Abstract
An intercalator-linked macrocycle 3,6-bis(dimethylamino)-10-(6-[1,4,7-triazacyclononane]hexyl) acridinium bromide was synthesized, its copper(ii) complex was prepared, and the DNA cleavage activity of this metal complex was compared to that of the intercalator-free (Cu[9]aneN3)2+ complex.
There has been considerable interest in developing new synthetic reagents to manipulate DNA, with selectivity different from the naturally occurring systems. Artificial nucleases, employing metal ions as catalysts, are one focus of this research.1 Highly selective cleavage of large DNA molecules has been achieved using a PNA-based strand invasion strategy to induce local single-stranded structures that are highly susceptible to cleavage by Ce(iv)EDTA.2 Simple metal complexes have been successfully employed to accelerate the rate of double-stranded DNA hydrolysis, and those metal complexes with intrinsically high affinity for DNA are the most effective reagents.3–7 To increase the affinity for DNA, various recognition groups have been appended to metal catalysts.8–11 These bifunctional molecules are not consistently more reactive in DNA cleavage; the nature and length of the tether has a strong influence on the reactivity of the compound.3,12,13 A consistent result is that greater flexibility in the linker affords greater reactivity. In the case of intercalator-driven DNA binding, the tether length allows flexibility in the orientation of the appended metal complex but does not influence DNA binding.14
We have previously studied mononuclear copper(ii) complexes of the macrocycle triazacyclononane, and showed these complexes to be effective DNA cleavage agents.5,15 Herein, we report the synthesis of the intercalator-linked macrocycle 3,6-bis(dimethylamino)-10-(6-[1,4,7-triazacyclononane]hexyl) acridinium bromide, 1, and the ability of this ligand to support metal-mediated DNA cleavage. The most efficient cleavage is obtained with the copper complex, 2-Cu, which cleaved DNA under both aerobic and anaerobic conditions. The efficiency of this intercalator-linked complex is compared to the free copper(ii) macrocycle, Cu([9]aneN3)Cl2, 3. Interestingly, the intercalator-linked complex is not consistently more efficient in cleaving DNA than the simple metal complex.

The intercalator-linked macrocycle 1 was prepared following literature precedent. The 6-carbon linker was selected because optimal metal-mediated DNA cleavage was observed with other bifunctional chelates with linkers of 5–8 carbon atoms.3,12,13 Furthermore, Cu([9]aneN3)2+ appended to an acridine intercalator via a 1-carbon linker bound to and cleaved DNA.16 Bowler et al.17,18 previously prepared an analogous compound to 1, in which the chemotherapeutic metal complex cisplatinum was appended to acridine orange. This bifunctional complex bound to DNA via intercalation of the acridine orange moiety and via coordination of platinum, demonstrating sufficient flexibility for both functional groups to interact with the DNA. We adapted the method of Bowler to prepare 1, as shown in Scheme 1. The functionalized intercalator 10-(6-bromohexyl)-3,6-bis(dimethylamino)-acridinium iodide17,18 4 was reacted with 1,4,7-triazacyclo[5.2.1.04.10]decane19 5 under basic conditions, and the protected bifunctional macrocycle 6 was formed. Removal of the methine carbon under strongly basic conditions, and neutralization with HBr, formed the HBr salt 1, which was isolated by crystallization. 1H NMR, high-resolution FAB mass spectrometry, and elemental analysis were used to characterize 1. Elemental analysis yielded the following mass percentages: Found: C 36.1%, H 5.8%, N 8.8%, Br 43.4%. Calc. for C29H45N6Br·5HBr: C 36.2%, H 5.8%, N 8.7%, Br 41.5%.
Scheme 1.
Reagents and conditions: (i) CH3CN, 25 °C, 3 h; (ii) H2O, reflux, 3 h; (iii) NaOH, reflux, 4 h; (iv) 48% HBr, 25 °C.
