Abstract
Salmonella ranks high among the pathogens causing foodborne disease outbreaks. According to the Centers for Disease Control and Prevention, Salmonella contributed to about 53.4% of all foodborne disease outbreaks from 2006 to 2017, and approximately 32.7% of these foodborne Salmonella outbreaks were associated with consumption of produce. Trace-back investigations have suggested that irrigation water may be a source of Salmonella contamination of produce and a vehicle for transmission. Presence and persistence of Salmonella have been reported in surface waters such as rivers, lakes, and ponds, while ground water in general offers better microbial quality for irrigation. To date, culture methods are still the gold standard for detection, isolation and identification of Salmonella in foods and water. In addition to culture, other methods for the detection of Salmonella in water include most probable number, immunoassay, and PCR. The U.S. Food and Drug Administration (FDA) issued the Produce Safety Rule (PSR) in January 2013 based on the Food Safety Modernization Act (FSMA), which calls for more efforts toward enhancing and improving approaches for the prevention of foodborne outbreaks. In the PSR, agricultural water is defined as water used for in a way that is intended to, or likely to, contact covered produce, such as spray, wash, or irrigation. In summary, Salmonella is frequently present in surface water, an important source of water for irrigation. An increasing evidence indicates irrigation water as a source (or a vehicle) for transmission of Salmonella. This pathogen can survive in aquatic environments by a number of mechanisms, including entry into the viable but nonculturable (VBNC) state and/or residing within free-living protozoa. As such, assurance of microbial quality of irrigation water is critical to curtail the produce-related foodborne outbreaks and thus enhance the food safety. In this review, we will discuss the presence and persistence of Salmonella in water and the mechanisms Salmonella uses to persist in the aquatic environment, particularly irrigation water, to better understand the impact on the microbial quality of water and food safety due to the presence of Salmonella in the water environment.
Keywords: irrigation water, agricultural water, Salmonella, produce safety, prevalence, pathogen detection, foodborne outbreaks
Introduction
Salmonella is a natural inhabitant in the gastrointestinal tract of many animals, including birds, reptiles, livestock, and humans (1–7). Salmonellosis caused by nontyphoidal Salmonella ranks among the highest in all gastroenteritis cases linked to food consumption, affecting the health of approximately one million people annually in the United States alone (8, 9), resulting in medical costs of $3.7 billion. It is estimated that Salmonella species causes 93.8 million cases of gastroenteritis worldwide annually with 155,000 deaths (5−95th percentile, 39,000–303,000) (10). The causative source for salmonellosis has traditionally been attributed to animal origin (11, 12), including meat, eggs, and other poultry products, which has attracted considerable regulatory attention and enormous mitigation efforts (2, 9). In recent years, the number of foodborne outbreaks due to nontraditional sources of the pathogen such as domestic or imported fresh fruits, vegetables, spices, and nuts has been increasing (9, 13–15). A recent report from the Centers for Disease Control and Prevention (CDC) suggested nearly half (46%) of foodborne illnesses and 23% of deaths were associated with produce consumption (16). For instance, during July 2015 to February 2016, a Salmonella outbreak associated with consumption of imported Mexican cucumbers caused illness in at least 907 people, with six deaths in 40 states within the United States (17).
Consumption of more fresh fruits and vegetables has been advocated as a healthier diet habit because raw, or less processed, fruits and vegetables are good sources of vitamins, fiber, and other beneficial nutrients (18, 19). Since the early 1970s, the demand for fresh produce in the US has been on the rise continuously, and the estimated increase of per capita consumption of fresh fruits and vegetables from 1982 to 1997 reached approximately 32% (13). The consumption of vegetables grew more rapidly than fruits from 1976 to 2009 (20). A recent investigation by the US Department of Agriculture also suggested Americans consumed more fresh produce in 2015 than in 1970 (21) As a result, it is expected that more produce-related outbreaks of disease will occur even if the contamination sources and rates stay at the present levels. This has attracted the attention of the legislative branch and food safety regulatory agencies in the United States. The Food Safety Modernization Act (FSMA), which was enacted in 2011, emphasizes the significance of produce safety. In response to that, the US Food and Drug Administration (FDA) issued the Produce Safety Rule (PSR) in January 2013 to establish science-based standards for the growing, harvesting, packing, and holding by domestic and foreign farms of produce consumed in the United States (22).
The causative pathogens for produce contaminations include viruses, bacteria, and parasites. Contamination may occur at various stages during the production process (2). Salmonella has been regarded as the primary pathogen for causing produce-related foodborne outbreaks. Pathogenic bacteria carried by produce that can lead to an outbreak may involve multiple external sources and production stages. For instance, at the pre-harvest stage, Salmonella can come from specific agricultural practices such as using animal manure as fertilizer (1) and others may include using contaminated water for irrigation, pesticide spraying, or anti-frost spraying (2, 23, 24).
Water has been shown as a source of microbial contamination of fresh produce and a vehicle for pathogen transmission (23, 25). FSMA defines “agricultural water” partially as water that is “intended to, or is likely to contact covered produce or food contact surfaces” (22). Irrigation water consists of a major component of agricultural water. The sources of irrigation water can come from ground water, surface water, or municipal water. Irrigation water can be applied to produce through various ways including drip tape, furrows, and overhead sprinklers (26). In this review, we will primarily focus on the presence, survival, persistence, and source of Salmonella in surface water, particularly irrigation water, to help us better understand the impact on the microbial quality of water and food safety due to the presence of Salmonella in the water environment.
Produce as a vehicle for transmission of Salmonella
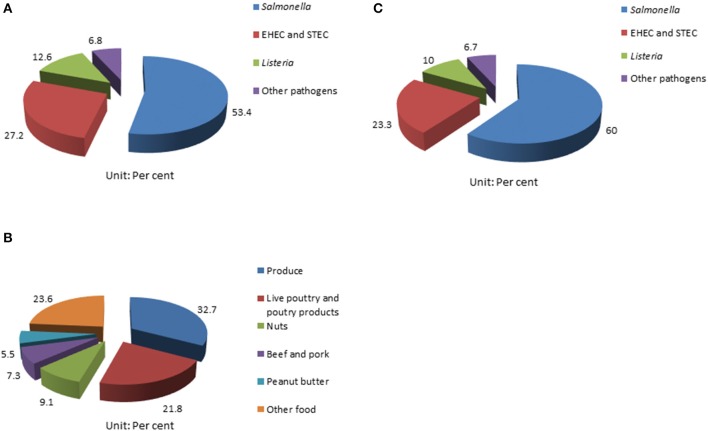
Foodborne outbreaks associated with fresh produce in the United States have been on the rise in the last few decades; and Salmonella has been recognized as the primary causative pathogen (1, 11, 13, 27). According to a report from the CDC, 31 Salmonella outbreaks from 2002 to 2003 were associated with fresh produce; while 29 were poultry related (2, 28). Also, Salmonella contributes to 53.4% (55/103) of the foodborne disease outbreaks documented by CDC among all pathogens investigated from 2006 to 2017 (Figure 1A); 32.7% (18/55) of the multistate foodborne Salmonella outbreaks were associated with produce (Figure 1B); and 60% (18/60) of the fresh produce-related outbreaks were attributed to Salmonella among all the pathogens involved (Figure 1C) (29). Many common types of produce have been implicated in Salmonella-related outbreaks, including beans, alfalfa sprouts, tomatoes, hot peppers, lettuce, cucumbers, cantaloupes, water melons, papayas, and mangoes (1, 2, 29–32).
Figure 1.
Summary of surveillance of foodborne pathogen outbreaks from CDC (29). (A) Multistate foodborne outbreaks caused by Salmonella rank highest in all the pathogens surveilled; (B) the percentage of multistate Salmonella outbreaks linked to produce from 2006 to 2017; and (C) and Salmonella contributes to 60% of all produce-related outbreaks caused by different pathogens surveilled.
Salmonella is originally from, and has adapted to, the microenvironment of the gastrointestinal tract of animals. Once released from the animal gastrointestinal tract or other sources with feces or exudates, Salmonella can be carried to surface waters through rainfall and surface runoffs, survive many challenges such as ultraviolet (UV) radiation from sunlight, poor nutrients, the changes in pH, and temperature (33–35). After attaching to produce, Salmonella must persist under these adverse environmental conditions at a sufficient concentration to cause human illness (13). The surface of various fruits and vegetables provides a niche for numerous bacterial species, such as epiphytes, plant pathogens, and opportunistic human pathogens, such as Salmonella (36). Interestingly, some plant pathogenic bacteria, such as Pectobacterium carotovorum, can promote the growth of Salmonella by macerating plant tissue and providing nutrients (9, 37), whereas other bacterial species such as Panebacillus spp. on tomatoes and cilantro may inhibit or even kill Salmonella (38, 39). Thus Salmonella has to survive the inhibition from these microbes and outcompete them for the acquisition of nutrients and space (33).
The mechanisms for how Salmonella are carried in irrigation water or from other sources interact with plants, survive or persist on these intermediate hosts have become an interesting topic in recent years. Studies indicate Salmonella can be internalized into tomato plants through different routes and may possibly colonize and employ the plant as an alternative host (2, 9, 36). The formation of biofilm-like structures on the surface of roots, the colonizing regions of emerging lateral roots and wounded tissues has been reported (40, 41). Moreover, Salmonella inoculated into soil or blossoms can be recovered from the fruit at low internal levels, suggesting its ability to colonize and internalize tomato plants, and however, survival ability with different serovars varies in the soil and in different parts of the plants (42). Salmonella was also recovered from tomato fruits after it was introduced into the plant by stem injection or by flower inoculation (43). Stine et al. demonstrated that Salmonella was still detectable for at least 14 days after inoculation, indicating the bacteria probably developed a mechanism to adapt and survive within this hostile environment (44).
Recent studies have further suggested Salmonella may not only passively use plants as a “shelter” for survival, but a growing body of evidence has indicated it also may have evolved mechanisms for active adhesion and escape of plant immunity systems and actively internalize and proliferate in some plants, such as the tomato (9, 42, 45–47). It was found that Salmonella populations inoculated onto the exterior of tomatoes can increase in numbers at suitable humidity and temperature and such bacterial growth is serovar dependent (48). This may explain why some serovars, such as Newport, Montevideo, and Saintpaul, are more frequently linked to Salmonella outbreaks (2). A latest study suggested that the mechanisms Salmonella requires to colonize tomato plants are similar to phytopathogens, such as biosynthesis of amino acids, lipopolysaccharides (LPS), and nucleotides, indicating the flexibility of this pathogen to fit different hosts (49). The studies on Salmonella entry into inner leaves indicated that while trichomes are postulated as preferential colonization sites (50, 51), stomata are shown to be entry points Salmonella utilize for penetration of lettuce leaves (52, 53). The interaction of Salmonella with Arabdopsis, potato, sprouts and other plants are also reported and reviewed by Schikora et al. (47).
