Abstract
The collected data have revealed the beneficial effects of dipeptidyl peptidase-4 (DPP-4) inhibitors on the vascular endothelium, including vildagliptin. However, the involved mechanisms are not yet clear. In this study, Sprague-Dawley rats were randomly divided into the following four groups: control, diabetic, diabetic + low-dose vildagliptin (10 mg/kg/d), and diabetic + high-dose vildagliptin (20 mg/kg/d). The diabetic model was created by feeding a high-fat diet for four weeks and injection of streptozotocin. Then, vildagliptin groups were given oral vildagliptin for twelve weeks, and the control and diabetic groups were given the same volume of saline. The metabolic parameters, endothelial function, and whole genome expression in the aorta were examined. After 12 weeks of treatment, vildagliptin groups showed significantly reduced blood glucose, blood total cholesterol, and attenuated endothelial dysfunction. Notably, vildagliptin may inhibit angiopoietin-like 3 (Angptl3) and betaine-homocysteine S-methyltransferase (Bhmt) expression and activated paraoxonase-1 (Pon1) in the aorta of diabetic rats. These findings may demonstrate the vasoprotective pathway of vildagliptin in vivo.
1. Introduction
The incidence and prevalence of diabetes mellitus are dramatically increasing worldwide [1]. Epidemiological research shows that diabetic patients have a higher risk of cardiovascular diseases [2]. The main cause of morbidity and mortality in diabetic patients is cardiovascular diseases.
Glucagon-like peptide-1 (GLP-1) is produced in gut L-cells. It contains 30 amino acids. The main physiological function of GLP-1 is stimulation of insulin secretion from pancreatic β cells when glucose is orally taken up in the human body. GLP-1 can also inhibit glucose production and appetite, activate adipose and muscle glucose uptake and storage, and thus moderate insulin sensitivity. However, GLP-1 is quickly hydrolyzed by dipeptidyl peptidase-4 (DPP-4).
DPP-4 inhibitors are a new class of GLP-1 based antidiabetic drugs. As one type of DPP-4 inhibitors, vildagliptin controls blood glucose by inhibiting the enzymatic activity of DPP-4. DPP-4 is also found on endothelial cells in the cardiovascular system, and increasing research has focused on the benefit of DPP-4 inhibitors on cardiovascular function. Sitagliptin (one type of DPP-4 inhibitor) has been proven to significantly attenuate heart failure-related left ventricular (LV) end-diastolic pressure, systolic performance, and chamber stiffness in an ablation-induced cardiac dysfunction rat model [3]. In a clinical trial, sitagliptin enhanced global and regional LV function in type 2 diabetes mellitus (T2DM) patients with coronary artery [4]. Vildagliptin exerts cardioprotective effects in obesity-based insulin resistance [5, 6], myocardial infarction (MI) [7], and ischemia-reperfusion (I/R) injury rat models [8]. However, the exact mechanism of the beneficial effect of vildagliptin on the aorta in diabetic rats remains to be elucidated.
In this study, we hypothesized that vildagliptin improved aorta function through multiple pathways. We employed a whole genomic expression array and bioinformatics method to explore the pathway involved in aorta vascular function moderation in diabetic rats.
2. Materials and Methods
2.1. Animal Treatments and Diets
Five-week-old male Sprague-Dawley rats were obtained from the Institute of Laboratory Animal Science, Chinese Academy of Medical Sciences, and Peking Union Medical College (Beijing, China, SCXK-2014-0013). The animal protocol was approved by the Animal Care Committee of the Peking Union Medical Hospital Animal Ethics Committee (Project XHDW-2015-0051, 15 Feb 2015), and all efforts were made to minimize suffering. All the rats were fed in a light/dark cycle (12 hours : 12 hours) environment and were free to drink water. Three days after arrival, rats were randomly divided into four groups (n = 6 per group): normal control group, diabetic group, low-dose vildagliptin (vil-low), and high-dose vildagliptin (vil-high). The normal control group was fed a standard rodent diet (kcal%: 10% fat, 20% protein, and 70% carbohydrate; 3.85 kcal/gm). Other groups were fed a high-fat diet (kcal%: 45% fat, 20% protein, and 35% carbohydrate; 4.73 kcal/gm, Research Diet, New Brunswick, NJ, USA). After 4 weeks, diabetic, low-dose vildagliptin, and high-dose vildagliptin groups were given a single injection of streptozotocin (STZ, 30 mg/kg body weight, i.p., Sigma-Aldrich, St. Louis, MO, USA). Fasting blood glucose > 11.1 mmol/L was the standard for the diabetic model. Then, vil-low and vil-high groups were treated with 10 mg or 20 mg vildagliptin (Novartis Pharma AG, Basel, Switzerland)/kg of body weight by daily gavage for 12 weeks. Normal control and diabetic groups were given normal saline. After 12 weeks of treatment, the rats were anesthetized using ketamine (100 mg/kg i.p., Pharmacia and Upjohn Ltd., Crawley, UK), followed by withdrawal of food overnight. Blood samples were obtained from the abdominal aorta. Then, the rats were sacrificed by decapitation. The thoracic aorta was quickly removed. Some aortas were placed in Krebs solution (120 mmol/L of NaCl, 4.7 mmol/L of KCl, 1.18 mmol/L of KH2PO4, 2.25 mmol/L of CaCl2, 24.5 mmol/L of NaHCO3, 1.2 mmol/L of MgSO4·7H2O, 11.1 mmol/L of glucose, and 0.03 mmol/L of EDTA) and aerated with 95% O2 and 5% CO2. Other aortas were frozen in liquid nitrogen and stored at −80°C for a gene microarray experiment.
2.2. Body Weight and Fasting Blood Glucose Measurements
The rats were weighed every 4 weeks. Fasting blood glucose levels were measured by Bayer Contour TS glucometer (Hamburg, Germany).
2.3. Oral Glucose Tolerance Test (OGTT)
An OGTT was performed after 12 weeks of treatment. Blood glucose levels were measured at 30, 60, and 120 min after an oral administration of 20% glucose at a dose of 2 g/kg. The area under the curve (AUC) was calculated by the linear trapezoid method [9].
2.4. Serum Insulin and Lipid Panel Measurements
Serum fasting insulin was analyzed using an ELISA kit (Millipore, Billerica, MA, USA). The homeostasis model assessment of insulin resistance (HOMA-IR) was calculated by the following formula: FBG (mmol/L) × fasting insulin (μIU/mL)/22.5. Serum total cholesterol (TC), triglyceride (TG), high-density lipoprotein (HDL), and low-density lipoprotein (LDL) were measured using an enzyme end-point kit (Roche Diagnostics GmbH, Mannheim, Germany).
2.5. Isometric Contractile Tension Assay
Isometric contractile tension was determined as follows and described previously [10]. The aortic ring segments (3 mm width) were incubated with Krebs solution (120 mmol/L of NaCl, 4.7 mmol/L of KCl, 1.18 mmol/L of KH2PO4, 2.25 mmol/L of CaCl2, 24.5 mmol/L of NaHCO3, 1.2 mmol/L of MgSO4·7H2O, 11.1 mmol/L of glucose, and 0.03 mmol/L of EDTA), aerated with 95% O2 and 5% CO2, and then preconstricted with phenylephrine (Phe, 10−7 mmol/L). Isometric contractile tension was measured by a BL-410 Biological function system (Chengdu Tai Meng Science and Technology Co., Ltd., Chengdu, China). Once a basal contraction was obtained, 10−10 to 10−4 mol/L of acetylcholine (Ach) was cumulatively added to the solution.
2.6. RNA Extraction, Amplification, Labeling, and Hybridization
To look for the differentially expressed genes under vildagliptin treatment in diabetic rats, we performed gene whole transcript-based array in vil-high group and diabetic group (n = 3 in each group). Total RNA was extracted from aortas using a mirVana™ RNA Isolation Kit (Ambion, San Paulo, SP, Brazil). The RNA was purified using an RNeasy Kit (Qiagen, Hilden, Germany), quantified by NanoDrop ND-2000 spectrophotometry (Nanodrop Tech, Rockland, Del, Wilmington, DE, USA), and qualified by agarose gel electrophoresis. Total RNA (100 ng) was used for cDNA synthesis. cRNA was synthesized followed by two-strand cDNA. Biotin-labeled cRNA was hybridized to an Affymetrix GeneChip Rat Gene 2.0 ST whole transcript-based array (Affymetrix Technologies, Santa Clara, CA, USA). After hybridization, the microarrays were washed, stained, and scanned with an Affymetrix Scanner 3000 7G (Santa Clara, CA, USA).
