Abstract
Yeast Cdc13 protein (related to human CTC1) maintains telomere stability by preventing 5′-3′ end resection. While Cdc13 and Yku70/Yku80 proteins appear to prevent excessive resection, their combined contribution to maintenance of telomere ends across the genome and their relative roles at specific ends of different chromosomes have not been addressable because Cdc13 and Yku70/Yku80 double mutants are sickly. Using our PFGE-shift approach where large resected molecules have slower pulse field gel electrophoresis mobilities, along with methods for maintaining viable double mutants, we address end-resection on most chromosomes as well as telomere end differences. In this global approach to looking at ends of most chromosomes, we identify chromosomes with 1-end resections and end-preferences. We also identify chromosomes with resection at both ends, previously not possible. 10–20% of chromosomes exhibit PFGE-shift when cdc13-1 cells are switched to restrictive temperature (37 °C). In yku70Δ cdc13-1 mutants, there is a telomere resection “storm” with approximately half the chromosomes experiencing at least 1-end resection, ~10 kb/telomere, due to exonuclease1 and many exhibiting 2-end resection. Unlike for random internal chromosome breaks, resection of telomere ends is not coordinated. Telomere restitution at permissive temperature is rapid (< 1 h) in yku70Δ cdc13-1 cells. Surprisingly, survival can be high although strain background dependent. Given large amount of resected telomeres, we examined associated proteins. Up to 90% of cells have ≥1 Rfa1 (RPA) focus and 60% have multiple foci when ~30–40 telomeres/cell are resected. The ends are dispersed in the nucleus suggesting wide distribution of resected telomeres across nuclear space. The previously reported Rad52 nuclear centers of repair for random DSBs also appear in cells with many resected telomere ends, suggesting a Rad52 commonality to the organization of single strand ends and/or limitation on interactions of single-strand ends with Rad52.
Keywords: Telomere, Resection, Yku, Rad52, Yeast, Foci
1. Introduction
Telomeres of all organisms provide protection of chromosomal ends from inappropriate degradation, cell stress responses and they provide the opportunity for lengthening to counteract inability to replicate completely both ends in a semi-conservative fashion. While they are essential to organismal longevity, regeneration of telomeres is associated with unlimited mitotic division and cancer [1]. The chromosomal ends of budding yeast Saccharomyces cerevisiae have provided a structural model for understanding telomere biology [2]. The telomeres consist of TG1-3 repeats (unlike the TTAGGG repeats in mammalian cells) that range in size from around 250–350 bases. At the extreme ends, there is a small, approximately 10–20 base 3′ single-strand DNA (ssDNA) tail [3]. Several proteins protect the ends from resection including Cdc13 (related to human CTC1) that binds to ssDNA and coordinates several telomere activities (summarized in [4]), the Yku heterodimer and Rap1, which bind to the TG repeats. Given that telomere DNA has features that resemble DNA double-strand breaks (DSBs) and that many of the same proteins such as the MRX complex and the Yku70/80 heterodimer associate with the two types of structures, there has been a long-term interest in the metabolism of the two types of ends [5–7]. Interestingly, the resected end of the telomere resembles the early 5′ to 3′ resection step first proposed for recombinational repair of DSBs [8].
While a great deal is known from a variety of organisms about telomere organization, structure and protein functions, there is relatively little known about differences in protection between the ends of individual chromosomes or between chromosomes (i.e., global changes). Also, there is no information about possible coordination of events between ends of chromosomes. In the present study, we have extended our recent approaches for addressing resection events at random DSBs induced by ionizing radiation to characterizing global changes at telomeres in yeast. Since IR-induced DSBs are produced randomly in the genome, the opportunity to characterize them in a population was challenging, especially compared to molecular analysis of a single defined DSB. For this we developed the pulse field gel electrophoresis shift assay (PFGE-shift) [8]. We showed that single-strand DNA at either or both ends of large, double-stranded DNA molecules significantly reduces their electrophoretic mobility during PFGE resulting in an apparent increase in size. Mung bean nuclease treatment of plug samples was used to demonstrate that the PFGE-shift effect is indeed due to extensive resection at the ends of large DNA molecules. For the PFGE conditions used in this study, short resection tracks of ~200 bases or less are barely detectable [9], but increasing the length of single-stranded ends up to ~3 kb progressively increases the mobility shift to a maximum apparent increase in chromosome size of about 40 kb (for 1-end resections) and about 140 kb (for 2-end resections). Extensive resection lengths greater than ~3 kb do not cause further increase in the apparent size of resected chromosomes [8,10]. Importantly, this difference between the maximum PFGE-shift for 1-end vs 2-end resections enables the assay to distinguish between 1-end and 2-end extensive resection events. With this system we were able to demonstrate that resection at the two ends of a single, randomly produced DSB in a circular chromosome was coordinated and that we could distinguish 1- end vs 2-end resection in various mutants.
Based on these findings that enabled us to examine events and coordination at the two ends of a chromosomal size linear molecule, we have turned our attention to events at the two ends of telomeres. This is particularly interesting in light of possible associations between telomere ends of chromosomes [11]. Since many of the linear 16 chromosomes of budding yeast can be distinguished on a PFGE gel, we anticipated being able to address similarities and differences between chromosomes in terms of 0, 1 or 2 ends of individual chromosomes as well as a global opportunity to compare changes between chromosomes. Previous approaches to addressing telomere resection were focused on small defined regions of individual telomere ends using various amplification PCR-based approaches or to amplification of single-strand regions with random primers (QAOS [5,12]).
In this study, we address the impact of the Yku70 and Cdc13 proteins in protecting the telomeres across the yeast genome and potential differences between chromosomes. Previously, we demonstrated that in a cdc13-1 mutant, the resulting resection could lead to PFGE-shift in some of the chromosomes in a population of cells [8]. However, we did not address 1- vs 2-end events as we had done for IR-induced DSBs [10]. Notably, we have subsequently developed conditions and identified genetic backgrounds in which yku70Δ cdc13-1 double mutants were not synthetically lethal, allowing assessment of the total contribution of each component to end resection of individual chromosomes as well as globally. Earlier studies have reported incompatability between double mutants [13,14]. We found that considerable protection is provided by the combined Yku70 and Cdc13 proteins. In the absence of both proteins nearly 50–75% of chromatids experience at least 1-end resection and about 25% are resected at both ends. Remarkably, in spite of such a high level of resection in yku70Δ cdc13-1 double mutants at the restrictive temperature (37°), there was nearly complete recovery of double-strand telomeres in less than one hour when cells were returned to permissive temperature and, depending on the strain background, there was little loss of survival.
With this system we also investigated associated appearance in single cells of Rad52 and Rfa1 (subunit of RPA) foci that are expected to be associated with the resected ends. We found that there is a corresponding increase in Rfa1 single-stranded DNA binding protein foci with the large increase in resected telomere ends. However, while many cells have a Rad52 focus there is typically only one per cell. Thus, using a combination of our PFGE-shift approaches and single-cell protein analysis, we have been able to address resection of telomeres globally and associated changes in protein associations with resected telomeres.
2. Materials and methods
2.1. Yeast strains, media, growth conditions and probes
Genotypes of all strains used in this study are listed in Supplementary Table S1.
Construction of CG379 background strains DAG635 (MATα ade5-1 his7-2 leu2-3,112 trp1-289 ura3Δ can1Δ) and its cdc13-1 derivative DAG760 are described in [15]. From DAG635 and DAG760, the dominant selectable marker hygromycin cassette (hphMX4) [16] was used for deletion of EXO1, and the nourseothricin (natMX4) cassette for deletion of the YKU70 gene to generate double and triple mutants in the CG379 strain background.
For W303 background strains [17,18], cells endogenously expressing Rad52-YFP and Rfa1-CFP were crossed to yku70Δ and cdc13-1 to obtain KBY583-1D, KBY647-2D, and KBY823-6C. (All the W303 cells were RAD5+ and isogenic except for indicated genotypes.) The cdc13-F684S allele (abbreviated in this study as cdc13-FS, [13]) was introduced into KBY583-1D using plasmid pVL5439 to generate JWW2024.1. The natMX4 cassette was used for deletion of the YKU70 gene for JWW2037.1 and JWW2027.2.