Preservation of taste, nutrients, and other desirable qualities of produce demand minimum processing, including avoidance of heating and sanitation with disinfectant to leafy vegetables and sprouts (2, 54), if washing is inadequate, the contamination acquired in field or production stage and postharvest easily transit from field to the table. Washing with chlorinated water, water-dip disinfection procedures may be applied to fruits such as melons, mango, and papaya. But if the disinfection efficiency of chlorinated water is not adequately monitored, the contaminated Salmonella can still be attached; and the rough surfaces for some types of melon like Cantaloupes can increase the adherence of bacteria and compromise the effect of washing (15, 30, 31, 55). Nowadays, the precut ready-to-eat vegetables and fruits are widely available to consumers in grocery stores; and poor hygiene and sanitary practice in preparation of these foods can bring additional chances for bacterial contamination (15, 56, 57). Therefore, the elimination of Salmonella contamination of fresh produce from fields to forks is almost impossible because of the above-mentioned loopholes. In general, the frequent occurrence of Salmonella outbreaks associated with produce and the scarcity of approaches available for sanitizing contaminated produce underscore the urgency for development of more efficient preventive control measures that can be applied at an earlier stage in the produce production.
Irrigation water as a source or a vehicle for transmission of Salmonella
The original source of Salmonella on produce may come from soil, manure, irrigation water, and contact with reptiles, birds, or other small animals (23, 24, 58). Irrigation water has drawn considerable attention in recent years, and studies have implicated irrigation water as a source of Salmonella contamination (23, 24, 59). This hypothesis has been previously reviewed (5, 23, 24, 27, 60, 61) and appears to be supported by trace-back investigations of produce-related Salmonella outbreaks (62, 63). The 2005 multistate Salmonella outbreak involving tomatoes was caused by Newport serotype, and a trace-back investigation linked this strain to an outbreak associated with tomatoes that occurred 3 years earlier. Furthermore, the isolates from these two outbreaks were shown to be genotypically identical by pulsed-field gel electrophoresis (PFGE), and the same bacterial strains were found present in a pond the farm used for irrigation (62). A case control study of a 2008 multistate outbreak identified Salmonella Saintpaul on serrano and jalapeño peppers and was also present in irrigation ponds (63). Another study indicated that the PFGE patterns of some of the isolates from irrigation ponds of produce farms in southern Georgia were indistinguishable from strains that were associated with the Salmonella Thompson outbreaks in 2010, 2012, and 2013, Salmonella Enteritidis outbreaks in 2011 and 2013, and a Salmonella Javiana outbreak in 2012 (61). The investigation of papaya Salmonella outbreak happened from 2006 to 2007 in Australia also found that the river water used for washing papayas was contaminated with Salmonella (64).
Prevalence and source of Salmonella in irrigation water
The fresh produce consumption over the last 40 years and number of foodborne disease outbreaks associated with fresh produce has been increasing (15, 18). Irrigation is a critical factor for the production of fruits and vegetables, Salmonella present in irrigation waters has been regarded as one of the major source for fresh produce contamination (2, 23, 24), and this has become a public health concern and drawn more attention of food safety regulatory agencies. Irrigation is a critical factor for the production of fruits and vegetables. As identified above, the source of irrigation water can include groundwater from wells, surface water (rivers and irrigation ponds), and treated wastewater (24, 27, 65, 66). Groundwater in wells is naturally filtered by soil and generally has a higher microbial quality (i.e., less microbes present). But it may be compromised by inferior construction or insufficient depth of the well, and may be contaminated from nearby latrines, septic tanks leaching fields, land application of waste water, and rainfalls (27, 67). Incidences of Salmonella contamination of ground water is mainly a concern of developing countries, especially in the rural areas, due to poor hygienic conditions, deficiently-structured water supply systems, and inadequate disinfection treatment (68), but occasionally occurs in developed countries as well (69, 70).
Surface waters, which include ponds, lakes, rivers, and streams, account for nearly half of the water used for irrigation in the United States (26). They are more exposed to environmental events such as discharge of sewage, rainfall, animal husbandry, and wildlife, and thus are more susceptible to contamination as compared to groundwater (23, 67). Rivers have been widely used as an irrigation source for agricultural practice (71, 72); river water, however, has been shown to be one of the largest reservoirs of viable Salmonella (2, 73). An estimation of Salmonella loads from a coastal Mediterranean river of a 16-month period (74) indicated Salmonella occurrence up to 95% during high waterflow (21% of the year) such as storm events. The subsequent study discovered S. Typhimurium accounted for 33.1% of all isolates recovered from the river in that period (73). In another study (12), Salmonella were detected from 57 of 72 (79.2%) water samples monthly collected from six stations of Little River in upper Suwannee Bain of Southern Georgia State of the United States. The recovery of Salmonella from rivers exhibited seasonality pattern, with summer time being highest, which is similar to the prevalence of Salmonella in irrigation ponds (6, 61). This increased prevalence of Salmonella during summer time may be related to multiple environmental factors, such as host shedding, enhanced persistence of Salmonella in warm temperature, and increase of storm events (12). In the Suwannee River watershed of southern Georgia, open surface pond waters are the main source of irrigation water. The case rates in this region have been observed to be 1.5-fold higher than the national average (6, 12). Presence of Salmonella in irrigation ponds within this region has been surveyed by Li et al. and showed that Salmonella was recovered from 50 of 170 (29.4%) water samples collected monthly over 27 months from 10 selected irrigation ponds that serve as water sources for irrigation of vegetables (61). Furthermore, more than half of the isolates were identified as S. Newport and antimicrobial susceptibility testing confirmed 16 S. Newport isolates were multidrug resistant (MDR), exhibiting resistance to ampicillin, chloramphenicol, streptomycin, sulfamethoxazole, and tetracycline (ACSSuT) and to the 1st, 2nd, and 3rd generations of cephalosporins (cephalothin, amoxicillin-clavulanic acid ceftriaxone) (61). Another monthly survey by Luo et al. on 10 ponds in the same area isolated Salmonella from 28.2% of all samples (n = 635) and the most common serotypes were Hadar, Montevideo, and Newport. In addition, 98.9% of the strains were reported to be resistant to streptomycin and about 20% were MDR strains (6). The contamination of fresh produce by these nontyphi Salmonella species present in irrigation waters could generate detrimental clinical and public health consequence because the increasing antibiotic resistance limits options of treatment after microbial diagnosis. In foodborne disease outbreak settings, the infection of MDR strains may bring excess of mortality and morbidity (75, 76). Moreover, the encodings genes of MDR may be horizontally transferred to other pathogenic bacteria resulting the spreading of antibiotic resistance (75).
The microbial quality of treated municipal water depends on the efficiency of treatments to remove pathogenic enteric bacteria, viruses, and parasites. Prevalence of Salmonella in treated effluent has been reported (77). In the United States, only a limited scale of treated municipal water is used for irrigation of crops because of the concern of potential bacterial contamination (24). However, in countries or regions with a shortage of fresh water, wastewater treated to a suitable level can be used as a substitute for ground water or surface water for irrigation purposes, and the microbial quality guidelines adopted for treated wastewater must be the strictest (23).
It is generally accepted that Salmonella present in water can be traced back to its animal origins. This pathogen may directly be transported from feces or exudates of wild animals by rain water runoff to rivers or ponds used for irrigation (12). Manure of domesticated animals has long been used to fertilize soil because it is economical and beneficial to the environment. However, studies have indicated that Salmonella in manure can survive as long as 231 days and may eventually contaminate produce by rain water splashing and/or by surface irrigation water (78). Sewage effluents contain waste water from human toilets with a high concentration of bacteria and other pathogens (79). The average concentration of Salmonella can reach as high as 2.7 × 102 CFU/100 ml (80, 81), which could become a major source of contamination if discharged directly or with inadequate treatment. Salmonella and other bacteria in sewage water can be effectively reduced to very low levels with modern treatment methods, but it is not practical to eliminate all the bacteria. When discharged, this will pose another contamination source of Salmonella to surface waters (79, 82) as illustrated in the proposed model of Salmonella transmission in the environment (5).
Salmonella survival and persistence in irrigation water
The gastrointestinal tract of vertebrates is generally regarded as the natural habitat for Salmonella (83), and bacteria released from feces could be transported into aquatic systems by sewage discharges, rainfall events or associated surface runoffs (84). Salmonella may survive in an environment with a broad range of pH (4.05–9.5) and can multiply in a broad range of temperatures (7–48°C) (34). In a closed environment at room temperature (25°C), it has been shown that Salmonella can survive for up to 5 years in sterile water or a phosphate buffered solution (85). However, the eco-environment of irrigation water, such as river or pond water, is harsher and more complicated and dynamic. Survival and persistence of Salmonella in water depends on multiple environmental factors, such as temperature, pH, salt, dissolved oxygen concentration, nutrient availability, interaction with other microorganisms, and exposure to UV light radiation (23, 27, 86, 87). As a result, Salmonella viability will decrease in water over time and could survive generally less than 30 days (23). However, many studies now show that biofilms can facilitate the survival of Salmonella in water and invertebrates such as free-living protozoa and various vertebrate animals can serve as reservoirs for Salmonella and other pathogenic bacteria. For example, Gaertner et al. detected Salmonella from water biofilms and crayfish samples from the headwater spring of Spring Lake, Texas and found that Salmonella isolates from biofilms collected 23 days apart shared the same Rep-PCR profile, suggesting Salmonella infrequently washed into an aquatic system might take up water biofilms as dwellings for long term persistence (88). Sha et al. further demonstrated Salmonella isolated from biofilms on a concrete surface exhibited significant microheterogeneity, but remained pathogenic (89). Additionally, Salmonella were recovered from sediments in water, and the concentration was higher than in the overlying water. Similar phenomena have also been observed in survival of other bacteria in water. This might be caused by sedimentation and absorption (90–92), which may help explain why continuous prevalence of Salmonella over a long period of time was found in river and surface ponds in multiple studies (5, 12).
