Abstract
Many genetic studies for Alzheimer’s Disease (AD) have been focused on the identification of common genetic variants associated with AD risk and not on other aspects of the disease, such as age at onset or rate of dementia progression. There are multiple approaches to untangling the genetic architecture of these phenotypes. We hypothesized that the genetic architecture of rate of progression is different than the risk for developing AD dementia. To test this hypothesis, we used longitudinal clinical data from ADNI and the Knight-ADRC at Washington University and we calculated PRS (polygenic risk score) based on the IGAP study to compare the genetic architecture of AD risk and dementia progression. Dementia progression was measured by the change of Clinical Dementia Rating Sum of Boxes (CDR)-SB per year. Out of the 21 loci for AD risk, no association with the rate of dementia progression was found. The PRS rate was significantly associated with the rate of dementia progression (β=0.146, p.value= 0.03). In the case of rare variants, TREM2 (β=0.309, p.value=0.02) was also associated with the rate of dementia progression. TREM2 variant carriers showed a 23% faster rate of dementia compared with non-variant carriers. In conclusion, our results indicate that the recently identified common and rare variants for AD susceptibility have a limited impact on the rate of dementia progression in AD patients.
Keywords: progression, AD risk, Polygenic risk score, Clinical Dementia Rating sum of boxes, International Genomics of Alzheimer’s Project (IGAP)
Introduction
Alzheimer’s disease (AD) is a common and incurable form of dementia [1]. There is significant heterogeneity in the progression and in the symptomatology of the disease, but little understanding about the causes of this variability. Most genetic studies of AD have focused on the identification of common genetic variants associated with AD risk. The largest GWAS for AD risk included more than 74,000 AD cases and controls and found more than 21 loci associated with AD [2]. Some genes under these loci are involved in inflammation and the immune response [3]. Each locus only accounts for a small proportion of the variance in AD susceptibility [4], indicating that there are still additional genetic variants and genes associated with AD risk remaining to be identified.
Most studies identified variants that are associated with AD risk and no other aspect of the disease, such as age at onset or rate of dementia progression. There are multiple approaches to untangle the genetic architecture of these phenotypes. One approach is to use alternative endophenotypes such as imaging traits or biochemical biomarkers [5]. These intermediate phenotypes, such as levels of cerebrospinal fluid (CSF) phospho-tau (ptau) can identify genetic variants associated with rate of progression. Thus far no genetic study of a relative large population focused on rate of dementia progression has been performed. We defined rate of dementia progression as the change in the Clinical Dementia Rating Sum of Boxes (CDR-SB) in patients diagnosed with AD [6]. We hypothesized that the genetic architecture of rate of progression is different to that of AD risk. This is supported by our previous studies in which we found that genetic variants associated with AD risk are also associated with age at onset and CSF Aβ level; however, the variants associated with CSF tau levels were associated with disease progression but not onset [6, 7].
It has been proposed that low frequency variants could explain some of the missing hereditability. As a result, some recent findings have identified low frequency variants with large effects in some of the genes under the identified loci that are independent of the GWAS signal, as in the case of SORL1 and ABCA7 [8–12]. In addition, low frequency coding variants in TREM2 [13–20] and PLD3 associated with AD risk have been reported. Polygenic Risk Scores (PRS) have been successfully used to collapse the effects of common variants and are useful to calculate the overall risk of an individual or to identify individuals at risk [21]. Even though the predictive power and accuracy of PRS are still insufficient to be applied in a clinical setting [22], it has been suggested that a PRS created from the 21 genome-wide loci capture the overall genetic architecture of late-onset AD and may help to predict AD risk [23].
In this study, we used longitudinal clinical data from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) and the Knight Alzheimer’s Disease Research Center (Knight-ADRC) coupled with GWAS data in order to elucidate the genetic architecture of rate of dementia progression, which may help to identify additional pathways and genes implicated in AD.
Methods
Participants
Participants from this study were enrolled in two different longitudinal studies: the Knight-ADRC at Washington University and ADNI. The recruitment, assessment and exclusion criteria methods for the Knight-ADRC study have been published previously [24]. Written informed consent was obtained from participants and their family members by the Clinical and Genetics Core of the Knight ADRC. The ADNI study has been previously described [25]. The IRB approval number for this study is 201104178.
Cognitive assessment
Individuals in the Knight-ADRC cohort were evaluated by personnel at Washington University [24, 26]. For ADNI, multiple sites evaluated the participants [25, 27]. Briefly, participants were evaluated in accordance with the Clinical Dementia Rating (CDR), where 0 indicates cognitive normality, 0.5 is very mild dementia, 1 is mild dementia, 2 is moderate dementia, and 3 is severe dementia [28, 29]. The scores in each of the six areas are summed to yield a sum of box scores ranging from 0 (no impairment) to 18 (maximal impairment).
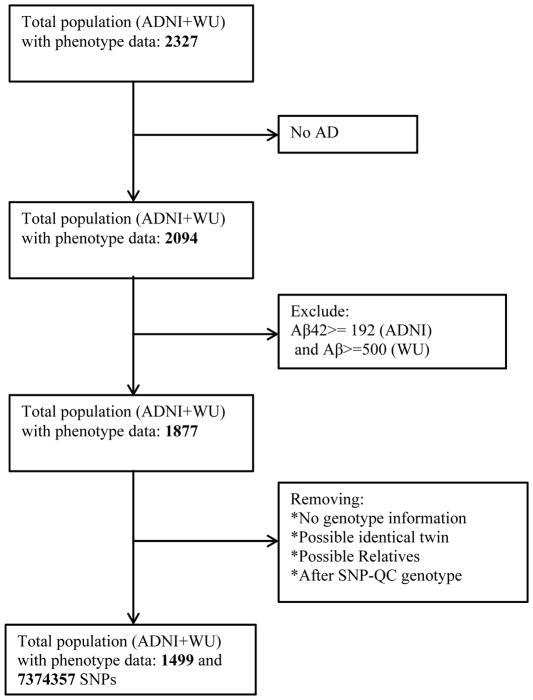
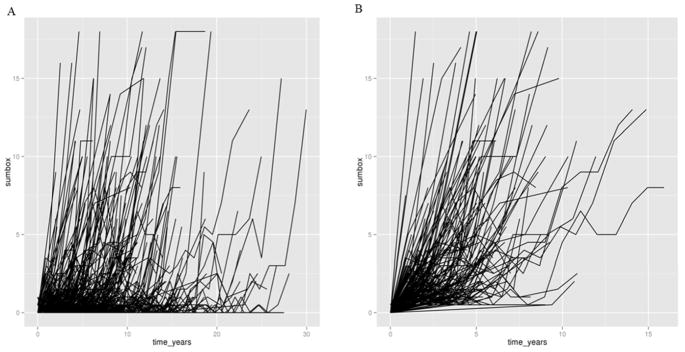
Figure 1 describes the inclusion and exclusion criteria for this study. Briefly, only participants with an AD diagnosis and CDR > 0 at their last visit were included in our analyses. Individuals with dementia caused by neurological diseases other than AD were excluded. For those samples with data available, individuals with cerebrospinal fluid (CSF) amyloid β peptide42 (Aβ42) with values equal or greater than 192pg/mL (ADNI) [30] or 500pg/mL (Knight-ADRC) [31] were also excluded. Individuals with CSF Aβ42 levels below these thresholds have been shown to have fibrillar Aβ deposits in brain [32, 33], and individuals with higher levels do not shown Aβ brain pathology and are likely not have Alzheimer’s Disease. Therefore by removing individuals with high Aβ levels we are removing individuals that have very high likelihood of having a non-AD dementia. Non-informative longitudinally measured CDR-SB was removed for each participant. Non-informative longitudinal data is defined as data in which the CDR-SB is either 0 or 18 and remains constant over a period of time (Figure 2). Figure 2 shows spaghetti plots of CDR-SB before and after QC of our data, when non-informative data was removed. A clear trend of progression is shown after QC. Only individuals with at least two visits and 1.5 year of follow-up were included.
