Abstract
Shigella are ranked among the most prevalent aetiologies of diarrhoeal disease worldwide, disproportionately affecting young children in developing countries and high-risk communities in developed settings. Antimicrobial treatment, most commonly with fluoroquinolones, is currently recommended for Shigella infections to alleviate symptoms and control disease transmission. Resistance to fluoroquinolones has emerged in differing Shigella species (S. dysenteriae, flexneri and sonnei) since the turn of the 21st century, originating in endemic areas, and latterly spreading into non-endemic regions. Despite occurring independently, the emergence of fluoroquinolone resistance in these different Shigella species shares striking similarities regarding their epidemiology and resistance mechanisms. Here, we review and discuss the current epidemiology of fluoroquinolone-resistant Shigella species, particularly in the light of recent genomic insights.
Keywords: Shigella, fluoroquinolone resistance, Asia, quinolone resistance determining region (QRDR), epidemiology, genomic
Impact Statement.
Shigella is a genus of human-adapted bacterial pathogens that cause dysenteric diarrhoea (shigellosis) in developing and developed countries. A specific class of antimicrobials, known as the fluoroquinolones, is recommended for the treatment of shigellosis, but resistance to this group of antimicrobials is rising rapidly within this genus. Here, we have combined available epidemiological and high-resolution genomic data to outline common themes that define the emergence and circulation of fluoroquinolone-resistant Shigella species. The information gathered in this review will be useful for determining optimal shigellosis treatment regimens and to tailor public health measures for alerting, containing and preventing the future spread of these antimicrobial-resistant enteric pathogens.
Introduction
Shigella, a pathogenic genus within the extensive Gram-negative family Enterobacteriaceae, is a major cause of diarrhoeal disease worldwide [1, 2]. The global burden of shigellosis is estimated to be 125 million cases per year, of which 160 000 lead to death [2, 3]. The disease disproportionately affects young children in low-income tropical settings, where malnutrition, inadequate sanitation and limited access to clean water appear to facilitate the transmission of the infecting organisms. The genus Shigella does not comprise a monophyletic group of organisms but is formed of multiple discrete Escherichia coli lineages, all of which harbour a signature virulence plasmid responsible for the distinctive invasive pathogenesis [4, 5]. Current serology classifies the genus into four species or serogroups (S. dysenteriae, boydii, flexneri and sonnei), which differ significantly in their epidemiology. Toxigenic S. dysenteriae serotype 1 (Sd1) is the causative agent of the now rare, often fatal, epidemic bacillary dysentery. S. boydii is only sporadically isolated from diarrhoeal cases in the Indian subcontinent [6–8]. The overwhelming majority of shigellosis cases are presently attributed to S. flexneri and S. sonnei, which predominantly circulate in developing and developed regions, respectively [9].
Shigellosis usually results in profuse diarrhoea, often accompanied by mucous or bloody discharge. This clinical presentation is associated with disruption of the intestinal epithelium, which is mediated by intracellular proliferation of the infecting Shigella. Although the disease is self-limiting, antimicrobial treatment is recommended to prevent further complications, assist recovery and restrict faecal shedding [10, 11]. One of the most commonly prescribed groups of antimicrobials for shigellosis is the fluoroquinolones, which directly interact with the bacterial DNA gyrase (encoded by gyrA and gyrB) and topoisomerase IV (encoded by parC and parE) to inhibit functional replication and induce bacterial cell death [12]. Routine surveillance has documented dramatic increases in the frequency of fluoroquinolone-resistant (FQR) Shigella, estimating that resistance increased from 0.6 % in 1998–2000 to 29 % in 2007–2009 of the endemic shigellosis in Asia and Africa [9, 13] (Fig. 1). The genetic mechanism(s) underlying resistance is commonly attributed to mutations in the quinolone resistance determining region (QRDR), ultimately diminishing the interaction between the antimicrobial and its target proteins [14]. Resistance to fluoroquinolones narrows ever-dwindling treatment options, placing those who are vulnerable at the increased risk of complications and hampering the efficient management of outbreaks. These factors have placed FQR Shigella on the list of global priority pathogens that urgently need focused development of novel antimicrobials [15]. This review aims to summarize the epidemiology of various FQR Shigella species, highlighting insights provided through genome sequencing and phylogenetic reconstruction. Due to its low prevalence and research focus, S. boydii will be excluded from this discussion.