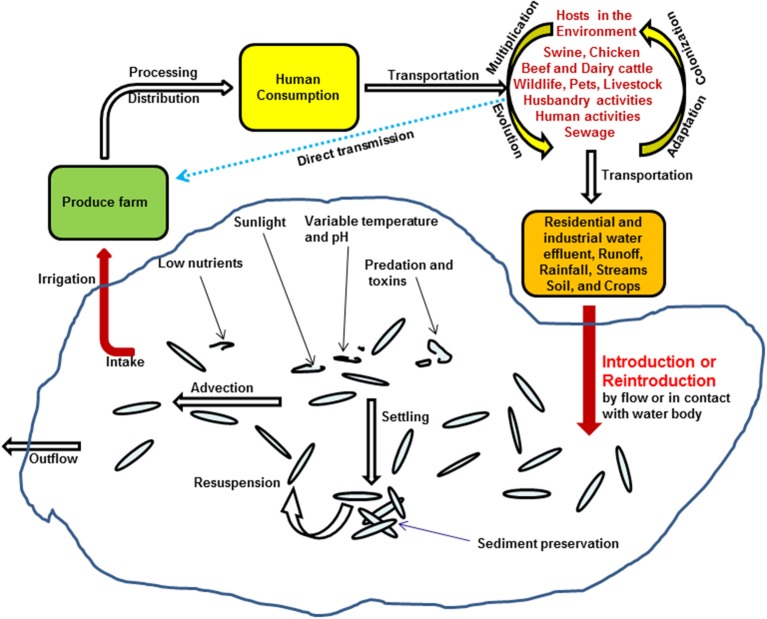
Besides the elongation of Salmonella viability and survivability in water environment, recently, Li et al. also suggested that frequent detection of Salmonella in irrigation water might be due to numerous reintroduction events associated with several different hosts in the environment. This was supported by genomic microarray analysis on Salmonella Newport isolates (5). On the one hand, produces growing in the field can be directly contaminated by Salmonella excreted from proximate environmental hosts, such as swine, chicken, beef, dairy cattle, husbandry or human, other wild lives, or inadequately composited manures applied in the fields (4, 5, 9); on the other hand, these bacteria could also be repeatedly introduced to the irrigation ponds through storm events and rainfalls, survive in water, and reach fresh produce through irrigation, and circulate back to human and other animals (Figure 2) (5).
Figure 2.
Proposed model of Salmonella transmission in irrigation water in the environment, adapted from Li et al. (5).
Furthermore, the prevalence and distribution of Salmonella in rivers, streams, or ponds exhibit seasonality, which has been documented in multiple studies (12, 61, 93). Thus, running water formed by rainfall may act as an important vehicle for Salmonella transportation. In addition, a number of studies suggest that bacteria can enter a viable but nonculturable (VBNC) state under stress conditions (94), such as low temperature (95), osmotic stress (96), pH changes (97), or nutrient starvation (98). Bacteria in the VBNC state cannot be recovered with routine growth media, but are alive (i.e., they metabolic activity) and these cells can become culturable again on resuscitation (99). Salmonella is one of the bacteria reported using VNBC as a survival strategy in harsh environments, including aquatic systems (100–102). The existence of VBNC has been controversial, since although it was reported 30 years ago, there is still little direct conclusive information on the molecular mechanisms underlying VNBC state induction and resuscitation (92, 103).
As mentioned above, many bacterial pathogens are known to survive within free-living protozoa, particularly amoebae (104). In fact, more than 20 species of pathogenic bacteria have been reported to associate with a single species (i.e., Acanthamoeba) of amoebae (105). This provides a potential mechanism for bacterial survival in the aquatic environment with the amoebae serving as an environmental reservoir. Salmonella Typhimurium was shown to replicate and survive in Acathamoeba spp.; however, it was cytotoxic and killed the amoebae (106–108). Douesnard-Malo and Daigle investigated the interactions between Acathamoeba castellanii and Salmonella Typhi, the etiologic agent of typhoid fever (109). They showed that S. Typhi could survive at least 3 weeks when grown in coculture with A. castellanii as opposed to less than 10 days when grown as singly cultured bacteria under the same conditions. Additionally, growth rates of amoebae after 14 days were similar in cocultures or when amoebae were singly cultured, suggesting that S. Typhi was not cytotoxic to A. castellanii (109). These studies suggest that certain species of free-living amoebae can serve as an environmental reservoir for pathogenic Salmonella species.
With an increasing number of outbreaks associated with consumption of fresh produce, understanding the mechanisms of produce contamination by foodborne pathogens is useful to develop preventative and processing measures to curtail the microbial populations in produce. A relatively new topic regarding the mechanisms of produce contamination is the concept of internalization of produce by pathogens, which has given rise to debate in the field in the last two decades (43, 110–113). Internalization is defined as the uptake of pathogens through the roots into the intercellular spaces between plant cells and in the plant vasculature tissues, xylem and phloem (114). Internalized pathogens cannot be removed by washing or disinfection and thus can pose a risk for human health if contaminated produce is eaten uncooked (115). The uptake of different foodborne microbes including bacteria (Salmonella and E. coli) (42, 116–121), viruses (norovirus) (122–124), fungi (125), and protozoa (126) through roots into produce has been reported. However, the presence of pathogens including Salmonella inside plant cells remains controversial (35), and further comprehensive and in-depth study is needed on the internalization of produce by pathogens.
Indirect detection of Salmonella in irrigation water
Rapid and accurate detection/estimation of the levels of Salmonella and other pathogens is prerequisite for understanding the dynamics of microbial populations and determination of microbial quality of irrigation water (102). The major source of microbial contamination of irrigation water is from fecal origin (4, 127, 128), and thus the microbial quality guidelines of irrigation water are through testing total counts of coliforms, fecal coliforms, E. coli, fecal streptococci, and nematode eggs (23, 129). Because total counts of coliforms and fecal coliforms cannot exclude bacteria from nonfecal origin, the presence of E. coli is now regarded as a better indicator of microbial quality of irrigation water (23, 129–131). The PSR establishes microbial quality criteria for various uses of agricultural water using generic E. coli as an indicator for fecal pollution. However, the use of generic E. coli as an index organism for the presence of human pathogens in water sources has been discussed in the literature as well. The PSR also requires that agricultural water must be safe and of adequate sanitary quality for its intended use. It has been shown that the concentration of E. coli can predict the level of Salmonella present in water (6, 132). However, Salmonella could be identified even if E. coli counts were below the actionable levels, raising a concern that the satisfaction of the current limit of generic E. coli may not necessarily represent the absence of Salmonella (6). Accurate estimation/enumeration and isolation of Salmonella in water are still indispensable in source-tracking investigations of produce associated outbreaks.
Direct detection of Salmonella in irrigation water
Traditional methods for detection, isolation, and identification of Salmonella in water involve nonselective and selective pre-enrichment in liquid culture followed by isolation using selective and differential agar plates. Such methods are laborious and time-consuming, which may take 4–5 days to complete (133). To overcome these limitations, immunoassays such as enzyme-linked immunosorbent assay (ELISA) have been combined with culture methods for detection of Salmonella (134), but the application of ELISA is greatly hampered by its poor performance in sensitivity and specificity. Fortunately, a combination of culture-based methods with DNA-based technologies has given a great boost for Salmonella detection in various food matrices. DNA-based methods, including conventional and real-time PCR (qPCR), have become the most common methods for the detection of Salmonella (135–137), with qPCR being more advantageous due to its specificity, sensitivity, and short turn-round time.
There are two different approaches used in qPCR detection. One approach uses SYBR Green dye to nonspecifically bind to synthesized double DNA (dsDNA). This dye only fluoresces when bound to dsDNA, thus the intensity of fluorescence quantitatively reflects the amount of the newly synthesized dsDNA (138, 139). The other approach is specific, employing a DNA probe with a fluorescent reporter incorporated at one end and a quencher of fluorescence at the other end to prevent detection of fluorescence. Degradation of the probe by 5'−3' nuclease activity of TaqMan polymerase will allow the unquenched emission of fluorescence, and the probe is also complementary to the DNA target and can anneal in each cycle. Thus, the increase of fluorescence can proportionately reflect the amplification of the DNA product. The advantage of this method is that it can be used to detect multiple targets simultaneously with high specificity and sensitivity and is widely used in detection and identification of microorganisms (140–142). The Loop-Mediated Isothermal Amplification (LAMP) can be carried out at a constant temperature and the amplicons are detected by measuring turbidity or fluorescence (143–147). It is a good option for detection of Salmonella from waters in rural area or developing countries where thermocycler is not equipped or budget is limited.
Most conventional PCR and qPCR assays for Salmonella target the invasive gene (invA) because it is an important virulence factor gene (148). This locus is considered to be present in all Salmonella spp., including a wide range of Salmonella serotypes and absent in other closely related bacterial species and genera (149–151). The biological confirmation of positive PCR results of irrigation water samples with convention methods should proceed, especially when the results are to serve the regulatory purpose. This can be achieved by plating on conventionally-used selective growth agars, including XLD (Xylose Lysine Deoxycholate) (152), HE (Hektoen eteric) (153), and BS (Bismuth sulfite, a modification of Wilson and Blair agar) (154) agar. The presumed Salmonella colonies on these plates based on morphology may undergo further biochemical or molecular identification process. Salmonella is urease-negative, hence, this trait can also be used for differentiation from some urease-positive bacteria such as Proteus and Citrobacter species on urea medium with phenol red as in indicator (155).
Enumeration of Salmonella in irrigation water
In general, the level of Salmonella cells in food, poultry, produce, or other food matrices is low, and direct enumeration of Salmonella cell number has always been a challenge (156). A variety of methods have been established, including direct plating, fluorescence in situ hybridization (157), most probable number (MPN) (158), modified MPN methods (132, 156), and qPCR (135, 159, 160). These methods can be applied for enumeration of Salmonella populations in irrigation water, however, direct plating sometimes is inefficient due to low level of Salmonella cells in water and competitiveness from natural microbiota (161). Direct counting by immunofluorescence-based methods are also not widely used due to low sensitivity in enumeration, problems of antibody quality and linkage of fluorochrome (162). The MPN method is still widely used, in particular, for determination of low concentration of Salmonella samples (156), and it has been improved by combination with serology or multiplex PCR in the confirmation step in the MPN method (158, 163). McEgan et al. have developed a modified MPN method for irrigation water. This modified MPN includes a three-by-three MPN dilution test, selective enrichment, plating, biological confirmation, and PCR confirmation, and it normally takes about a week to complete the entire process (132).