Figure 1.
General flowchart for subject inclusion criteria and SNP selection for longitudinal analysis.
Figure 2. Longitudinal change in CDR-SB.
(A) This plot represents the longitudinal values before QC. (B) This plot represents the longitudinal values after QC, when non-informative data was removed. The increasing CDR-SB is shown clearly following QC.
Genotype data
Participants were genotyped with the Illumina 610 or Omniexpress chip (Illumina, San Diego, CA, USA). As part of routine quality control steps, single-nucleotide polymorphisms (SNPs) with minor allele frequency < 1%, call rates < 98%, Hardy–Weinberg equilibrium p-values >10-6 and individuals with > 2% missing genotypes were removed from the analysis. Imputation was performed as described previously [34]. Briefly IMPUT2 v2.3.2 software and the 1,000 genome (phase3 NCBI build 37) data were used as reference to impute up to 6 million SNPs. To avoid the possibility of spurious association of population structure [35] and to confirm ethnicity of each sample, the two principal components (PCs) scores were used as covariates in the analysis. These PCs were obtained using PLINK v1.9 (http://www.cog-genomics.org/plink2) [36] and Hapmap2 [37, 38] reference population data. Only individuals that clustered with the European-American cluster were included in the study. PLINK v1.9 was also used to find duplicate and related individuals which were eliminated from the analysis. There were 5,986,883 imputed and genotyped SNPs after removing SNPs with a call rate <95% or r2≤0.3.
Both, PLD3 and TREM2, genes were sequenced using pooled-DNA sequencing designed as described previously [17, 39]. All polymorphisms were validated by Sequenom and KASPar genotype in each individual included in the pool.
Neuropsychological data
A subset of individuals (n=460) from the Knight-ADRC cohort who met the criteria in Figure 1 underwent neuropsychological testing within one year of a cognitive assessment where the CDR had been assigned. Once individuals progress to CDR= 2 they typically have difficulty performing neuropsychological testing, so rarely a subject is tested after being rated CDR= 2. Therefore, the neuropsychological data captured early (CDR=0 to CDR=2) but not late changes (CDR=2 to CDR=3). Within these 450 participants, all underwent neuropsychological testing at least twice with a minimal interval between first and last testing of 1.5 years. We evaluated the following three episodic memory measures that are routinely collected as part of the Knight-ADRC cognitive battery: the picture version of the Free and Cued Selective Reminding Test with immediate recall, free recall portion (FCSRT-free)[40]. The Wechsler Memory Scale-III (WMS-III) Logical Memory Immediate and Delayed Recall (logical memory); and Verbal Paired Associates (associate learning)[41].
Statistical Analyses
Longitudinal Regression
A linear mixed-model repeated measure framework was used to account for correlation between repeated measures in the same individual. The linear mixed model analysis was carried out using R statistical software [42] and the package nlme [43]. Change in CDR Sum of Boxes per year (CDR-SB) was set as the independent variable with the following covariates: baseline CDR, baseline age, sex, time of follow-up, years of education, the interaction between baseline CDR and time, and to avoid the possibility of spurious association due to population substructure, the two first principal components scores were included as covariates. A random effect for time and individual was included in the model with an AR(1) covariance structure. We are testing the interaction term SNP*times against CDR-SB, as well as the interaction term PRS*time. When we are testing the former, SNP and time are also included as covariates, in case of the latter; PRS and time are added in the model.
We also performed a gene-based CMC method using PLD3 and TREM2. In this analysis all the rare variants from each gene region was coded as “0” if the locus was a wildtype and “1” if the participant carried a coding variant. In this way we can gain power in the analysis when we test for the null hypothesis that the gene is not associated with the change in CDR-SB. The model was the same as described above. Similar models were used to evaluate whether TREM2 variants were associated with decline on cognitive tests.
Association of GWAS hits with AD risk and Disease Progression
From the longitudinal regression analysis, we explored the relationship between the 21 susceptibility loci with high risk of LOAD [2] which included SNPs in the SLC24A4-RIN3, SORL1, BIN1, PICALM, CASS4, PTK2B, CELF1, CD2AP, INPP5D, EPHA1, MS4A6A, CD33, FERMT2, ZCWPW1, NME8, DSG2, ABCA7, CR1, CLU, HLA-DRB5-HLA-DRB1 and MEF2C gene regions.
Computation of Polygenic Risk Score
We derived a weighted PRS [44], as described previously. In short, the odds ratios were modelled as reported in IGAP [2] using a logarithm of base 2 transformation. SNPs utilized for the score either have a high genotyping rate (around 95%) or were a proxy to the IGAP hits. PLINK v1.9 was used to calculate the PRS choosing the score function and the no-mean-imputation option to ensure that no scores would be imputed. The resulting mean was corrected by multiplying the allele count (Log OR Score). The original score did not include APOE, to analyze whether there is a genetic overlap between risk and progression beyond APOE. However, we also calculated another PRS that included APOE. APOE was modeled as previously, using a model that captures the entire variability of APOE 2 and 4 alleles [45]. Weights (OR) for each APOE allele was based on previous studies (i.e. ε2/ε2=0.6, ε2/ε3=0.6, ε2/ε4 =2.6, ε3/ε4=3.2, ε4/ε4=14.9)[45].
The association results of the longitudinal regression models for Polygenic Risk Scored derived for Lambert hits was calculated by using the following equation, we also calculated the a Pseudo-R2 using the MuMIn package in R
ADNI material and methods
Data used in the preparation of this article were obtained from the ADNI database (www.loni.ucla.edu\ADNI). The ADNI was launched in 2003 by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, the Food and Drug Administration, private pharmaceutical companies and non-profit organizations, as a $60 million, 5-year public-private partnership. The Principal Investigator of this initiative is Michael W. Weiner, M.D. ADNI is the result of efforts of many co-investigators from a broad range of academic institutions and private corporations, and subjects have been recruited from over 50 sites across the U.S. and Canada. The initial goal of ADNI was to recruit 800 adults, ages 55 to 90, to participate in the research -approximately 200 cognitively normal older individuals to be followed for 3 years, 400 people with MCI to be followed for 3 years, and 200 people with early AD to be followed for 2 years.” For up-to-date information see www.adni-info.org.
Results
Participants
The number of participants, after passing our inclusion criteria, with longitudinal cognitive assessment is presented in Table 1; they were grouped into 2 cohorts, ADNI (721) and Knight-ADRC (778). Briefly, a total of 1499 individuals with AD were included in the analysis after passing our inclusion criteria (Figure 1). The combined cohort was 52% male, the Knight-ADRC cohort was 44% males and the ADNI cohort and 60% male. The average year of education in the combined cohort was 14.5 years. Participants in the ADNI cohort had an average of 15.6 years of education and participants in the Knight-ADRC cohort had an average of 13.6 years of education. The follow-up time was longer in Knight-ADRC (4.83 years) than ADNI (3.23 years). In both cohorts, most participants had very mild dementia (CDR 0.5) at baseline. Likely because of the longer follow up time in the Knight-ADRC cohort, the percentage of moderate dementia and severe dementia cases at the final follow-up assessment was higher in the Knight-ADRC (61.2%) than the ADNI (21%) cohorts.
