605639103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holmberg N, Bulow L. Improving stress tolerance in plants by gene transfer. Trends Plant Sci. 1998;3:61–66. doi: 10.1016/S1360-1385(97)01163-1. [DOI] [Google Scholar]
- 24.Rosenow DT, Quisenberry JE, Wendt CW, Clark LE. Drought Tolerant Sorghum and Cotton Germplasm. Agr. Water Manage. 1983;7:207–222. doi: 10.1016/0378-3774(83)90084-7. [DOI] [Google Scholar]
- 25.Ngara R, Ndimba BK. Model plant systems in salinity and drought stress proteomics studies: a perspective on Arabidopsis and Sorghum. Plant Biology. 2014;16:1029–1032. doi: 10.1111/plb.12247. [DOI] [PubMed] [Google Scholar]
- 26.Paterson AH, et al. The Sorghum bicolor genome and the diversification of grasses. Nature. 2009;457:551–556. doi: 10.1038/nature07723. [DOI] [PubMed] [Google Scholar]
- 27.Johnson SM, et al. Transcriptomic analysis of Sorghum bicolor responding to combined heat and drought stress. BMC Genomics. 2014;15:456. doi: 10.1186/1471-2164-15-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jedmowski C, et al. Comparative Analysis of Sorghum bicolor Proteome in Response to Drought Stress and following Recovery. Int. J. Proteomics. 2014;2014:395905. doi: 10.1155/2014/395905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ngara R, Ndimba BK. Mapping and characterisation of the sorghum cell suspension culture secretome. Afr. J. Biotechnol. 2011;10:253–266. [Google Scholar]
- 30.Chivasa S, Slabas AR. Plant extracellular ATP signalling: new insight from proteomics. Molecular Biosyst. 2012;8:445–452. doi: 10.1039/C1MB05278K. [DOI] [PubMed] [Google Scholar]
- 31.Ndimba BK, Chivasa S, Simon WJ, Slabas AR. Identification of Arabidopsis salt and osmotic stress responsive proteins using two-dimensional difference gel electrophoresis and mass spectrometry. Proteomics. 2005;5:4185–4196. doi: 10.1002/pmic.200401282. [DOI] [PubMed] [Google Scholar]
- 32.Brisson LF, Tenhaken R, Lamb C. Function of oxidative cross-linking of cell wall structural proteins in plant disease resistance. Plant Cell. 1994;6:1703–1712. doi: 10.1105/tpc.6.12.1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Delledonne M, Xia Y, Dixon RA, Lamb C. Nitric oxide functions as a signal in plant disease resistance. Nature. 1998;394:585–588. doi: 10.1038/29087. [DOI] [PubMed] [Google Scholar]
- 34.Delledonne M, Zeier J, Marocco A, Lamb C. Signal interactions between nitric oxide and reactive oxygen intermediates in the plant hypersensitive disease resistance response. P. Natl. Acad. Sci. USA. 2001;98:13454–13459. doi: 10.1073/pnas.231178298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gupta S, et al. Secretome analysis of chickpea reveals dynamic extracellular remodeling and identifies a Bet v1-like protein, CaRRP1 that participates in stress response. Sci. Rep. 2015;5:18427. doi: 10.1038/srep18427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kwon C, Bednarek P, Schulze-Lefert P. Secretory pathways in plant immune responses. Plant Physiol. 2008;147:1575–1583. doi: 10.1104/pp.108.121566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kwon C, et al. Co-option of a default secretory pathway for plant immune responses. Nature. 2008;451:835–840. doi: 10.1038/nature06545. [DOI] [PubMed] [Google Scholar]
- 38.Duan Y, et al. An endoplasmic reticulum response pathway mediates programmed cell death of root tip induced by water stress in Arabidopsis. New Phytol. 2010;186:681–695. doi: 10.1111/j.1469-8137.2010.03207.x. [DOI] [PubMed] [Google Scholar]
- 39.Wang D, Dong X. A highway for war and peace: the secretory pathway in plant-microbe interactions. Mol. Plant. 2011;4:581–587. doi: 10.1093/mp/ssr053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Davies G, Henrissat B. Structures and mechanisms of glycosyl hydrolases. Structure. 1995;3:853–859. doi: 10.1016/S0969-2126(01)00220-9. [DOI] [PubMed] [Google Scholar]
- 41.Henrissat B, Davies G. Structural and sequence-based classification of glycoside hydrolases. Curr. Opin. Struc. Biol. 1997;7:637–644. doi: 10.1016/S0959-440X(97)80072-3. [DOI] [PubMed] [Google Scholar]
- 42.Garrett, R., Grisham, C. M. & Sabat, M. Biochemistry. 4th. edn, (Brooks/Cole Pub Co, 2010).