Detection of low level of Salmonella in food or water samples with molecular methods typically requires a pre-enrichment step to increase the number of the target cells, but the high background microbiota in some foods, such as fresh produce, will also multiply in this process and consequentially complicates the subsequent steps (164). Methods for quantification of Salmonella in foods have been developed by using qPCR to detect the DNA of the samples in order to estimate the copy number of a target gene that reflects the quantity of bacterial cells (135, 165). Elimination of the enrichment process presumably may improve the accuracy of the quantification, but this approach is not without drawbacks. First, only a very small amount of DNA template can be added to the PCR reaction mixture, and DNA purity and proportion of target DNA in the total DNA may affect the accuracy of the results. Therefore, it becomes critical to select the appropriate preparation method for separation of bacteria in water or food. The other issue involves the inability of PCR to discriminate DNA from viable cells and dead cells, which may undermine the reliability of the assay. One way to overcome this issue is to detect the presence of RNA (166, 167), which is laborious and can result in false-negative results due to degradation of RNA. The other approach involves using a photoreactive dye propidium monoazide (PMA), which has been incorporated into the PCR to differentiate dead from live bacterial cells in foods (142, 168–170). The principle for PMA's selectivity in detection is based on its ability to penetrate only cell membranes of dead cells, covalently bind to DNA upon light exposure, and subsequently inhibit the modified DNA from amplification by PCR. But caution should be exercised because in some cases inhibition of amplification of PMA-bound DNA from dead cells was found incomplete (171–174).
Reducing Salmonella in irrigation water
Irrigation water is extensively applied during the produce growth stage, and thus the assurance of microbial quality of irrigation water is critical in the protection of produce safety. In the United States, the standards for drinking water (the Safe Drinking Water Act, SDWA) was enacted in 1974, and subsequently the Environmental Protection Agency set the drinking water standards, which requires total coliforms (including the fecal coliforms and E. coli) to be 0 per 100 ml water (EPA) (https://www.epa.gov/ground-water-and-drinking-water/national-primary-drinking-water-regulations). Developing general standards for water used for irrigation is rather complicated because the source of water for irrigation can vary greatly with region, season, and climate. Even within the same region, water microbial quality can be quite dynamic. Precipitation, the distance to domesticated animal raising facilities, and the number of wild or domestic animals in the proximity of the water can also constitute substantial changes to the microbial quality of the water (61). However, with the passage of FSMA, and more specifically, the PSR, many growers producing covered crops must now meet certain minimum requirements for the safe use of agricultural water. This may serve as a good solution for improving microbial quality of irrigation water reduction of foodbonre outbreaks assocaited to fresh produces.
Conclusions
Salmonella is frequently detected in surface water, which accounts for nearly half of the water used for irrigation. Trace-back investigations of outbreaks often implicate irrigation water as a source (or a vehicle) for transmission of Salmonella. In addition, the bacterium can survive in these aquatic environments by a number of mechanisms, including entry into the VBNC state and/or residing within free-living protozoa. As such, assurance of microbial quality of irrigation water is vital to the mitigation of produce-related foodborne outbreaks.
Author contributions
BL conceived this project. HL, CW, BL wrote the manuscript. All authors have read and approved the manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The reviewers LB and AH and handling Editor declared their shared affiliation.
The views expressed in this article are those of the authors and do not necessarily reflect the official policy of the Department of Health and Human Services, the U.S. Food and Drug Administration (FDA), or the U.S. Government. Reference to any commercial materials, equipment, or process does not in any way constitute approval, endorsement, or recommendation by the FDA.
References
- 1.Tauxe R, Kruse H, Hedberg C, Potter M, Madden J, Wachsmuth K. Microbial hazards and emerging issues associated with produce - a preliminary report to the National Advisory Committee on Microbiologic Criteria for Foods. J Food Prot. (1997) 60:1400–8. 10.4315/0362-028X-60.11.1400 [DOI] [PubMed] [Google Scholar]
- 2.Hanning IB, Nutt JD, Ricke SC. Salmonellosis outbreaks in the United States due to fresh produce: sources and potential intervention measures. Foodborne Pathog Dis. (2009) 6:635–48. 10.1089/fpd.2008.0232 [DOI] [PubMed] [Google Scholar]
- 3.Ruby T, McLaughlin L, Gopinath S, Monack D. Salmonella's long-term relationship with its host. FEMS Microbiol Rev. (2012) 36:600–15. 10.1111/j.1574-6976.2012.00332.x [DOI] [PubMed] [Google Scholar]
- 4.Andino A, Hanning I. Salmonella enterica: survival, colonization, and virulence differences among serovars. Scientific World Journal. (2015) 2015:520179. 10.1155/2015/520179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li B, Jackson SA, Gangiredla J, Wang W, Liu H, Tall BD, et al. Genomic evidence reveals numerous Salmonella enterica serovar Newport reintroduction events in Suwannee watershed irrigation ponds. Appl Environ Microbiol. (2015) 81:8243–53. 10.1128/AEM.02179-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luo ZY, Gu GY, Ginn A, Giurcanu MC, Adams P, Vellidis G, et al. Distribution and characterization of Salmonella enterica isolates from irrigation ponds in the Southeastern United States. Appl Environ Microbiol. (2015) 81:4376–87. 10.1128/AEM.04086-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Whiley H, Gardner MG, Ross K. A review of Salmonella and squamates (lizards, snakes and amphisbians): implications for public health. Pathogens (2017) 6:E38. 10.3390/pathogens6030038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Batz MB, Hoffman S, Morris JG. Ranking the Risks: The 10 Pathogen-Food Combinations with the Greatest Burden on Public Health. Gainesville, FL: University of Florida, Emerging Pathogens Institute; (2011). [Google Scholar]
- 9.Brandl MT, Cox CE, Teplitski M. Salmonella interactions with plants and their associated microbiota. Phytopathology (2013) 103:316–25. 10.1094/PHYTO-11-12-0295-RVW [DOI] [PubMed] [Google Scholar]
- 10.Majowicz SE, Musto J, Scallan E, Angulo FJ, Kirk M, O'Brien SJ, et al. The global burden of nontyphoidal Salmonella gastroenteritis. Clin Infect Dis. (2010) 50:882–9. 10.1086/650733 [DOI] [PubMed] [Google Scholar]
- 11.Sivapalasingam S, Friedman CR, Cohen L, Tauxe RV. Fresh produce: a growing cause of outbreaks of foodborne illness in the United States, 1973 through 1997. J Food Prot. (2004) 67:2342–53. 10.4315/0362-028X-67.10.2342 [DOI] [PubMed] [Google Scholar]
- 12.Haley BJ, Cole DJ, Lipp EK. Distribution, diversity, and seasonality of waterborne Salmonellae in a rural watershed. Appl Environ Microbiol. (2009) 75:1248–55. 10.1128/AEM.01648-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harris L, Farber JN, Beuchat LR, Parish ME, Suslow TV, Garrett EH, et al. Outbreaks associated with fresh produce: incidence, growth, and survival of pathogens in fresh and fresh-cut produce. Comp Rev Food Sci Food Safety (2003) 2:78–141. 10.1111/j.1541-4337.2003.tb00031.x [DOI] [Google Scholar]
- 14.Walsh C, Duffy G, Sheridan JJ, Fanning S, Blair IS, McDowell DA. Thermal resistance of antibiotic-resistant and antibiotic-sensitive Salmonella spp. on chicken meat. J Food Safety (2005) 25:288–302. 10.1111/j.1745-4565.2005.00021.x [DOI] [Google Scholar]
- 15.Walsh KA, Bennett SD, Mahovic M, Gould LH. Outbreaks associated with cantaloupe, watermelon, and honeydew in the United States, 1973-2011. Foodborne Pathog. Dis. (2014) 11:945–52. 10.1089/fpd.2014.1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Painter JA, Hoekstra RM, Ayers T, Tauxe RV, Braden CR, Angulo FJ, et al. Attribution of foodborne illnesses, hospitalizations, and deaths to food commodities by using outbreak data, United States, 1998-2008. Emerging Infect Dis. (2013) 19:407–15. 10.3201/eid1903.111866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang YT, Swinburne M. New produce safety regulations: promises and challenges. Public Health Rep. (2016) 131:754–7. 10.1177/0033354916669495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lynch MF, Tauxe RV, Hedberg CW. The growing burden of foodborne outbreaks due to contaminated fresh produce: risks and opportunities. Epidemiol Infect. (2009) 137:307–15. 10.1017/S0950268808001969 [DOI] [PubMed] [Google Scholar]
- 19.Denis N, Zhang H, Leroux A, Trudel R, Bietlot H. Prevalence and trends of bacterial contamination in fresh fruits and vegetables sold at retail in Canada. Food Control (2016) 67:225–34. 10.1016/j.foodcont.2016.02.047 [DOI] [Google Scholar]
- 20.Cook R, Tracking Demographics US Fruit Vegetable Consumption Patterns (2011). Available online at: https://arefiles.ucdavis.edu/uploads/filer_public/2014/05/19/blueprintseoeconsumptioncookfinaljan2012figures.pdf
- 21.Usda-Ers Economic Research Service, United States Department of Agriculture. (2015). Available online at: https://www.ers.usda.gov/data-products/ag-and-food-statistics-charting-the-essentials/food-availability-and-consumption/