Table 1.
Demographic Characteristics and Baseline and Last measurement of CDR
| Variables | TOTAL | ADNI | WU |
|---|---|---|---|
| Total of Participants | 1499 | 721 | 778 |
| Male (%) | 51.7% | 60.2% | 43.8% |
| Baseline age, mean(SD) y | 75.0(7.8) | 74.5(7.1) | 75.5(8.4) |
| Age onset, mean(SD), y | 73.0(8.9) | 71.9(7.7) | 73.3(9.3) |
| Educational level, mean(SD),y | 14.6(3.3) | 15.6(2.9) | 13.6(3.4) |
| Sum of box, mean(SD) | 7.7(5.1) | 5.7(4.1) | 9.8(5.1) |
| Follow-up time, y | 4.1(2.5) | 3.23(2.0) | 4.8(2.6) |
| Baseline CDR 0 (%) | 11.4% | 4.8% | 17.6% |
| Last CDR | 1.4(0.9) [0.5–3] | 1.0(0.7) [0.5–3] | 1.8(0.9) [0.5–3] |
| APOE e4+ % | 62.3% | 65.9% | 59.0% |
Abbreviations: CDR, Clinical Dementia Rating. APOE, Apolipoprotein E. Y, years
Sex and disease progression
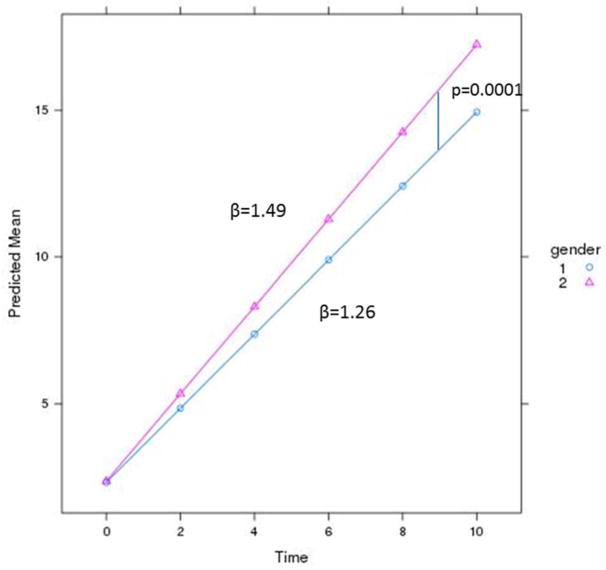
Before running the association analyses with the genetic variants, we wanted to identify the most important covariates for rate of progression. After adjusting for CDR at baseline, follow up time, age, education and the first two principal components, we found a very significant association of sex with rate of progression, in which females progressed 20.8% faster than males (β=0.225, p= 8.62×10−05; Table 2 and Figure 3). Both datasets, the Knight-ADRC and the ADNI dataset contributed to the association of sex with disease progression (β=0.21, p= 0.004 and β=0.15, p= 0.09, respectively).
Table 2.
Association results of the longitudinal regression models for Sex
|
|
|||
|---|---|---|---|
| CDR-SB (N:1,499) | |||
|
| |||
| Beta | P | Male (%) | |
| Sex*time | 0.225 | 8.62×10−05 | 51.6 |
The model for longitudinal regression was adjusted for CDR baseline, Follow-up time, sex, age, education, the first two principal components (PC1, PC2) and the interaction term CDR baseline* follow-up time.
Figure 3. Sex is associated with higher change in CDR-SB.
Females (2) have a faster dementia progression, β= 1.49 than Males (1), β=1.26.
Role of APOE in rate of dementia
APOE is the strongest genetic risk factor for AD, Dementia with Lewy Bodies (DBL) and progression from mild cognitive impairment (CDR 0.5) to AD dementia. Individuals with memory complaints not caused by AD may not present develop progressive memory decline and should have a lower APOE ε4 frequency. To test this hypothesis, we analyzed the association of APOE genotype for any individual with longitudinal data, as well as, the individuals from each step of the inclusion criteria filter (Figure S1). We found a very strong association of APOE with change in CDR-SB/year when every individual with clinical longitudinal data was included (p=2.94×10−11; Figure S1). This association is still strong even after non-AD subjects are removed Figure S1). This association was mainly driven by ADNI in both cases; this was expected because ADNI, by study design, recruit a large number of MCI, not all with AD. When individuals with high CSF Aβ42 levels were removed (>192pg/mL (ADNI) or >500pg/mL (Knight-ADRC)), the strength of association dropped again (β=0.124, p=2.58×10−03; Figure S1). To show the effect of APOE in rate of progression we did a comparison between the betas of 1) APOE4− vs APOE4+ and 2) APOE4− vs APOE 4 vs APOE 44 Figure S2. As noted in the tables having one APOE4 only increase the rate of progression in 1.16 times (FigureS2A) and having two APOE 4 alleles in 1.25 times (FigureS2B). However, the effect of one APOE4 allele in AD risk is either 2.6 or 3.2 (ε2/ε4, ε3/ε4) and two alleles is 14.9(ε4/ε4) [45]. These results suggest that the effect of APOE in rate of progression is lower than for AD risk.
Overlap between the genetic architecture of AD risk and progression
None of 21 GWAS loci associated with AD risk were associated with change in CDR-SB, even at nominal level. Table 4 shows the p-value for rate of progression for each of the 21 most significant SNPs reported as AD risk by the International Genomics of Alzheimer’s Project (IGAP). It is important to note that in this analysis, for APOE we are reporting the p-value for rs769449, which is the SNP reported on the original manuscript. rs769449 is a proxy of rs429358 (r2=0.81), the SNPs that codes for APOE4 allele. Therefore, rs769449 does not capture all the information as APOE 4, neither the entire APOE genotype.
Table 4.