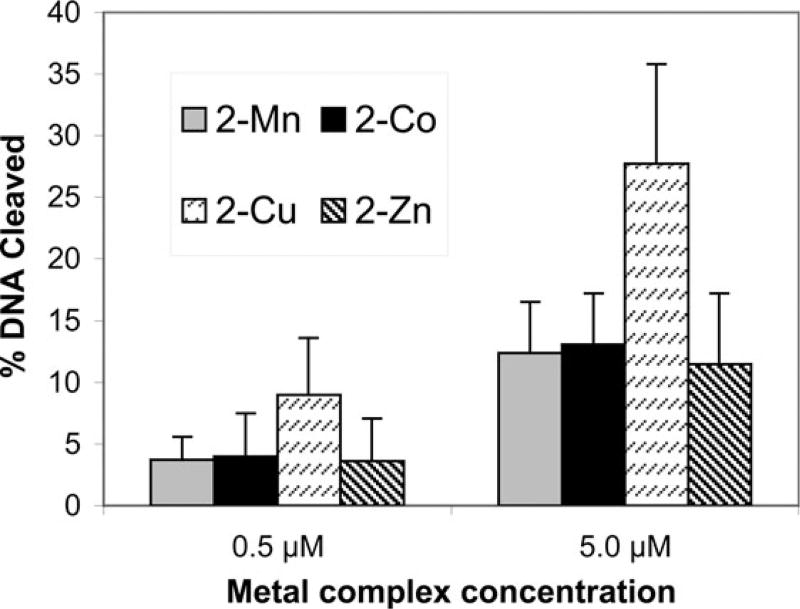
Divalent metal complexes of 1 promoted cleavage of plasmid DNA, with the greatest efficiency noted for Cu2+. All manipulations with ligand 1 and DNA were carried out under scrupulous exclusion of light in a sealed laboratory under safe light illumination. For detailed experimental methods see ESI †. Ligand 1 was combined with 1 equiv. metal salt (MnBr2, Co(NO3)2, CuBr2, or Zn(NO3)2) in water to prepare stock solutions of the metal complexes 2–M. Metal complex 2–M (0.5 µM or 5.0 µM) and supercoiled plasmid DNA (0.05 mg cm−3, 0.15 mM in phosphate groups) were combined in HEPPSO buffer (40 mM, pH 7.8). The samples were incubated in the dark in air for 12 h at 50 °C. Reactions were quenched in ice, and the extent of conversion of supercoiled DNA to nicked and linear DNA was assessed by ethidium bromide-containing agarose gel electrophoresis. Densitometry was used to quantitate the cleavage as previously described (see ESI†).15 Control experiments at a high concentration of 1, and lacking any metal ion, were performed: when ligand 1 (50 µM) was incubated with supercoiled plasmid DNA (0.05 mg cm−3, 0.15 mM in phosphate groups) in the dark, at 50 °C for 12 h, a maximum of 2% cleavage was observed. Fig. 1 illustrates that all the metal complexes 2–M cleaved supercoiled DNA. Complexes 2–Mn, 2–Co, and 2–Zn all cleaved the DNA to the same extent, with approximately 12% cleavage at the higher concentration. In contrast, 2–Cu cleaved DNA at more than twice the efficiency of the other metals, with 28% cleavage at the higher concentration. This result is reasonable given that Cu2+ is the strongest Lewis acid among the divalent transition metals.20 Furthermore, Cu2+ has the highest affinity for nitrogen donor groups among these metal ions, and thus Cu2+ is expected to bind stoichiometrically to the ligand even at the low concentrations used.
Fig. 1.
Aerobic DNA cleavage activity of 2 with varying metal ions. Shown is the percentage of supercoiled plasmid DNA that was converted to nicked and linear forms, averaged from three experiments. DNA, pBluescript II KS(−) (0.05 mg cm−3), was incubated with metal complexes 2–Mn, 2–Co, 2–Cu and 2–Zn in HEPPSO (40 mM, pH 7.8) for 12 h at 50 °C in air and in the dark. The extent of cleavage was normalized to the control reaction, and the error bars plotted are standard deviation calculated using the formula for a small number of data points.
Complex 2-Cu cleaved DNA effectively under anaerobic conditions, as illustrated in Fig. 2. For anaerobic experiments, all reagents were prepared in a nitrogen-filled glove box, which was contained within the light-tight laboratory. Individual reaction vials were sealed in a dessicator filled with argon gas, and placed in an incubator at 50 °C. Complex 2-Cu (0.5 µM or 5.0 µM) was incubated with supercoiled plasmid DNA (0.05 mg cm−3, 0.15 mM in phosphate groups) in either HEPPSO or HEPES buffers (40 mM, pH 7.8). No effect of buffer was observed and data from both buffers are included in Fig. 2. The DNA cleavage behavior of 2-Cu is similar to that which we previously observed for complex 3, where the efficiency of DNA cleavage decreased somewhat in the absence of air.5 This observation suggests that two different pathways are operative, one that is O2-dependent and one that is O2-independent. Since complex 2-Cu exhibits approximately 80% cleavage activity under anaerobic conditions, O2-dependent cleavage accounts for a modest fraction of the total DNA cleaving ability.