The microarray signals were analyzed using Expression Console software (version 1.4.1, Affymetrix, Santa Clara, CA, USA). The significance of the difference in genes was determined by one-way ANOVA. Differentially expressed genes were defined as genes with a fold change > 2.0 and P < 0.05. The microarray raw data were submitted to the Gene Expression Omnibus (GEO) repository (GSE102196).
2.7. Bioinformatics Analysis for Microarray
DAVID (Database for Annotation, Visualization and Integrated Discovery) software (https://david.abcc.ncifcrf.gov/) [11] was used to perform gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses. The Search Tool for the Retrieval of Interacting Genes (STRING) database (https://string-db.org/) [12] was used to analyze the interaction network for differentially expressed genes.
2.8. Validation of Differentially Expressed Genes by Quantitative PCR
The expression levels of three differentially expressed genes were measured by qPCR to validate the results of the microarray. All primers are listed in Table 1. Actin was used as an internal control.
Table 1.
Oligonucleotide sequences for qPCR analysis.
| Gene symbol | GenBank ID | Forward primer | Reverse Primer | Product size |
|---|---|---|---|---|
| Angptl3 | NM_001025065 | AAAGGGTTTTGGGAGGCTTGA | CCCAAAAGCGCTATGGTCTC | 117 |
| Bhmt | NM_030850 | GATGCTTGGGGAGTGACGAA | TGTGGCTACTGTGCGGATTT | 119 |
| Pon1 | NM_032077 | AAGCTGGCTACACCCACATC | CAACATTCGTTGGTGAGCGG | 103 |
Angptl3: angiopoietin-like 3; Bhmt: betaine-homocysteine S-methyltransferase; Pon1: paraoxonase 1.
2.9. Statistical Analyses
Data are presented as the means ± standard deviation (SD). One-way ANOVA analysis followed by Student's t-test was used to compare differences among groups. P < 0.05 was considered significant.
3. Results
3.1. Vildagliptin Moderated Blood Glucose and Insulin Resistance
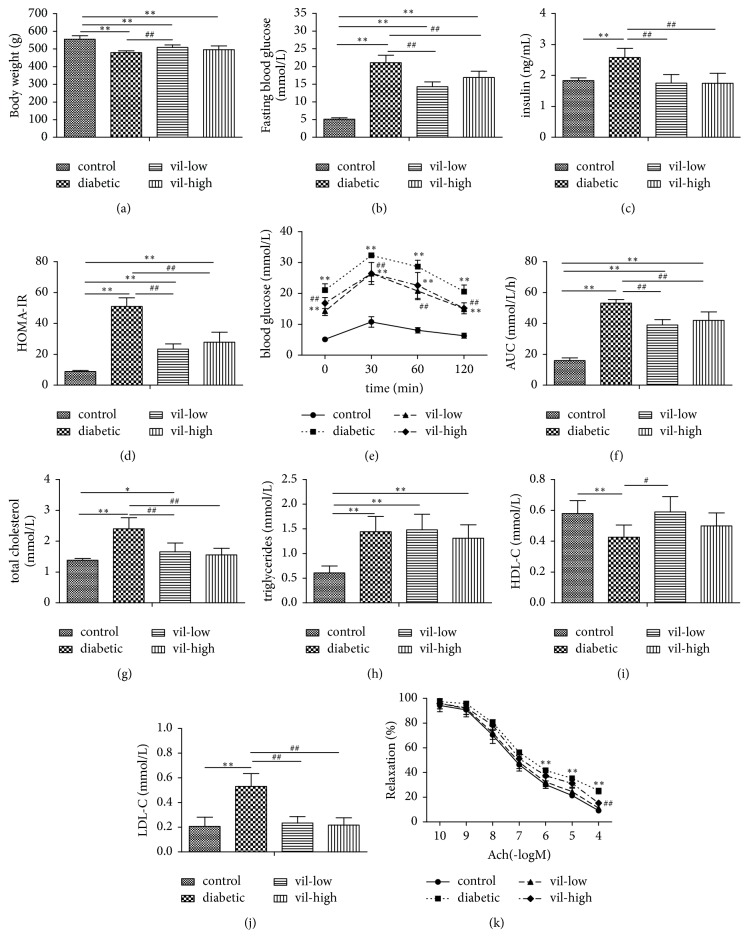
After 12 weeks of treatment, vildagliptin significantly reduced fasting blood glucose AUC in OGTT (P < 0.05, Figures 1(b) and 1(f)). Additionally, fasting serum insulin and HOMA-IR index in the vildagliptin-treated group were lower than in the diabetic group (P < 0.05, Figures 1(c) and 1(d)).
Figure 1.
Effect of vildagliptin on metabolic parameters in rats. (a) Body weight, (b) fasting blood glucose, (c) insulin, (d) homeostasis model assessment (HOMA-IR), (e) blood glucose in oral glucose tolerance test (OGTT), (f) area under the curve (AUC) in OGTT, (g) total cholesterol, (h) triglyceride, (i) high-density lipoprotein cholesterol (HDL-C), (j) low-density lipoprotein cholesterol (LDL-C), and (k) relaxation responses to Ach in aortic rings. Values are mean ± SD (n = 6), ∗P < 0.05, ∗∗P < 0.01, versus control group; #P < 0.05, ##P < 0.01 versus diabetic group. vil-low: low dose of vildagliptin; vil-high: high dose of vildagliptin.
3.2. Vildagliptin Decreased Blood Lipid Levels
Vildagliptin decreased TC and LDL-C levels in diabetic rats (P < 0.01, Figures 1(g) and 1(j)).
3.3. Vildagliptin Depressed Vasorelaxation Responses of Aortic Rings to Ach
As shown in Figure 1(k), endothelium-dependent vasodilation was impaired in diabetic rats compared with normal rats. Treatment with vildagliptin ameliorated this impairment (P < 0.01).
3.4. Identification of Differentially Expressed Genes
After filtering genes that had a cutoff of a 1.5-fold change or greater, we found that 150 genes were upregulated and 120 genes were downregulated in the vil-high group compared with the diabetic group (P < 0.05).
3.5. GO Term Enrichment Analysis of Differentially Expressed Genes
In general, 91 GO terms were significantly enriched (P < 0.05, Table 2). The top 20 most significant biological process (BP) terms are shown in Figure 2 (P < 0.01). They were negative regulation of endopeptidase activity, epoxygenase P450 pathway, acute-phase response, blood coagulation, oxidation-reduction processes, cytoskeleton organization, negative regulation of fibrinolysis, microtubule-based processes, phosphatidylcholine metabolic processes, fibrinolysis, organ regeneration, complement activation classical pathway, inflammatory response, positive regulation of lipid catabolic process, triglyceride homeostasis, aging, cellular response to interferon gamma, cholesterol metabolic process, cholesterol homeostasis, and tissue regeneration.
Table 2.
The enriched GO terms with differentially expressed genes (P < 0.05).
