Abstract
The order Sphingomonadales is a taxon of bacteria with a variety of physiological features and carotenoid pigments. Some of the coloured strains within this order are known to be aerobic anoxygenic phototrophs that contain characteristic photosynthesis gene clusters (PGCs). Previous work has shown that majority of the ORFs putatively involved in the biosynthesis of C40 carotenoids are located outside the PGCs in these strains. The main purpose of this study was to understand the genetic basis for the various colour/carotenoid phenotypes of the strains of Sphingomonadales. Comparative analyses of the genomes of 41 strains of this order revealed that there were different patterns of clustering of carotenoid biosynthesis (crt) ORFs, with four ORF clusters being the most common. The analyses also revealed that co-occurrence of crtY and crtI is an evolutionarily conserved feature in Sphingomonadales and other carotenogenic bacteria. The comparisons facilitated the categorisation of bacteria of this order into four groups based on the presence of different crt ORFs. Yellow coloured strains most likely accumulate nostoxanthin, and contain six ORFs (group I: crtE, crtB, crtI, crtY, crtZ, crtG). Orange coloured strains may produce adonixanthin, astaxanthin, canthaxanthin and erythroxanthin, and contain seven ORFs (group II: crtE, crtB, crtI, crtY, crtZ, crtG, crtW). Red coloured strains may accumulate astaxanthin, and contain six ORFs (group III: crtE, crtB, crtI, crtY, crtZ, crtW). Non-pigmented strains may contain a smaller subset of crt ORFs, and thus fail to produce any carotenoids (group IV). The functions of many of these ORFs remain to be characterised.
Keywords: Sphingomonadales, Sphingomonadaceae, Erythrobacteraceae, carotenogenesis, LOG, DUF2141
Data Summary
A full list of accession numbers for the whole-genome sequence data and locus tags of protein sequences used in this study are provided in Tables 1 and S1 (available with the online version of this article), respectively. Additional data is provided in Table S2 and Fig. S1.
Table 1. Features of the strains of Sphingomonadales and their genomes used in this study.
| Serial no. | Species (strain) | Chromosome size (bp) [genome status] | G+C (mol%) | GenBank/RefSeq accession no. | crt ORFs identified | Group based on crt genotype | Colour of the strain | Carotenoids produced | Reference (for colour and/or carotenoid)* |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Sphingopyxis alaskensis (RB2256T) | 3345170 [complete] | 65.50 | CP000356 | E, B, I, Y, Z, G | I | Yellow to beige | NostoxanthinP, caloxanthinP |
[77] |
| 2 | Sphingopyxis macrogoltabida (EY-1) | 4757879 [complete] | 64.90 | CP012700 | E, B, I, Y, Z, G | I | Unknown | Unknown | None |
| 3 | Sphingopyxis fribergensis (Kp5.2T) | 4993584 [complete] | 63.90 | CP009122 | E, B, I, Y, Z, G | I | Yellow | NostoxanthinP, caloxanthinP |
[78] |
| 4 | Sphingomonas sanxanigenens (DSM 19645T) | 6205897PGC[complete] | 66.80 | CP006644 | E, B, I, Y, Z, G | I | Colourless/white | Phytoene/noneP | [64] |