- 43.Kaffarnik FA, Jones AM, Rathjen JP, Peck SC. Effector proteins of the bacterial pathogen Pseudomonas syringae alter the extracellular proteome of the host plant, Arabidopsis thaliana. Mol. Cell. Proteomics. 2009;8:145–156. doi: 10.1074/mcp.M800043-MCP200. [DOI] [PubMed] [Google Scholar]
- 44.Tran HT, Plaxton WC. Proteomic analysis of alterations in the secretome of Arabidopsis thaliana suspension cells subjected to nutritional phosphate deficiency. Proteomics. 2008;8:4317–4326. doi: 10.1002/pmic.200800292. [DOI] [PubMed] [Google Scholar]
- 45.Sekhwal MK, Sharma V, Sarin R. Annotation of glycoside hydrolases in Sorghum bicolor using proteins interaction approach. J. Proteome Sci. Comput. Biol. 2013;2:2. doi: 10.7243/2050-2273-2-2. [DOI] [Google Scholar]
- 46.Cosgrove DJ. New genes and new biological roles for expansins. Curr. Opin. Plant Biol. 2000;3:73–78. doi: 10.1016/S1369-5266(99)00039-4. [DOI] [PubMed] [Google Scholar]
- 47.Johnson KL, Jones BJ, Bacic A, Schultz CJ. The fasciclin-like arabinogalactan proteins of Arabidopsis. A multigene family of putative cell adhesion molecules. Plant Physiol. 2003;133:1911–1925. doi: 10.1104/pp.103.031237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim ST, et al. Secretome analysis of differentially induced proteins in rice suspension-cultured cells triggered by rice blast fungus and elicitor. Proteomics. 2009;9:1302–1313. doi: 10.1002/pmic.200800589. [DOI] [PubMed] [Google Scholar]
- 49.Vierstra RD. Proteolysis in plants: mechanisms and functions. Plant Mol. Biol. 1996;32:275–302. doi: 10.1007/BF00039386. [DOI] [PubMed] [Google Scholar]
- 50.McGurl B, Pearce G, Orozco-Cardenas M, Ryan CA. Structure, expression, and antisense inhibition of the systemin precursor gene. Science. 1992;255:1570–1573. doi: 10.1126/science.1549783. [DOI] [PubMed] [Google Scholar]
- 51.Lindsey K. Plant peptide hormones: The long and the short of it. Curr. Biol. CB. 2001;11:R741–743. doi: 10.1016/S0960-9822(01)00435-3. [DOI] [PubMed] [Google Scholar]
- 52.Liu H, et al. A rice serine carboxypeptidase-like gene OsBISCPL1 is involved in regulation of defense responses against biotic and oxidative stress. Gene. 2008;420:57–65. doi: 10.1016/j.gene.2008.05.006. [DOI] [PubMed] [Google Scholar]
- 53.Hiraga S, Sasaki K, Ito H, Ohashi Y, Matsui H. A large family of class III plant peroxidases. Plant Cell Physiol. 2001;42:462–468. doi: 10.1093/pcp/pce061. [DOI] [PubMed] [Google Scholar]
- 54.Hancock JT, Desikan R, Neill SJ. Role of reactive oxygen species in cell signalling pathways. Biochemical Soc. Trans. 2001;29:345–350. doi: 10.1042/bst0290345. [DOI] [PubMed] [Google Scholar]
- 55.Desikan R, Reynolds A, Hancock JT, Neill SJ. Harpin and hydrogen peroxide both initiate programmed cell death but have differential effects on defence gene expression in Arabidopsis suspension cultures. Biochemical J. 1998;330:115–120. doi: 10.1042/bj3300115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Levine A, Pennell RI, Alvarez ME, Palmer R, Lamb C. Calcium-mediated apoptosis in a plant hypersensitive disease resistance response. Curr. Biol. 1996;6:427–437. doi: 10.1016/S0960-9822(02)00510-9. [DOI] [PubMed] [Google Scholar]
- 57.Arner ES, Holmgren A. Physiological functions of thioredoxin and thioredoxin reductase. Eur. J. Biochem. 2000;267:6102–6109. doi: 10.1046/j.1432-1327.2000.01701.x. [DOI] [PubMed] [Google Scholar]
- 58.Meyer Y, et al. Glutaredoxins and thioredoxins in plants. Biochim. Biophys. Acta. 2008;1783:589–600. doi: 10.1016/j.bbamcr.2007.10.017. [DOI] [PubMed] [Google Scholar]
- 59.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 60.Krause C, Richter S, Knoll C, Jurgens G. Plant secretome - from cellular process to biological activity. Biochim. Biophys. Acta. 2013;1834:2429–2441. doi: 10.1016/j.bbapap.2013.03.024. [DOI] [PubMed] [Google Scholar]
- 61.Ding Y, Robinson DG, Jiang L. Unconventional protein secretion (UPS) pathways in plants. Curr. Opin. Cell Biol. 2014;29:107–115. doi: 10.1016/j.ceb.2014.05.008. [DOI] [PubMed] [Google Scholar]
- 62.Ngara R, Rees J, Ndimba BK. Establishment of sorghum cell suspension culture system for proteomics studies. Afr. J. Biotechnol. 2008;7:744–749. [Google Scholar]
- 63.Smith SJ, Kroon JT, Simon WJ, Slabas AR, Chivasa S. A Novel Function for Arabidopsis CYCLASE1 in Programmed Cell Death Revealed by Isobaric Tags for Relative and Absolute Quantitation (iTRAQ) Analysis of Extracellular Matrix Proteins. Mol. Cell. Proteomics. 2015;14:1556–1568. doi: 10.1074/mcp.M114.045054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nature Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 65.Mulder, N. J. et al. New developments in the InterPro database. Nucleic Acids Res. 35 (2007). [DOI] [PMC free article] [PubMed]
- 66.Wilson, D. et al. SUPERFAMILY–sophisticated comparative genomics, data mining, visualization and phylogeny. Nucleic Acids Res. 37 (2009). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analysed during the current study are available from the corresponding author on request.