For most PFGE-shift experiments, yeast cultures were grown at the 20 °C permissive temperature to late log phase in YPDA media (1% yeast extract, 2% Bacto-Peptone, 2% dextrose, 60 μg/mL adenine sulfate) containing 1M sorbitol (Amresco, Solon, OH). Exceptions are found in the experiments described in Supplementary Figs. S2 and S3, which test effects of 20 °C vs 23 °C with (YPD+SRB) and without 1M sorbitol, and also in Fig. 4 and Supplementary Fig. S5, for cells grown in synthetic complete (SC) medium [19] containing 100 μg/mL adenine (AD) and 1M sorbitol (SRB) [SC+AD+SRB] as indicated in the figure legends. For all experiments using SC, nocadazole was added at final concentration of 15 μg/mL just before shifting from permissive temperature to 37 °C. Additional 7.5 μg/mL nocodazole was added at 1 h intervals during the remainder of the time course to compensate for degradation of nocodazole at 37 °C. Growth conditions for live cell florescent microscopy are described below.
Fig. 4.
The influence of the cdc13-FS and cdc13-1 in global resection of telomeres. Experiments with the mutants in the W303 background were comparable to those in Fig. 1C and D except that cells were cultured in synthetic defined media.
2.2. PFGE procedures
PFGE sample plugs were prepared as described previously [8]. NotI restriction enzyme treatments of sample plugs in Supplementary Figs. S4E and S4F were described in [10]. All PFGE separations were performed with a CHEF Mapper XA system (Bio-Rad, Hercules, CA). Except for the 2D CHEF in Fig. 3, the CHEF Mapper was programmed in autoalgorithm mode, 250–1400 kb, 24 h total run time, using 1/2 X TBE (44.5mM Tris, 44.5mM boric acid, 2mM EDTA) as running buffer. Gels were stained in Sybr® Gold (Invitrogen) and images were photographed using a Kodak GelLogic 200 digital imaging system. For the 2D CHEF shown in Fig. 3, the first dimension PFGE was performed as in [10] except that the CHEF mapper was programmed in autoalgorithm mode, 250–2000 kb, 22 h. First dimension gel slices were removed and equilibrated in TE [10]. Each gel slice was digested with 180 units of mung bean nuclease (Promega Corporation, Madison, WI) in 9 ml of reaction buffer (10X buffer provided by supplier) at 37 °C for 1 h. Reactions were stopped on ice with 100mM EDTA, pH 8.0, and equilibrated in 100 ml TE (10mM Tris, pH 8.0, 1mM EDTA) before running the 2nd dimension CHEF for 22 h, 250–2000 kb autoalgorithm.
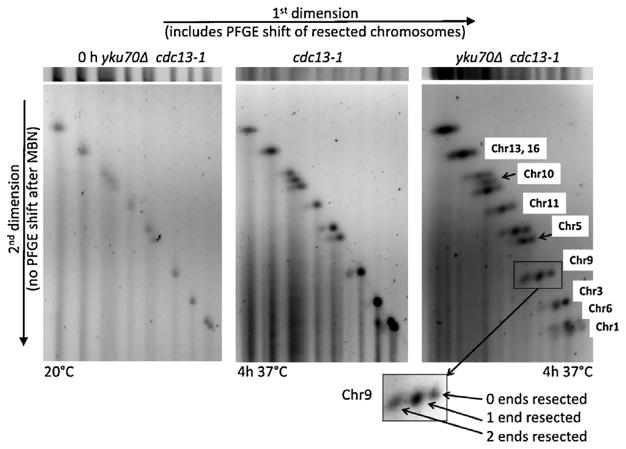
Fig. 3.
2-D PFGE analysis along with nuclease treatment identifies gobal 0-, 1-, and 2-end resection along with chromosomal differences. Lanes (horizontal slices) in the first dimension were cut out from the PFGE gel and exposed to mung bean nuclease to remove ssDNA tails. The lane is then subjected to PFGE in the second dimension. The bands of chromosomes with ssDNA tails that were PFGE-shifted in the first dimension do not undergo PFGE-shift in the second dimension. Because of the nuclease digestion, they are shortened compared to the chromosome with no resection and will have greater mobility. Because the 1-end and 2-end resected chromosomes were separated from the 0-end chromosomes, the relative amounts of resected chromosomes can be approximately quantitated, as indicated in the inset.
2.3. Southern transfer hybridization
Southern transfers, probe preparations, 32P labeling and hybridizations were carried out as described in [8]. Probes were prepared by PCR amplification of genomic DNA using primers listed in Supplementary Table S2.
2.4. Brightfield microscopy
For visualization of yeast cells (see Supplementary Fig. S2) bright field images were captured on a Zeiss AxioObserver Z1 fluorescence microscope (Carl Zeiss Inc, Oberkochen, Germany) using a Plan-Apochromat 63x/1.40 Oil DIC M27. A 100W halogen lamp was used for illumination and a Zeiss Axiocam MRm camera with the Zen Blue Software (Carl Zeiss) was used to collect the images.
2.5. Live-cell fluorescent microscopy methods
W303 yeast cultures with genotypes indicated in the figure legends were grown in 5 ml SC medium containing adenine (SC+AD; 100 μg/mL) and sorbitol (1 M) overnight at 23 °C. After 24 h, the cultures were diluted 100-fold into (SC+AD+SRB) and incubated at 23 °C for an additional 24 h to OD600 0.3–0.4. Nocodazole was added to the culture at a final concentration of 15 μg/mL. The cells were shifted to 37 °C for either 0, 3, 4, or 5 h and prepared for microscopic imaging as described in [20]. Cells were visualized with a Nikon TiE inverted live-cell system using a 100x oil immersion objective (1.45 numerical aperture), a Prior Z-stage and a Photometrics HQ2 camera. Eleven Z-stack sections of 0.3 μm were captured with the following exposure times: 800 ms for Rad52-YFP; 500 ms for Rfa1-CFP; and 60 ms for differential interference contrast. All images were processed identically and analyzed using Nikon Elements software suite. The images were adjusted for brightness and contrast using Adobe Photoshop.
3. Results
3.1. Cdc13, but not Yku, prevents extensive global telomere resection
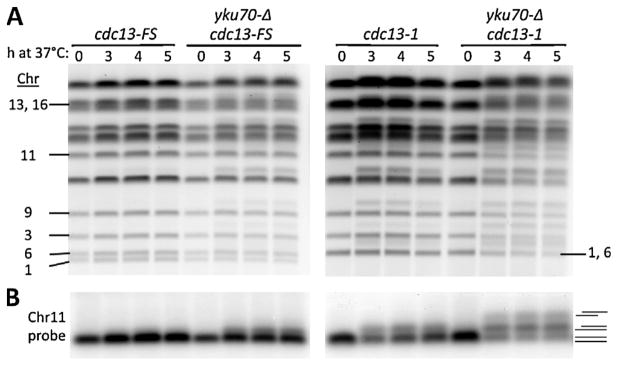
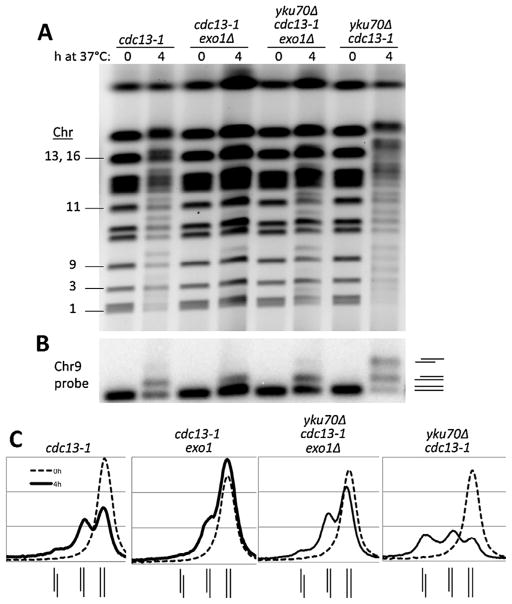
Previously, we showed that for all chromosomes that could be distinguished by PFGE, a shift of~10% of the molecules could be detected within 3 h after shifting the cdc13-1 mutant to the 37 °C restrictive temperature [8]. The frequency of shifted molecules in the population appeared to be chromosome specific with about 10–20% of the ChrIII molecules showing an “apparent” 40 kb shift. The previous findings are extended here to specific probing of Chr9 and Chr11 for cells held at 37 °C for 4 h where the frequency of resected Chr9 or Chr11 molecules in the population is also ~10–20%, as described in Fig. 1A and 1B. (Chr1, −3 and −13 are included in experiments presented below.)