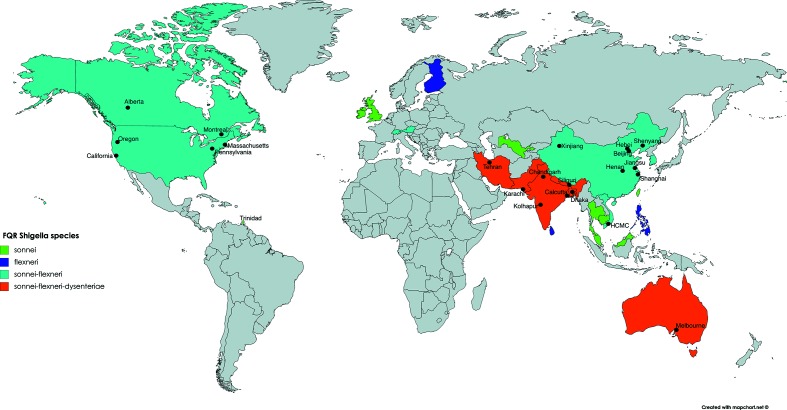
Fig. 1.
Worldwide distribution of FQR Shigella. Countries are coloured where different FQR Shigella species have been reported in the literature (see key). Black filled circles indicate specific regions where FQR Shigella have been isolated. Countries with no information or have not reported isolation of FQR Shigella are coloured grey. HCMC, Ho Chi Minh city.
Shigella dysenteriae
Resistance against fluoroquinolones had not been previously observed in Sd1 until FQR organisms were isolated during a dysentery outbreak arising in India and Bangladesh in 2002–2003 [16, 17] (Fig. 2). Molecular characterization by pulse field gel electrophoresis (PFGE) revealed that all contemporaneous FQR Sd1 isolates, causing either the outbreak or sporadic episodes across South Asia, belonged to a single clone [18–20]. However, fluoroquinolone resistance was attributed to two different QRDR mutation profiles: gyrA-S83L/D87G and S83L/D87N, which were associated with different geographical distributions [19]. These data suggested that the clone may have first acquired a gyrA-S83L mutation as early as 1994, later diverging into two FQR sub-populations, which were then characterized by differing secondary mutations. Indeed, a genomic investigation of the global phylogeny of Sd1 concluded that resistance to fluoroquinolones was acquired only once during the species’ evolutionary history, conferred by the co-occurrence of parC-S80I, gyrA-S83L and a secondary gyrA-D87 mutation between 1995 and 2002 [21]. This FQR clone belonged to the internationally successful lineage IV, which has witnessed at least nine independent single QRDR mutational events since the 1970s. The emergence of FQR Sd1 was followed by an abrupt decline after the outbreak, hampering routine monitoring and making the prospect of future FQR Sd1 outbreaks unpredictable [20, 22]. Information regarding resistance in alternative S. dysenteriae serotypes is limited, probably due to their low prevalence, even in regions where the disease was once highly endemic.
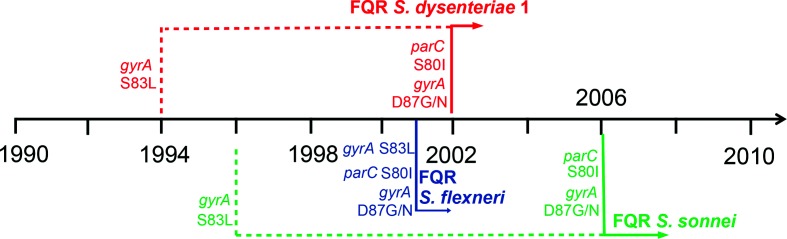
Fig. 2.
Timeline detailing the emergences of FQR Shigella species. The dashed lines represent the first occurrences of the initial QRDR mutation in the FQR clone if known, as described by epidemiological or genomic data. The solid lines indicate the first reports of FQR Shigella species as well as the QRDR mutations that became incorporated into these clones by this designated time. The presumed order of occurrence for these mutations is from top to bottom.
Shigella flexneri
The majority of epidemiological research on S. flexneri has been conducted in South Asia and China, where the pathogen’s burden remains significant. The first incidences of FQR in S. flexneri were documented in eastern and northern China in 2001–2002 [23, 24], and a detailed genetic screen revealed that the majority of these organisms possessed gyrA-S83L, gyrA-D87G and parC-S80I QRDR mutations [22]. Subsequently, a thorough examination of >2000 Bangladeshi S. flexneri underlined a worrying rising trend of fluoroquinolone resistance, which was almost exclusively found in serotype 2a [22]. Although it initially appeared in 2005, fluoroquinolone resistance escalated rapidly and its prevalence was >40 % of all native S. flexneri by 2010. These Bangladeshi isolates differed from their Chinese counterparts by a secondary QRDR mutation, harbouring gyrA-D87N instead of G. Furthermore, FQR S. flexneri with identical mutation profiles were recovered during a decade-long surveillance in Switzerland, highlighting the occurrence of fluoroquinolone resistance in non-endemic regions [25].