Hits from Lambert that could be related to progression in AD for CDR-SB
| Lambert et al | Progression CDR_SB Analysis | |||||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| SNP | CHR | Gene | Major/Minor | MAF | OR/Beta | P.Value | Beta | P.Value |
| rs10498633 | 14 | SLC24A4-RIN3 | G/T | 0.203 | 0.91/−0.09 | 5.50×10−09 | −0.01 | 0.07 |
| rs11218343 | 11 | SORL1 | T/C | 0.040 | 0.77/−0.26 | 9.70×10−15 | −0.19 | 0.06 |
| rs6733839 | 2 | BIN1 | C/T | 0.409 | 1.22/0.19 | 6.90×10−44 | 0.28 | 0.48 |
| rs10792832 | 11 | PICALM | G/A | 0.335 | 0.87/−0.14 | 9.30×10−26 | −0.001 | 0.97 |
| rs7274581 | 20 | CASS4 | T/C | 0.099 | 0.88/−0.12 | 2.50×10−08 | 0.09 | 0.21 |
| rs28834970 | 8 | PTK2B | T/C | 0.364 | 1.1/0.09 | 7.40×10−14 | 0.08 | 0.06 |
| rs10838725 | 11 | CELF1 | T/C | 0.296 | 1.08/0.08 | 1.10×10−08 | −0.04 | 0.39 |
| rs10948363 | 6 | CD2AP | A/G | 0.279 | 1.1/0.09 | 5.20×10−11 | 0.04 | 0.34 |
| rs35349669 | 2 | INPP5D | C/T | 0.476 | 1.08/0.08 | 3.20×10−08 | 0.06 | 0.12 |
| rs11771145 | 7 | EPHA1 | G/A | 0.353 | 0.9/−0.11 | 1.10×10−13 | −0.04 | 0.35 |
| rs983392 | 11 | MS4A6A | A/G | 0.358 | 0.9/−0.11 | 6.10×10−16 | −0.06 | 0.12 |
| rs3865444 | 19 | CD33 | C/A | 0.279 | 0.94/−0.06 | 3.00×10−06 | −0.04 | 0.37 |
| rs17125944 | 14 | FERMT2 | T/C | 0.104 | 1.14/0.13 | 7.90×10−09 | −0.001 | 0.89 |
| rs1476679 | 7 | ZCWPW1 | T/C | 0.263 | 0.91/−0.09 | 5.60×10−10 | 0.004 | 0.92 |
| rs2718058 | 7 | NME8 | A/G | 0.361 | 0.93/−0.07 | 4.80×10−09 | −0.04 | 0.38 |
| rs8093731 | 18 | DSG2 | C/T | 0.029 | 0.73/−0.31 | 1.00×10−04 | −0.05 | 0.71 |
| rs4147929 | 19 | ABCA7 | G/A | 0.177 | 1.15/0.14 | 1.10×10−15 | 0.03 | 0.61 |
| rs6656401 | 1 | CR1 | G/A | 0.197 | 1.18/0.17 | 5.70×10−24 | −0.006 | 0.93 |
| rs9331896 | 8 | CLU | T/C | 0.379 | 0.86/−0.15 | 2.80×10−25 | −0.43 | 0.28 |
| rs9271192 | 6 | HLA-DRB5-HLA-DRB1 | A/C | 0.276 | 1.11/0.14 | 2.90×10−12 | NA | NA |
| rs190982 | 5 | MEF2C | A/G | 0.408 | 0.93/−0.07 | 3.20×10−08 | 0.04 | 0.59 |
| rs769449 | 19 | APOE | G/A | - | - | - | 0.06 | 0.21 |
SNP, based on Build 37 of reference genome. MAF, Minor allele frequency. NA no data available.
Polygenic Risk Scores (PRS) aggregates the effects that multiple genetic markers (both protective and risk variants) confer to individuals for a specific complex trait. PRS can be employed as a measure to identify the extent of overlap between the genetic architecture of co-morbid complex traits. For this reason, we decided to calculate a PRS based on the IGAP results to test the potential overlap in the genetic architecture between AD risk and progression. The PRS rate reached a nominal association with cognitive decline (β=0.146, p=0.03) as shown on Table 5; this association became more significant when APOE genotype was added (β=0.054, p=9.51×10−03), as expected. It is important to notice that the amount of variability explained by the PRS or APOE or APOE+PRS (Table S1) is lower than the variability explained by these variants for case-control studies [23].
Table 5.
Association results of the longitudinal regression models for Polygenic Risk Scored derived for Lambert hits
|
|
||
|---|---|---|
| CDR-SB (N=1,499) | ||
|
| ||
| Beta? | P | |
| PRS*time | 0.146 | 0.029 |
| APOE*time | 0.045 | 0.041 |
| PRS+APOE*time | 0.054 | 9.51×10−03 |
The model for longitudinal regression was adjusted for CDR baseline, follow-up time, sex, age, education, the first two principal components (PC1, PC2) and the interaction term CDR baseline* follow-up time. The beta and p values are the results for the interaction terms of PRS, APOE and PRS+APOE and time in the model. Abbreviations: PRS, Polygenetic Risk Score. APOE, Apolipoprotein E. P, p-value.
The effects of APOE genotype on AD risk are not additive. APOE is added to the model the scale of PRS is totally different to the scale without APOE, and therefore the effect size can not be compared directly.
TREM2 and PLD3
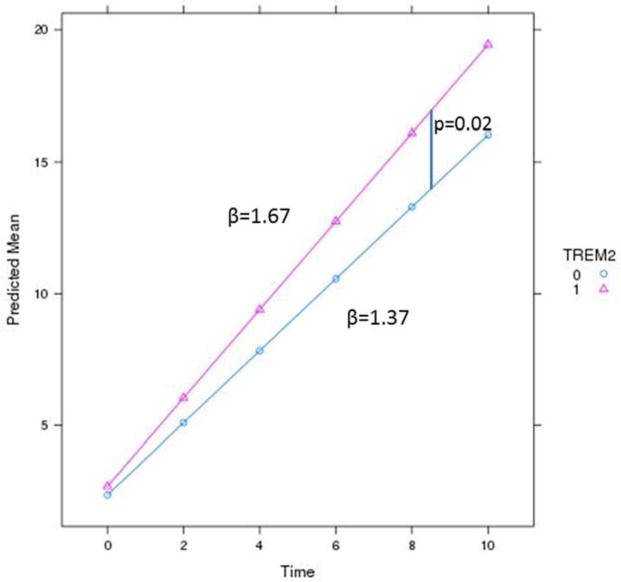
In our low frequency variant analyses for PLD3, only one variant was associated with faster progression in the carriers (Table S2). The gene-based analysis for PLD3 did not reach a statistically significant association with rate of progression (Table 3). On the other hand, individuals carrying TREM2 coding variants experienced faster progression than non-carriers, the carriers declined at 1.67 CDR-SB/year and non-carriers by 1.37 CDR-SB/yr, (p=0.02) (Table 3 and Figure 4). There was one TREM2 variant in exon 2, rs143332484 (p.R62H; Table S3), that reached nominal significance at p=0.027. This variant was already associated with AD as reported previously in the European-Americans [17] population.
Table 3.
Association results of the longitudinal regression models for PLD3 and TREM2
The model for longitudinal regression was adjusted for CDR baseline, Follow up time, sex, age, education, the first two principal components (PC1, PC2), the interaction term CDR baseline* follow-up time, and PLD3 or TREM2 status (presence or absent).
The number of individuals included in these analyses is lower than in the overall study because the sequence data was not available for all participants.
Figure 4. TREM2 variant carriers have a faster increase in CDR-SB than non-carriers.
TREM2 carriers (1) have a faster rate of dementia progression, β=1.67 compared to non-carriers (0), β=1.37.
Using a subset of 460 individuals from the Knight-ADRC cohort who had sufficient longitudinal neuropsychological data, we found that individuals carrying TREM2 variants declined at a faster rate on the FCSRT-Free Recall measure of episodic memory (p=0.02). Carriers of TREM2 variants also trended towards a faster decline on logical memory (p=0.07). There was no effect of TREM2 variant status on the rate of decline for associate learning. Overall, this data suggests that TREM2 variant carriers with AD have a faster rate of decline in episodic memory
Discussion
In order to understand the genetic architecture of AD, it is important to study multiple aspects of the diseases [46]. Here, we present a genetic study of rate of progression of AD. An interesting finding in this study is that in our data set, women have a twofold (22.5%) faster progression for cognitive decline, consistent with other recent reports [47, 48]. This suggests that the more rapid progression of AD in women is not due to longevity, higher rates of obesity, diabetes and other conditions [47] which increase the likelihood of developing AD, but rather to a specific pathogenic mechanisms that affects women.
However, the main hypothesis of this study was to investigate whether the genetic architecture of the rate of progression is different than AD risk. We ran analyses using common variant markers from two different cohorts with longitudinal data. Among the known 21 common susceptibility loci associated with risk of LOAD, no association with our progression analysis with CDR-SB was found. To increase the power in our analysis, we calculated, from the same 21 loci, a PRS. The main idea behind PRS is that it sums the additive effects of several variants with small effects individually in both directions (protective and risk) [44]. Therefore the best PRS will capture all the genetic architecture of a disease in one value or clinical test. Our study showed that even though the results for the PRS rate were nominally significant, without (p=0.03) and with APOE genotype (p=9.51×10−03), it did not reach the strength of association observed in previous studies in which the PRS showed a stronger association with risk for disease in a similar size dataset (p-value of the PRS without APOE = 1.21×10−03 and with APOE the 5.29×10−34 [49]. These results indicate that overall genetic architecture for rate of progression is different to that of AD risk, although the AD risk has a subtle effect in rate of progression.