Fig. 1.
Global resection of telomeres detected by PFGE-shift and protection provided by Cdc13 and Yku70. (A) Cells in the CG379 strain background grown to late log at 20 °C (YPDA+SRB) were diluted to fresh medium and shifted to 37 °C. Chromosomal DNA was examined using the PFGE protocols described in the Material and Methods. The PFGE-shift was detected as bands above the main chromosomal bands obtained with cells that were not raised to the higher temperature. (B) Southerns of Chr9 and Chr11 from the gel of 1A probed with respective 32P labeled probes. The line diagrams correspond to the relative positions of the bands. For example, the lowest band for the double mutant is unresected DNA, the next band up is for molecules with one or the other ends resected, and the upper band corresponds to molecules with both telomeres resected. (C) and (D) comparable experiments except with cells with the W303 background. Note the absence of PFGE-shifted chromosomes in the WT and yku70Δ single mutant.
The Yku complex protects telomeres from limited resection and prevents telomere shortening over multiple cell divisions [14]. However, unlike for cdc13-1, we found no observable resection based on the lack of PFGE-shift, as shown in the gel in Fig. 1A and the Southerns in Fig. 1B. Since we previously showed that the limit of detectable resection by PFGE-shift is in the range of ~200 bases [9], these global results along with previous findings that the extent of resection in Yku mutants is small [21] suggest that in the absence of Yku, there is little if any extended telomere resection across any of the chromosomes. (Note: because of their large sizes, PFGE-shift of Chr4 and Chr12 was not detectable under the conditions used.)
3.2. Yku prevents extensive telomere resection across the genome in the absence of Cdc13
Results with the single mutants led us to address the combined role of Yku and Cdc13 in stabilizing telomere DNAs across the genome. Previous attempts to address the combined role of these telomere end associated proteins have generally been hampered by the poor growth of the double mutants at permissive temperatures (for example, [14]). However, we discovered that double mutants in the CG379 genetic background that we had used previously [8] exhibited good growth (described in Fig. 6 below) at permissive temperature, thereby providing the opportunity to address the consequences of a combined Yku70 Cdc13 deficiency. As shown in Fig. 1A, there is a dramatic difference between the cdc13-1 single mutant and the yku70Δ cdc13-1 double mutant. For Chr9 and Chr11 nearly ¾ of the molecules in the double mutant, population exhibit PFGE-shift as detected with Southern probing (Fig. 1B). The appearance of a band is due to a maximum shift as the resection increases (which was previously described in [8]). If the distribution is random then the large majority of molecules have at least one extensively resected end. Similar results were found for Chr1 in a separate experiment described in Supplementary Figs. S1A and B. (We note that a slight amount of shifted molecules are detected occasionally prior to changing temperature suggesting some capability for global resection at the permissive temperature in the double mutant. Also, see results below for cells with the W303 background.)
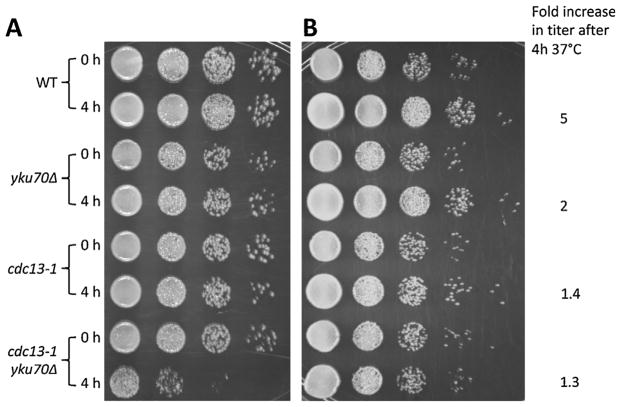
Fig. 6.
Impact of telomere resection on survival of cdc13-1 and yku70Δ cdc13-1 cells with W303 or CG379 background. W303 (in A) and CG379 (in B) Cells were grown to stationary (3 d) in YPDA+SRB at 20 °C, diluted to 1×107 cells/ml in YPDA+SRB. A portion of each culture was cultured at 37 °C for 4 h while the other portion was held on ice to be used as the 0 h samples. At the end of the time course, 10-fold serial dilutions of all samples were done in 96-well plates, followed by pronging to YPD+SRB agar plates. Titers were not adjusted after the 4 h incubation. (A) W303 background strain set. (B) CG379 background strain set. The WT and yku70 CG379 strains continued to grow at the higher temperature, and the relative increase based on hemacytometer counts are described.
Unlike, for the single cdc13-1 mutant, there was a second region of much slower PFGE-shifted molecules for Chr9 and Chr11, as seen in Fig. 1B and for Chr1 in Fig. S1B. We previously showed [10] that for the case of an IR or I-Sce I induced DSB in a circular chromosome, the second band of slower moving molecules is due to molecules that are resected at both ends (this is addressed further below in Fig. 3). However, a second band of PFGE-shifted molecule was not found for Chr3 (Supplementary Figs. S1C and S1D), which we suggest may be due to differences between telomere ends or subtelomeric regions (addressed in the Discussion section). As described in Supplementary Fig. S1C, global resection in the double mutant can be detected within 1 h after shifting to 37 °C. The resected chromosomes appear stable up to 8 h (Supplementary Fig. S1B).
From these results, we conclude that most of the protection from extensive resection at telomeres across the genome is due to Cdc13 and associated proteins. However, in the absence of Cdc13, Yku has a major role in preventing global telomere resection. Furthermore, our study provides the first demonstration that most telomere resection at the ends of a chromosome is independent based on the absence of 2-ended events in cdc13-1 mutants and the minority of 2-ended events in the double yku70Δ cdc13-1 mutant. This differs from our previous findings for resection at an induced DSB where resection between the two ends is coordinated, giving rise primarily to 2-ended PFGE-shift patterns [10].
3.3. Global telomere resection is also extensive in yku70Δ cdc13-1 cells with a W303 background
Many telomere studies have been done in the W303 background as well as studies on DNA repair/recombination protein associations [17,18,22] However, unlike for the CG379 background, the yku70Δ cdc13-1 double mutants of W303 grow poorly even at permissive temperature, which led us to identify conditions that would enable healthy growth of the double mutants. As shown in Supplementary Fig. S2, we found that the poor growth of freshly isolated double mutants on YPDA medium at 23 °C was associated with budded cells that had a stressed appearance (including lysis) as compared to WT or either single mutant. This led us to try sorbitol, commonly used to prevent lysis during preparation of spheroplasts. The presence of sorbitol resulted in normal growth rather than accumulation of cells with the G2 budding phenotype. (Since the yku70Δ cdc13-1 cells retained their lethal interaction when returned to medium without sorbitol, the permissive growth conditions were not due to accumulation of suppressor factors.)
As shown in Supplementary Figs. S3A and S3B, when yku70Δ cdc13-1 cells with the W303 background that had been maintained on YPDA+sorbitol were incubated overnight in medium lacking sorbitol, even at 20 °C, there was a small amount of resected chromosomes for all that could be identified on PFGE and specifically Chr9 and Chr11 (see scan in Supplementary Fig. 3C). Shifting to 37 °C for 4 h resulted in large amounts global telomere resection. Based on these results we chose to include sorbitol in all experiments including those above (Fig. 1A and B) with the CG379 strain background, even though the CG379 cells did not show stress. These findings suggest additional factors, which could be pursued in future studies, that can influence the impact of a combined Yku70 Cdc13 deficiency on cell growth and survival.
As shown in Fig. 1C and D and Supplementary Figs. S4A and S4B, the results in the W303 cells with yku70Δ, cdc13-1 or yku70Δ cdc13-1 mutations were comparable to those for the CG379 background cells including the 2-end resection for Chr9 and Chr11 (also see Supplementary Figs. S5A and S5B for additional 2-end resection examples of Chr9 and Chr11). The maximum resection was achieved by 3 h. The Chr3 (Supplementary Figs. S4C and S4D) in the W303 yku70Δ cdc13-1 strain exhibited the 2-end resection PFGE-shift pattern observed for Chr 9 and Chr11 (Fig. 1C and D). However, since Chr3 in the CG379 strain showed only 1-end resection (Supplementary Fig. S1D), telomere differences between chromosomes and between strains can be detected with the PFGE-shift analysis.