Routine dysentery surveillance in China has reported a steady increase of FQR S. flexneri of various serotypes, including 1a, 1c, 2a, 2b, 2av, 4a, 4c and X [26–31]. This observation suggests that the FQR phenotype has either emerged on numerous independent occasions across several serotypes or was acquired once, prior to subsequent intensive serotype switching events. Previous genomic studies reported that serotype conversion within a lineage is a commonly observed phenomenon for S. flexneri [32, 33]. Available literature provides greater support for the role of serotype switching on creating multiple FQR S. flexneri serotypes. Despite being present in a wide range of serotypes and locations, identical QRDR mutations have been frequently encountered in S. flexneri in China, encompassing gyrA-S83L, gyrA-D87G/N and parC-S80I. Furthermore, these mutations are commonly accompanied by an unusual mutation (gyrA-H211Y), which was also present in the aforementioned Bangladeshi FQR isolates [22, 28, 31, 34]. FQR S. flexneri from these two countries were also found to exhibit a close genetic relationship via PFGE [22]. This combined evidence indicates that spatially dispersed FQR S. flexneri probably belong to one dominant widespread clone, where the gyrA-H211Y, gyrA-S83L and parC-S80I mutations arose prior to geographical divergence. Later, a secondary mutation in gyrA delineated the Bangladeshi (gyrA-D87N) and the Chinese (gyrA-D87G) FQR isolates. However, the increasing isolation frequency of the gyrA-D87N variant in parts of China may be the result of a higher degree of trans-border dissemination from South Asia and/or the separate, indigenous emergence of a competent FQR subclone [31]. Due to the degree of genetic diversity and the complex population structures within S. flexneri, the true nature of such events cannot be easily measured using low-resolution molecular typing methods.
Shigella sonnei
A shift in species dominance (from S. flexneri to S. sonnei) has been observed concurrently in multiple Asian countries as they undergo rapid economic transition; this has been recorded in Bangladesh, China, Thailand and Vietnam [35–39]. This intriguing trend greatly increases the burden of S. sonnei worldwide, making antimicrobial resistance in this species a focal target for monitoring. Surveillance studies in developed countries have identified strong epidemiological links between FQR S. sonnei and a travel history to India [40, 41]. Moreover, despite disparate spatial distributions, these isolates share the same pulsotype (via PFGE) with FQR S. sonnei recovered in South Asia [36, 42–44]. These results suggest that contemporaneous FQR S. sonnei are clonal and have evolved and spread in the region before disseminating intercontinentally. Indeed, phylogenetic analysis on representative extant FQR S. sonnei confirmed this hypothesis, concluding that South Asia was the most likely origin of these organisms [45]. Furthermore, this study identified two distinct regional diversifications of the FQR clone out of South Asia, with one circulating in Southeast Asia and another appearing to instigate sustained transmission within Europe and America. These observations concur with frequent reports of native FQR S. sonnei circulating in Cambodia, Vietnam and California [46–48]. Fluoroquinolone resistance in S. sonnei is generally determined by the sequential accumulation of three mutations: gyrA-S83L, parC-S80I and gyrA-D87G [45, 49]. However, other resistance mechanisms, including differing mutations in QRDR (gyrA-D87N instead of D87G) and the synergy between the plasmid-mediated qnrB gene and gyrA mutations, have also been identified [50, 51]. It is of particular concern that the transmission of the FQR clone is intensified in high-risk contact networks, such as those reported among MSM (men who have sex with men) communities in non-endemic Canada and Taiwan [52, 53]. Therefore, the propagation of FQR Shigella should be closely monitored in MSM networks, especially in the wake of increased shigellosis incidence, HIV infection and resistance to other antimicrobials, such as azithromycin, within this high-risk group [54].
Outlook
The presented evidence reveals striking similarities between the emergence of fluoroquinolone resistance among the discrete Shigella species. (1) FQR is almost exclusively determined by sequential QRDR mutations in the following order: gyrA-S83L, parC-S80I and gyrA-D87G/N. (2) To date, the majority of FQR isolates identified within an individual species are clonal despite their wide geographical distribution. (3) South Asia, and potentially China, serve as likely reservoirs for the rise and spread of resistant clones. These interpretations are currently deduced from genomic insights into Sd1 and S. sonnei, and are subjected to various confounders, including geographical bias in sample collection. However, the extensive genetic diversity within S. flexneri may present an alternative scenario, which will benefit from large-scale molecular epidemiology data generated through whole-genome sequencing.