Fig. 2.
Comparison of the extent of DNA cleavage by 2-Cu under aerobic and anaerobic conditions. Shown is the percentage of supercoiled plasmid DNA that was converted to nicked and linear forms. DNA, pBluescript II KS(−) (0.05 mg cm−3), was incubated with 2-Cu in HEPPSO or HEPES (40 mM, pH 7.8) for 12 h at 50°C in the dark. Aerobic samples were handled in air; anaerobic samples were prepared in a nitrogen-filled glove box and incubated under argon.
A comparison of the anaerobic cleavage activities of the simple macrocyclic complex 3 and the intercalator-linked complex 2–Cu reveals differences in the concentration-dependent behavior of these two complexes. Fig. 3 shows a comparison of the cleavage activity of 3 and 2–Cu at varied concentrations of metal complex. Supercoiled plasmid DNA (0.05 mg cm−3, 0.15 mM in phosphate groups) was incubated with 2–Cu in HEPES buffer (40 mM, pH 7.8) at the concentrations indicated in Fig. 3. Reactions were carried out under anaerobic conditions, as described above, for 24 h at 50 °C. As previously indicated, all manipulations with 2–Cu were carried out in the dark. The DNA cleavage efficiency of 2–Cu as a function of concentration was compared to that previously obtained for 3.15 Interestingly, 2–Cu is less effective in cleaving DNA at low concentrations: at 5.0 µM 3 is approximately three times more efficient than 2–Cu.
Fig. 3.
Comparison of DNA cleavage by the simple copper(ii) complex 3 and the intercalator-linked complex 2–Cu. Shown is the percentage of supercoiled plasmid DNA that was converted to nicked and linear forms. Reactions of 3 and 2–Cu with DNA pBluescript II KS(−) (0.05 mg cm−3) were carried in HEPES or HEPPSO (40 mM, pH 7.8) for 24 h at 50 °C under inert atmosphere conditions. Reactions with 2–Cu were carried out in the dark. The extent of cleavage was normalized to control reactions, and the error bars are standard deviation calculated using formula for a small number of data points.
The two complexes exhibit very different dose response behaviors. Complex 2–Cu exhibited consistent time and concentration dependent increases in the extent of DNA cleavage under anaerobic conditions, behavior that is different from that observed previously for 3.15 Complex 3 is maximally efficient at 25 µM, and at higher concentrations it becomes less effective at cleaving DNA. This decrease in DNA cleavage activity at higher concentrations was attributed to the formation of inactive hydroxo-bridged dimers.15 In contrast, complex 2–Cu appears to increase in effectiveness toward DNA cleavage as the concentration is increased, and at 100 µM 2–Cu is approximately 2.4 times more effective than 3. This difference in concentration-dependent behavior may be attributed to the effect of the intercalator on the metal complex. Neighbor exclusion effects make it unlikely that two molecules of 2–Cu will be in close contact; therefore, the formation of inactive hydroxo-bridged dimers is inhibited at the higher concentrations. The fact that 2–Cu is less effective at lower concentrations than the free complex 3, suggests that there may be cooperativity between the monomeric complexes in the cleavage process and the neighbor exclusion effect operates to decrease the likelihood that two Cu([9]aneN3)2+ units will cooperate.
The data presented herein reinforce the conclusion that simply tethering a DNA-cleaving metal complex to a DNA-binding functionality does not necessarily result in an increased rate of DNA cleavage. A surprising result of our prior work was the observation that simple copper(ii) complexes of [9]aneN3 and i-Pr3[9]aneN3 were more reactive toward DNA hydrolysis than their reactivity toward an activated model phosphodiester would predict.15 This study produced another unanticipated result: at low concentrations intercalator-linked (Cu[9]aneN3)2+ was less effective as an DNA cleavage agent than the free metal complex. A plausible explanation for both unexpected observations is that on polyanionic DNA, individual cationic metal complexes collaborate to promote DNA hydrolysis without forming inactive, hydroxide-bridged dimers. One known difference between free and intercalator-linked (Cu[9]aneN3)2+ species is the metal ion mobility when bound to DNA. EPR analysis suggested that the free complex was highly mobile while a 1-carbon intercalator-linked complex was rigidly held in two distinct conformations.16 Naturally occurring metallonucleases frequently rely on cooperativity among two or more metal ions, and in some synthetic systems, multiple metal ions induce substantially improved reactivity toward DNA and model phosphate esters.21 The challenge remains to effectively tailor the structure of synthetic multinuclear complexes to consistently promote efficient hydrolysis of DNA.