Given the robustness of the PFGE-shift assay, we also established that we could address events at the individual ends of a chromosome. Although PFGE-shift analysis of whole chromosomes enables 1-end and 2-end extensive resection events to be distinguished, it does not reveal if there is preferential protection from extensive resection at either the left or the right telomere. To address this issue, we digested DNA in agarose plugs with NotI restriction endonuclease followed by PFGE and Southern transfer and then hybridized the blot with probes specific for NotI fragments containing either the left or right telomere of Chr11. Based on the Southerns and density analysis (Supplementary Fig. S4E and S4F, respectively), there is no clear preference for resection of either the left or right telomere of Chr11 in the cdc13-1 or yku70Δ cdc13-1strains (W303 background).
3.4. Most of the global resection at telomeres of cdc13-1 and yku70Δ cdc13-1 mutants is due to Exo1
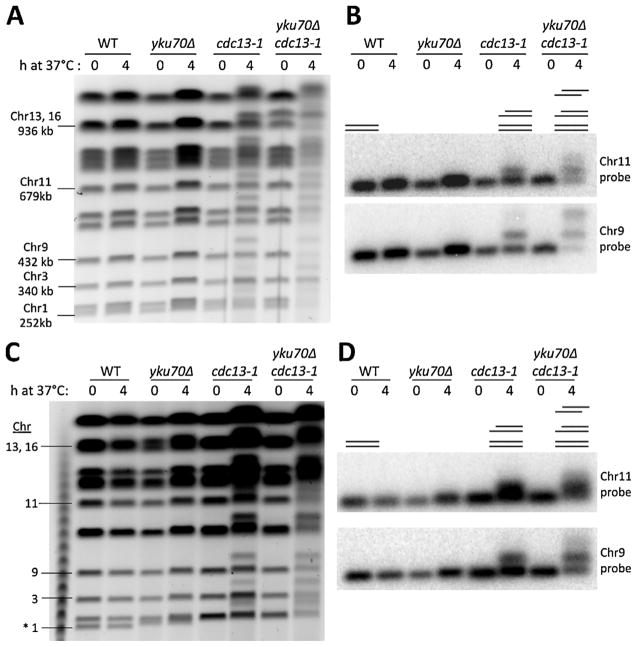
Much of the resection at individual telomeres has been reported to be due to Exo1 [23,24], unlike for radiation-induced DSBs where there is considerable redundancy between Exo1 and Sgs1/Dna2. We chose to examine specifically the global role of Exo1 since lack of PFGE-shift in the nuclease deficient cells would be diagnostic of a prominent role in resection. As shown in Fig. 2A for genomic analysis (SYBR gold stained gel) and Fig. 2B that is specific for Chr9, the frequency of molecules that are shifted in the cdc13-1 strain after 4 h at 37 °C is dramatically reduced by the exo1Δ mutation. The results are even more dramatic for the yku70Δ cdc13-1 double mutant vs the yku70Δ cdc13-1 exo1Δ triple mutant. Visually, along with a scan of the gels probed for Chr9 (Fig. 2C), the band indicative of the 2-end resection in the double mutant is almost absent in the triple mutant. There is a corresponding increase in molecules with no apparent resection (~20% EXO1+ vs ~80% exo1Δ−). Thus, these findings extend previous reports on the role of Exo1 in telomere resection to show that there are 1-end and 2-end resected chromosomes identified by PFGE-shift and these are primarily due to Exo1. The remaining, Exo1 indepedent resection is likely due to Sgs1, which has also been shown to play a role in resection at uncapped telomeres, especially in the absence of Rad9 [25].
Fig. 2.
The role of Exo1 in global resection at telomeres of cdc13-1 and yku70Δ cdc13-1 mutants. (A) Cells with indicated genotypes in the CG379 strain background grown to late log at 20 °C (YPDA+SRB) were diluted to fresh medium and shifted to 37 °C. Chromosomal DNA was examined using the PFGE protocols (see Material and Methods). (B) Southern of the gel shown in panel (A) using a 32P labeled Chr 9 probe. (C) The signal intensity profiles of 0 h and 4 h timepoints from the Southern from panel (B) indicate the distribution of 0-, 1-, and 2-end resected forms of Chr 9 for each strain. The line diagrams beneath each graph indicate the positions (left to right) of the 2-end, 1-end, and 0-end resection peaks.
3.5. Global chromosome differences in resection between 1-end and 2-end can be detected by 2D PFGE-shift approaches
We developed a 2D PFGE approach to address further the global resection at the individual chromosome level and to characterize the extent of 1-end and 2-end resected chromosomes. Briefly, lanes in the first dimension were cut out (slices are above the 2D gels in Fig. 3) and exposed to mung bean nuclease to remove ssDNA tails. The lane slices were then subjected to PFGE in the second dimension. The bands of chromosomes with ssDNA tails that were PFGE-shifted in the first dimension migrate only according to molecular weight in the second dimension. Because of the nuclease digestion of the first dimension gel slice, they will be shortened (compared to their chromosome counterparts that were not resected during the 37 °C temperature shift) and will have greater mobility. In addition, because the 1-end and 2-end resected chromosomes were separated from the 0-end chromosomes, this provides the unique opportunity to quantitate the relative amounts of resected chromosomes and make comparisons between chromosomes on the same gel.
As shown in the left image of Fig. 3, chromosomes from yku70Δ cdc13-1 cells (CG379 background) that were grown at the permissive temperature (20 °C) separated in both the first and second dimension only according to size, generating a single diagonal of unresected chromosomal spots in the second dimension separation. For the cdc13-1 cells shifted to 37 °C for 4 h (middle image of Fig. 3), all the chromosomes had a major spot and a minor spot, which generated two diagonals, corresponding to 0-end and 1-end resection chromosomal spots, respectively. These results contrast with the yku70Δ cdc13-1 double mutant cells that were shifted to 37 °C for 4 h (see right image of Fig. 3), where there are three diagonals of chromosomal spots. Most chromosomes showed three spots with the slowest (PFGE-shift of 2-end resections) in the first dimension migrating the fastest (i.e., without PFGE-shift) in the second dimension, as expected, following mung bean nuclease treatment of 2-end resected molecules from the 1st dimension lane slice. There were individual chromosome differences in the relative amounts of 0-, 1- and 2-end resections. In all cases there were more molecules with 1-end than 0-end resected chromosomes. For Chr1 and Chr6 most of the chromosomes were in the 1-end band in an approximate ratio of 1:2:1 (0-end; 1-end; 2-end), suggesting that around 75% of these chromosomes in the population were resected. Consistent with results in Supplementary Fig. S1D, there was relatively little 2-end resection of Chr3. We also found that there was only a small amount, at most, of 2-end resection of Chr5 and Chr10. As indicated in Supplementary Fig. 4B, there appeared to be more Chr3 molecules resected in the W303 background than in the CG379 background, and approximately 25% of them showed 2-end resection.
While the proportions of 1-end and 2-end resected chromosomes from the yku70Δ cdc13-1 cells varied, the PFGE-shift patterns indicate that the fraction of all chromosomes with at least one resected end was greater than 50%. This corresponds to a minimum of 32 resected ends per cell based on 32 chromatids and 2 ends each in the G2-arrested cells. When 2-end resections are included and assuming random end-resection, over 40 ends out of 64 telomere ends per G2 cell are resected. Overall, this study provides the first measurement of the number of resected ends and comparisons between mutants. (By way of comparison, a dose of 12.5 krad to a G2 haploid would be expected based on our previous findings to result in a similar number of DSB resected ends: 20 DSBs times 2 ends.)
As described in Supplementary Fig. S5, we were able to assess the lengths of resection of the yku70Δ cdc13-1 chromosomes in Fig. 3 and Supplementary Fig. S5A. We found that there is a linear relation between migration distance and molecular weight from Chr2 (825 kb) down to Chr1 (250 kb) as shown in Supplementary Fig. S5B. This enabled us to determine the size of the two-end resected molecules (orange dots) after mung bean nuclease digestion of the single strand tails and PFGE in the 2nd dimension. For the seven chromosomes with 2-end resections, the total amount of resection varied between 15–30 kb, with an average of ~20 kb, corresponding to 10 kb/telomere end. Thus, in the absence of protection by both Yku70 and Cdc13 proteins, most of the chromosomes experience extended lengths of resection.