| Term ID | Term name | Count | P-value | Fold Enrichment | catalogue | Genes |
|---|---|---|---|---|---|---|
| GO:0010951 | negative regulation of endopeptidase activity | 16 | 7.22 × 10−10 | 8.425 | Biology Process | KNG1, MUG2, MUG1, SERPINA10, PZP, C5, AHSG, AMBP, SERPINA3N, SERPINF2, SERPINA4, SERPINC1, ITIH4, HRG, ITIH2, SERPIND1 |
| GO:0019373 | epoxygenase P450 pathway | 7 | 5.88 × 10−7 | 21.821 | Biology Process | CYP2C6V1, CYP2B3, CYP2J4, CYP2C23, CYP2C13, CYP2A1, CYP2A2 |
| GO:0006953 | acute-phase response | 7 | 7.97 × 10−6 | 14.356 | Biology Process | KNG1, MUG1, CRP, ITIH4, SAA4, SERPINA1, AHSG |
| GO:0007596 | blood coagulation | 8 | 2.08 × 10−5 | 9.446 | Biology Process | F13B, F12, SERPINA10, C9, F9, SERPIND1, CPB2, PLG |
| GO:0055114 | oxidation-reduction process | 23 | 3.29 × 10−5 | 2.753 | Biology Process | ALDH8A1, CYP2B3, CYP2J4, HSD3B5, DECR2, RGD1304810, CYP2D3, EGLN1, KMO, P4HTM, VAT1, IYD, CYP2C6V1, TDO2, CYP3A23/3A1, HIF1AN, CYP4F4, CYP2C23, CYP2C13, SH3BGRL3, RDH16, CYP2A1, CYP2A2 |
| GO:0007010 | cytoskeleton organization | 10 | 3.95 × 10−5 | 6.089 | Biology Process | CCL24, TNIK, CAP2, TUBB2A, SVIL, CAMSAP1, STRIP2, TUBB6, TUBA1A, TUBB4B |
| GO:0051918 | negative regulation of fibrinolysis | 4 | 6.93 × 10−5 | 44.533 | Biology Process | SERPINF2, HRG, CPB2, PLG |
| GO:0007017 | microtubule-based process | 6 | 1.68 × 10−4 | 11.405 | Biology Process | TUBB2A, TUBB5, TUBB6, TUBA1A, DCTN1, TUBB4B |
| GO:0046470 | phosphatidylcholine metabolic process | 4 | 6.75 × 10−4 | 22.267 | Biology Process | APOA5, PON1, GPLD1, LIPC |
| GO:0042730 | fibrinolysis | 4 | 8.35 × 10−4 | 20.782 | Biology Process | F12, HRG, CPB2, PLG |
| GO:0031100 | organ regeneration | 7 | 0.00110 | 5.995 | Biology Process | APOA2, BAAT, ADH1, ALDOC, APOA5, PRPS2, AHSG |
| GO:0006958 | complement activation, classical pathway | 5 | 0.00123 | 10.532 | Biology Process | C9, C5, CRP, C4BPB, C4BPA |
| GO:0006954 | inflammatory response | 12 | 0.00158 | 3.149 | Biology Process | CCL24, KNG1, TLR10, SERPINA3N, TNFRSF10B, MUG1, CCL21, C5, HRH4, CCL9, SERPINA1, CXCR3 |
| GO:0050996 | positive regulation of lipid catabolic process | 3 | 0.00236 | 38.967 | Biology Process | APOA2, APOA5, ANGPTL3 |
| GO:0070328 | triglyceride homeostasis | 4 | 0.00431 | 11.990 | Biology Process | APOC4, APOA5, LIPC, ANGPTL3 |
| GO:0007568 | aging | 11 | 0.00745 | 2.721 | Biology Process | ADRB3, CYP3A23/3A1, CRYAB, ENDOG, ALDOC, CRP, SPINK1, IGFBP1, ATP5G3, SREBF2, HTR2A |
| GO:0071346 | cellular response to interferon-gamma | 5 | 0.00766 | 6.388 | Biology Process | CCL24, SERPINA3N, CCL21, CCL9, CFH |
| GO:0008203 | cholesterol metabolic process | 5 | 0.00811 | 6.285 | Biology Process | APOA2, PON1, LIPC, ANGPTL3, SREBF2 |
| GO:0042632 | cholesterol homeostasis | 5 | 0.00906 | 6.089 | Biology Process | CAV3, APOA2, APOA5, LIPC, ANGPTL3 |
| GO:0042246 | tissue regeneration | 4 | 0.00996 | 8.907 | Biology Process | SERPINA10, APOA5, IGFBP1, PLG |
| GO:0046330 | positive regulation of JNK cascade | 5 | 0.0112 | 5.730 | Biology Process | DIXDC1, CCL21, SERPINF2, MAP3K10, TPD52L1 |
| GO:0050921 | positive regulation of chemotaxis | 3 | 0.0134 | 16.700 | Biology Process | CCL21, C5, CXCR3 |
| GO:0006956 | complement activation | 3 | 0.0153 | 15.587 | Biology Process | C8A, C5, CFH |
| GO:2000649 | regulation of sodium ion transmembrane transporter activity | 3 | 0.0153 | 15.587 | Biology Process | CAV3, SCN1B, SCN2B |
| GO:0042573 | retinoic acid metabolic process | 3 | 0.0173 | 14.613 | Biology Process | ALDH8A1, ADH1, RDH16 |
| GO:0009395 | phospholipid catabolic process | 3 | 0.0217 | 12.989 | Biology Process | APOA2, ENPP2, ANGPTL3 |
| GO:0001666 | response to hypoxia | 9 | 0.0230 | 2.598 | Biology Process | CRYAB, ANG, CAMK2G, ALDOC, CRP, SERPINA1, EGLN1, PAK1, LIPC |
| GO:0055117 | regulation of cardiac muscle contraction | 3 | 0.0241 | 12.305 | Biology Process | CAV3, P2RX4, CALM3 |
| GO:0055090 | acylglycerol homeostasis | 2 | 0.0254 | 77.933 | Biology Process | APOA5, ANGPTL3 |
| GO:0034370 | triglyceride-rich lipoprotein particle remodeling | 2 | 0.0254 | 77.933 | Biology Process | APOA2, APOA5 |
| GO:0031116 | positive regulation of microtubule polymerization | 3 | 0.0265 | 11.690 | Biology Process | CAV3, MAP1B, DCTN1 |
| GO:0032956 | regulation of actin cytoskeleton organization | 4 | 0.0287 | 5.995 | Biology Process | DIXDC1, HRG, PAK1, SH3BGRL3 |
| GO:0048247 | lymphocyte chemotaxis | 3 | 0.0317 | 10.627 | Biology Process | CCL24, CCL21, CCL9 |
| GO:0070098 | chemokine-mediated signaling pathway | 4 | 0.0363 | 5.469 | Biology Process | CCL24, CCL21, CCL9, CXCR3 |
| GO:0045959 | negative regulation of complement activation, classical pathway | 2 | 0.0378 | 51.956 | Biology Process | C4BPB, C4BPA |
| GO:0002542 | Factor XII activation | 2 | 0.0378 | 51.956 | Biology Process | KNG1, F12 |
| GO:0009725 | response to hormone | 5 | 0.0381 | 3.936 | Biology Process | ANG, APOA5, LIPC, ANGPTL3, SREBF2 |
| GO:0042311 | vasodilation | 3 | 0.0402 | 9.352 | Biology Process | KNG1, P2RX4, ALB |
| GO:0006631 | fatty acid metabolic process | 4 | 0.0413 | 5.196 | Biology Process | BAAT, DECR2, LIPC, ANGPTL3 |
| GO:0007399 | nervous system development | 7 | 0.0428 | 2.741 | Biology Process | PLXNA4, SCN2B, CAMK2G, KREMEN1, MAP1B, DPYSL3, NUMBL |
| GO:0048675 | axon extension | 3 | 0.0431 | 8.992 | Biology Process | SEMA7A, MAP1B, POU4F3 |
| GO:0072659 | protein localization to plasma membrane | 4 | 0.0485 | 4.871 | Biology Process | CAV3, TNIK, FGF13, CDH1 |
| GO:0006935 | chemotaxis | 4 | 0.0485 | 4.871 | Biology Process | CCL24, ENPP2, C5, CXCR3 |
| GO:0006810 | transport | 6 | 0.0489 | 3.017 | Biology Process | P2RX4, DYNC1LI1, AFM, LOC360919, ALB, LOC500473 |
| GO:0072562 | blood microparticle | 22 | 1.84 × 10−18 | 14.631 | Cellular Components | KNG1, GC, C9, MUG1, APCS, C4BPA, PLG, AHSG, C8A, AMBP, APOA2, AFM, SERPINF2, ALB, HPX, APOA5, PON1, CFH, SERPINC1, ITIH4, HRG, ITIH2 |
| GO:0005615 | extracellular space | 49 | 2.19 × 10−11 | 2.922 | Cellular Components | MUG2, MUG1, PZP, CRP, GPLD1, SPINK1, ANPEP, AZGP1, APOA2, ANG, SEMA7A, SERPINA4, APOA5, CFH, SERPINA1, SEMA3B, KNG1, F12, APCS, LOC360919, F9, C8A, AMBP, SERPINA3N, SERPINF2, SCGB2A2, GC, SERPINA10, ENPP2, C5, CCL9, AHSG, CCL24, ALB, CCL21, SERPINC1, ANGPTL3, C4BPB, DPYSL3, C4BPA, PLG, AFM, HPX, PON1, METRNL, SERPIND1, IGFBP1, LIPC, CPB2 |
| GO:0034364 | high-density lipoprotein particle | 7 | 4.31 × 10−8 | 32.313 | Cellular Components | APOA2, APOC4, APOA5, PON1, GPLD1, SAA4, LIPC |