| 5 | Sphingorhabdus sp. (M41) | 3339521 [complete] | 56.70 | CP014545 | E, B, I, Y, Z, G | I | Colourless/white | Phytoene/noneP | Personal communication1 |
| 6 | Sphingopyxis sp. (113P3) | 4420776 [complete] | 64.00 | CP009452 | E, B, I, Y, Z, G | I | Yellowish brown | NostoxanthinP, caloxanthinP |
[79] |
| 7 | Sphingopyxis terrae (NBRC 15098T) | 3979087 [complete] | 64.60 | CP013342 | E, B, I, Y, Z, G | I | Light or deep-yellow | NostoxanthinP, caloxanthinP |
[80] |
| 8 | Sphingobium sp. (SYK-6) | 4199332 [complete] | 65.60 | NC_015976 | E, B, I, Y, Z, G | I | Yellow | NostoxanthinP, caloxanthinP |
Personal communication2 |
| 9 | Sphingomonas hengshuiensis (WHSC-8T) | 5191536PGC [complete] | 66.70 | CP010836 | E, B, I, Y, Z, G | I | Yellow | NostoxanthinP, caloxanthinP |
[65] |
| 10 | Sphingomonas sp. (Root241) | 4212322 [draft] | 66.00 | NZ_LMIV00000000 | E, B, I, Y, Z, G | I | Unknown | Unknown | None |
| 11 | Sphingomonas sp. (ATCC 31555) | 4046117 [draft] | 65.90 | NZ_ALBQ00000000 | E, B, I, Y, Z, G, W | II | Red | Canthaxanthin and othersP | [81] |
| 12 | Sphingomonas paucimobilis (NBRC 13935T) | 4327402 [draft] | 65.70 | NZ_BBJS00000000 | E, B, I, Y, Z, G | I | Yellow | NostoxanthinC, caloxanthinP |
[82] |
| 13 | Sphingomonas astaxanthinifaciens (DSM 22298T) | 2533034PGC [draft] | 68.40 | NZ_JONN00000000 | E, B, I, Y, Z, W, X | III | Red | Astaxanthin and its derivativesC | [58] |
| 14 | Sphingobium chlorophenolicum (L-1) | 3080818 [chromosome 1, complete] | 63.90 | CP002798 | E, B, I, Y, Z, G | I | Yellow | NostoxanthinP, caloxanthinP |
[83] |
| 15 | Sphingobium sp. (MI1205) | 3351250 [chromosome 1, complete] | 62.30 | CP005188 | E, B, I, Y, Z, G | I | Brownish–yellow | NostoxanthinP, caloxanthinP |
[84] |
| 16 | Sphingobium sp. (EP60837) | 2669660 [chromosome 1, complete] | 62.40 | CP015986 | E, B, I, Y, Z, G | I | Colourless/white | Phytoene/noneP | Personal communication3 |
| 17 | Sphingobium sp. (YBL2) | 4766421 [complete] | 64.80 | CP010954 | E, B, I, Y, Z, G | I | Yellow | NostoxanthinP, caloxanthinP |
[85] |
| 18 | Novosphingobium aromaticivorans (DSM 12444T) | 3561584 [complete] | 65.20 | CP000248 | E, B, I, Y, Z, G | I | Yellow | NostoxanthinP, caloxanthinP |
[86] |
| 19 | Citromicrobium sp. (JL477) | 3258499PGC [complete] | 65.00 | CP011344 | E, B, I, Y, Z, G | I | Yellow | NostoxanthinP, caloxanthinP |
Personal communication4 |
| 20 | Altererythrobacter dongtanensis (KCTC 22672T) | 3009495 [complete] | 65.80 | CP016591 | E, B, I, Y, Z, G | I | Yellow | NostoxanthinP, caloxanthinP |
[87] |
| 21 | Altererythrobacter namhicola (JCM 16345T) | 2591679 [complete] | 65.00 | CP016545 | E, B, I, Y, Z, G, W | II | Orange | Canthaxanthin and othersP | [88] |
| 22 | Altererythrobacter epoxidivorans (CGMCC 1.7731T) | 2786256 [complete] | 61.50 | CP012669 | E, B, I, Y, Z, G | I | Yellow | NostoxanthinP, caloxanthinP |
[89] |
| 23 | Altererythrobacter atlanticus (26DY36T) | 3386291 [complete] | 61.90 | CP011452 | E, B, I, Y, Z, G | I | Yellow | NostoxanthinP, caloxanthinP |
[90] |
| 24 | Altererythrobacter ishigakiensis (NBRC 107699T) | 2673978 PGC[complete] | 56.90 | CP015963 | E, B, I, Y, Z, G, W | II | Orange–red | Canthaxanthin and othersC | [63] |