3.6. Allelic difference in resection between cdc13 mutant alleles
While the cdc13-1 temperature sensitive (ts) mutant has been commonly used to address telomere stability and protection in yeast, the ts phenotype was proposed to be due to a combination of natural high temperature sensitivity and the cdc13-1 defect [26]. Additional cdc13 mutants were isolated by the Lundblad lab that could specifically prevent growth at lower temperatures. Among the mutants were those that had reduced amount of Cdc13 protein at nonpermissive temperatures. Since such mutants might have altered levels of resection, we investigated one of them using our PFGE approach. Presented in Fig. 4A & B are results for the cdc13-FS allele (which corresponds to the cdc13-F684S allele from [26]) and the cdc13-1 allele either as a single mutation or in combination with a yku70Δ mutation. (These experiments with W303 cells, which were done in the defined SC+AD+SRB medium used in the foci experiments below, yielded results similar to those in YPDA+SRB, as shown in Supplementary Figs. S6A and S6B.) As compared to the cdc13-1 mutants, we found less molecules that were extensively resected, possibly suggesting differences in retained binding or telomere complex formation between the Cdc13-FS and the Cdc13-1 proteins. This was useful for later experiments below where we wanted to address the consequences of a reduced number of resected molecules. The findings also provide new approaches to addressing allelic differences in genes that influence telomere stability.
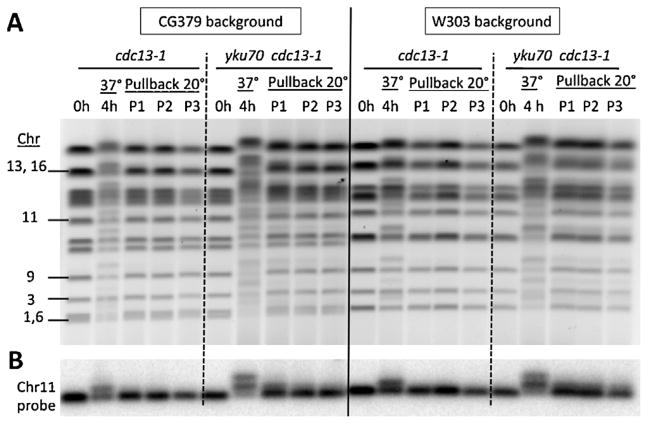
3.7. Efficient telomere restoration after extensive telomere resection
Since over 50% of the chromatids in the yku70Δ cdc13-1 strains had extensively resected telomere ends and many had resection at both ends by 4 h after shifting to 37 °C (Figs. 1 and 3), we investigated the ability to restore the resected molecules across the genome to the double-strand state. The CG379 and the W303 cells growing at 20 °C in YPDA+SRB were shifted to 37 °C for 4 h and then returned to 20 °C. As shown in Fig. 5, there was considerable resection by 4 h at 37 °C with results comparable to those for the cdc13-1 and yku70Δ cdc13-1 mutants in Figs. 1, 2 and 4. For the cdc13-1 single mutants in both genetic backgrounds the restitution appeared to be complete by 1 h after pullback to 20 °C, at least at the level that could be detected with the stained PFGE gel. For the double mutant, it appeared that restitution was 80–90% complete within 1 h, possibly somewhat less in the W303 background. Thus, in spite of the high frequency of extensively resected molecules, there was efficient restitution across the genome. These findings are expected to provide opportunities to address the nature of restitution and the role of various polymerases as well as the possible role of recombination.
Fig. 5.
Efficient and rapid restoration of telomeres following resection in cdc13-1 and yku70Δ cdc13-1 cells having a W303 or CG379 background. Cells were grown to late log at 20 °C in YPDA+SRB. They were diluted in fresh media and shifted to 37 °C for 4 h and then incubated for 1–3 h (P1, P2 and P3) at 20 °C. Presented in (A) are the stained pulse field gels and in (B) Southerns using Chr11 specific 32P labeled probe.
3.8. Effect of resection on survival in the CG379 and the W303 cells
The high frequency of chromosomes with at least 1-end resection in the yku70Δ cdc13-1 strains after 4 h at 37 °C and subsequent efficient restitution when cells are returned to 20 °C led us to investigate the impact on survival. Cells of the cdc13-1 and yku70Δ cdc13-1 in both genetic backgrounds were grown to stationary phase in YPDA+SRB at 20 °C and were diluted in fresh media and adjusted to a common titer of 107 cells/ml before shifting the temperature to 37 °C for 4 h. (For the 0 h samples, a portion of each diluted culture was held on ice during the time course.) Survival was assessed by 10-fold serial dilution prongings to YPDA+SRB agar plates.
As shown in Fig. 6A and B, the plating efficiencies prior to raising the temperature were comparable for all strains: WT, yku70Δ, cdc13-1 and cdc13-1 yku70Δ. After raising the temperature to 37 °C for 4 h, there was an increase in titer and corresponding survival for the WT and the yku70Δ single mutants. There was no apparent decrease in survival for the cdc13-1 or cdc13-1 yku70Δ strains in the CG379 background. This contrasts with results for the W303 background where there was a > 20-fold reduction in survival for the yku70Δ cdc13-1 but little change in the WT, yku70Δ or cdc13-1cells. Based on the results with yku70Δ cdc13-1 double mutant cells in the CG379 background, we conclude that short term extensive global resection of telomeres is not per se a major threat to genome integrity. The > 20-fold reduction in survival of the corresponding double mutant in the W303 background is due to unknown differences between the strain backgrounds, since there were comparable levels of resection as well as restitution after return to permissive temperature. These findings of high frequencies of resected ends and rapid restitution along with high survival (as well as strain differences) can provide opportunities to address genetic factors that can influence telomere maintenance.
3.9. Rad52 and Rfa1 protein associations with resected telomeres
The Cdc13 protein along with RPA and other proteins bind to telomere ends to protect them and to coordinate end replication. Since we show here that there is considerable resection in yku70Δ cdc13 double mutants involving most telomeres and that there is efficient restitution at permissive temperature, we investigated the relationship between Rad52, Rfa1 (the largest subunit of RPA) and resected telomeres. RPA associates strongly with ssDNA and facilitates DNA synthesis. The Rad52 protein also associates with ssDNA and initiates early steps in recombination leading to Rad51 filament intermediates [27–29]. Given the high frequencies of ssDNA we asked whether there was a corresponding change in Rad52 and Rfa1 associations in single cells by examining foci formation. When multiple DSBs are induced by radiation, there is primarily only one Rad52 focus per cell, or at most a few cells with multiple Rad52 foci [30]. We assessed whether similar events occurred when many telomeres are resected. As described above, there can be a large number of telomere resection events per cell; however, Cdc13-dependent resection events at the ends of the same chromosome are independent, unlike for IR-induced DSBs, where resection at the two ends of the DSB is coordinated [10].
To address Rad52 and RPA foci formation in relation to telomere resection, we used W303 cells expressing YFP tagged Rad52 and CFP tagged Rfa1 that have been utilized in previous studies that addressed induction and processing of DSBs and other DNA lesions [22]. To log phase cells grown in SC+AD+SRB at 23 °C nocodazole (15 ug/ml final concentration) was added and cells were transferred to 37 °C. The nocodazole assured all cells would arrest at G2/M since WT and yku70Δ might continue growing at the high temperature. We used SC+AD+SRB to better discern foci, although there were somewhat less resected molecules compared to YPDA+SRB (Supplementary Fig. S6A).