In the gene-based collapsing analysis, we observed a nominal association for TREM2 (β=0.309, p=0.02) but not for PLD3 (β= 0.268, p=0.06) in the decline of AD patients. TREM2 variant carriers have a faster rate of dementia progression. This finding suggests that TREM2 plays a role in rate of progression. The role of TREM2 in the biology of AD is not clear, but some studies suggest that TREM2 may be involved in the clearance of Aβ [50, 51]. If TREM2 helps clear Aβ, it is possible that variants that reduce TREM2 activity could accelerate AD pathogenesis and lead to faster AD progression, congruent with our findings.
Limitations of our study include the relatively small sample sizes and variability in time of follow-up of the cases for cognitive decline between ADNI and Knight-ADRC. Furthermore, CDR-SB is a one measure, but certainly not the only measure, of dementia progression. Finally, because of the lack of a validation cohort, our GWAS findings are meant for hypothesis generation for future larger studies.
In conclusion, common and rare markers are associated with AD progression, which indicates that they play a role in the development of disease over time, but their effect size is relatively small. These suggest that there is a different genetic architecture for AD risk and AD progression.
Supplementary Material
Acknowledgments
We thank all participants and their families for their commitment and dedication to helping advance research into the early detection and causation of AD. This work was supported by grants from the National Institutes of Health (R01-AG044546, P01-AG003991, RF1AG053303, R01-AG035083, and R01-NS085419), and the Alzheimer Association (BAND-14-338165). The recruitment and clinical characterization of research participants at Washington University were supported by NIH P50-AG05681, P01-AG03991, and P01-AG026276. This work was supported by access to equipment made possible by the Hope Center for Neurological Disorders, and the Departments of Neurology and Psychiatry at Washington University School of Medicine.
Data collection and sharing for this project was funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol-Myers Squibb Company; CereSpir, Inc.; Cogstate; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Lumosity; Lundbeck; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Therapeutic Research Institute at the University of Southern California. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest
References
- 1.Holtzman DM, Morris JC, Goate AM. Alzheimer’s disease: the challenge of the second century. Sci Transl Med. 2011;3:77sr71. doi: 10.1126/scitranslmed.3002369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, DeStafano AL, Bis JC, Beecham GW, Grenier-Boley B, Russo G, Thorton-Wells TA, Jones N, Smith AV, Chouraki V, Thomas C, Ikram MA, Zelenika D, Vardarajan BN, Kamatani Y, Lin CF, Gerrish A, Schmidt H, Kunkle B, Dunstan ML, Ruiz A, Bihoreau MT, Choi SH, Reitz C, Pasquier F, Cruchaga C, Craig D, Amin N, Berr C, Lopez OL, De Jager PL, Deramecourt V, Johnston JA, Evans D, Lovestone S, Letenneur L, Moron FJ, Rubinsztein DC, Eiriksdottir G, Sleegers K, Goate AM, Fievet N, Huentelman MW, Gill M, Brown K, Kamboh MI, Keller L, Barberger-Gateau P, McGuiness B, Larson EB, Green R, Myers AJ, Dufouil C, Todd S, Wallon D, Love S, Rogaeva E, Gallacher J, St George-Hyslop P, Clarimon J, Lleo A, Bayer A, Tsuang DW, Yu L, Tsolaki M, Bossu P, Spalletta G, Proitsi P, Collinge J, Sorbi S, Sanchez-Garcia F, Fox NC, Hardy J, Deniz Naranjo MC, Bosco P, Clarke R, Brayne C, Galimberti D, Mancuso M, Matthews F, Moebus S, Mecocci P, Del Zompo M, Maier W, Hampel H, Pilotto A, Bullido M, Panza F, Caffarra P, Nacmias B, Gilbert JR, Mayhaus M, Lannefelt L, Hakonarson H, Pichler S, Carrasquillo MM, Ingelsson M, Beekly D, Alvarez V, Zou F, Valladares O, Younkin SG, Coto E, Hamilton-Nelson KL, Gu W, Razquin C, Pastor P, Mateo I, Owen MJ, Faber KM, Jonsson PV, Combarros O, O’Donovan MC, Cantwell LB, Soininen H, Blacker D, Mead S, Mosley TH, Jr, Bennett DA, Harris TB, Fratiglioni L, Holmes C, de Bruijn RF, Passmore P, Montine TJ, Bettens K, Rotter JI, Brice A, Morgan K, Foroud TM, Kukull WA, Hannequin D, Powell JF, Nalls MA, Ritchie K, Lunetta KL, Kauwe JS, Boerwinkle E, Riemenschneider M, Boada M, Hiltuenen M, Martin ER, Schmidt R, Rujescu D, Wang LS, Dartigues JF, Mayeux R, Tzourio C, Hofman A, Nothen MM, Graff C, Psaty BM, Jones L, Haines JL, Holmans PA, Lathrop M, Pericak-Vance MA, Launer LJ, Farrer LA, van Duijn CM, Van Broeckhoven C, Moskvina V, Seshadri S, Williams J, Schellenberg GD, Amouyel P European Alzheimer’s Disease I, Genetic, Environmental Risk in Alzheimer’s D, Alzheimer’s Disease Genetic C, Cohorts for H, Aging Research in Genomic E. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet. 2013;45:1452–1458. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.International Genomics of Alzheimer’s Disease C. Convergent genetic and expression data implicate immunity in Alzheimer’s disease. Alzheimers Dement. 2015;11:658–671. doi: 10.1016/j.jalz.2014.05.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ertekin-Taner N. Genetics of Alzheimer disease in the pre- and post-GWAS era. Alzheimers Res Ther. 2010;2:3. doi: 10.1186/alzrt26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deming Y, Xia J, Cai Y, Lord J, Del-Aguila JL, Fernandez MV, Carrell D, Black K, Budde J, Ma S, Saef B, Howells B, Bertelsen S, Bailey M, Ridge PG, Holtzman D, Morris JC, Bales K, Pickering EH, Lee J-M, Heitsch L, Kauwe J, Goate A, Piccio L, Cruchaga C. Genetic studies of plasma analytes identify novel potential biomarkers for several complex traits. Scientific Reports. 2016;6:18092. doi: 10.1038/srep18092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cruchaga C, Kauwe JS, Mayo K, Spiegel N, Bertelsen S, Nowotny P, Shah AR, Abraham R, Hollingworth P, Harold D, Owen MM, Williams J, Lovestone S, Peskind ER, Li G, Leverenz JB, Galasko D, Morris JC, Fagan AM, Holtzman DM, Goate AM Alzheimer’s Disease Neuroimaging I. SNPs associated with cerebrospinal fluid phospho-tau levels influence rate of decline in Alzheimer’s disease. PLoS Genet. 