| GO:0070062 | extracellular exosome | 66 | 6.06 × 10−8 | 1.957 | Cellular Components | ALDH8A1, CYP2J4, MUG1, TUBB2A, CRP, GPLD1, ANPEP, CKB, AZGP1, APOA2, DES, PACSIN3, ANG, SERPINA4, ITIH4, TUBB5, CFH, TUBB6, ITIH2, SEMA3B, SERPINA1, TUBA1A, KNG1, F12, TNIK, APCS, CRYAB, F9, METTL7A, VAT1, AMBP, C8A, RPS16, SERPINF2, BHMT, PRNP, SLC27A2, PRPS2, GC, C9, SERPINA10, ALDOC, C5, CDH1, KMO, AHSG, ALCAM, ALB, SERPINC1, DOPEY2, HRG, TUBB4B, COTL1, PLG, P2RX4, AFM, HPX, ARF3, PON1, CALM3, PAPPA2, METRNL, SERPIND1, SH3BGRL3, MYH14, CPB2 |
| GO:0031090 | organelle membrane | 11 | 2.10 × 10−7 | 9.591 | Cellular Components | CYP2C6V1, CYP2B3, UGT2B37, UGT2B17, CYP3A23/3A1, CYP4F4, CYP2C23, CYP2D3, CYP2C13, CYP2A1, CYP2A2 |
| GO:0030018 | Z disc | 9 | 1.24 × 10−4 | 6.037 | Cellular Components | CAV3, JPH2, DES, CRYAB, BAG3, PDLIM3, PAK1, HOMER1, FLNC |
| GO:0014704 | intercalated disc | 6 | 4.60 × 10−4 | 9.232 | Cellular Components | CAV3, SCN1B, DES, ATP2A2, FGF13, PAK1 |
| GO:0030426 | growth cone | 9 | 6.13 × 10−4 | 4.772 | Cellular Components | ANG, CRP, MAP1B, CALM3, DPYSL3, FGF13, MYH14, PAK1, HAP1 |
| GO:0005874 | microtubule | 11 | 8.97 × 10−4 | 3.658 | Cellular Components | DYNC1LI1, TUBB2A, CAMSAP1, MAP1B, TUBB5, TUBB6, FGF13, TUBA1A, DCTN1, TUBB4B, GLYATL2 |
| GO:0043034 | costamere | 4 | 0.00196 | 15.695 | Cellular Components | SVIL, SYNM, HOMER1, FLNC |
| GO:0005576 | extracellular region | 19 | 0.00456 | 2.088 | Cellular Components | APCS, C9, CRP, NTN4, GPLD1, SAA4, C4BPB, FGF13, PTH2, C4BPA, PLG, LOC500473, CCL24, C8A, SERPINA3N, ALB, APOA5, HRG, LIPC |
| GO:0005789 | endoplasmic reticulum membrane | 16 | 0.00504 | 2.258 | Cellular Components | CYP2B3, CDIPT, HSD3B5, CYP2D3, SREBF2, CYP2C6V1, UGT2B37, UGT2B17, CYP3A23/3A1, ATP2A2, CYP4F4, CYP2C23, CYP2C13, CYP2A1, CYP2A2, SLC27A2 |
| GO:0030424 | axon | 12 | 0.00657 | 2.609 | Cellular Components | GC, ALCAM, P2RX4, CRYAB, ALDOC, MAP1B, FGF13, CDH1, MYH14, PAK1, HOMER1, HTR2A |
| GO:0034361 | very-low-density lipoprotein particle | 3 | 0.0192 | 13.848 | Cellular Components | APOA2, APOC4, APOA5 |
| GO:0043231 | intracellular membrane-bounded organelle | 16 | 0.0302 | 1.825 | Cellular Components | KNG1, CYP2B3, CAV3, CYP2J4, HSD3B5, GPLD1, PLG, SREBF2, AMBP, UGT2B17, CYP3A23/3A1, PON1, CYP2C23, UGT2A3, CYP2A1, SLC27A2 |
| GO:0005856 | cytoskeleton | 8 | 0.0345 | 2.605 | Cellular Components | TNIK, DES, FILIP1, MAP1B, FLNC, COTL1, TPM2, HAP1 |
| GO:0044216 | other organism cell | 2 | 0.0376 | 52.316 | Cellular Components | C4BPB, C4BPA |
| GO:0033017 | sarcoplasmic reticulum membrane | 3 | 0.0457 | 8.719 | Cellular Components | JPH2, ATP2A2, CAMK2G |
| GO:0004867 | serine-type endopeptidase inhibitor activity | 15 | 6.34 × 10−11 | 11.167 | Molecular Function | MUG2, PZP, MUG1, SERPINA10, SPINK1, AMBP, SERPINA3N, SERPINF2, SERPINA4, SERPINC1, ITIH4, HRG, ITIH2, SERPINA1, SERPIND1 |
| GO:0070330 | aromatase activity | 9 | 2.65 × 10−8 | 18.039 | Molecular Function | CYP2C6V1, CYP2B3, CYP3A23/3A1, CYP4F4, CYP2C23, CYP2D3, CYP2C13, CYP2A1, CYP2A2 |
| GO:0008392 | arachidonic acid epoxygenase activity | 8 | 1.97 × 10−7 | 18.393 | Molecular Function | CYP2C6V1, CYP2B3, CYP2J4, CYP2C23, CYP2D3, CYP2C13, CYP2A1, CYP2A2 |
| GO:0008395 | steroid hydroxylase activity | 8 | 5.35 × 10−7 | 16.035 | Molecular Function | CYP2C6V1, CYP2B3, CYP2J4, CYP2C23, CYP2D3, CYP2C13, CYP2A1, CYP2A2 |
| GO:0020037 | heme binding | 13 | 1.46 × 10−6 | 6.122 | Molecular Function | CYP2B3, CYP2J4, CYP2D3, AMBP, CYP2C6V1, TDO2, CYP3A23/3A1, CYP4F4, CYP2C23, HRG, CYP2C13, CYP2A1, CYP2A2 |
| GO:0008092 | cytoskeletal protein binding | 8 | 1.07 × 10−5 | 10.423 | Molecular Function | DES, PACSIN3, CRYAB, ALDOC, PDLIM3, CDH1, FLNC, ABCG2 |
| GO:0005506 | iron ion binding | 13 | 1.11 × 10−5 | 5.031 | Molecular Function | CYP2B3, CYP2J4, CYP2D3, EGLN1, P4HTM, CYP2C6V1, CYP3A23/3A1, HIF1AN, CYP4F4, CYP2C23, CYP2C13, CYP2A1, CYP2A2 |
| GO:0004866 | endopeptidase inhibitor activity | 5 | 5.40 × 10−4 | 13.028 | Molecular Function | MUG2, MUG1, C5, SERPINA1, AHSG |
| GO:0017080 | sodium channel regulator activity | 5 | 6.95 × 10−4 | 12.214 | Molecular Function | CAV3, SCN1B, SCN2B, GPLD1, FGF13 |
| GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen | 5 | 8.78 × 10−4 | 11.495 | Molecular Function | CYP2B3, CYP2J4, CYP3A23/3A1, CYP2D3, CYP2A1 |
| GO:0005200 | structural constituent of cytoskeleton | 6 | 0.00223 | 6.514 | Molecular Function | TUBB2A, TUBB5, TUBB6, SYNM, TUBA1A, TUBB4B |
| GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors | 4 | 0.00262 | 14.213 | Molecular Function | HIF1AN, RGD1304810, EGLN1, P4HTM |
| GO:0008201 | heparin binding | 8 | 0.00268 | 4.283 | Molecular Function | SERPINA10, ANG, APOA5, SERPINC1, CFH, HRG, SERPIND1, LIPC |
| GO:0016705 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen | 5 | 0.00297 | 8.316 | Molecular Function | CYP2C6V1, CYP2B3, CYP2C23, EGLN1, CYP2C13 |
| GO:0005102 | receptor binding | 12 | 0.00666 | 2.598 | Molecular Function | KNG1, HAO1, P2RX4, BAAT, ANG, LRRC4B, DECR2, HRG, HOMER1, SLC27A2, HAP1, PLG |
| GO:0004497 | monooxygenase activity | 5 | 0.00944 | 6.013 | Molecular Function | CYP2C6V1, CYP2B3, CYP2C23, CYP2D3, CYP2C13 |
| GO:0004622 | lysophospholipase activity | 3 | 0.01328 | 16.751 | Molecular Function | ENPP2, LIPC, GDPD1 |
| GO:0042803 | protein homodimerization activity | 19 | 0.01474 | 1.845 | Molecular Function | RBPMS2, CRYAB, GIMAP7, CAMK2G, CRP, NR4A1, TPD52L1, ABCG2, AMBP, ADRB3, APOA2, HIF1AN, ANG, SERPINF2, ADH1, TENM3, PON1, RDH16, PRPS2 |
| GO:0005543 | phospholipid binding | 5 | 0.0198 | 4.825 | Molecular Function | APOA2, APOA5, MAP1B, PON1, SYTL3 |
| GO:0044325 | ion channel binding | 6 | 0.0233 | 3.693 | Molecular Function | CAV3, CALM3, FGF13, PRNP, HOMER1, HAP1 |
| GO:0048020 | CCR chemokine receptor binding | 3 | 0.0239 | 12.342 | Molecular Function | CCL24, CCL21, CCL9 |
| GO:0043395 | heparan sulfate proteoglycan binding | 3 | 0.0289 | 11.167 | Molecular Function | CFH, HRG, LIPC |
| GO:0030215 | semaphorin receptor binding | 3 | 0.0289 | 11.167 | Molecular Function | SEMA7A, SEMA3B, SH3BGRL3 |
| GO:0005509 | calcium ion binding | 16 | 0.0330 | 1.797 | Molecular Function | MATN2, F12, APCS, CALR4, TBC1D9, ENPP2, CRP, DECR2, F9, CDH1, P4HTM, ATP2A2, PON1, ITIH4, CALM3, SYTL3 |
| GO:0008307 | structural constituent of muscle | 3 | 0.0371 | 9.771 | Molecular Function | PDLIM3, SYNM, TPM2 |
| GO:0003779 | actin binding | 8 | 0.0406 | 2.511 | Molecular Function | GC, DIXDC1, CAP2, ANG, DIAPH3, MAP1B, COTL1, TPM2 |
| GO:0015020 | glucuronosyltransferase activity | 3 | 0.0460 | 8.685 | Molecular Function | UGT2B37, UGT2B17, UGT2A3 |
| GO:0005507 | copper ion binding | 4 | 0.0463 | 4.963 | Molecular Function | P2RX4, ANG, PRNP, ATP7B |
| GO:0005515 | protein binding | 29 | 0.0484 | 1.427 | Molecular Function | CAV3, GC, SCN1B, ALDOC, CRP, CDH1, RHOV, CKB, A1CF, TUBB5, SERPINC1, SERPINA1, PAK1, TUBA1A, HAP1, TUBB4B, SCN2B, CRYAB, MAP1B, HRK, NR4A1, HOMER1, NUMBL, P2RX4, ATP2A2, CALM3, SLC27A2, PRPS2, HTR2A |
Figure 2.