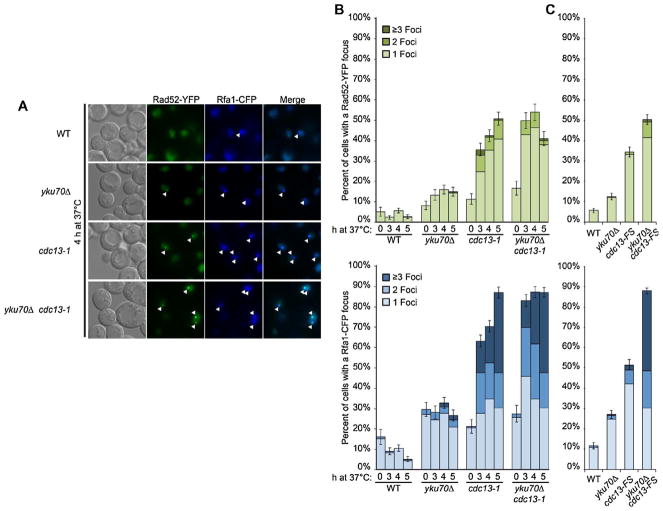
Presented in Fig. 7A are pictures of the Rad52-YFP and Rfa1-CFP foci after shifting cells from 23 °C to 37 °C for WT and the various mutants. The frequencies are summarized in Fig. 7B and C. We note that under all conditions, regardless of the mutant, Rad52 foci were always merged with the Rfa1 foci, consistent with Rad52 interactions with RPA on ssDNA for stable Rad52 binding [31]. Prior to the shift, ~5% or less of the WT cells had a Rad52 focus and 5–10% had an Rfa1 focus. There were no multiple Rad52 foci and at most a few percent of cells had multiple Rfa1 foci. The nocadazole arrested WT cells exhibited no increase in the frequency of Rad52 foci after 3–5 h at 37 °C. There did appear to be some decrease in Rfa1 foci, which might be due to completion of unfinished replication.
Fig. 7.
Appearance of Rad52-YFP and Rfa1-CFP foci in WT and various cdc13-1 and cdc13-FS mutants experiencing global telomere resection. Logarithmically growing (W303 background) cells in SC+AD+SRB that express Rad52-YFP and Rfa1-CFP were switched from 23 °C to 37 °C and nocadazole (15 micrograms/ml) was added to arrest cells in G2 since WT and yku70Δ cells would continue growing. Nocadazole was added hourly at half the amount to assure arrest. (These microscopy experiments correspond to the PFGE-shift experiments in the right half of gel of Supplementary Fig. S6.) At the times indicated, cells were harvested by centrifugation and large budded cells were examined for focus formation by fluorescent microscopy with 11 z-stacks corresponding to 3 μm examined. A focus is counted if the focus is observed in a minimum of 3 consecutive Z-planes. (A) Examples of a single Z-stack showing cells with Rad52 and Rfa1 foci at 4 h after the switch to 37 °C. Note that a Rad52 focus is always detected in association with a Rfa1 focus and typically only one Rad52 focus is observed per cell. (B) Includes cells with the cdc13-1 allele. Presented are results from two independent experiments. Total cells examined at “0” time was between 100 and 200. For 3–5 h the totals were between 200 and 300. (C) Rad52 and Rfa1 foci determined in cells with the cdc13-FS allele at 4 h. Presented are results from three independent experiments and a total of 400–500 cells examined.
The frequency of yku70Δ mutant cells with Rad52 and Rfa1 foci was ~2-fold more than for WT cells prior to temperature shift. There was no change in Rfa1 following the shift to 37 °C suggesting no additional lesions that would lead to resection. This is consistent with the absence of PFGE-shifted molecules (Fig. 1). The temperature increase did lead to a small increase in cells with Rad52 foci reaching ~15% of the cells.
The results for the cdc13-1 mutant were markedly different from the WT and the yku70Δ strains, although the incidence of foci at “0” time was similar for the yku70Δ and the cdc13-1 mutants. By 3 h, ~35% and ~65% of cdc13-1 cells had at least one Rad52 or Rfa1 focus. At 5 h the percent of cells with a Rad52 focus increased to ~50%, with ~10% of the cells exhibiting two Rad52 foci and few multiple foci (≥3 foci). This contrasts with Rfa1, where the frequency of cells with at least 1 focus at 5 h increased to ~90% with nearly ~60% cells having ≥2 foci and 40% of the cells having ≥3 foci.
The cdc13-1 single mutant and the double mutant yku70Δ cdc13-1 cells had similar frequencies of Rad52 foci at 5 h after incubation at 37 °C, as was also found for the Rfa1 foci although the number of extensive resection events is much larger in the double mutant. However, while the frequencies of cells with foci were high at 3, 4, and 5 h for the double mutant, it took until 5 h to reach the corresponding high levels of Rad52 and Rfa1 foci with the single mutant. The number of cells with a Rad52 focus is far less than the number of 1- or 2-end resected chromosome molecules in cells (Fig. 5). At most only 10% of the cdc13-1 or yku70Δ cdc13-1 mutants had two Rad52 foci. This contrasted with Rfa1 foci where the frequencies of cells with multiple (≥3) foci increased to nearly 40% for both the cdc13-1 and the yku70Δ cdc13-1 strains. However, the number of Rfa1 foci per cell is still much less than the expected frequency of extensively resected molecules in the yku70Δ cdc13-1: ~30–40/cell, based on the frequencies of molecules that do not exhibit PFGE-shift.
Although similar frequencies of Rad52 and Rfa1 foci were found, respectively, in the cdc13-1 and yku70Δ cdc13-1 mutants, the frequencies of resected molecules were ~3-fold less for the single cdc13-1 mutant for the chromosomes examined by Southern blot and there were almost no 2-end resected molecules as shown in Supplementary Figs. S6A and S6B. This led us to investigate cells with the cdc13-FS allele, since the incidence of resected molecules at 37 °C was much less than for the cdc13-1 cells (Fig. 4). Based on the stained gels and Southern analysis of Chr11, < 5% and ~10–20% molecules (i.e., 6–12 of 64 chromatids) were resected in the cdc13-FS and the yku70Δ cdc13-FS strains, respectively. In spite of the much lower frequencies of resected molecules, ~35% of the single mutant cdc13-FS cells had a Rad52 focus and ~50% had at least one Rfa1 focus after 4 h at 37 °C compared with ~40% and 70%, respectively, for the cells containing the cdc13-1 allele, as described in Fig. 7B and C. The incidence of multiple Rfa1 foci was much less for the single mutant cdc13-FS cells, consistent with the fewer resected molecules and, therefore, fewer opportunities per cell for RPA/ssDNA formation. For the yku70Δ cdc13-FS cells, ~50% had a Rad52 focus, which is comparable to that for the yku70Δ cdc13-1 double mutant, in spite of fewer resected molecules and few cells had 2 or more Rad52 foci. The low numbers of Rad52 foci and the few multiple foci for both double mutants, yku70Δ cdc13-1 and yku70Δ cdc13-FS, is consistent with our proposed central location for repair of resected telomeres. For the Rfa1 foci, the frequencies of cells with Rfa1 foci were ~85 to 90% for both double mutants, with two thirds of them having multiple foci and 4–10% of cells with 7 or 8 Rfa1 foci (Supplementary Table S3). Among the cells with 7 or 8 Rfa1 foci, none had 2 or more Rad52 foci.
In summary, both the single cdc13-1 and cdc13-FS defects can result in at least one third of the cells accumulating Rad52/Rfa1 foci. This increases to nearly one half of the cells having Rad52/Rfa1 foci for both the yku70Δ cdc13-1 and the yku70Δ cdc13-FS mutants where nearly 90% of cells have at least one Rfa1 focus and 60% have multiple Rfa1 foci. Therefore, while the appearance of large numbers of Rfa1 foci is prevented by Cdc13, the incidence of Rfa1 foci is not directly related to the number of extensively resected molecules in the cdc13 mutants.
4. Discussion
The protection of chromosomal DNA is essential in all eukaryotes to assure genome stability within and between generations. The ends of chromosomes are continual potential threats that have been reckoned with by elaborate protein complexes that both deal with the intricacies of end replication and prevent access to nucleases [32,33,21]. Here, using our PFGE-shift approach and modifications of growth conditions we addressed globally the extent of telomere protection provided by Cdc13 and Yku.
In WT and yku70 single mutants, we did not detect PFGE-shifted chromosomes. Although Yku has been shown to protect ends from resection and to assure telomere length over many generations [14], in yku70 single mutants the extent of resection is small, limited to the region near the telomere tips [21]. This is consistent with our previous report that 300 kb Chr3 molecules with less than a few hundred bases of resection are at the threshold of detection by PFGE-shift, On the other hand, our global analyses of resection in cdc13-1 mutants reflects previous measurements on individual chromosomes in that the lengths of resection can be substantial. The extent of PFGE-shift is consistent with resection lengths in the range of thousands of bases based on our previous findings with resection at DSB ends [8] and also nuclease digestion of ends (Fig. 3). Based on Southern analysis and comparisons with the stained gels, we are able to estimate that ~20–30% or ~6–9 chromatids per G2 cdc13-1 cell experience long telomere end resection.