| 25 | Erythrobacter litoralis (HTCC2594) | 3052398 [complete] | 63.10 | CP000157 | E, B, I, Y, Z, G, W | II | Pink | Unknown | [91] |
| 26 | Erythrobacter atlanticus (s21-N3T) | 3012400 [complete] | 58.20 | CP011310 | E, B, I, Y, Z, G | I | Yellow–brown | NostoxanthinP, caloxanthinP |
[92] |
| 27 | Sphingobium yanoikuyae (ATCC 51230T) | 5500358 [draft)] | 64.40 | NZ_AGZU00000000 | E, B, I, Y, Z, G | I | Creamy white | Phytoene/noneP | [82] |
| 28 | Novosphingobium sp. (PP1Y) | 3911486 [complete] | 63.70 | FR856862 | E, B, I, Y, Z, G | I | Yellow | NostoxanthinP, caloxanthinP |
[93] |
| 29 | Novosphingobium pentaromativorans (US6-1T) | 3979506 [complete] | 63.50 | CP009291 | E, B, I, Y, Z, G | I | Yellow | NostoxanthinP, caloxanthinP |
[94] |
| 30 | Porphyrobacter neustonensis (DSM 9434T) | 3090363PGC[complete] | 65.30 | CP016033 | E, B, I, Y, Z, G, W | II | Orange | Canthaxanthin and othersP | [10] |
| 31 | Sphingomonas sp. (NIC1) | 3408545 [complete] | 67.40 | CP015521 | E, B, I, Y, Z, G | I | Unknown | Unknown | None |
| 32 | Sphingomonas melonis (TY) | 4100783 [draft] | 67.10 | NZ_LQCK00000000 | E, B, I, Y, Z, G | I | Yellow | NostoxanthinP, caloxanthinP |
[95] |
| 33 | Altererythrobacter marensis (KCTC 22370T) | 2885033 [complete] | 64.70 | CP011805 | E, B, I, Y, Z, G | I | Yellow | NostoxanthinP, caloxanthinP |
[96] |
| 34 | Croceicoccus naphthovorans (PQ-2T) | 3543806 [complete] | 62.60 | CP011770 | E, B, I, Y, Z, G | I | Yellow | NostoxanthinP, caloxanthinP |
[97] |
| 35 | Sphingomonas taxi (ATCC 55669) | 3859099 [complete] | 68.00 | CP009571 | E, B, I, Y, Z, G, W | II | Yellow to orange | Canthaxanthin and othersP | [98] |
| 36 | Sphingomonas sp. (RIT328) | 4343511 [draft] | 68.30 | NZ_JFYV00000000 | E, B, I, Y, Z, G, W | II | Unknown | Unknown | None |
| 37 | Sphingobium japonicum (UT26S) | 3514822 [chromosome 1, complete] | 64.80 | NC_014006 | E, B, I, Y, Z, G | I | Yellow | NostoxanthinP, caloxanthinP |
[99] |
| 38 | Sphingomonas sp. (MM-1) | 4054833 [complete] | 67.20 | CP004036 | E, I, Y | IV | Colourless/white | None | Personal communication5 |
| 39 | Sphingobium baderi (DE-13) | 4107398 [complete] | 62.40 | CP013264 | E, I | IV | Colourless/white | None | [100] |
| 40 | Sphingomonas wittichii (RW1T) | 5382261 [complete] | 68.40 | CP000699 | E, Y | IV | Greyish-white (faintly yellow) | None | [101] |
| 41 | Sphingopyxis granuli (TFA) | 4679853 [complete] | 66.20 | CP012199 | E | IV | Yellowish | None | [102] |
C, Confirmed in one or more studies published previously; P, presumptive based on the colour of the strain reported and the crt ORFs identified; PGC, genome contains a putative photosynthesis gene cluster – production of spirilloxanthin (using CrtC, CrtD and CrtF) and Bchl a from this cluster can potentially affect the colour of the host strains; T, type strain.
1, Dr Che Ok Jeon, Chung-Ang University, Republic of Korea; 2, Dr Eiji Masai, Nagaoka University of Technology, Japan; 3, Dr Byung-Yong Kim, ChunLab, Inc., Republic of Korea; 4, Dr Qiang Zheng, Xiamen University, People's Republic of China; 5, Dr Yuji Nagata, Tohoku University, Japan.
Impact Statement.