The first widely used fluoroquinolone, ciprofloxacin, was introduced to clinical practice in 1987. However, resistance in Shigella only began to emerge in the early 2000s. The intervening period witnessed the emergence of Shigella exhibiting resistance to multiple antimicrobials including co-trimoxazole, ampicillin and nalidixic acid [55]. Therefore, fluoroquinolones, such as ciprofloxacin, began to be deployed more commonly to manage drug-resistant shigellosis, and its use became routine, as recommended by the World Health Organization in 2005 [11, 56]. Recent experimental and modelling work into the evolution of fluoroquinolone resistance could offer explanations for the observed pattern between the various Shigella species. The ordered QRDR mutations are selected in favour of those in efflux regulatory machinery due to their co-optimization for non-susceptibility (MIC levels) and fitness cost [57, 58]. For both in vitro generated and clinical isolates, resistance to fluoroquinolones almost exclusively commences with an initial mutation, gyrA-S83L. This mutation has arisen independently on multiple occasions for different Shigella lineages, possibly as an adaptive strategy for resistance against the first-generation quinolone, nalidixic acid [21, 32, 59]. However, the key determining factor in this stepwise evolution is the subsequent mutation, parC-S80I, which occurs much less frequently during evolution but gives the bacterium a significant increase in fluoroquinolone MIC and potentially a non-inferior fitness. Such a mutation is suggested to be favourably selected in abundance of mutation supply, fulfilled either by a large population size or a high mutation rate, when antimicrobial pressure is high [58]. Given that the mutation rate of the bacterium Shigella is relatively stable, the first scenario appears to be more plausible [21, 32, 49, 59]. High population densities in South Asia could promote extensive and sustained Shigella transmission, resulting in a large bacterial population. Suboptimal public health measures in the region, exemplified by the fact that only 40 % of the Indian population has access to improved sanitation [60], further amplify the transmission cycle of Shigella. This expansion has arisen on a backdrop of rapidly increasing fluoroquinolone use for treatment of multiple enteric and febrile diseases since the turn of this century [56, 61]. Indeed, India, with 12.9 billion units, was ranked as the world’s largest antimicrobial consumer in 2010 [62]. These contributing factors might render South Asia a unique focal point for the emergence of human-restricted FQR enteric bacteria, including Shigella and Salmonella Typhi [63].
Conclusion
The emergence of FQR Shigella has been quickly followed by the expansion and, for S. sonnei, rapid international spread. Furthermore, co-resistance to other first-line antimicrobials, such as the macrolides and third-generation cephalosporins, is frequently identified among these bacteria [47, 50]. These antimicrobial resistance combinations present a serious public health threat for the effective treatment and management of shigellosis. It has been experimentally demonstrated that the described QRDR mutations may carry no detrimental or limited fitness cost to the resistant Enterobacteriaceae, even in the absence of fluoroquinolone pressure [57, 64]. Alternatively, fluoroquinolone resistance has been coupled with the successful clonal propagation of several multi-drug-resistant pathogens, including Staphylococcus aureus, Klebsiella pneumoniae, E. coli, Clostridium difficile and Neisseria gonorrhoeae [65, 66]. All these major FQR clones were found to harbour specific QRDR mutation combinations, indicating that these resistance genotypes induce a minimal fitness disadvantage. Although little is known about the impact of alleviating fluoroquinolone pressure on the clonal dominance of FQR bacteria in nature, we speculate that withdrawal of such pressure in clinical settings is unlikely to discontinue the dominance of FQR Shigella in the transmission chain. However, future research is warranted to challenge this hypothesis, as well as to develop the best practices for controlling and treating new emerging antimicrobial-resistant clones.
Funding information
HCT received a DPhil scholarship from the Tropical Network Fund, Nuffield Department of Medicine, University of Oxford. SB is a Sir Henry Dale Fellow, jointly funded by the Wellcome Trust and the Royal Society (100087/Z/12/Z).
Conflicts of interest
The authors declare that there are no conflicts of interest.
Ethical statement
No ethical approval was required for the research in this study.
Footnotes
Abbreviations: FQR, fluoroquinolone-resistant; PFGE, pulse field gel electrophoresis; QRDR, quinolone resistance determining region; Sd1, Shigella dysenteriae serotype 1; MSM, men who have sex with men.