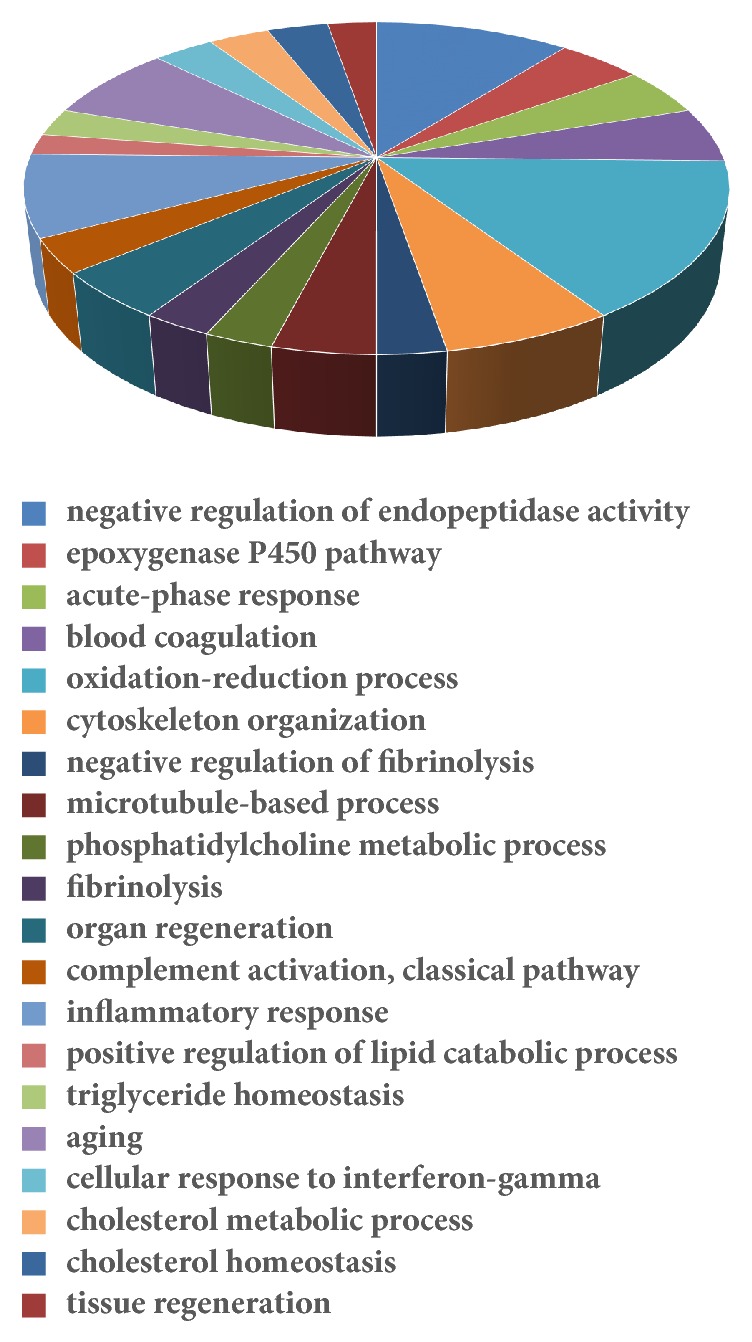
Enriched biological process for the differentially expressed genes between vil-high group and diabetic group.
3.6. Pathway Enrichment Analysis of Differentially Expressed Genes
In the pathway enrichment analysis, 12 pathway terms were significantly enriched (P < 0.05, Table 3). The top 10 most significant pathway terms were complement and coagulation cascades, retinol metabolism, steroid hormone biosynthesis, chemical carcinogenesis, linoleic acid metabolism, inflammatory mediator regulation of TRP channels, Gap junction, prion diseases, metabolic pathways, and arachidonic acid metabolism.
Table 3.
The enriched Kegg pathway with differentially expressed genes (P < 0.05).
| Pathway ID | Pathway name | Count | P-value | Fold Enrichment | Genes |
|---|---|---|---|---|---|
| rno04610 | Complement and coagulation cascades | 16 | 7.92 × 10−14 | 14.904 | KNG1, F12, C9, C5, F9, C4BPB, C4BPA, PLG, F13B, C8A, SERPINF2, CFH, SERPINC1, SERPINA1, SERPIND1, CPB2 |
| rno00830 | Retinol metabolism | 12 | 3.00 × 10−8 | 9.697 | CYP2C6V1, CYP2B3, UGT2B37, UGT2B17, CYP3A23/3A1, ADH1, CYP2C23, CYP2C13, UGT2A3, CYP2A1, RDH16, CYP2A2 |
| rno00140 | Steroid hormone biosynthesis | 10 | 2.63 × 10−6 | 8.280 | CYP2C6V1, CYP2B3, UGT2B37, UGT2B17, CYP3A23/3A1, HSD3B5, CYP2C23, CYP2D3, CYP2C13, UGT2A3 |
| rno05204 | Chemical carcinogenesis | 9 | 5.47 × 10−5 | 6.633 | CYP2C6V1, CYP2B3, UGT2B37, UGT2B17, CYP3A23/3A1, ADH1, CYP2C23, CYP2C13, UGT2A3 |
| rno00591 | Linoleic acid metabolism | 5 | 3.01 × 10−3 | 8.179 | CYP2C6V1, CYP2J4, CYP3A23/3A1, CYP2C23, CYP2C13 |
| rno04750 | Inflammatory mediator regulation of TRP channels | 7 | 6.98 × 10−3 | 4.082 | CYP2C6V1, CYP2J4, CAMK2G, CYP2C23, CALM3, CYP2C13, HTR2A |
| rno04540 | Gap junction | 6 | 9.57 × 10−3 | 4.573 | TUBB2A, TUBB5, TUBB6, TUBA1A, TUBB4B, HTR2A |
| rno05020 | Prion diseases | 4 | 1.14 × 10−2 | 8.384 | C8A, C9, C5, PRNP |
| rno01100 | Metabolic pathways | 30 | 1.17 × 10−2 | 1.556 | CYP2B3, CYP2J4, PGS1, CDIPT, TUSC3, ALDOC, HSD3B5, KMO, ANPEP, ATP5G3, CKB, TDO2, ADH1, CYP2C13, UGT2A3, CYP2C6V1, MAN2A2, HAO1, UGT2B37, UGT2B17, BAAT, CYP3A23/3A1, BHMT, PON1, CYP2C23, LIPC, RDH16, CYP2A1, CYP2A2, PRPS2 |
| rno00590 | Arachidonic acid metabolism | 5 | 3.16 × 10−2 | 4.140 | CYP2C6V1, CYP2B3, CYP2J4, CYP2C23, CYP2C13 |
| rno04726 | Serotonergic synapse | 6 | 3.63 × 10−2 | 3.245 | CYP2C6V1, CYP2J4, CYP2C23, CYP2D3, CYP2C13, HTR2A |
| rno04360 | Axon guidance | 6 | 4.08 × 10−2 | 3.144 | PLXNA4, SEMA7A, NTN4, SEMA3B, PAK1, UNC5C |
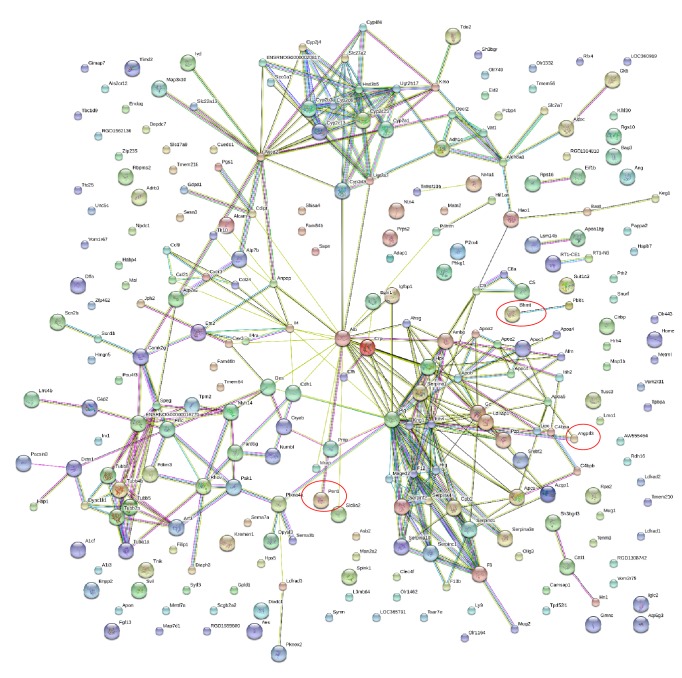
3.7. Gene Interaction Network
Based on 270 differentially expressed genes, a gene interaction network was analyzed by the String software. We found that 257 nodes and 338 edges were constructed (Figure 3). Twenty-five genes had more than 5 edges. The top 10 genes by degree are listed in Table 4.