2010;6:e1001101. doi: 10.1371/journal.pgen.1001101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cruchaga C, Kauwe JS, Harari O, Jin SC, Cai Y, Karch CM, Benitez BA, Jeng AT, Skorupa T, Carrell D, Bertelsen S, Bailey M, McKean D, Shulman JM, De Jager PL, Chibnik L, Bennett DA, Arnold SE, Harold D, Sims R, Gerrish A, Williams J, Van Deerlin VM, Lee VM, Shaw LM, Trojanowski JQ, Haines JL, Mayeux R, Pericak-Vance MA, Farrer LA, Schellenberg GD, Peskind ER, Galasko D, Fagan AM, Holtzman DM, Morris JC, Consortium G, Goate AM Alzheimer’s Disease Neuroimaging I, Alzheimer Disease Genetic C. GWAS of cerebrospinal fluid tau levels identifies risk variants for Alzheimer’s disease. Neuron. 2013;78:256–268. doi: 10.1016/j.neuron.2013.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rogaeva E, Meng Y, Lee JH, Gu Y, Kawarai T, Zou F, Katayama T, Baldwin CT, Cheng R, Hasegawa H, Chen F, Shibata N, Lunetta KL, Pardossi-Piquard R, Bohm C, Wakutani Y, Cupples LA, Cuenco KT, Green RC, Pinessi L, Rainero I, Sorbi S, Bruni A, Duara R, Friedland RP, Inzelberg R, Hampe W, Bujo H, Song YQ, Andersen OM, Willnow TE, Graff-Radford N, Petersen RC, Dickson D, Der SD, Fraser PE, Schmitt-Ulms G, Younkin S, Mayeux R, Farrer LA, St George-Hyslop P. The neuronal sortilin-related receptor SORL1 is genetically associated with Alzheimer disease. Nat Genet. 2007;39:168–177. doi: 10.1038/ng1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Louwersheimer E, Ramirez A, Cruchaga C, Becker T, Kornhuber J, Peters O, Heilmann S, Wiltfang J, Jessen F, Visser PJ, Scheltens P, Pijnenburg YA, Teunissen CE, Barkhof F, van Swieten JC, Holstege H, Van der Flier WM. Influence of genetic variants in SORL1 gene on the manifestation of Alzheimer’s disease. Neurobiol Aging. 2015;36:1605e1613–1620. doi: 10.1016/j.neurobiolaging.2014.12.007. [DOI] [PubMed] [Google Scholar]
- 10.Del-Aguila JL, Fernandez MV, Jimenez J, Black K, Ma S, Deming Y, Carrell D, Saef B, Howells B, Budde J, Cruchaga C. Role of ABCA7 loss-of-function variant in Alzheimer’s disease: a replication study in European-Americans. Alzheimers Res Ther. 2015;7:73. doi: 10.1186/s13195-015-0154-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hollingworth P, Harold D, Sims R, Gerrish A, Lambert JC, Carrasquillo MM, Abraham R, Hamshere ML, Pahwa JS, Moskvina V, Dowzell K, Jones N, Stretton A, Thomas C, Richards A, Ivanov D, Widdowson C, Chapman J, Lovestone S, Powell J, Proitsi P, Lupton MK, Brayne C, Rubinsztein DC, Gill M, Lawlor B, Lynch A, Brown KS, Passmore PA, Craig D, McGuinness B, Todd S, Holmes C, Mann D, Smith AD, Beaumont H, Warden D, Wilcock G, Love S, Kehoe PG, Hooper NM, Vardy ER, Hardy J, Mead S, Fox NC, Rossor M, Collinge J, Maier W, Jessen F, Ruther E, Schurmann B, Heun R, Kolsch H, van den Bussche H, Heuser I, Kornhuber J, Wiltfang J, Dichgans M, Frolich L, Hampel H, Gallacher J, Hull M, Rujescu D, Giegling I, Goate AM, Kauwe JS, Cruchaga C, Nowotny P, Morris JC, Mayo K, Sleegers K, Bettens K, Engelborghs S, De Deyn PP, Van Broeckhoven C, Livingston G, Bass NJ, Gurling H, McQuillin A, Gwilliam R, Deloukas P, Al-Chalabi A, Shaw CE, Tsolaki M, Singleton AB, Guerreiro R, Muhleisen TW, Nothen MM, Moebus S, Jockel KH, Klopp N, Wichmann HE, Pankratz VS, Sando SB, Aasly JO, Barcikowska M, Wszolek ZK, Dickson DW, Graff-Radford NR, Petersen RC, van Duijn CM, Breteler MM, Ikram MA, DeStefano AL, Fitzpatrick AL, Lopez O, Launer LJ, Seshadri S, Berr C, Campion D, Epelbaum J, Dartigues JF, Tzourio C, Alperovitch A, Lathrop M, Feulner TM, Friedrich P, Riehle C, Krawczak M, Schreiber S, Mayhaus M, Nicolhaus S, Wagenpfeil S, Steinberg S, Stefansson H, Stefansson K, Snaedal J, Bjornsson S, Jonsson PV, Chouraki V, Genier-Boley B, Hiltunen M, Soininen H, Combarros O, Zelenika D, Delepine M, Bullido MJ, Pasquier F, Mateo I, Frank-Garcia A, Porcellini E, Hanon O, Coto E, Alvarez V, Bosco P, Siciliano G, Mancuso M, Panza F, Solfrizzi V, Nacmias B, Sorbi S, Bossu P, Piccardi P, Arosio B, Annoni G, Seripa D, Pilotto A, Scarpini E, Galimberti D, Brice A, Hannequin D, Licastro F, Jones L, Holmans PA, Jonsson T, Riemenschneider M, Morgan K, Younkin SG, Owen MJ, O’Donovan M, Amouyel P, Williams J. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer’s disease. Nat Genet. 2011;43:429–435. doi: 10.1038/ng.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sassi C, Nalls MA, Ridge PG, Gibbs JR, Ding J, Lupton MK, Troakes C, Lunnon K, Al-Sarraj S, Brown KS, Medway C, Clement N, Lord J, Turton J, Bras J, Almeida MR, Holstege H, Louwersheimer E, van der Flier WM, Scheltens P, Van Swieten JC, Santana I, Oliveira C, Morgan K, Powell JF, Kauwe JS, Cruchaga C, Goate AM, Singleton AB, Guerreiro R, Hardy J. ABCA7 p.G215S as potential protective factor for Alzheimer’s disease. Neurobiol Aging. 2016 doi: 10.1016/j.neurobiolaging.2016.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Benitez BA, Cooper B, Pastor P, Jin SC, Lorenzo E, Cervantes S, Cruchaga C. TREM2 is associated with the risk of Alzheimer’s disease in Spanish population. Neurobiol Aging. 2013;34:1711e1715–1717. doi: 10.1016/j.neurobiolaging.2012.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Benitez BA, Cruchaga C. TREM2 and neurodegenerative disease. N Engl J Med. 2013;369:1567–1568. doi: 10.1056/NEJMc1306509#SA4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cady J, Koval ED, Benitez BA, Zaidman C, Jockel-Balsarotti J, Allred P, Baloh RH, Ravits J, Simpson E, Appel SH, Pestronk A, Goate AM, Miller TM, Cruchaga C, Harms MB. TREM2 variant p.R47H as a risk factor for sporadic amyotrophic lateral sclerosis. JAMA Neurol. 2014;71:449–453. doi: 10.1001/jamaneurol.2013.6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guerreiro R, Wojtas A, Bras J, Carrasquillo M, Rogaeva E, Majounie E, Cruchaga C, Sassi C, Kauwe JS, Younkin S, Hazrati L, Collinge J, Pocock J, Lashley T, Williams J, Lambert JC, Amouyel P, Goate A, Rademakers R, Morgan K, Powell J, St George-Hyslop P, Singleton A, Hardy J. TREM2 variants in Alzheimer’s disease. N Engl J Med. 2013;368:117–127. doi: 10.1056/NEJMoa1211851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jin SC, Benitez BA, Karch CM, Cooper B, Skorupa T, Carrell D, Norton JB, Hsu S, Harari O, Cai Y, Bertelsen S, Goate AM, Cruchaga C. Coding variants in TREM2 increase risk for Alzheimer’s disease. Hum Mol Genet. 