References
- 1.Kotloff KL, Nataro JP, Blackwelder WC, Nasrin D, Farag TH, et al. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet. 2013;382:209–222. doi: 10.1016/S0140-6736(13)60844-2. [DOI] [PubMed] [Google Scholar]
- 2.Troeger C, Forouzanfar M, Rao PC, Khalil I, Brown A, et al. Estimates of global, regional, and national morbidity, mortality, and aetiologies of diarrhoeal diseases: a systematic analysis for the Global Burden of Disease Study 2015. Lancet Infect Dis. 2017;17:909–948. doi: 10.1016/S1473-3099(17)30276-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bardhan P, Faruque AS, Naheed A, Sack DA. Decrease in shigellosis-related deaths without Shigella spp.-specific interventions, Asia. Emerg Infect Dis. 2010;16:1718–1723. doi: 10.3201/eid1611.090934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sahl JW, Morris CR, Emberger J, Fraser CM, Ochieng JB, et al. Defining the phylogenomics of Shigella species: a pathway to diagnostics. J Clin Microbiol. 2015;53:951–960. doi: 10.1128/JCM.03527-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chung The H, Thanh DP, Holt KE, Thomson NR, Baker S. The genomic signatures of Shigella evolution, adaptation and geographical spread. Nat Rev Microbiol. 2016;14:235–250. doi: 10.1038/nrmicro.2016.10. [DOI] [PubMed] [Google Scholar]
- 6.Shiga K. Ueber den erreger der dysenterie in Japan (vorläufige mitteilung) Zentralbl Bakteriol Mikrobiol. 1898;23:599–600. [Google Scholar]
- 7.Pal SC. Epidemic bacillary dysentery in West Bengal, India, 1984. The Lancet. 1984;323:1462. doi: 10.1016/S0140-6736(84)91948-2. [DOI] [PubMed] [Google Scholar]
- 8.Barceloux DG. Medical Toxicology of Natural Substances. Hoboken, New Jersey, USA: John Wiley & Sons, Inc; 2008. Shigella species (Shiga Enterotoxins) pp. 150–155. [Google Scholar]
- 9.Gu B, Cao Y, Pan S, Zhuang L, Yu R, et al. Comparison of the prevalence and changing resistance to nalidixic acid and ciprofloxacin of Shigella between Europe-America and Asia-Africa from 1998 to 2009. Int J Antimicrob Agents. 2012;40:9–17. doi: 10.1016/j.ijantimicag.2012.02.005. [DOI] [PubMed] [Google Scholar]
- 10.Vinh H, Wain J, Chinh MT, Tam CT, Trang PT, et al. Treatment of bacillary dysentery in Vietnamese children: two doses of ofloxacin versus 5-days nalidixic acid. Trans R Soc Trop Med Hyg. 2000;94:323–326. doi: 10.1016/S0035-9203(00)90343-2. [DOI] [PubMed] [Google Scholar]
- 11.WHO Guidelines for the control of shigellosis, including epidemics due to Shigella dysenteriae type 1. 2005. World Health Organization.
- 12.Redgrave LS, Sutton SB, Webber MA, Piddock LJ. Fluoroquinolone resistance: mechanisms, impact on bacteria, and role in evolutionary success. Trends Microbiol. 2014;22:438–445. doi: 10.1016/j.tim.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 13.Kahsay AG, Muthupandian S. A review on Sero diversity and antimicrobial resistance patterns of Shigella species in Africa, Asia and South America, 2001–2014. BMC Res Notes. 2016;9:422. doi: 10.1186/s13104-016-2236-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tamanna, Ramana J. Structural insights into the fluoroquinolone resistance mechanism of Shigella flexneri DNA gyrase and topoisomerase IV. Microb Drug Resist. 2016;22:404–411. doi: 10.1089/mdr.2015.0018. [DOI] [PubMed] [Google Scholar]
- 15.Tacconelli E, Magrini N, Kahlmeter G, Singh N. Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. World Heal Organ. 2017:1–7. [Google Scholar]
- 16.Bhattacharya SK, Sarkar K, Balakrish Nair G, Faruque AS, Sack DA. Multidrug-resistant Shigella dysenteriae type 1 in south Asia. Lancet Infect Dis. 2003;3:755. doi: 10.1016/S1473-3099(03)00829-6. [DOI] [PubMed] [Google Scholar]
- 17.Naheed A, Kalluri P, Talukder KA, Faruque AS, Khatun F, et al. Fluoroquinolone-resistant Shigella dysenteriae type 1 in northeastern Bangladesh. Lancet Infect Dis. 2004;4:607–608. doi: 10.1016/S1473-3099(04)01143-0. [DOI] [PubMed] [Google Scholar]
- 18.Pazhani GP, Sarkar B, Ramamurthy T, Bhattacharya SK, Takeda Y, et al. Clonal multidrug-resistant Shigella dysenteriae type 1 strains associated with epidemic and sporadic dysenteries in eastern India. Antimicrob Agents Chemother. 2004;48:681–684. doi: 10.1128/AAC.48.2.681-684.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Talukder KA, Khajanchi BK, Islam MA, Dutta DK, Islam Z, et al. Genetic relatedness of ciprofloxacin-resistant Shigella dysenteriae type 1 strains isolated in south Asia. J Antimicrob Chemother. 2004;54:730–734. doi: 10.1093/jac/dkh425. [DOI] [PubMed] [Google Scholar]