Figure 3.
Interaction of differentially expressed genes between vil-high group and diabetic group from String software. Angptl3, Bhmt, and Pon1 are circled in red.
Table 4.
The top 10 genes from gene interaction analysis.
| Gene Accession | Gene Symbol | gene name | degree |
|---|---|---|---|
| NM_019369 | Itih4 | inter-alpha-trypsin inhibitor heavy chain family, member 4 | 17 |
| NM_053491 | Plg | plasminogen | 16 |
| NM_001014006 | F12 | coagulation factor XII (Hageman factor) | 12 |
| NM_001012027 | Serpinc1 | serpin peptidase inhibitor, clade C (antithrombin), member 1 | 11 |
| NM_138514 | Cyp2c13 | cytochrome P450, family 2, subfamily c, polypeptide 13 | 9 |
| NM_019287 | Apob | apolipoprotein B | 8 |
| ENSRNOT00000074103 | Cyp2c6 | cytochrome P450, family 2, subfamily C, polypeptide 6, variant 1 | 8 |
| NM_053318 | Hpx | hemopexin | 8 |
| NM_022519 | Serpina1 | clade A (alpha-1 antiproteinase, antitrypsin), member 1 | 8 |
| NM_001108802 | Speg | SPEG complex locus | 7 |
3.8. Confirmation with qPCR
From the gene expression array results, we found that Pon1 and Bhmt were in “metabolic pathway” (KEGG ID: rno01100), and Angptl3 and Pon1 were in several GO terms such as “positive regulation of lipid catabolic process” (GO: 0050996), “triglyceride homeostasis” (GO: 0070328), “cholesterol metabolic process” (GO: 0008203), “cholesterol homeostasis” (GO: 0042632), “response to hypoxia” (GO: 0001666), and “acylglycerol homeostasis” (GO: 0055090). So, we focused on these three genes in further study. As shown in Figure 4, the relative mRNA levels of angiopoietin-like 3 (Angptl3) and betaine-homocysteine S-methyltransferase (Bhmt) in diabetic rats were significantly higher than those of normal rats (P < 0.01), whereas the expression of these two genes was significantly reduced in vildagliptin-treated groups compared with those in diabetic rats (P < 0.01). Conversely, paraoxonase 1 (Pon1) decreased in the diabetic group. However, Pon1 increased in vildagliptin-treated rats (P < 0.01). These results were consistent with the microarray results.
Figure 4.
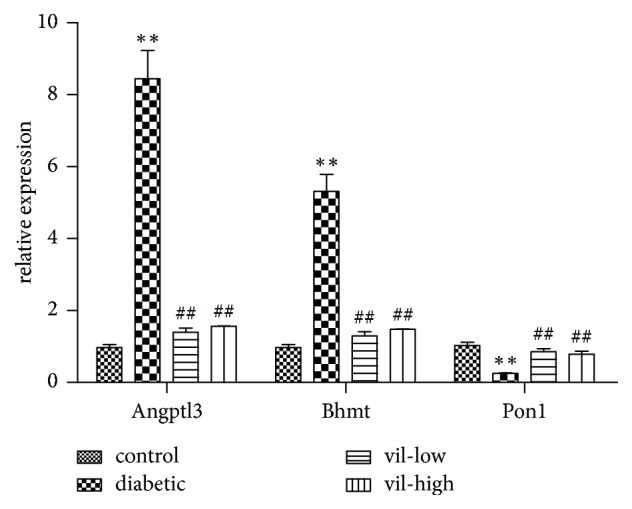
Confirmation of three representative differentially expressed genes by qPCR. Values are mean ± SD (n = 6). Angptl3: angiopoietin-like 3; Bhmt: betaine-homocysteine S-methyltransferase; Pon1: paraoxonase 1. ∗∗P < 0.01 versus control group; ##P < 0.01 versus diabetic group. vil-low: low dose of vildagliptin; vil-high: high dose of vildagliptin.
4. Discussion
In our study, we found that vildagliptin reduced blood glucose, TC, and LDL-C. In a clinical trial, sitagliptin-combined metformin add-on therapy led to greater improvement in HbA1c than the metformin monotherapy group after 18 weeks of treatment. Moreover, sitagliptin combined with metformin significantly reduced TG, TC, and LDL-C and increased HDL-C compared with the metformin group [13].
We used aortic rings to test the Ach-induced endothelium-dependent vasodilation. In diabetic rats, Ach-induced endothelium-dependent vasodilation was impaired. Vildagliptin augmented endothelial function. Other DDP-4 inhibitors also had similar vasoprotective effects. Saxagliptin treatment for 8 weeks increased aortic nitric oxide (NO) release by 22% and reduced mean arterial pressure in spontaneously hypertensive rats [14]. Moreover, sitagliptin treatment for 2 weeks protected endothelial function and reduced systolic blood pressure in spontaneously hypertensive rats through GLP-1 signaling [15].
In our research, the expression of Angptl3 was downregulated in vildagliptin-treated rats. Angptl3 is a key regulator, which can inhibit lipoprotein lipase (LPL) activity [16, 17]. The enzyme LPL hydrolyzes TG to free fatty acids (FFA). ANGPTL3 overexpression mice had high plasma TG levels [18]. Patients with a loss-of-function mutation of Angptl3 are characterized by low plasma TC, TG, HDL-C, and LDL-C [19, 20]. In 2008, Kathiresan et al. first identified an SNP site near ANGPTL3 that was associated with plasma TG levels in a genome-wide association study [21]. In obesity and T2D subjects, the ANGPTL3 level was higher than that in healthy subjects [22]. Moreover, the level of ANGPTL3 was increased in the livers of insulin-deficient and insulin-resistant diabetic mice [23]. In mice and monkeys, using ANGPTL3-specific antibodies led to reduced plasma TG [24–26]. The therapeutic targets of the inhibition activity of ANGPTL require further research to treat dyslipidemia [16, 17].