While the absence of Yku did not lead to PFGE-shift detectable resected chromosomes, there was a rapid increase in the frequency of resected chromosomes reaching 60–70% by 5 h in the yku70Δ cdc13-1 cells after shifting to the nonpermissive temperature. Thus, Yku70/80 provides considerable protection from extensive resection in the absence of Cdc13, suggesting it can still associate with telomere ends in a cdc13-1 mutant.
The PFGE-shift approach also provided a unique opportunity to identify 1- and 2-end resected molecules. Previously, we had shown that resection at the 2-ends of a DSB end was coordinated and that in the absence of Rad50/Mre11 most resected molecules had only 1- end resected [10]. In the present experiments we found that nearly all telomere resection in the cdc13-1 mutant was 1-end, suggesting independent resection of telomere ends of chromosomes although there may be associations between telomeres [11]. The PFGE-system allowed us to address the length of resection for 1-end and 2-end events. We found that on average, there was about 10 kb resection per end within 4 h, which did not increase over an additional hour. This is similar to what we found for resection at radiation-induced DSBs. It would be interesting to explore factors affecting resection and impacts on different chromosomes.
Nearly all chromosomes had the potential to undergo at least 1-end resection. (The large sizes of Chr4 and Chr12 precluded addressing PFGE-shift under the conditions used.) The greatly reduced or lack of 2-end events for Chr3 in the CG379 strains could be explained by reduced access to one of the two ends, an issue that would be interesting to pursue in future studies. Based on close examination in Figs. 1 and 3, there appears to be a higher percentage of nonresected Chr3 molecules than other chromosomes. As noted above, resection of telomere ends differs between the CG379 and the W303 background in that for the W303 strain there is the appearance of 2-end events for Chr3 and there are less nonresected Chr3 molecules (compare Fig. 1A and C). Overall, these results demonstrate for the first time that protection against telomere end resection can differ between ends, which has implications for telomere metabolism along with individual telomere and chromosome stability. Factors that might contribute to telomere differences in protection include variability in the telomere TG1-3 and the X and Y′ subtelomeric repeat regions as well as differences in Rap1 binding and associated Rif1/Rif2 proteins [21,34].
Taking into account 1- and 2-end resected molecules, over 40 telomere ends per cell are resected in the yku70Δ cdc13-1 at restrictive temperature. The large differences in survival of yku70Δ cdc13-1 double mutants in the CG379 and W303 backgrounds despite efficient, rapid restoration of double-strand telomere ends could be exploited to address factors that influence potential biological consequences of resection, such as various repair mutants and genes that affect telomere length [35]. For example, previously we had found that specific telomere resection leads to increased regional mutation [15]. Also, there might be differences in telomere recombinational activities. Using PFGE to address chromosomal changes (cf. [36]), it would be interesting to address chromosome instability including rearrangements and loss in diploid yku70Δ cdc13-1 mutants [8]. This system also provides a good opportunity to address mechanisms and limitations of DNA synthesis at resected telomeres including polymerases and nucleotide pools.
By combining measurements of foci formation with our results on global resection we were able to relate the underlying Rad52 and RPA processes to frequencies of resected molecules. The Rad52 and the RPA proteins associate with ssDNA to provide opportunities for recombination and, in the case of RPA, it is important in resection [37] and for protecting ssDNA. As expected, the Rad52 foci are always seen in association with Rfa1 foci, but not vice versa, since the Rad52 is stabilized at RPA covered ssDNA substrates [31]. While multiple radiation-induced DSBs lead to the appearance of Rad52 foci, typically there are only 1 or 2 per cell [30]. A similar phenomenon of primarily just one focus was found in our present study although the number of resected molecules differed considerably between the cdc13-1 and the yku70Δ cdc13-1 mutants, even when the frequency of resected molecules was much lower in cells with the cdc13-FS allele. However, the average number of Rfa1 foci is at least 2-fold higher than Rad52 foci with as many as 7 or more foci/cell (Supplementary Table S3). Based on the average number of Rfa1 foci, the limitation on Rad52 foci is not due to limited amounts of RPA at ssDNA. Our results suggest that there may be a Rad52 determined center that associates with the resected ends. Given our findings on the distribution of the Rad52 and RPA proteins, it would be interesting to address the influence of these genes on survival when cells are returned to permissive conditions.
The finding of multiple Rfa1 foci (Fig. 7 and Supplementary Table 3) when there was only a small per cent of extensively resected chromosomes per cell as found for the yku70Δ cdc13-FS and the single cdc13-1 mutants (Fig. 4) indicates that resected ends may not be closely clustered in the nucleus. Previous findings with foci in normal cells have indicated that telomeres assemble into multiple clusters near nuclear pores [38]. Since telomeres are anchored at nuclear pores, it would be interesting to address the nuclear organization of the highly resected telomere regions and our observed single Rad52 focus in light of studies with eroding telomeres in cells lacking telomerase [39].
Thus, in our global approach to addressing the roles of Yku and Cdc13 in protecting yeast telomeres, we establish that most telomeres are subject to resection in their absence and that the resection is not coordinated at individual chromosome ends, unlike for damage induced DSBs. On its own, Yku appears to protect against localized [21] but not extended telomere resection. However, it does protect against the potential for large-scale global resection in cdc13 mutants. We show for the first time that even large numbers of resected telomeres, as found in the yku70Δ cdc13-1 double mutants, can be rapidly repaired, resulting in high survival and suggesting highly robust mechanisms for telomere protection.
Supplementary Material
Acknowledgments
Funding
This study was supported by the Intramural Research Program of the National Institute of Environmental Health Sciences, NIH (project 1 Z01 ES065073 to M.A.R.) and the National Institutes of Health grant (ES024872 to K.A.B.) and the American Cancer Society (129182-RSG-16-043-01-DMC to K.A.B.).
We thank Dr. Vicki Lundbland for providing plasmids to create various CDC13 mutants. We also thank Dr. Dmitry Gordenin for providing the DAG635 and DAG760 strains, from which all other CG379 background strains used in this study were derived. The authors would like to thank Cynthia Sakofsky and Jessica Williams for critical comments on the manuscript. The authors appreciate the efforts of the Fluorescence Microscopy and Imaging Center, NIEHS, especially Erica Scappini and Jeff Tucker.
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at https://doi.org/10.1016/j.dnarep.2017.11.010.