- 20.Taneja N. Changing epidemiology of shigellosis and emergence of ciprofloxacin-resistant Shigellae in India. J Clin Microbiol. 2007;45:678–679. doi: 10.1128/JCM.02247-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Njamkepo E, Fawal N, Tran-Dien A, Hawkey J, Strockbine N, et al. Global phylogeography and evolutionary history of Shigella dysenteriae type 1. Nat Microbiol. 2016;1:16027. doi: 10.1038/nmicrobiol.2016.27. [DOI] [PubMed] [Google Scholar]
- 22.Azmi IJ, Khajanchi BK, Akter F, Hasan TN, Shahnaij M, et al. Fluoroquinolone resistance mechanisms of Shigella flexneri isolated in Bangladesh. PLoS One. 2014;9:e102533. doi: 10.1371/journal.pone.0102533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang XY, Du L, von Seidlein L, Xu ZY, Zhang YL, et al. Occurrence of shigellosis in the young and elderly in rural China: results of a 12-month population-based surveillance study. Am J Trop Med Hyg. 2005;73:416–422. [PubMed] [Google Scholar]
- 24.Qin T, Bi R, Fan W, Kang H, Ma P, et al. Novel mutations in quinolone resistance-determining regions of gyrA, gyrB, parC and parE in Shigella flexneri clinical isolates from eastern Chinese populations between 2001 and 2011. Eur J Clin Microbiol Infect Dis. 2016;35:2037–2045. doi: 10.1007/s10096-016-2761-2. [DOI] [PubMed] [Google Scholar]
- 25.Nüesch-Inderbinen M, Heini N, Zurfluh K, Althaus D, Hächler H, et al. Shigella antimicrobial drug resistance mechanisms, 2004–2014. Emerg Infect Dis. 2016;22:1083–1085. doi: 10.3201/eid2206.152088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang J, Jin H, Hu J, Yuan Z, Shi W, et al. Antimicrobial resistance of Shigella spp. from humans in Shanghai, China, 2004–2011. Diagn Microbiol Infect Dis. 2014;78:282–286. doi: 10.1016/j.diagmicrobio.2013.11.023. [DOI] [PubMed] [Google Scholar]
- 27.Qiu S, Xu X, Wang Y, Yang G, Wang Z, et al. Emergence of resistance to fluoroquinolones and third-generation cephalosporins in Shigella flexneri subserotype 1c isolates from China. Clin Microbiol Infect. 2012;18:E95. doi: 10.1111/j.1469-0691.2012.03768.x. [DOI] [PubMed] [Google Scholar]
- 28.Cui X, Wang J, Yang C, Liang B, Ma Q, et al. Prevalence and antimicrobial resistance of Shigella flexneri serotype 2 variant in China. Front Microbiol. 2015;6:435. doi: 10.3389/fmicb.2015.00435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pu XY, Pan JC, Wang HQ, Zhang W, Huang ZC, et al. Characterization of fluoroquinolone-resistant Shigella flexneri in Hangzhou area of China. J Antimicrob Chemother. 2009;63:917–920. doi: 10.1093/jac/dkp087. [DOI] [PubMed] [Google Scholar]
- 30.Zhang W, Luo Y, Li J, Lin L, Ma Y, et al. Wide dissemination of multidrug-resistant Shigella isolates in China. J Antimicrob Chemother. 2011;66:2527–2535. doi: 10.1093/jac/dkr341. [DOI] [PubMed] [Google Scholar]
- 31.Qin T, Qian H, Fan W, Ma P, Zhou L, et al. Newest data on fluoroquinolone resistance mechanism of Shigella flexneri isolates in Jiangsu Province of China. Antimicrob Resist Infect Control. 2017;6:1–8. doi: 10.1186/s13756-017-0249-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Connor TR, Barker CR, Baker KS, Weill FX, Talukder KA, et al. Species-wide whole genome sequencing reveals historical global spread and recent local persistence in Shigella flexneri. Elife. 2015;4:1–16. doi: 10.7554/eLife.07335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang N, Lan R, Sun Q, Wang J, Wang Y, et al. Genomic portrait of the evolution and epidemic spread of a recently emerged multidrug-resistant Shigella flexneri clone in China. J Clin Microbiol. 