- 22.FDA Food Safety Modernization Act: Standards for the Growing, Harvesting, Packing, and Holding of Produce for Human Consumption; Proposed Rule, 2013 (Docket No. FDA-2011-N-0921). Washington, DC: U.S. Food and Drug Administration; (2013). [Google Scholar]
- 23.Steele M, Odumeru J. Irrigation water as source of foodborne pathogens on fruit and vegetables. J Food Prot. (2004) 67:2839–49. 10.4315/0362-028X-67.12.2839 [DOI] [PubMed] [Google Scholar]
- 24.Pachepsky Y, Shelton DR, McLain JET, Patel J, Mandrell RE. Irrigation waters as a source of pathogenic microorganisms in produce: a review. Adv Agron. (2011) 113:73–138. 10.1016/B978-0-12-386473-4.00002-6 [DOI] [Google Scholar]
- 25.Raudales R, Parke JL, Guy CL, Fisher PR. Control of waterborne microbes in irrigation:A review. Agric Water Manage. (2014) 143:9–28. 10.1016/j.agwat.2014.06.007 [DOI] [Google Scholar]
- 26.Bihn E, Smart CD, Hoepting CA, Worobo RW. Use of surface water in the production of fresh fruits and vegetables: a survey of fresh produce growers and their water management practices. Food Protection Trends (2013) 33:307–14. Available online at: http://www.foodprotection.org/files/food-protection-trends/Sep-Oct-13-bihn.pdf [Google Scholar]
- 27.CPS Agricultural Water: Center for Produce Safety 5 -Year Research Review, Version 1.1. (2014). Available online at: http://www.pma.com/~/media/pma-files/food-safety/cps/cps-research-reportag-water-200813version-11final.pdf
- 28.CDC Centers for Disease Control and Prevention. Annual Listing of Foodborne Disease Outbreaks. Atlanta, GA: CDC; (2008). Available online at: http://www.cdc.gov/foodborneoutbreaks/outbreak_data.htm [Google Scholar]
- 29.CDC Centers for Disease Control and Prevention: List of Selected Multistate Foodborne Outbreaks and Investigations. (2017). Available online at: https://www.cdc.gov/foodsafety/outbreaks/multistate-outbreaks/outbreaks-list.html
- 30.Sivapalasingam S, Barrett E, Kimura A, Van Duyne S, De Witt W, Ying M, et al. A multistate outbreak of Salmonella enterica serotype Newport infection linked to mango consumption: impact of water-dip disinfestation technology. Clin Infect Dis. (2003) 37:1585–90. 10.1086/379710 [DOI] [PubMed] [Google Scholar]
- 31.Castillo A, Mercado I, Lucia LM, Martinez-Ruiz Y, De Leon JP, Murano EA, et al. Salmonella contamination during production of Cantaloupe: a binational study. J Food Prot. (2004) 67:713–20. 10.4315/0362-028X-67.4.713 [DOI] [PubMed] [Google Scholar]
- 32.CDC Centers for Disease Control and Prevention. List of Selected Outbreaks. 2006-2015. (2006-2015). Available online at: http://www.cdc.gov/salmonella/outbreaks.html
- 33.Fett WF. Inhibition of Salmonella enterica by plant-associated pseudomonads in vitro and on sprouting alfalfa seed. J Food Prot. (2006) 69:719–28. 10.4315/0362-028X-69.4.719 [DOI] [PubMed] [Google Scholar]
- 34.Fatica MK, Schneider KR. Salmonella and produce: survival in the plant environment and implications in food safety. Virulence (2011) 2:573–9. 10.4161/viru.2.6.17880 [DOI] [PubMed] [Google Scholar]
- 35.Wiedemann A, Virlogeux-Payant I, Chausse AM, Schikora A, Velge P. Interactions of Salmonella with animals and plants. Front Microbiol. (2015) 5:791. 10.3389/fmicb.2014.00791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leff JW, Fierer N. Bacterial communities associated with the surfaces of fresh fruits and vegetables. PLoS ONE (2013) 8:e59310. 10.1371/journal.pone.0059310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goudeau DM, Parker CT, Zhou Y, Sela S, Kroupitski Y, Brandl MT. The salmonella transcriptome in lettuce and cilantro soft rot reveals a niche overlap with the animal host intestine. Appl Environ Microbiol. (2013) 79:250–62. 10.1128/AEM.02290-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ottesen AR, Gonzalez A, Bell R, Arce C, Rideout S, Allard M, et al. Co-enriching microflora associated with culture based methods to detect Salmonella from tomato phyllosphere. PLoS ONE (2013) 8:e73079. 10.1371/journal.pone.0073079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jarvis KG, White JR, Grim CJ, Ewing L, Ottesen AR, Beaubrun JJG, et al. Cilantro microbiome before and after nonselective pre-enrichment for Salmonella using 16S rRNA and metagenomic sequencing. BMC Microbiol. (2015) 15:160. 10.1186/s12866-015-0497-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Iniguez AL, Dong Y, Carter HD, Ahmer BM, Stone JM, Triplett EW. Regulation of enteric endophytic bacterial colonization by plant defenses. Mol Plant Microbe Interact. (2005) 18:169–78. 10.1094/MPMI-18-0169 [DOI] [PubMed] [Google Scholar]
- 41.Schikora A, Carreri A, Charpentier E, Hirt H. The dark side of the salad: Salmonella typhimurium overcomes the innate immune response of Arabidopsis thaliana and shows an endopathogenic lifestyle. PLoS ONE (2008) 3:e2279. 10.1371/journal.pone.0002279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zheng J, Allard S, Reynolds S, Millner P, Arce G, Blodgett RJ, et al. Colonization and internalization of Salmonella enterica in tomato plants. Appl Environ Microbiol. (2013) 79:2494–502. 10.1128/AEM.03704-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guo X, Chen JR, Brackett RE, Beuchat LR. Survival of Salmonellae on and in tomato plants from the time of inoculation at flowering and early stages of fruit development through fruit ripening. Appl Environ Microbiol. (2001) 67:4760–4. 10.1128/AEM.67.10.4760-4764.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stine SW, Song I, Choi CY, Gerba CP. Effect of relative humidity on preharvest survival of bacterial and viral pathogens on the surface of cantaloupe, lettuce, and bell peppers. J Food Prot. (2005) 68:1352–8. 10.4315/0362-028X-68.7.1352 [DOI] [PubMed] [Google Scholar]
- 45.Barak JD, Gorski L, Naraghi-Arani P, Charkowski AO. Salmonella enterica virulence genes are required for bacterial attachment to plant tissue. Appl Environ Microbiol. (2005) 71:5685–91. 10.1128/AEM.71.10.5685-5691.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gu G, Hu J, Cevallos-Cevallos JM, Richardson SM, Bartz JA, Van Bruggen AH. Internal colonization of Salmonella enterica serovar Typhimurium in tomato plants. PLoS ONE (2011) 6:e27340. 10.1371/journal.pone.0027340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schikora A, Garcia AV, Hirt H. Plants as alternative hosts for Salmonella. Trends Plant Sci. (2012) 17:245–9. 10.1016/j.tplants.2012.03.007 [DOI] [PubMed] [Google Scholar]
- 48.Iturriaga MH, Tamplin ML, Escartin EF. Colonization of tomatoes by Salmonella Montevideo is affected by relative and storage temperature. J Food Prot. (2007) 70:30–4. 10.4315/0362-028X-70.1.30 [DOI] [PubMed] [Google Scholar]
- 49.De Moraes MH, Desai P, Porwollik S, Canals R, Perez DR, Chu W, et al. Salmonella persistence in tomatoes requires a distinct set of metabolic functions identified by transposon insertion sequencing. Appl Environ Microbiol. (2017) 83:e03028-16. 10.1128/AEM.03028-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Barak JD, Kramer LC, Hao LY. Colonization of tomato plants by Salmonella enterica is cultivar dependent, and type 1 trichomes are preferred colonization sites. Appl Environ Microbiol. (2011) 77:498–504. 10.1128/AEM.01661-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Golberg D, Kroupitski Y, Belausov E, Pinto R, Sela S. Salmonella Typhimurium internalization is variable in leafy vegetables and fresh herbs. Int J Food Microbiol. (2011) 145:250–7. 10.1016/j.ijfoodmicro.2010.12.031 [DOI] [PubMed] [Google Scholar]
- 52.Kroupitski Y, Golberg D, Belausov E, Pinto R, Swartzberg D, Granot D, et al. Internalization of Salmonella enterica in leaves is induced by light and involves chemotaxis and penetration through open stomata. Appl Environ Microbiol. (2009) 75:6076–86. 10.1128/AEM.01084-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kroupitski Y, Pinto R, Brandl MT, Belausov E, Sela S. Interactions of Salmonella enterica with lettuce leaves. J Appl Microbiol. (2009) 106:1876–85. 10.1111/j.1365-2672.2009.04152.x [DOI] [PubMed] [Google Scholar]
- 54.Herman KM, Hall AJ, Gould LH. Outbreaks attributed to fresh leafy vegetables, United States, 1973-2012. Epidemiol Infect. (2015) 143:3011–21. 10.1017/S0950268815000047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Parnell TL, Harris LJ, Suslow TV. Reducing Salmonella on cantaloupes and honeydew melons using wash practices applicable to postharvest handling, foodservice, and consumer preparation. Int J Food Microbiol. (2005) 99:59–70. 10.1016/j.ijfoodmicro.2004.07.014 [DOI] [PubMed] [Google Scholar]
- 56.Boriss H, Brunke H, Kreith M. Commodity Profile: Melons. Davis, CA: University of California Agricultural Issues Center; (2006). [Google Scholar]
- 57.Patil R. TJ, Ryser E. Extent of Listeria monocytogenes transfer during cutting of cantaloupe and honeydew melon. J Food Prot. (2013) 76(Suppl. A):3–99. Available online at: https://iafp.confex.com/iafp/2013/webprogram/Paper4931.html [Google Scholar]
- 58.Jacobsen CS, Bech TB. Soil survival of Salmonella and transfer to freshwater and fresh produce. Food Res Int. (2012) 45:557–66. 10.1016/j.foodres.2011.07.026 [DOI] [Google Scholar]
- 59.Jones LA, Worobo RW, Smart CD. Plant-pathogenic oomycetes, Escherichia coli strains, and Salmonella spp. Frequently found in surface water used for irrigation of fruit and vegetable crops in New York State. Appl Environ Microbiol. (2014) 80:4814–20. 10.1128/AEM.01012-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jablasone J, Brovko LY, Griffiths MW. A research note: the potential for transfer of Salmonella from irrigation water to tomatoes. J Sci Food Agric. (2004) 84:287–9. 10.1002/jsfa.1646 [DOI] [Google Scholar]