Footnotes
Conflict of interest statement
The authors declare that there are no conflicts of interest
References
- 1.Maciejowski J, de Lange T. Telomeres in cancer: tumour suppression and genome instability. Nat Rev Mol Cell Biol. 2017;18:175–186. doi: 10.1038/nrm.2016.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kupiec M. Biology of telomeres: lessons from budding yeast. FEMS Microbiol Rev. 2014;38:144–171. doi: 10.1111/1574-6976.12054. [DOI] [PubMed] [Google Scholar]
- 3.Larrivee M, LeBel C, Wellinger RJ. The generation of proper constitutive G-tails on yeast telomeres is dependent on the MRX complex. Genes Dev. 2004;18:1391–1396. doi: 10.1101/gad.1199404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garvik B, Carson M, Hartwell L. Single-stranded DNA arising at telomeres in cdc13 mutants may constitute a specific signal for the RAD9 checkpoint. Mol Cell Biol. 1995;15:6128–6138. doi: 10.1128/mcb.15.11.6128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dewar JM, Lydall D. Similarities and differences between uncapped telomeres and DNA double-strand breaks. Chromosoma. 2012;121:117–130. doi: 10.1007/s00412-011-0357-2. [DOI] [PubMed] [Google Scholar]
- 6.Lydall D. Hiding at the ends of yeast chromosomes: telomeres, nucleases and checkpoint pathways. J Cell Sci. 2003;116:4057–4065. doi: 10.1242/jcs.00765. [DOI] [PubMed] [Google Scholar]
- 7.Marcomini I, Gasser SM. Nuclear organization in DNA end processing: telomeres vs double-strand breaks. DNA Repair (Amst) 2015;32:134–140. doi: 10.1016/j.dnarep.2015.04.024. [DOI] [PubMed] [Google Scholar]
- 8.Westmoreland J, Ma W, Yan Y, Van Hulle K, Malkova A, Resnick MA. RAD50 is required for efficient initiation of resection and recombinational repair at random, gamma-induced double-strand break ends. PLoS Genet. 2009;5:e1000656. doi: 10.1371/journal.pgen.1000656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Westmoreland JW, Resnick MA. Recombinational repair of radiation-induced double-strand breaks occurs in the absence of extensive resection. Nucleic Acids Res. 2016;44:695–704. doi: 10.1093/nar/gkv1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Westmoreland JW, Resnick MA. Coincident resection at both ends of random, gamma-induced double-strand breaks requires MRX (MRN), Sae2 (Ctp1), and Mre11-nuclease. PLoS Genet. 2013;9:e1003420. doi: 10.1371/journal.pgen.1003420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bystricky K, Laroche T, van Houwe G, Blaszczyk M, Gasser SM. Chromosome looping in yeast: telomere pairing and coordinated movement reflect anchoring efficiency and territorial organization. J Cell Biol. 2005;168:375–387. doi: 10.1083/jcb.200409091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dewar JM, Lydall D. Simple, non-radioactive measurement of single-stranded DNA at telomeric, sub-telomeric, and genomic loci in budding yeast. Methods Mol Biol. 2012;920:341–348. doi: 10.1007/978-1-61779-998-3_24. [DOI] [PubMed] [Google Scholar]
- 13.Paschini M, Mandell EK, Lundblad V. Structure prediction-driven genetics in Saccharomyces cerevisiae identifies an interface between the t-RPA proteins Stn1 and Ten1. Genetics. 2010;185:11–21. doi: 10.1534/genetics.109.111922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Polotnianka RM, Li J, Lustig AJ. The yeast Ku heterodimer is essential for protection of the telomere against nucleolytic and recombinational activities. Curr Biol. 1998;8:831–834. doi: 10.1016/s0960-9822(98)70325-2. [DOI] [PubMed] [Google Scholar]
- 15.Yang Y, Sterling J, Storici F, Resnick MA, Gordenin DA. Hypermutability of damaged single-strand DNA formed at double-strand breaks and uncapped telomeres in yeast Saccharomyces cerevisiae. PLoS Genet. 2008;4:e1000264. doi: 10.1371/journal.pgen.1000264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goldstein AL, McCusker JH. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast. 1999;15:1541–1553. doi: 10.1002/(SICI)1097-0061(199910)15:14<1541::AID-YEA476>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 17.Thomas BJ, Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell. 1989;56:619–630. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- 18.Zhao X, Muller EG, Rothstein R. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol Cell. 1998;2:329–340. doi: 10.1016/s1097-2765(00)80277-4. [DOI] [PubMed] [Google Scholar]
- 19.Sherman FG, Fink GR, Hicks JB. Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press; Plainview NY: 1986. [Google Scholar]
- 20.Lisby M, Rothstein R, Mortensen UH. Rad52 forms DNA repair and recombination centers during S phase. Proc Natl Acad Sci U S A. 2001;98:8276–8282. doi: 10.1073/pnas.121006298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bonetti D, Clerici M, Anbalagan S, Martina M, Lucchini G, Longhese MP. Shelterin-like proteins and Yku inhibit nucleolytic processing of Saccharomyces cerevisiae telomeres. PLoS Genet. 2010;6:e1000966. doi: 10.1371/journal.pgen.1000966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lisby M, Barlow JH, Burgess RC, Rothstein R. Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell. 2004;118:699–713. doi: 10.1016/j.cell.2004.08.015. [DOI] [PubMed] [Google Scholar]
- 23.Zubko MK, Guillard S, Lydall D. Exo1 and Rad24 differentially regulate generation of ssDNA at telomeres of Saccharomyces cerevisiae cdc13-1 mutants. Genetics. 2004;168:103–115. doi: 10.1534/genetics.104.027904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maringele L, Lydall D. EXO1-dependent single-stranded DNA at telomeres activates subsets of DNA damage and spindle checkpoint pathways in budding yeast yku70Delta mutants. Genes Dev. 2002;16:1919–1933. doi: 10.1101/gad.225102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ngo HP, Lydall D. Survival and growth of yeast without telomere capping by Cdc13 in the absence of Sgs1, Exo1, and Rad9. PLoS Genet. 2010;6:e1001072. doi: 10.1371/journal.pgen.1001072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paschini M, Toro TB, Lubin JW, Braunstein-Ballew B, Morris DK, Lundblad V. A naturally thermolabile activity compromises genetic analysis of telomere function in Saccharomyces cerevisiae. Genetics. 2012;191:79–93. doi: 10.1534/genetics.111.137869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Benson FE, Baumann P, West SC. Synergistic actions of Rad51 and Rad52 in recombination and DNA repair. Nature. 1998;391:401–404. doi: 10.1038/34937. [DOI] [PubMed] [Google Scholar]
- 28.Shinohara A, Ogawa T. Stimulation by Rad52 of yeast Rad51-mediated recombination. Nature. 1998;391:404–407. doi: 10.1038/34943. [DOI] [PubMed] [Google Scholar]
- 29.New JH, Sugiyama T, Zaitseva E, Kowalczykowski SC. Rad52 protein stimulates DNA strand exchange by Rad51 and replication protein A. Nature. 1998;391:407–410. doi: 10.1038/34950. [DOI] [PubMed] [Google Scholar]
- 30.Lisby M, Mortensen UH, Rothstein R. Colocalization of multiple DNA double-strand breaks at a single Rad52 repair centre. Nat Cell Biol. 2003;5:572–577. doi: 10.1038/ncb997. [DOI] [PubMed] [Google Scholar]
- 31.Sugiyama T, Kantake N. Dynamic regulatory interactions of rad51, rad52, and replication protein-a in recombination intermediates. J Mol Biol. 2009;390:45–55. doi: 10.1016/j.jmb.2009.05.009. [DOI] [PubMed] [Google Scholar]
- 32.Greetham M, Skordalakes E, Lydall D, Connolly BA. The telomere binding protein cdc13 and the single-stranded DNA binding protein RPA protect telomeric DNA from resection by exonucleases. J Mol Biol. 2015;427:3023–3030. doi: 10.1016/j.jmb.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Booth C, Griffith E, Brady G, Lydall D. Quantitative amplification of single-stranded DNA (QAOS) demonstrates that cdc13-1 mutants generate ssDNA in a telomere to centromere direction. Nucleic Acids Res. 2001;29:4414–4422. doi: 10.1093/nar/29.21.4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vodenicharov MD, Laterreur N, Wellinger RJ. Telomere capping in non-dividing yeast cells requires Yku and Rap1. EMBO J. 2010;29:3007–3019. doi: 10.1038/emboj.2010.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Askree SH, Yehuda T, Smolikov S, Gurevich R, Hawk J, Coker C, Krauskopf A, Kupiec M, McEachern MJ. A genome-wide screen for Saccharomyces cerevisiae deletion mutants that affect telomere length. Proc Natl Acad Sci U S A. 2004;101:8658–8663. doi: 10.1073/pnas.0401263101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Argueso JL, Westmoreland J, Mieczkowski PA, Gawel M, Petes TD, Resnick MA. Double-strand breaks associated with repetitive DNA can reshape the genome. Proc Natl Acad Sci U S A. 2008;105:11845–11850. doi: 10.1073/pnas.0804529105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen H, Lisby M, Symington LS. RPA coordinates DNA end resection and prevents formation of DNA hairpins. Mol Cell. 2013;50:589–600. doi: 10.1016/j.molcel.2013.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gotta M, Laroche T, Formenton A, Maillet L, Scherthan H, Gasser SM. The clustering of telomeres and colocalization with Rap1, Sir3, and Sir4 proteins in wild-type Saccharomyces cerevisiae. J Cell Biol. 1996;134:1349–1363. doi: 10.1083/jcb.134.6.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Khadaroo B, Teixeira MT, Luciano P, Eckert-Boulet N, Germann SM, Simon MN, Gallina I, Abdallah P, Gilson E, Geli V, et al. The DNA damage response at eroded telomeres and tethering to the nuclear pore complex. Nat Cell Biol. 2009;11:980–987. doi: 10.1038/ncb1910. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.