2014;52:1119–1126. doi: 10.1128/JCM.02669-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang C, Li P, Zhang X, Ma Q, Cui X, et al. Molecular characterization and analysis of high-level multidrug-resistance of Shigella flexneri serotype 4s strains from China. Sci Rep. 2016;6:29124. doi: 10.1038/srep29124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thompson CN, Duy PT, Baker S. The rising dominance of Shigella sonnei: an intercontinental shift in the etiology of bacillary dysentery. PLoS Negl Trop Dis. 2015;9:e0003708. doi: 10.1371/journal.pntd.0003708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ud-Din AI, Wahid SU, Latif HA, Shahnaij M, Akter M, et al. Changing trends in the prevalence of Shigella species: emergence of multi-drug resistant Shigella sonnei biotype g in Bangladesh. PLoS One. 2013;8:e82601. doi: 10.1371/journal.pone.0082601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qiu S, Xu X, Yang C, Wang J, Liang B, et al. Shift in serotype distribution of Shigella species in China, 2003-2013. Clin Microbiol Infect. 2015;21:252.e5–252.e8. doi: 10.1016/j.cmi.2014.10.019. [DOI] [PubMed] [Google Scholar]
- 38.Bangtrakulnonth A, Vieira AR, Lo Fo Wong DM, Pornreongwong S, Pulsrikarn C, et al. Shigella from humans in Thailand during 1993 to 2006: spatial-time trends in species and serotype distribution. Foodborne Pathog Dis. 2008;5:773–784. doi: 10.1089/fpd.2008.0109. [DOI] [PubMed] [Google Scholar]
- 39.Vinh H, Nhu NT, Nga TV, Duy PT, Campbell JI, et al. A changing picture of shigellosis in southern Vietnam: shifting species dominance, antimicrobial susceptibility and clinical presentation. BMC Infect Dis. 2009;9:204. doi: 10.1186/1471-2334-9-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.De Lappe N, O'Connor J, Garvey P, Mckeown P, Cormican M. Ciprofloxacin-resistant Shigella sonnei associated with travel to India. Emerg Infect Dis. 2015;21:894–896. doi: 10.3201/eid2105.141184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bowen A, Hurd J, Hoover C, Khachadourian Y, Traphagen E, et al. Importation and domestic transmission of Shigella sonnei resistant to ciprofloxacin – United States, May 2014-February 2015. MMWR Morb Mortal Wkly Rep. 2015;64:318–320. [PMC free article] [PubMed] [Google Scholar]
- 42.Nandy S, Dutta S, Ghosh S, Ganai A, Rajahamsan J, et al. Foodborne-associated Shigella sonnei, India, 2009 and 2010. Emerg Infect Dis. 2011;17:2072–2073. doi: 10.3201/eid1711.110403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ghosh S, Pazhani GP, Chowdhury G, Guin S, Dutta S, et al. Genetic characteristics and changing antimicrobial resistance among Shigella spp. isolated from hospitalized diarrhoeal patients in Kolkata, India. J Med Microbiol. 2011;60:1460–1466. doi: 10.1099/jmm.0.032920-0. [DOI] [PubMed] [Google Scholar]
- 44.Ruekit S, Wangchuk S, Dorji T, Tshering KP, Pootong P, et al. Molecular characterization and PCR-based replicon typing of multidrug resistant Shigella sonnei isolates from an outbreak in Thimphu, Bhutan. BMC Res Notes. 2014;7:95–99. doi: 10.1186/1756-0500-7-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chung The H, Rabaa MA, Pham Thanh D, De Lappe N, Cormican M, et al. South Asia as a reservoir for the global spread of ciprofloxacin-resistant Shigella sonnei: a cross-sectional study. PLoS Med. 2016;13:e1002055. doi: 10.1371/journal.pmed.1002055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Poramathikul K, Bodhidatta L, Chiek S, Oransathid W, Ruekit S, et al. Multidrug-resistant Shigella infections in patients with diarrhea, Cambodia, 2014–2015. Emerg Infect Dis. 2016;22:1640–1643. doi: 10.3201/eid2209.152058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim JS, Kim JJ, Kim SJ, Jeon S, Seo KY, et al. Shigella sonnei associated with travel to Vietnam, Republic of Korea. Emerg Infect Dis. 2015;21:1247–1250. doi: 10.3201/eid2107.150363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kozyreva VK, Jospin G, Greninger AL, Watt JP, Eisen JA, et al. Recent outbreaks of Shigellosis in California caused by two distinct populations of Shigella sonnei with either increased virulence or fluoroquinolone resistance. mSphere. 2016;1:e00344-16. doi: 10.1128/mSphere.00344-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chung The H, Rabaa MA, Thanh DP, Ruekit S, Wangchuk S, et al. Introduction and establishment of fluoroquinolone-resistant Shigella sonnei into Bhutan. Microb Genom. 2015;1:1–11. doi: 10.1099/mgen.0.000042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sadouki Z, Day MR, Doumith M, Chattaway MA, Dallman TJ, et al. Comparison of phenotypic and WGS-derived antimicrobial resistance profiles of Shigella sonnei isolated from cases of diarrhoeal disease in England and Wales, 2015. J Antimicrob Chemother. 2017;72:2496–2502. doi: 10.1093/jac/dkx170. [DOI] [PubMed] [Google Scholar]