2014;23:5838–5846. doi: 10.1093/hmg/ddu277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jin SC, Carrasquillo MM, Benitez BA, Skorupa T, Carrell D, Patel D, Lincoln S, Krishnan S, Kachadoorian M, Reitz C, Mayeux R, Wingo TS, Lah JJ, Levey AI, Murrell J, Hendrie H, Foroud T, Graff-Radford NR, Goate AM, Cruchaga C, Ertekin-Taner N. TREM2 is associated with increased risk for Alzheimer’s disease in African Americans. Mol Neurodegener. 2015;10:19. doi: 10.1186/s13024-015-0016-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luis EO, Ortega-Cubero S, Lamet I, Razquin C, Cruchaga C, Benitez BA, Lorenzo E, Irigoyen J, Pastor MA, Pastor P. Frontobasal gray matter loss is associated with the TREM2 p.R47H variant. Neurobiol Aging. 2014;35:2681–2690. doi: 10.1016/j.neurobiolaging.2014.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Piccio L, Deming Y, Del-Aguila JL, Ghezzi L, Holtzman DM, Fagan AM, Fenoglio C, Galimberti D, Borroni B, Cruchaga C. Cerebrospinal fluid soluble TREM2 is higher in Alzheimer disease and associated with mutation status. Acta Neuropathol. 2016;131:925–933. doi: 10.1007/s00401-016-1533-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vassos E, Di Forti M, Coleman J, Iyegbe C, Prata D, Euesden J, O’Reilly P, Curtis C, Kolliakou A, Patel H, Newhouse S, Traylor M, Ajnakina O, Mondelli V, Marques TR, Gardner-Sood P, Aitchison KJ, Powell J, Atakan Z, Greenwood KE, Smith S, Ismail K, Pariante C, Gaughran F, Dazzan P, Markus HS, David AS, Lewis CM, Murray RM, Breen G. An Examination of Polygenic Score Risk Prediction in Individuals With First-Episode Psychosis. Biol Psychiatry. 2016 doi: 10.1016/j.biopsych.2016.06.028. [DOI] [PubMed] [Google Scholar]
- 22.Dudbridge F. Power and predictive accuracy of polygenic risk scores. PLoS Genet. 2013;9:e1003348. doi: 10.1371/journal.pgen.1003348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Escott-Price V, Shoai M, Pither R, Williams J, Hardy J. Polygenic score prediction captures nearly all common genetic risk for Alzheimer’s disease. Neurobiol Aging. 2017;49:214e217–214e211. doi: 10.1016/j.neurobiolaging.2016.07.018. [DOI] [PubMed] [Google Scholar]
- 24.Berg L, McKeel DW, Jr, Miller JP, Storandt M, Rubin EH, Morris JC, Baty J, Coats M, Norton J, Goate AM, Price JL, Gearing M, Mirra SS, Saunders AM. Clinicopathologic studies in cognitively healthy aging and Alzheimer’s disease: relation of histologic markers to dementia severity, age, sex, and apolipoprotein E genotype. Arch Neurol. 1998;55:326–335. doi: 10.1001/archneur.55.3.326. [DOI] [PubMed] [Google Scholar]
- 25.Petersen RC, Aisen PS, Beckett LA, Donohue MC, Gamst AC, Harvey DJ, Jack CR, Jr, Jagust WJ, Shaw LM, Toga AW, Trojanowski JQ, Weiner MW. Alzheimer’s Disease Neuroimaging Initiative (ADNI): clinical characterization. Neurology. 2010;74:201–209. doi: 10.1212/WNL.0b013e3181cb3e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Williams MM, Roe CM, Morris JC. Stability of the Clinical Dementia Rating, 1979–2007. Arch Neurol. 2009;66:773–777. doi: 10.1001/archneurol.2009.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saykin AJ, Shen L, Foroud TM, Potkin SG, Swaminathan S, Kim S, Risacher SL, Nho K, Huentelman MJ, Craig DW, Thompson PM, Stein JL, Moore JH, Farrer LA, Green RC, Bertram L, Jack CR, Jr, Weiner MW Alzheimer’s Disease Neuroimaging I. Alzheimer’s Disease Neuroimaging Initiative biomarkers as quantitative phenotypes: Genetics core aims, progress, and plans. Alzheimers Dement. 2010;6:265–273. doi: 10.1016/j.jalz.2010.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morris JC. The Clinical Dementia Rating (CDR): current version and scoring rules. Neurology. 1993;43:2412–2414. doi: 10.1212/wnl.43.11.2412-a. [DOI] [PubMed] [Google Scholar]
- 29.Morris JC, McKeel DW, Jr, Fulling K, Torack RM, Berg L. Validation of clinical diagnostic criteria for Alzheimer’s disease. Ann Neurol. 1988;24:17–22. doi: 10.1002/ana.410240105. [DOI] [PubMed] [Google Scholar]
- 30.Shaw LM, Vanderstichele H, Knapik-Czajka M, Clark CM, Aisen PS, Petersen RC, Blennow K, Soares H, Simon A, Lewczuk P, Dean R, Siemers E, Potter W, Lee VM, Trojanowski JQ Alzheimer’s Disease Neuroimaging I. Cerebrospinal fluid biomarker signature in Alzheimer’s disease neuroimaging initiative subjects. Ann Neurol. 2009;65:403–413. doi: 10.1002/ana.21610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morris JC, Roe CM, Xiong C, Fagan AM, Goate AM, Holtzman DM, Mintun MA. APOE predicts amyloid-beta but not tau Alzheimer pathology in cognitively normal aging. Ann Neurol. 2010;67:122–131. doi: 10.1002/ana.21843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fagan AM, Mintun MA, Mach RH, Lee SY, Dence CS, Shah AR, LaRossa GN, Spinner ML, Klunk WE, Mathis CA, DeKosky ST, Morris JC, Holtzman DM. Inverse relation between in vivo amyloid imaging load and cerebrospinal fluid Abeta42 in humans. Ann Neurol. 2006;59:512–519. doi: 10.1002/ana.20730. [DOI] [PubMed] [Google Scholar]