- 61.Li B, Vellidis G, Liu H, Jay-Russell M, Zhao S, Hu Z, et al. Diversity and antimicrobial resistance of Salmonella enterica isolates from surface water in Southeastern United States. Appl Environ Microbiol. (2014) 80:6355–65. 10.1128/AEM.02063-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Greene SK, Daly ER, Talbot EA, Demma LJ, Holzbauer S, Patel NJ, et al. Recurrent multistate outbreak of Salmonella Newport associated with tomatoes from contaminated fields, 2005. Epidemiol Infect. (2008) 136:157–65. 10.1017/S095026880700859X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Klontz KC, Klontz JC, Mody RK, Hoekstra RM. Analysis of tomato and jalapeno and Serrano pepper imports into the United States from Mexico before and during a National Outbreak of Salmonella serotype Saintpaul infections in 2008. J Food Prot. (2010) 73:1967–74. 10.4315/0362-028X-73.11.1967 [DOI] [PubMed] [Google Scholar]
- 64.Gibbs R, Pingault N, Mazzucchelli T, O'reilly L, Mackenzie B, Green J, et al. An outbreak of Salmonella enterica serotype Litchfield infection in Australia linked to consumption of contaminated papaya. J Food Prot. (2009) 72:1094–8. 10.4315/0362-028X-72.5.1094 [DOI] [PubMed] [Google Scholar]
- 65.Leifert C, Ball K, Volakakis N, Cooper JM. Control of enteric pathogens in ready-to-eat vegetable crops in organic and ‘low input’ production systems: a HACCP-based approach. J Appl Microbiol. (2008) 105:931–50. 10.1111/j.1365-2672.2008.03794.x [DOI] [PubMed] [Google Scholar]
- 66.Santiago P, Jimenez-Belenguer A, Garcia-Hernandez J, Estelles RM, Hernandez Perez M, Castillo Lopez MA, et al. High prevalence of Salmonella spp. in wastewater reused for irrigation assessed by molecular methods. Int J Hyg Environ Health (2018) 221:95–101. 10.1016/j.ijheh.2017.10.007 [DOI] [PubMed] [Google Scholar]
- 67.Gerba The role of water and water testing in produce safety, In: Fan X, Niemira BA, Doona CJ, Feeherry FE, Gravani RB. editors. Microbial Safety of Fresh Produce. Oxford, UK: Blackwell-Wiley; IFT Press; (2009). [Google Scholar]
- 68.Levantesi C, Bonadonna L, Briancesco R, Grohmann E, Toze S, Tandoi V. Salmonella in surface and drinking water: occurrence and water-mediated transmission. Food Res Int. (2012) 45:587–602. 10.1016/j.foodres.2011.06.037 [DOI] [Google Scholar]
- 69.Goss M, Richards C. Development of a risk-based index for source water protection planning, which supports the reduction of pathogens from agricultural activity entering water resources. J Environ Manage. (2008) 87:623–32. 10.1016/j.jenvman.2006.12.048 [DOI] [PubMed] [Google Scholar]
- 70.Bockelmann U, Dorries HH, Ayuso-Gabella MN, De Marcay MS, Tandoi V, Levantesi C, et al. Quantitative PCR monitoring of antibiotic resistance genes and bacterial pathogens in three european artificial groundwater recharge systems. Appl Environ Microbiol. (2009) 75:154–63. 10.1128/AEM.01649-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Assadian NW, Fenn LB, Flores-Ortiz MA, Ali AS. Spatial variability of solutes in a pecan orchard surface-irrigated with untreated effluents in the upper Rio Grande River basin. Agric Water Manage. (1999) 42:143–56. 10.1016/S0378-3774(99)00037-2 [DOI] [Google Scholar]
- 72.Garcia SS, Ake C, Clement B, Huebner HJ, Donnelly KC, Shalat SL. Initial results of environmental monitoring in the Texas Rio Grande Valley. Environ Int. (2001) 26:465–74. 10.1016/S0160-4120(01)00027-7 [DOI] [PubMed] [Google Scholar]
- 73.Baudart J, Lemarchand K, Brisabois A, Lebaron P. Diversity of Salmonella strains isolated from the aquatic environment as determined by serotyping and amplification of the ribosomal DNA spacer regions. Appl Environ Microbiol. (2000) 66:1544–52. 10.1128/AEM.66.4.1544-1552.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Baudart J, Grabulos J, Barusseau JP, Lebaron P. Salmonella spp. and fecal coliform loads in coastal waters from a point vs. nonpoint source of pollution. J Environ Qual. (2000) 29:241–50. 10.2134/jeq2000.00472425002900010031x [DOI] [Google Scholar]
- 75.Su LH, Chiu CH, Chu C, Ou JT. Antimicrobial resistance in nontyphoid Salmonella serotypes: a global challenge. Clin Infect Dis. (2004) 39:546–51. 10.1086/422726 [DOI] [PubMed] [Google Scholar]
- 76.Molbak K. Human health consequences of antimicrobial drug-resistant Salmonella and other foodborne pathogens. Clin Infect Dis. (2005) 41:1613–20. 10.1086/497599 [DOI] [PubMed] [Google Scholar]
- 77.Odjadjare EC, Olaniran AO. Prevalence of antimicrobial resistant and virulent Salmonella spp. in treated effluent and receiving aquatic milieu of wastewater treatment plants in Durban, South Africa. Int J Environ Res Public Health (2015) 12:9692–713. 10.3390/ijerph120809692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Islam M, Morgan J, Doyle MP, Phatak SC, Millner P, Jiang XP. Fate of Salmonella enterica serovar Typhimurium on carrots and radishes grown in fields treated with contaminated manure composts or irrigation water. Appl Environ Microbiol. (2004) 70:2497–502. 10.1128/AEM.70.4.2497-2502.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kay D, Crowther J, Stapleton CM, Wyer MD, Fewtrell L, Edwards A, et al. Faecal indicator organism concentrations in sewage and treated effluents. Water Res. (2008) 42:442–54. 10.1016/j.watres.2007.07.036 [DOI] [PubMed] [Google Scholar]
- 80.Rose JB, Huffman DE, Riley K, Farrah SR, Lukasik JO, Hamann CL. Reduction of enteric microorganisms at the Upper Occoquan Sewage Authority Water Reclamation Plant. Water Environ Res. (2001) 73:711–20. 10.2175/106143001X143457 [DOI] [PubMed] [Google Scholar]
- 81.Howard I, Espigares E, Lardelli P, Martin JL, Espigares M. Evaluation of microbiological and physicochemical indicators for wastewater treatment. Environ Toxicol. (2004) 19:241–9. 10.1002/tox.20016 [DOI] [PubMed] [Google Scholar]
- 82.Naidoo S, Olaniran AO. Treated wastewater effluent as a source of microbial pollution of surface water resources. Int J Environ Res Public Health (2014) 11:249–70. 10.3390/ijerph110100249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Woodward DL, Khakhria R, Johnson WM. Human salmonellosis associated with exotic pets. J Clin Microbiol. (1997) 35:2786–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sha Q, Forstner MR, Hahn D. Diversity of Salmonella in biofilms and water in a headwater ecosystem. FEMS Microbiol Ecol. (2013) 83:642–9. 10.1111/1574-6941.12021 [DOI] [PubMed] [Google Scholar]
- 85.Liao CH, Shollenberger LM. Survivability and long-term preservation of bacteria in water and in phosphate-buffered saline. Lett Appl Microbiol. (2003) 37:45–50. 10.1046/j.1472-765X.2003.01345.x [DOI] [PubMed] [Google Scholar]
- 86.Wilkes G, Edge TA, Gannon VPJ, Jokinen C, Lyautey E, Neumann NF, et al. Associations among pathogenic bacteria, parasites, and environmental and land use factors in multiple mixed-use watersheds. Water Res. (2011) 45:5807–25. 10.1016/j.watres.2011.06.021 [DOI] [PubMed] [Google Scholar]
- 87.Wanjugi P, Harwood VJ. The influence of predation and competition on the survival of commensal and pathogenic fecal bacteria in aquatic habitats. Environ Microbiol. (2013) 15:517–26. 10.1111/j.1462-2920.2012.02877.x [DOI] [PubMed] [Google Scholar]
- 88.Gaertner JP, Mendoza JA, Forstner MRJ, Hahn D. Recovery of Salmonella from biofilms in a headwater spring ecosystem. J Water Health (2011) 9:458–66. 10.2166/wh.2011.173 [DOI] [PubMed] [Google Scholar]
- 89.Sha Q, Gunathilake A, Forstner MR, Hahn D. Temporal analyses of the distribution and diversity of Salmonella in natural biofilms. Syst Appl Microbiol. (2011) 34:353–9. 10.1016/j.syapm.2011.01.005 [DOI] [PubMed] [Google Scholar]
- 90.Hendricks CW. Increased recovery rate of Salmonellae from stream bottom sediments versus surface waters. Appl Microbiol. (1971) 21:379–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Morinigo MA, Borrego JJ, Romero P. Comparative study of different methods for detection and enumeration of Salmonella spp. in natural waters. J Appl Bacteriol. (1986) 61:169–76. 10.1111/j.1365-2672.1986.tb04272.x [DOI] [PubMed] [Google Scholar]
- 92.Moore BC, Martinez E, Gay JM, Rice DH. Survival of Salmonella enterica in freshwater and sediments and transmission by the aquatic midge Chironomus tentans (Chironomidae: Diptera). Appl Environ Microbiol. (2003) 69:4556–60. 10.1128/AEM.69.8.4556-4560.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Huang KH, Hsu BM, Chou MY, Tsai HL, Kao PM, Wang HJ, et al. Application of molecular biological techniques to analyze Salmonella seasonal distribution in stream water. FEMS Microbiol Lett. (2014) 352:87–96. 10.1111/1574-6968.12381 [DOI] [PubMed] [Google Scholar]
- 94.Xu HS, Roberts N, Singleton FL, Attwell RW, Grimes DJ, Colwell RR. Survival and viability of nonculturable escherichia-coli and vibrio-cholerae in the estuarine and marine-environment. Microb Ecol. (1982) 8:313–23. 10.1007/BF02010671 [DOI] [PubMed] [Google Scholar]
- 95.Cook KL, Bolster CH. Survival of Campylobacter jejuni and Escherichia coli in groundwater during prolonged starvation at low temperatures. J Appl Microbiol. (2007) 103:573–83. 10.1111/j.1365-2672.2006.03285.x [DOI] [PubMed] [Google Scholar]
- 96.Kusumoto A, Asakura H, Kawamoto K. General stress sigma factor RpoS influences time required to enter the viable but non-culturable state in Salmonella enterica. Microbiol Immunol. (2012) 56:228–37. 10.1111/j.1348-0421.2012.00428.x [DOI] [PubMed] [Google Scholar]