Supplementary Material
Acknowledgments
Support of the Wisconsin Alumni Research Foundation through the Technology Innovation Fund is gratefully acknowledged. T.A.T. acknowledges support of the Chemistry-Biology Interface Training Grant, GM-08505. Grants NSF CHE-9208463 and NIH 1 S10 RR08389-01 provided support for NMR instrumentation.
Footnotes
Electronic supplementary information (ESI) available: Experimental methods and characterization of all new compounds. See DOI: 10.1039/b812183d
References
- 1.Mancin F, Scrimin P, Tecilla P, Tonellato U. Chem. Commun. 2005:2540–2548. doi: 10.1039/b418164f. [DOI] [PubMed] [Google Scholar]
- 2.Yamamoto Y, Miura K, Komiyama M. Chem. Lett. 2006;35:594–595. [Google Scholar]
- 3.Hettich R, Schneider H-J. J. Am. Chem. Soc. 1997;119:5638–5647. [Google Scholar]
- 4.Schnaith LMT, Hanson RS, Que L., Jr Proc. Natl. Acad. Sci. U. S. A. 1994;91:569–573. doi: 10.1073/pnas.91.2.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hegg EL, Burstyn JN. Inorg. Chem. 1996;35:7474–7481. [Google Scholar]
- 6.Sreedhara A, Freed JD, Cowan JA. J. Am. Chem. Soc. 2000;122:8814–8824. [Google Scholar]
- 7.Sissi C, Mancin F, Gatos M, Palumbo M, Tecilla P, Tonellato U. Inorg. Chem. 2005;44:2310–2317. doi: 10.1021/ic049316o. [DOI] [PubMed] [Google Scholar]
- 8.Nomura A, Sugiura Y. J. Am. Chem. Soc. 2004;126:15374–15375. doi: 10.1021/ja045663l. [DOI] [PubMed] [Google Scholar]
- 9.Chen XQ, Peng XJ, Wang JY, Wang Y, Wu S, Zhang LZ, Wu T, Wu YK. Eur. J. Inorg. Chem. 2007:5400–5407. [Google Scholar]
- 10.Fitzsimons MP, Barton JK. J. Am. Chem. Soc. 1997;119:3379–3380. [Google Scholar]
- 11.Gupta T, Dhar S, Nethaji M, Chakravarty AR. Dalton Trans. 2004:1896–1900. doi: 10.1039/B404673K. [DOI] [PubMed] [Google Scholar]
- 12.Boseggia E, Gatos M, Lucatello L, Mancin F, Moro S, Palumbo M, Sissi C, Tecilla P, Tonellato U, Zagotto G. J. Am. Chem. Soc. 2004;126:4543–4549. doi: 10.1021/ja039465q. [DOI] [PubMed] [Google Scholar]
- 13.Hashimoto S, Nakamura Y. J. Chem. Soc., Perkin Trans. 1996;1:2623–2628. [Google Scholar]
- 14.Ellis LT, Perkins DF, Turner P, Hambley TW. Dalton Trans. 2003:2728–2736. [Google Scholar]
- 15.Deck KM, Tseng TA, Burstyn JN. Inorg. Chem. 2002;41:669–677. doi: 10.1021/ic0107025. [DOI] [PubMed] [Google Scholar]
- 16.Hirohama T, Arii H, Chikira M. J. Inorg. Biochem. 2004;98:1778–1786. doi: 10.1016/j.jinorgbio.2004.07.014. [DOI] [PubMed] [Google Scholar]
- 17.Bowler BE, Hollis LS, Lippard SJ. J. Am. Chem. Soc. 1984;106:6102–6104. [Google Scholar]
- 18.Bowler BE, Ahmed KJ, Sundquist WI, Hollis LS, Whang EE, Lippard SJ. J. Am. Chem. Soc. 1989;111:1299–1306. [Google Scholar]
- 19.Atkins TJ. J. Am. Chem. Soc. 1980;102:6364–6365. [Google Scholar]
- 20.Hegg EL, Burstyn JN. Coord. Chem. Rev. 1998;173:133–165. [Google Scholar]
- 21.Liu CL, Wang M, Zhang TL, Sun HZ. Coord. Chem. Rev. 2004;248:147–168. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.