- 33.Jagust WJ, Landau SM, Shaw LM, Trojanowski JQ, Koeppe RA, Reiman EM, Foster NL, Petersen RC, Weiner MW, Price JC, Mathis CA Alzheimer’s Disease Neuroimaging I. Relationships between biomarkers in aging and dementia. Neurology. 2009;73:1193–1199. doi: 10.1212/WNL.0b013e3181bc010c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 36.Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.International HapMap C. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.International HapMap C. Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, Gibbs RA, Belmont JW, Boudreau A, Hardenbol P, Leal SM, Pasternak S, Wheeler DA, Willis TD, Yu F, Yang H, Zeng C, Gao Y, Hu H, Hu W, Li C, Lin W, Liu S, Pan H, Tang X, Wang J, Wang W, Yu J, Zhang B, Zhang Q, Zhao H, Zhao H, Zhou J, Gabriel SB, Barry R, Blumenstiel B, Camargo A, Defelice M, Faggart M, Goyette M, Gupta S, Moore J, Nguyen H, Onofrio RC, Parkin M, Roy J, Stahl E, Winchester E, Ziaugra L, Altshuler D, Shen Y, Yao Z, Huang W, Chu X, He Y, Jin L, Liu Y, Shen Y, Sun W, Wang H, Wang Y, Wang Y, Xiong X, Xu L, Waye MM, Tsui SK, Xue H, Wong JT, Galver LM, Fan JB, Gunderson K, Murray SS, Oliphant AR, Chee MS, Montpetit A, Chagnon F, Ferretti V, Leboeuf M, Olivier JF, Phillips MS, Roumy S, Sallee C, Verner A, Hudson TJ, Kwok PY, Cai D, Koboldt DC, Miller RD, Pawlikowska L, Taillon-Miller P, Xiao M, Tsui LC, Mak W, Song YQ, Tam PK, Nakamura Y, Kawaguchi T, Kitamoto T, Morizono T, Nagashima A, Ohnishi Y, Sekine A, Tanaka T, Tsunoda T, Deloukas P, Bird CP, Delgado M, Dermitzakis ET, Gwilliam R, Hunt S, Morrison J, Powell D, Stranger BE, Whittaker P, Bentley DR, Daly MJ, de Bakker PI, Barrett J, Chretien YR, Maller J, McCarroll S, Patterson N, Pe’er I, Price A, Purcell S, Richter DJ, Sabeti P, Saxena R, Schaffner SF, Sham PC, Varilly P, Altshuler D, Stein LD, Krishnan L, Smith AV, Tello-Ruiz MK, Thorisson GA, Chakravarti A, Chen PE, Cutler DJ, Kashuk CS, Lin S, Abecasis GR, Guan W, Li Y, Munro HM, Qin ZS, Thomas DJ, McVean G, Auton A, Bottolo L, Cardin N, Eyheramendy S, Freeman C, Marchini J, Myers S, Spencer C, Stephens M, Donnelly P, Cardon LR, Clarke G, Evans DM, Morris AP, Weir BS, Tsunoda T, Mullikin JC, Sherry ST, Feolo M, Skol A, Zhang H, Zeng C, Zhao H, Matsuda I, Fukushima Y, Macer DR, Suda E, Rotimi CN, Adebamowo CA, Ajayi I, Aniagwu T, Marshall PA, Nkwodimmah C, Royal CD, Leppert MF, Dixon M, Peiffer A, Qiu R, Kent A, Kato K, Niikawa N, Adewole IF, Knoppers BM, Foster MW, Clayton EW, Watkin J, Gibbs RA, Belmont JW, Muzny D, Nazareth L, Sodergren E, Weinstock GM, Wheeler DA, Yakub I, Gabriel SB, Onofrio RC, Richter DJ, Ziaugra L, Birren BW, Daly MJ, Altshuler D, Wilson RK, Fulton LL, Rogers J, Burton J, Carter NP, Clee CM, Griffiths M, Jones MC, McLay K, Plumb RW, Ross MT, Sims SK, Willey DL, Chen Z, Han H, Kang L, Godbout M, Wallenburg JC, L’Archeveque P, Bellemare G, Saeki K, Wang H, An D, Fu H, Li Q, Wang Z, Wang R, Holden AL, Brooks LD, McEwen JE, Guyer MS, Wang VO, Peterson JL, Shi M, Spiegel J, Sung LM, Zacharia LF, Collins FS, Kennedy K, Jamieson R, Stewart J. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cruchaga C, Karch CM, Jin SC, Benitez BA, Cai Y, Guerreiro R, Harari O, Norton J, Budde J, Bertelsen S, Jeng AT, Cooper B, Skorupa T, Carrell D, Levitch D, Hsu S, Choi J, Ryten M, Consortium UKBE, Hardy J, Ryten M, Trabzuni D, Weale ME, Ramasamy A, Smith C, Sassi C, Bras J, Gibbs JR, Hernandez DG, Lupton MK, Powell J, Forabosco P, Ridge PG, Corcoran CD, Tschanz JT, Norton MC, Munger RG, Schmutz C, Leary M, Demirci FY, Bamne MN, Wang X, Lopez OL, Ganguli M, Medway C, Turton J, Lord J, Braae A, Barber I, Brown K, Passmore P, Craig D, Johnston J, McGuinness B, Todd S, Heun R, Kolsch H, Kehoe PG, Hooper NM, Vardy ER, Mann DM, Pickering-Brown S, Brown K, Kalsheker N, Lowe J, Morgan K, David Smith A, Wilcock G, Warden D, Holmes C, Pastor P, Lorenzo-Betancor O, Brkanac Z, Scott E, Topol E, Morgan K, Rogaeva E, Singleton AB, Hardy J, Kamboh MI, St George-Hyslop P, Cairns N, Morris JC, Kauwe JS, Goate AM Alzheimer’s Research UKC. Rare coding variants in the phospholipase D3 gene confer risk for Alzheimer’s disease. Nature. 2014;505:550–554. doi: 10.1038/nature12825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grober E, Buschke H, Crystal H, Bang S, Dresner R. Screening for dementia by memory testing. Neurology. 1988;38:900–903. doi: 10.1212/wnl.38.6.900. [DOI] [PubMed] [Google Scholar]
- 41.Wechsler DCS. Wechsler Memory Scale Manual. New York, NY: The Psychological Corporation; 1973. [Google Scholar]
- 42.Team RDC. R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2016. [Google Scholar]
- 43.Team JPaDBaSDaDSaRC. 2016 [Google Scholar]
- 44.International Schizophrenia C. Purcell SM, Wray NR, Stone JL, Visscher PM, O’Donovan MC, Sullivan PF, Sklar P. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Farrer LA, Cupples LA, Haines JL, Hyman B, Kukull WA, Mayeux R, Myers RH, Pericak-Vance MA, Risch N, van Duijn CM. Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease. A meta-analysis. APOE and Alzheimer Disease Meta Analysis Consortium. JAMA. 1997;278:1349–1356. [PubMed] [Google Scholar]
- 46.Schrodi SJ. Reflections on the Field of Human Genetics: A Call for Increased Disease Genetics Theory. Front Genet. 2016;7:106. doi: 10.3389/fgene.2016.00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lin KA, Choudhury KR, Rathakrishnan BG, Marks DM, Petrella JR, Doraiswamy PM Alzheimer’s Disease Neuroimaging I. Marked gender differences in progression of mild cognitive impairment over 8 years. Alzheimers Dement (N Y) 2015;1:103–110. doi: 10.1016/j.trci.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Seshadri S, Wolf PA, Beiser A, Au R, McNulty K, White R, D’Agostino RB. Lifetime risk of dementia and Alzheimer’s disease. The impact of mortality on risk estimates in the Framingham Study. Neurology. 1997;49:1498–1504. doi: 10.1212/wnl.49.6.1498. [DOI] [PubMed] [Google Scholar]
- 49.Cruchaga C, Del-Aguila JL, Saef B, Black K, Fernandez MV, Budde J, Ibanez L, Kapoor M, Tosto G, Mayeux RP, Holtzman DM, Fagan AM, Morris JC, Bateman RJ, Goate AM, Harari O Dominantly Inherited Alzheimer N Disease Neuroimaging I study N-Lf. Polygenic risk score of sporadic late-onset Alzheimer’s disease reveals a shared architecture with the familial and early-onset forms. Alzheimers Dement. 2017 doi: 10.1016/j.jalz.2017.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hickman SE, El Khoury J. TREM2 and the neuroimmunology of Alzheimer’s disease. Biochem Pharmacol. 2014;88:495–498. doi: 10.1016/j.bcp.2013.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ulrich JD, Holtzman DM. TREM2 Function in Alzheimer’s Disease and Neurodegeneration. ACS Chem Neurosci. 2016;7:420–427. doi: 10.1021/acschemneuro.5b00313. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.