- 51.Das A, Natarajan M, Mandal J. The emergence of quinolone resistant Shigella sonnei, Pondicherry, India. PLoS One. 2016;11:e0160290. doi: 10.1371/journal.pone.0160290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gaudreau C, Ratnayake R, Pilon PA, Gagnon S, Roger M, et al. Ciprofloxacin-resistant Shigella sonnei among men who have sex with men, Canada, 2010. Emerg Infect Dis. 2011;17:1747–1750. doi: 10.3201/eid1709.102034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chiou CS, Izumiya H, Kawamura M, Liao YS, Su YS, et al. The worldwide spread of ciprofloxacin-resistant Shigella sonnei among HIV-infected men who have sex with men, Taiwan. Clin Microbiol Infect. 2016;22:383.e11–383.e16. doi: 10.1016/j.cmi.2015.12.021. [DOI] [PubMed] [Google Scholar]
- 54.Mohan K, Hibbert M, Rooney G, Canvin M, Childs T, et al. What is the overlap between HIV and shigellosis epidemics in England: further evidence of MSM transmission? Sex Transm Infect. 2018;94:67–71. doi: 10.1136/sextrans-2016-052962. [DOI] [PubMed] [Google Scholar]
- 55.Niyogi SK. Increasing antimicrobial resistance–an emerging problem in the treatment of shigellosis. Clin Microbiol Infect. 2007;13:1141–1143. doi: 10.1111/j.1469-0691.2007.01829.x. [DOI] [PubMed] [Google Scholar]
- 56.Kotwani A, Chaudhury RR, Holloway K. Antibiotic-prescribing practices of primary care prescribers for acute diarrhea in New Delhi, India. Value Health. 2012;15:S116–S119. doi: 10.1016/j.jval.2011.11.008. [DOI] [PubMed] [Google Scholar]
- 57.Marcusson LL, Frimodt-Møller N, Hughes D. Interplay in the selection of fluoroquinolone resistance and bacterial fitness. PLoS Pathog. 2009;5:e1000541. doi: 10.1371/journal.ppat.1000541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huseby DL, Pietsch F, Brandis G, Garoff L, Tegehall A, et al. Mutation supply and relative fitness shape the genotypes of ciprofloxacin-resistant Escherichia coli. Mol Biol Evol. 2017;34:1029–1039. doi: 10.1093/molbev/msx052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Holt KE, Baker S, Weill FX, Holmes EC, Kitchen A, et al. Shigella sonnei genome sequencing and phylogenetic analysis indicate recent global dissemination from Europe. Nat Genet. 2012;44:1056–1059. doi: 10.1038/ng.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.World Bank World development indicators. 2015. https://data.worldbank.org/indicator/ Available from.
- 61.Laxminarayan R, Chaudhury RR. Antibiotic resistance in India: drivers and opportunities for action. PLoS Med. 2016;13:e1001974. doi: 10.1371/journal.pmed.1001974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.van Boeckel TP, Gandra S, Ashok A, Caudron Q, Grenfell BT, et al. Global antibiotic consumption 2000 to 2010: an analysis of national pharmaceutical sales data. Lancet Infect Dis. 2014;14:742–750. doi: 10.1016/S1473-3099(14)70780-7. [DOI] [PubMed] [Google Scholar]
- 63.Wong VK, Baker S, Pickard DJ, Parkhill J, Page AJ, et al. Phylogeographical analysis of the dominant multidrug-resistant H58 clade of Salmonella Typhi identifies inter- and intracontinental transmission events. Nat Genet. 2015;47:632–639. doi: 10.1038/ng.3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Baker S, Duy PT, Nga TV, Dung TT, Phat VV, et al. Fitness benefits in fluoroquinolone-resistant Salmonella Typhi in the absence of antimicrobial pressure. Elife. 2013;2:e01229. doi: 10.7554/eLife.01229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fuzi M, Szabo D, Csercsik R. Double-serine fluoroquinolone resistance mutations advance major international clones and lineages of various multi-drug resistant bacteria. Front Microbiol. 2017;8:1–14. doi: 10.3389/fmicb.2017.02261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Grad YH, Harris SR, Kirkcaldy RD, Green AG, Marks DS, et al. Genomic epidemiology of gonococcal resistance to extended-spectrum cephalosporins, macrolides, and fluoroquinolones in the United States, 2000–2013. J Infect Dis. 2016;214:1579–1587. doi: 10.1093/infdis/jiw420. [DOI] [PMC free article] [PubMed] [Google Scholar]