We also found that the expression of Bhmt was reduced in the vildagliptin-treated group. Methionine (Met) was produced from methylate homocysteine (Hcy) by BHMT enzymes with betaine. Thus, BHMT can reduce Hcy levels and increase Met levels. Met can then be converted to S-adenosylmethionine (SAM). In vivo, SAM is a main methyl donor to regulate methionine metabolism in many reactions. An increase in BHMT activity and SAM levels was observed in the livers of both type 1 diabetic rats [27] and the type 2 diabetic model [28, 29]. Insulin treatment in the rat hepatoma cell line and STZ-induced diabetic rats can inhibit the excess expression of BHMT and SAM [28, 30].
Moreover, our results showed that Pon1 was upregulated in the vildagliptin-treated group. Pon1 is an enzyme that has antioxidant functions. In serum, Pon1 is located in HDL. Pon1 protects LDL from oxidation and hydrolyzed oxidized LDL [31–34]. HDL levels are negatively associated with the risk of developing coronary artery disease (CAD), but high levels of oxidized LDL (oxLDL) in the aorta lead to cholesterol accumulation, foam cell formation, and atherosclerotic lesions [31, 35]. Low Pon1 expression accelerates aortic lesion development in mice [36]. In humans, serum PON level is reduced in patients with a history of myocardial infarction [32] and diabetic patients [37]. PON1 transgenic mice had decreased oxidative stress and atherosclerotic lesions [38, 39], whereas PON1 knockout mice had increased serum oxidative stress and were more susceptible to high-fat-diet-induced atherosclerosis [34, 40].
5. Conclusion
In conclusion, this study has confirmed that vildagliptin significantly attenuates endothelial dysfunction in diabetic rats. Importantly, our study indicates that vildagliptin may activate Pon1 expression and inhibit the expression of Bhmt and Angptl3 in the aorta. This study may increase the understanding of the pathways that contribute to which DPP-4 inhibitor attenuates endothelial dysfunction. More studies are needed to investigate whether or not vildagliptin treatment directly alters Agptl3, Bhmt, and Pon1 expression in the aorta of diabetic rats, independent of blood glucose (Figure 5).
Figure 5.
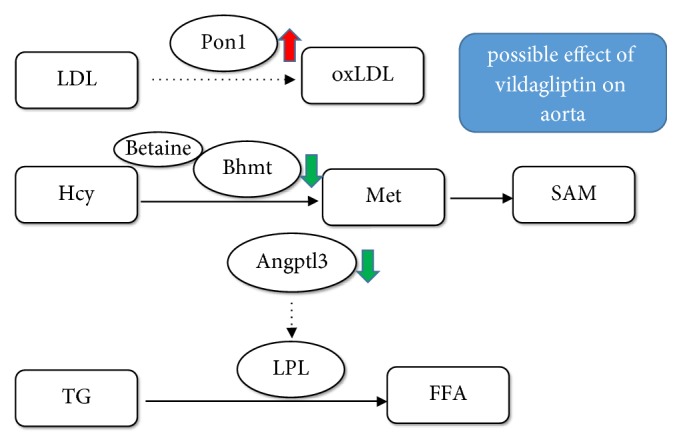
The possible mechanism of attenuation of vildagliptin on endothelial dysfunction. Vildagliptin attenuates endothelial dysfunction in diabetic rats through activating Pon1 expression in aorta to inhibit oxLDL production; inhibiting Bhmt expression to reduce Met and SAM production; inhibiting Angptl3 expression to activate LPL activity and thereby reduce plasma TG level. LDL: low-density lipoprotein; Pon1: paraoxonase 1; oxLDL: oxidized low-density lipoprotein; Hcy: homocysteine; Bhmt: betaine-homocysteine S-methyltransferase; Met: methionine, SAM: S-adenosylmethionine; TG: triglyceride.
Acknowledgments
This work was supported by grants from the National Key R&D Program of China (2017YFC1309603), National Key Research and Development Program of China (2016YFA0101002), National Natural Science Foundation of China (nos. 81170736 and 81570715), National Natural Science Foundation for Young Scholars of China (no. 81300649), China Scholarship Council foundation (201308110443), PUMC Youth Fund (33320140022) and Fundamental Research Funds for the Central Universities, Scientific Activities Foundation for Selected Returned Overseas Professionals of Human Resources and Social Security Ministry, and Novartis AG. The authors are very grateful to Beijing Compass Biotechnology Company for the excellent technical assistance with the microarray experiments.
Data Availability
The data used to support the findings of this study are included within the article.
Disclosure
The funding institutions had no role in study design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
Conflicts of Interest
The authors declare no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Authors' Contributions
Xinhua Xiao designed the experiments and contributed reagents and materials. Qian Zhang, Jia Zheng, Tong Wang, and Xiaojing Wang conducted the experiments. Miao Yu, Ming Li, and Fan Ping analyzed the data. Qian Zhang wrote the manuscript.
References
- 1.Ogurtsova K., da Rocha Fernandes J., Huang Y., et al. IDF Diabetes Atlas: Global estimates for the prevalence of diabetes for 2015 and 2040. Diabetes Research and Clinical Practice. 2017;128:40–50. doi: 10.1016/j.diabres.2017.03.024. [DOI] [PubMed] [Google Scholar]
- 2.Haffner S. M., Lehto S., Rönnemaa T., Pyörälä K., Laakso M. Mortality from coronary heart disease in subjects with type 2 diabetes and in nondiabetic subjects with and without prior myocardial infarction. The New England Journal of Medicine. 1998;339(4):229–234. doi: 10.1056/NEJM199807233390404. [DOI] [PubMed] [Google Scholar]
- 3.Dos Santos L., Salles T. A., Arruda-Junior D. F., et al. Circulating dipeptidyl peptidase IV activity correlates with cardiac dysfunction in human and experimental heart failure. Circulation: Heart Failure. 2013;6(5):1029–1038. doi: 10.1161/CIRCHEARTFAILURE.112.000057. [DOI] [PubMed] [Google Scholar]
- 4.McCormick L. M., Kydd A. C., Read P. A., et al. Chronic dipeptidyl peptidase-4 inhibition with sitagliptin is associated with sustained protection against ischemic left ventricular dysfunction in a pilot study of patients with type 2 diabetes mellitus and coronary artery disease. Circulation: Cardiovascular Imaging. 2014;7(2):274–281. doi: 10.1161/circimaging.113.000785. [DOI] [PubMed] [Google Scholar]
- 5.Apaijai N., Pintana H., Chattipakorn S. C., Chattipakorn N. Cardioprotective effects of metformin and vildagliptin in adult rats with insulin resistance induced by a high-fat diet. Endocrinology. 2012;153(8):3878–3885. doi: 10.1210/en.2012-1262. [DOI] [PubMed] [Google Scholar]
- 6.Apaijai N., Pintana H., Chattipakorn S. C., Chattipakorn N. Effects of vildagliptin versus sitagliptin, on cardiac function, heart rate variability and mitochondrial function in obese insulin-resistant rats. British Journal of Pharmacology. 2013;169(5):1048–1057. doi: 10.1111/bph.12176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Inthachai T., Lekawanvijit S., Kumfu S., et al. Dipeptidyl peptidase-4 inhibitor improves cardiac function by attenuating adverse cardiac remodelling in rats with chronic myocardial infarction. Experimental Physiology. 2015;100(6):667–679. doi: 10.1113/EP085108. [DOI] [PubMed] [Google Scholar]
- 8.Apaijai N., Chinda K., Palee S., Chattipakorn S., Chattipakorn N. Combined vildagliptin and metformin exert better cardioprotection than monotherapy against ischemia-reperfusion injury in obese-insulin resistant rats. PLoS ONE. 2014;9(7) doi: 10.1371/journal.pone.0102374.e102374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Q., Sun X., Xiao X., et al. Maternal Chromium Restriction Leads to Glucose Metabolism Imbalance in Mice Offspring through Insulin Signaling and Wnt Signaling Pathways. International Journal of Molecular Sciences. 2016;17(10):p. 1767. doi: 10.3390/ijms17101767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matsumoto S., Shimabukuro M., Fukuda D., et al. Azilsartan, an angiotensin II type 1 receptor blocker, restores endothelial function by reducing vascular inflammation and by increasing the phosphorylation ratio Ser1177/Thr497 of endothelial nitric oxide synthase in diabetic mice. Cardiovascular Diabetology. 2014;13(1, article no. 30) doi: 10.1186/1475-2840-13-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dennis G., Jr., Sherman B. T., Hosack D. A., Yang J., Gao W., Lane H. C. Database for Annotation, Visualization, and Integrated Discovery. Genome Biology. 2003;4(5)12734009 [PubMed] [Google Scholar]