- 97.Tholozan JL, Cappelier JM, Tissier JP, Delattre G, Federighi M. Physiological characterization of viable-but-nonculturable Campylobacter jejuni cells. Appl Environ Microbiol. (1999) 65:1110–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gupte AR, De Rezende CLE, Joseph SW. Induction and resuscitation of viable but nonculturable Salmonella enterica serovar typhimurium DT104. Appl Environ Microbiol. (2003) 69:6669–75. 10.1128/AEM.69.11.6669-6675.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Oliver JD. The viable but nonculturable state in bacteria. J Microbiol. (2005) 43:93–100. Available online at: https://pdfs.semanticscholar.org/e661/934dca6bb0dbb31a8781f3193232b7b5a8a4.pdf [PubMed] [Google Scholar]
- 100.Roszak DB, Grimes DJ, Colwell RR. Viable but nonrecoverable stage of Salmonella-enteritidis in aquatic systems. Can J Microbiol. (1984) 30:334–8. 10.1139/m84-049 [DOI] [PubMed] [Google Scholar]
- 101.Desmonts C, Minet J, Colwell R, Cormier M. Fluorescent-antibody method useful for detecting viable but nonculturable Salmonella spp. in chlorinated waste-water. Appl Environ Microbiol. (1990) 56:1448–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Morishige Y, Koike A, Tamura-Ueyama A, Amano F. Induction of viable but nonculturable Salmonella in exponentially grown cells by exposure to a low-humidity environment and their resuscitation by catalase. J Food Prot. (2017) 80:288–94. 10.4315/0362-028X.JFP-16-183 [DOI] [PubMed] [Google Scholar]
- 103.Pinto D, Santos MA, Chambel L. Thirty years of viable but nonculturable state research: unsolved molecular mechanisms. Crit Rev Microbiol. (2015) 41:61–76. 10.3109/1040841X.2013.794127 [DOI] [PubMed] [Google Scholar]
- 104.Molmeret M, Horn M, Wagner M, Santic M, Abu Kwaik Y. Amoebae as training grounds for intracellular bacterial pathogens. Appl Environ Microbiol. (2005) 71:20–8. 10.1128/AEM.71.1.20-28.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Winiecka-Krusnell J, Linder E. Bacterial infections of free-living amoebae. Res Microbiol. (2001) 152:613–9. 10.1016/S0923-2508(01)01240-2 [DOI] [PubMed] [Google Scholar]
- 106.Gaze WH, Burroughs N, Gallagher MP, Wellington EMH. Interactions between Salmonella typhimurium and Acanthamoeba polyphaga, and observation of a new mode of intracellular growth within contractile vacuoles. Microb Ecol. (2003) 46:358–69. 10.1007/s00248-003-1001-3 [DOI] [PubMed] [Google Scholar]
- 107.Tezcan-Merdol D, Ljungstrom M, Winiecka-Krusnell J, Linder E, Engstrand L, Rhen M. Uptake and replication of Salmonella enterica in Acanthamoeba rhysodes. Appl Environ Microbiol. (2004) 70:3706–14. 10.1128/AEM.70.6.3706-3714.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Feng Y, Hsiao YH, Chen HL, Chu CS, Tang P, Chiu CH. Apoptosis-like cell death induced by Salmonella in Acanthamoeba rhysodes. Genomics (2009) 94:132–7. 10.1016/j.ygeno.2009.05.004 [DOI] [PubMed] [Google Scholar]
- 109.Douesnard-Malo F, Daigle F. Increased persistence of Salmonella enterica serovar Typhi in the presence of Acanthamoeba castellanii. Appl Environ Microbiol. (2011) 77:7640–6. 10.1128/AEM.00699-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Guo X, Chen JR, Brackett RE, Beuchat LR. Survival of Salmonella on tomatoes stored at high relative humidity, in soil, and on tomatoes in contact with soil. J Food Prot. (2002) 65:274–9. 10.4315/0362-028X-65.2.274 [DOI] [PubMed] [Google Scholar]
- 111.Kutter S, Hartmann A, Schmid M. Colonization of barley (Hordeum vulgare) with Salmonella enterica and Listeria spp. FEMS Microbiol Ecol. (2006) 56:262–71. 10.1111/j.1574-6941.2005.00053.x [DOI] [PubMed] [Google Scholar]
- 112.Franz E, Visser AA, Van Diepeningen AD, Klerks MM, Termorshuizen AJ, Van Bruggen AH. Quantification of contamination of lettuce by GFP-expressing Escherichia coli O157:H7 and Salmonella enterica serovar Typhimurium. Food Microbiol. (2007) 24:106–12. 10.1016/j.fm.2006.03.002 [DOI] [PubMed] [Google Scholar]
- 113.Sharma M, Ingram DT, Patel JR, Millner PD, Wang X, Hull AE, et al. A novel approach to investigate the uptake and internalization of Escherichia coli O157:H7 in spinach cultivated in soil and hydroponic medium. J Food Prot. (2009) 72:1513–20. 10.4315/0362-028X-72.7.1513 [DOI] [PubMed] [Google Scholar]
- 114.Hirneisen KA, Sharma M, Kniel KE. Human enteric pathogen internalization by root uptake into food crops. Foodborne Pathog Dis. (2012) 9:396–405. 10.1089/fpd.2011.1044 [DOI] [PubMed] [Google Scholar]
- 115.Fornefeld E, Schierstaedt J, Jechalke S, Grosch R, Schikora A, Smalla K. Persistence of Salmonella Typhimurium LT2 in soil enhanced after growth in lettuce medium. Front Microbiol. (2017) 8:757. 10.3389/fmicb.2017.00757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bernstein N, Sela S, Neder-Lavon S. Assessment of contamination potential of lettuce by Salmonella enterica serovar Newport added to the plant growing medium. J Food Prot. (2007) 70:1717–22. 10.4315/0362-028X-70.7.1717 [DOI] [PubMed] [Google Scholar]
- 117.Bernstein N, Sela S, Pinto R, Ioffe M. Evidence for internalization of Escherichia coli into the aerial parts of maize via the root system. J Food Prot. (2007) 70:471–5. 10.4315/0362-028X-70.2.471 [DOI] [PubMed] [Google Scholar]
- 118.Brandl MT, Amundson R. Leaf age as a risk factor in contamination of lettuce with Escherichia coli O157:H7 and Salmonella enterica. Appl Environ Microbiol. (2008) 74:2298–306. 10.1128/AEM.02459-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lopez-Velasco G, Sbodio A, Tomas-Callejas A, Wei P, Tan KH, Suslow TV. Assessment of root uptake and systemic vine-transport of Salmonella enterica sv. Typhimurium by melon (Cucumis melo) during field production. Int J Food Microbiol. (2012) 158:65–72. 10.1016/j.ijfoodmicro.2012.07.005 [DOI] [PubMed] [Google Scholar]
- 120.Gomez-Lopez VM, Marin A, Allende A, Beuchat LR, Gil MI. Postharvest handling conditions affect internalization of Salmonella in baby spinach during washing. J Food Prot. (2013) 76:1145–51. 10.4315/0362-028X.JFP-12-539 [DOI] [PubMed] [Google Scholar]
- 121.Zhang Y, Sallach JB, Hodges L, Snow DD, Bartelt-Hunt SL, Eskridge KM, et al. Effects of soil texture and drought stress on the uptake of antibiotics and the internalization of Salmonella in lettuce following wastewater irrigation. Environ Pollut. (2016) 208:523–31. 10.1016/j.envpol.2015.10.025 [DOI] [PubMed] [Google Scholar]
- 122.Wei J, Jin Y, Sims T, Kniel KE. Manure- and biosolids-resident murine norovirus 1 attachment to and internalization by Romaine lettuce. Appl Environ Microbiol. (2010) 76:578–83. 10.1128/AEM.02088-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wang Q, Markland S, Kniel KE. Inactivation of human norovirus and its surrogates on alfalfa seeds by aqueous ozone. J Food Prot. (2015) 78:1586–91. 10.4315/0362-028X.JFP-15-029 [DOI] [PubMed] [Google Scholar]
- 124.Wang Q, Kniel KE. Survival and transfer of murine norovirus within a hydroponic system during kale and mustard microgreen harvesting. Appl Environ Microbiol. (2016) 82:705–13. 10.1128/AEM.02990-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Vorholt JA. Microbial life in the phyllosphere. Nat Rev Microbiol. (2012) 10:828–40. 10.1038/nrmicro2910 [DOI] [PubMed] [Google Scholar]
- 126.Deng W, Gibson KE. Interaction of microorganisms within leafy green phyllospheres: where do human noroviruses fit in? Int J Food Microbiol. (2017) 258:28–37. 10.1016/j.ijfoodmicro.2017.07.010 [DOI] [PubMed] [Google Scholar]
- 127.Yan T, Sadowsky MJ. Determining sources of fecal bacteria in waterways. Environ Monit Assess. (2007) 129:97–106. 10.1007/s10661-006-9426-z [DOI] [PubMed] [Google Scholar]
- 128.Aijuka M, Charimba G, Hugo CJ, Buys EM. Characterization of bacterial pathogens in rural and urban irrigation water. J Water Health (2015) 13:103–17. 10.2166/wh.2014.228 [DOI] [PubMed] [Google Scholar]
- 129.Edberg SC, Rice EW, Karlin RJ, Allen MJ. Escherichia coli: the best biological drinking water indicator for public health protection. Symp Ser Soc Appl Microbiol. (2000) 88:106S–16S. 10.1111/j.1365-2672.2000.tb05338.x [DOI] [PubMed] [Google Scholar]
- 130.Campbell LM, Michaels G, Klein RD, Roth IL. Isolation of Klebsiella pneumoniae from lake water. Can. J Microbiol. (1976) 22:1762–7. 10.1139/m76-260 [DOI] [PubMed] [Google Scholar]
- 131.Barrell RA, Hunter PR, Nichols G. Microbiological standards for water and their relationship to health risk. Commun Dis Public Health (2000) 3:8–13. Available online at: http://martechnicltd.com/sales/docs/LGL_Microbiological_Standards_forWater_HPA.pdf [PubMed] [Google Scholar]
- 132.McEgan R, Mootian G, Goodridge LD, Schaffner DW, Danyluk MD. Predicting Salmonella populations from biological, chemical, and physical indicators in Florida surface waters. Appl Environ Microbiol. (2013) 79:4094–105. 10.1128/AEM.00777-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Bell RL, Jarvis KG, Ottesen AR, McFarland MA, Brown EW. Recent and emerging innovations in Salmonella detection: a food and environmental perspective. Microb Biotechnol. (2016) 9:279–92. 10.1111/1751-7915.12359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Mansfield LP, Forsythe SJ. The detection of Salmonella using a combined immunomagnetic separation and ELISA end-detection procedure. Lett Appl Microbiol. (2000) 31:279–83. 10.1046/j.1472-765x.2000.00811.x [DOI] [PubMed] [Google Scholar]
- 135.Wolffs PFG, Glencross K, Thibaudeau R, Griffiths MW. Direct quantitation and detection of Salmonellae in biological samples without enrichment, using two-step filtration and real-time PCR. Appl Environ Microbiol. (2006) 72:3896–900. 10.1128/AEM.02112-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Eriksson E, Aspan A. Comparison of culture, ELISA and PCR techniques for Salmonella detection in faecal samples for cattle, pig and poultry. BMC Vet Res. (2007) 3:21. 10.1186/1746-6148-3-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Malorny B, Lofstrom C, Wagner M, Kramer N, Hoorfar J. Enumeration of Salmonella bacteria in food and feed samples by real-time PCR for quantitative microbial risk assessment. Appl Environ Microbiol. (2008) 74:1299–304. 10.1128/AEM.02489-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Fukushima H, Tsunomori Y, Seki R. Duplex real-time SYBR green PCR assays for detection of 17 species of food- or waterborne pathogens in stools. J Clin Microbiol. (2003) 41:5134–46. 10.1128/JCM.41.11.5134-5146.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Barbau-Piednoir E, Bertrand S, Mahillon J, Roosens NH, Botteldoorn N. SYBRA (R) Green qPCR Salmonella detection system allowing discrimination at the genus, species and subspecies levels. Appl Microbiol Biotechnol. (2013) 97:9811–24. 10.1007/s00253-013-5234-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Hoorfar J, Ahrens P, Radstrom P. Automated 5′ nuclease PCR assay for identification of Salmonella enterica. J Clin Microbiol. (2000) 38:3429–35. Available online at: http://jcm.asm.org/content/38/9/3429.full.pdf+html [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Malorny B, Paccassoni E, Fach P, Bunge C, Martin A, Helmuth R. Diagnostic real-time PCR for detection of Salmonella in food. Appl Environ Microbiol. (2004) 70:7046–52. 10.1128/AEM.70.12.7046-7052.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Li B, Chen JQ. Real-time PCR methodology for selective detection of viable Escherichia coli O157:H7 cells by targeting Z3276 as a genetic marker. Appl Environ Microbiol. (2012) 78:5297–304. 10.1128/AEM.00794-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Mori Y, Nagamine K, Tomita N, Notomi T. Detection of loop-mediated isothermal amplification reaction by turbidity derived from magnesium pyrophosphate formation. Biochem Biophys Res Commun. (2001) 289:150–4. 10.1006/bbrc.2001.5921 [DOI] [PubMed] [Google Scholar]
- 144.Hara-Kudo Y, Yoshino M, Kojima T, Ikedo M. Loop-mediated isothermal amplification for the rapid detection of Salmonella. FEMS Microbiol Lett. (2005) 253:155–61. 10.1016/j.femsle.2005.09.032 [DOI] [PubMed] [Google Scholar]
- 145.Tomita N, Mori Y, Kanda H, Notomi T. Loop-mediated isothermal amplification (LAMP) of gene sequences and simple visual detection of products. Nat Protoc. (2008) 3:877–82. 10.1038/nprot.2008.57 [DOI] [PubMed] [Google Scholar]
- 146.Chen S, Wang F, Beaulieu JC, Stein RE, Ge B. Rapid detection of viable Salmonellae in produce by coupling propidium monoazide with loop-mediated isothermal amplification. Appl Environ Microbiol. (2011) 77:4008–16. 10.1128/AEM.00354-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Oscorbin IP, Belousova EA, Zakabunin AI, Boyarskikh UA, Filipenko ML. Comparison of fluorescent intercalating dyes for quantitative loop-mediated isothermal amplification (qLAMP). BioTechniques (2016) 61:20–5. 10.2144/000114432 [DOI] [PubMed] [Google Scholar]
- 148.Galan JE, Ginocchio C, Costeas P. Molecular and functional characterization of the Salmonella invasion gene invA: homology of InvA to members of a new protein family. J Bacteriol. (1992) 174:4338–49. 10.1128/jb.174.13.4338-4349.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Rahn K, De Grandis SA, Clarke RC, McEwen SA, Galan JE, Ginocchio C, et al. Amplification of an invA gene sequence of Salmonella typhimurium by polymerase chain reaction as a specific method of detection of Salmonella. Mol Cell Probes (1992) 6:271–9. 10.1016/0890-8508(92)90002-F [DOI] [PubMed] [Google Scholar]
- 150.Malorny B, Hoorfar J, Bunge C, Helmuth R. Multicenter validation of the analytical accuracy of Salmonella PCR: towards an international standard. Appl Environ Microbiol. (2003) 69:290–6. 10.1128/AEM.69.1.290-296.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Mainar-Jaime RC, Andres S, Vico JP, San Roman B, Garrido V, Grillo MJ. Sensitivity of the ISO 6579:2002/Amd 1:2007 standard method for detection of Salmonella spp. on mesenteric lymph nodes from slaughter pigs. J Clin Microbiol. (2013) 51:89–94. 10.1128/JCM.02099-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Zajc-Satler J, Gragas AZ. Xylose lysine deoxycholate agar for the isolation of Salmonella and Shigella from clinical specimens. Zentralbl Bakteriol Orig. A (1977) 237:196–200. [PubMed] [Google Scholar]
- 153.Goo VY, Ching GQ, Gooch JM. Comparison of brilliant green agar and Hektoen enteric agar media in the isolation of Salmonellae from food products. Appl Microbiol. (1973) 26:288–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Wilson WJ, Blair EM. Use of a glucose bismuth sulphite iron medium for the isolation of B. typhosus and B. proteus. J Hyg (Lond.) (1927) 26:374–91. 10.1017/S0022172400009220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Christensen WB. Urea Decomposition as a Means of Differentiating Proteus and Paracolon Cultures from Each Other and from Salmonella and Shigella Types. J. Bacteriol. (1946) 52:461–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Fravalo P, Hascoet Y, Le Fellic M, Queguiner S, Petton J, Salvat G. Convenient method for rapid and quantitative assessment of Salmonella enterica contamination: the mini-MSRV MPN technique. J Rapid Methods Autom Microbiol. (2003) 11:81–8. 10.1111/j.1745-4581.2003.tb00031.x [DOI] [Google Scholar]
- 157.Stender H, Oliveira K, Rigby S, Bargoot F, Coull J. Rapid detection, identification, and enumeration of Escherichia coli by fluorescence in situ hybridization using an array scanner. J Microbiol Methods (2001) 45:31–9. 10.1016/S0167-7012(01)00218-4 [DOI] [PubMed] [Google Scholar]
- 158.Humbert FS, Salvat G, Lalande F, Colin P. Miniaturized most probable number and enrichment serology technique for the enumeration of Salmonella spp. on poultry carcasses. J Food Protect. (1997) 60:1306–11. 10.4315/0362-028X-60.11.1306 [DOI] [PubMed] [Google Scholar]
- 159.Malorny B, Tassios PT, Radstrom P, Cook N, Wagner M, Hoorfar J. Standardization of diagnostic PCR for the detection of foodborne pathogens. Int J Food Microbiol. (2003) 83:39–48. 10.1016/S0168-1605(02)00322-7 [DOI] [PubMed] [Google Scholar]
- 160.Mackay IM. Real-time PCR in the microbiology laboratory. Clin Microbiol Infect. (2004) 10:190–212. 10.1111/j.1198-743X.2004.00722.x [DOI] [PubMed] [Google Scholar]
- 161.Nye KJ, Fallon D, Frodsham D, Gee B, Graham C, Howe S, et al. An evaluation of the performance of XLD, DCA, MLCB, and ABC agars as direct plating media for the isolation of Salmonella enterica from faeces. J Clin Pathol. (2002) 55:286–8. 10.1136/jcp.55.4.286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Wang XL, Slavik MF. Rapid detection of Salmonella in chicken washes by immunomagnetic separation and flow cytometry. J Food Prot. (1999) 62:717–23. 10.4315/0362-028X-62.7.717 [DOI] [PubMed] [Google Scholar]
- 163.Soumet C, Ermel G, Rose V, Rose N, Drouin P, Salvat G, et al. Identification by a multiplex PCR-based assay of Salmonella typhimurium and Salmonella Enteritidis strains from environmental swabs of poultry houses. Lett Appl Microbiol. (1999) 29:1–6. 10.1046/j.1365-2672.1999.00559.x [DOI] [PubMed] [Google Scholar]
- 164.Malorny B, Junker E, Helmuth R. Multi-locus variable-number tandem repeat analysis for outbreak studies of Salmonella enterica serotype Enteritidis. BMC Microbiol. (2008) 8:84. 10.1186/1471-2180-8-84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Guy RA, Kapoor A, Holicka J, Shepherd D, Horgen PA. A rapid molecular-based assay for direct quantification of viable bacteria in slaughterhouses. J. Food Prot. (2006) 69:1265–72. 10.4315/0362-028X-69.6.1265 [DOI] [PubMed] [Google Scholar]
- 166.Miller ND, Draughon FA, D'souza DH. Real-time reverse-transcriptase-polymerase chain reaction for Salmonella enterica detection from jalapeno and serrano peppers. Foodborne Pathog Dis. (2010) 7:367–73. 10.1089/fpd.2009.0398 [DOI] [PubMed] [Google Scholar]
- 167.Techathuvanan C, Draughon FA, D'souza DH. Real-time reverse transcriptase PCR for the rapid and sensitive detection of Salmonella typhimurium from pork. J Food Prot. (2010) 73:507–514. 10.4315/0362-028X-73.3.507 [DOI] [PubMed] [Google Scholar]
- 168.Bohaychuk VM, Gensler GE, McFall ME, King RK, Renter DG. A real-time PCR assay for the detection of Salmonella in a wide variety of food and food-animal matricest. J Food Prot. (2007) 70:1080–7. 10.4315/0362-028X-70.5.1080 [DOI] [PubMed] [Google Scholar]
- 169.Nocker A, Sossa KE, Camper AK. Molecular monitoring of disinfection efficacy using propidium monoazide in combination with quantitative PCR. J Microbiol Methods (2007) 70:252–260. 10.1016/j.mimet.2007.04.014 [DOI] [PubMed] [Google Scholar]
- 170.Li BG, Chen JQ. Development of a sensitive and specific qPCR assay in conjunction with propidium monoazide for enhanced detection of live Salmonella spp. in food. BMC Microbiol. (2013) 13:273. 10.1186/1471-2180-13-273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Luo JF, Lin WT, Guo Y. Method to detect only viable cells in microbial ecology. Appl Microbiol Biotechnol. (2010) 86:377–84. 10.1007/s00253-009-2373-1 [DOI] [PubMed] [Google Scholar]
- 172.Contreras PJ, Urrutia H, Sossa K, Nocker A. Effect of PCR amplicon length on suppressing signals from membrane-compromised cells by propidium monoazide treatment. J Microbiol Methods (2011) 87:89–95. 10.1016/j.mimet.2011.07.016 [DOI] [PubMed] [Google Scholar]
- 173.Soejima T, Schlitt-Dittrich F, Yoshida S. Rapid detection of viable bacteria by nested polymerase chain reaction via long DNA amplification after ethidium monoazide treatment. Anal Biochem. (2011) 418:286–94. 10.1016/j.ab.2011.06.033 [DOI] [PubMed] [Google Scholar]
- 174.Schnetzinger F, Pan Y, Nocker A. Use of propidium monoazide and increased amplicon length reduce false-positive signals in quantitative PCR for bioburden analysis. Appl Microbiol Biotechnol. (2013) 97:2153–62. 10.1007/s00253-013-4711-6 [DOI] [PubMed] [Google Scholar]