- 12.Szklarczyk D., Franceschini A., Wyder S., et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Research. 2015;43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scott R., Loeys T., Davies M. J., Engel S. S. Efficacy and safety of sitagliptin when added to ongoing metformin therapy in patients with type 2 diabetes. Diabetes, Obesity and Metabolism. 2008;10(10):959–969. doi: 10.1111/j.1463-1326.2007.00839.x. [DOI] [PubMed] [Google Scholar]
- 14.Mason R. P., Jacob R. F., Kubant R., Ciszewski A., Corbalan J. J., Malinski T. Dipeptidyl peptidase-4 inhibition with saxagliptin enhanced nitric oxide release and reduced blood pressure and sICAM-1 levels in hypertensive rats. Journal of Cardiovascular Pharmacology. 2012;60(5):467–473. doi: 10.1097/FJC.0b013e31826be204. [DOI] [PubMed] [Google Scholar]
- 15.Liu L., Liu J., Wong W. T., et al. Dipeptidyl peptidase 4 inhibitor sitagliptin protects endothelial function in hypertension through a glucagon-like peptide 1-dependent mechanism. Hypertension. 2012;60(3):833–841. doi: 10.1161/HYPERTENSIONAHA.112.195115. [DOI] [PubMed] [Google Scholar]
- 16.Dijk W., Kersten S. Regulation of lipoprotein lipase by Angptl4. Trends in Endocrinology & Metabolism. 2014;25(3):146–155. doi: 10.1016/j.tem.2013.12.005. [DOI] [PubMed] [Google Scholar]
- 17.Kersten S. Physiological regulation of lipoprotein lipase. Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids. 2014;1841(7):919–933. doi: 10.1016/j.bbalip.2014.03.013. [DOI] [PubMed] [Google Scholar]
- 18.Shimizugawa T., Ono M., Shimamura M., et al. ANGPTL3 decreases very low density lipoprotein triglyceride clearance by inhibition of lipoprotein lipase. The Journal of Biological Chemistry. 2002;277(37):33742–33748. doi: 10.1074/jbc.m203215200. [DOI] [PubMed] [Google Scholar]
- 19.Musunuru K., Pirruccello J. P., Do R., et al. Exome sequencing, ANGPTL3 mutations, and familial combined hypolipidemia. The New England Journal of Medicine. 2010;363(23):2220–2227. doi: 10.1056/nejmoa1002926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Minicocci I., Montali A., Robciuc M. R., et al. Mutations in the ANGPTL3 gene and familial combined hypolipidemia: A clinical and biochemical characterization. The Journal of Clinical Endocrinology & Metabolism. 2012;97(7):E1266–E1275. doi: 10.1210/jc.2012-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kathiresan S., Melander O., Guiducci C., et al. Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nature Genetics. 2008;40(2):189–197. doi: 10.1038/ng.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abu-Farha M., Al-Khairi I., Cherian P., et al. Increased ANGPTL3, 4 and ANGPTL8/betatrophin expression levels in obesity and T2D. Lipids in Health and Disease. 2016;15(1) doi: 10.1186/s12944-016-0337-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Inukai K., Nakashima Y., Watanabe M., et al. ANGPTL3 is increased in both insulin-deficient and -resistant diabetic states. Biochemical and Biophysical Research Communications. 2004;317(4):1075–1079. doi: 10.1016/j.bbrc.2004.03.151. [DOI] [PubMed] [Google Scholar]
- 24.Wang Y., Gusarova V., Banfi S., Gromada J., Cohen J. C., Hobbs H. H. Inactivation of ANGPTL3 reduces hepatic VLDL-triglyceride secretion. Journal of Lipid Research. 2015;56(7):1296–1307. doi: 10.1194/jlr.M054882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gusarova V., Alexa C. A., Wang Y., et al. ANGPTL3 blockade with a human monoclonal antibody reduces plasma lipids in dyslipidemic mice and monkeys. Journal of Lipid Research. 2015;56(7):1308–1317. doi: 10.1194/jlr.M054890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dewey FE., Gusarova V., Dunbar RL., O'Dushlaine C., Schurmann C., Gottesman O., et al. Genetic and Pharmacologic Inactivation of ANGPTL3 and Cardiovascular Disease. The New England Journal of Medicine. 2017;377(3):211–221. doi: 10.1056/NEJMoa1612790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nieman K. M., Rowling M. J., Garrow T. A., Schalinske K. L. Modulation of methyl group metabolism by streptozotocin-induced diabetes and all-trans-retinoic acid. The Journal of Biological Chemistry. 2004;279(44):45708–45712. doi: 10.1074/jbc.M408664200. [DOI] [PubMed] [Google Scholar]
- 28.Ratnam S., Wijekoon E. P., Hall B., Garrow T. A., Brosnan M. E., Brosnan J. T. Effects of diabetes and insulin on betaine-homocysteine S-methyltransferase expression in rat liver. American Journal of Physiology-Endocrinology and Metabolism. 2006;290(5):E933–E939. doi: 10.1152/ajpendo.00498.2005. [DOI] [PubMed] [Google Scholar]
- 29.Wijekoon E. P., Hall B., Ratnam S., Brosnan M. E., Zeisel S. H., Brosnan J. T. Homocysteine metabolism in ZDF (Type 2) diabetic rats. Diabetes. 2005;54(11):3245–3251. doi: 10.2337/diabetes.54.11.3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jacobs R. L., House J. D., Brosnan M. E., Brosnan J. T. Effects of streptozotocin-induced diabetes and of insulin treatment on homocysteine metabolism in the rat. Diabetes. 1998;47(12):1967–1970. doi: 10.2337/diabetes.47.12.1967. [DOI] [PubMed] [Google Scholar]
- 31.Aviram M., Rosenblat M., Bisgaier C. L., Newton R. S., Primo-Parmo S. L., La Du B. N. Paraoxonase inhibits high-density lipoprotein oxidation and preserves its functions: a possible peroxidative role for paraoxonase. The Journal of Clinical Investigation. 1998;101(8):1581–1590. doi: 10.1172/JCI1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mackness B., Durrington P., McElduff P., et al. Low paraoxonase activity predicts coronary events in the Caerphilly Prospective study. Circulation. 2003;107(22):2775–2779. doi: 10.1161/01.cir.0000070954.00271.13. [DOI] [PubMed] [Google Scholar]
- 33.Navab M., Hama-Levy S., Van Lenten B. J., et al. Mildly oxidized LDL induces an increased apolipoprotein J/paraoxonase ratio. The Journal of Clinical Investigation. 1997;99(8):2005–2019. doi: 10.1172/JCI119369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shih D. M., Gu L., Xia Y.-R., et al. Mice lacking serum paraoxonase are susceptible to organophosphate toxicity and atherosclerosis. Nature. 1998;394(6690):284–287. doi: 10.1038/28406. [DOI] [PubMed] [Google Scholar]
- 35.Navab M., Berliner J. A., Subbanagounder G., et al. HDL and the Inflammatory Response Induced by LDL-Derived Oxidized Phospholipids. Arteriosclerosis, Thrombosis, and Vascular Biology. 2001;21(4):481–488. doi: 10.1161/01.ATV.21.4.481. [DOI] [PubMed] [Google Scholar]
- 36.Shih D. M., Gu L., Hama S., et al. Genetic-dietary regulation of serum paraoxonase expression and its role in atherogenesis in a mouse model. The Journal of Clinical Investigation. 1996;97(7):1630–1639. doi: 10.1172/JCI118589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mackness M. I., Harty D., Bhatnagar D., et al. Serum paraoxonase activity in familial hypercholesterolaemia and insulin-dependent diabetes mellitus. Atherosclerosis. 1991;86(2-3):193–199. doi: 10.1016/0021-9150(91)90215-O. [DOI] [PubMed] [Google Scholar]
- 38.Tward A., Xia Y.-R., Wang X.-P., et al. Decreased atherosclerotic lesion formation in human serum paraoxonase transgenic mice. Circulation. 2002;106(4):484–490. doi: 10.1161/01.CIR.0000023623.87083.4F. [DOI] [PubMed] [Google Scholar]
- 39.Oda M. N., Bielicki J. K., Ho T. T., Rubin E. M., Forte T. M., Berger T. Paraoxonase 1 overexpression in mice and its effect on high-density lipoproteins. Biochemical and Biophysical Research Communications. 2002;290(3):921–927. doi: 10.1006/bbrc.2001.6295. [DOI] [PubMed] [Google Scholar]
- 40.Shih D. M., Xia Y.-R., Wang X.-P., et al. Combined serum paraoxonase knockout/apolipoprotein E knockout mice exhibit increased lipoprotein oxidation and atherosclerosis. The Journal of Biological Chemistry. 2000;275(23):17527–17535. doi: 10.1074/jbc.m910376199. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data used to support the findings of this study are included within the article.