Despite pigmentation being the most obvious phenotype among members of the Sphingomonadales, not much is known about the genetic basis of its biosynthesis across different species and genera. The present study sought to fill this gap by leveraging the wealth of genomic data that has become available in the last 5 years. Through in-depth analyses of the genomes of 41 strains of Sphingomonadales, the genotypes (based on a combination of crtE, crtB, crtI, crtY, crtZ, crtW and/or crtG) that could engender the various colour/carotenoid phenotypes have been identified. This cataloguing effort would benefit not only bacteriologists interested in systematics, but also biologists investigating carotenoid biosynthesis and its evolution in different organisms. Although it is well known that carotenoid and cytokinin biosynthesis requires precursors derived from isoprenoids, the genetic association between these two pathways had not been identified hitherto. In this context, the discovery within crt loci of an ORF encoding a putative homologue of LOG has opened up novel frontiers in basic as well as applied research.
Introduction
The order Sphingomonadales was circumscribed within the hierarchical system of the second edition of the Bergey's Manual of Systematic Bacteriology as a distinct phylogenetic branch of the Alphaproteobacteria, which was a novel class of the phylum Proteobacteria [1–3]. Currently, Sphingomonadaceae and Erythrobacteraceae are the only two families within this order [4]. The class Alphaproteobacteria contains many well-characterised phototrophs, including the Bchl a-producing aerobic anoxygenic phototrophs (AAPs), which are believed to be incapable of using light as the sole source of energy [5, 6]. Thus, most of the AAPs are photoheterotrophs and inhabit a variety of nutrient-rich aquatic and terrestrial environments [6, 7]. It has been postulated that AAPs are descendants of anaerobic purple bacteria, and studies of these organisms may provide clues to understanding the evolution of non-photosynthetic aerobes [6]. Historically, Erythrobacter longus strain OCh101 is among the well-known AAPs [8, 9], and is incidentally a member of the Sphingomonadales [4]. Nevertheless, AAPs appear to be a minority within this order, because only a few species/strains containing Bchl a have been identified in the last four decades [10–13]. Comparative genomic analyses have shown that the photosynthesis gene clusters (PGCs) are conserved in Erythrobacter sp. NAP1 and Citromicrobium sp. JL354 [14]. A similar cluster has also been identified in the genome of Porphyrobacter neustonensis DSM 9434 [15]. These clusters contain ORFs encoding putative proteins of the photosynthetic reaction centre and light harvesting complexes. Not surprisingly, some ORFs within these PGCs were predicted to be involved in the biosynthesis of Bchl a [14, 15].
A feature that is more obvious and common than anoxygenic phototrophy, but equally fascinating, among members of the Sphingomonadales is their pigmentation [3]. While providing the description of Sphingomonadaceae fam. nov., Kosako et al. [11] reported the different colours (e.g. yellow, orange and brown) of the species. The description of Erythrobacteraceae fam. nov. also noted that the members produced yellow, orange and pink pigments [4]. Therefore, it appears that being ‘coloured’ is a common phenotypic feature among members of these two families, and the ‘non-coloured’ strains (e.g. Zymomonas spp.) are most likely an exception to the rule. The vivid colours are known to be due to different carotenoids [16, 17], some of which have been well documented in the Prokaryotic Carotenoid Database [18] and the Carotenoids Database [19]. Although the biosynthesis of carotenoid pigments by photoautotrophs is a physiological necessity [20], their presence in AAPs and non-photosynthetic aerobes is somewhat of a mystery. It has been proposed that carotenoid pigments in these bacteria may be involved in protecting the cell from reactive oxygen species and high-energy radiation [7, 20]. It has also been reported that the carotenoid compositions of anaerobic purple bacteria are different from those of AAPs, and that AAPs of the order Sphingomonadales contain a variety of carotenoids, including β-carotene, zeaxanthin, caloxanthin, nostoxanthin, spirilloxanthin, spheroidene, erythroxanthin sulfate and caloxanthin sulfate [20]. Interestingly, most of the ORFs putatively involved in the biosynthesis of carotenoids were located outside the PGCs in the genomes of AAPs of the order Sphingomonadales [14, 15]. Functional genomic studies have provided valuable insights into carotenogenesis in members of the Sphingomonadaceae, and have identified ORFs involved in the biosynthesis of various carotenoids [21–23]. The objectives of this study were to extend the current knowledge about carotenogenesis from a few species to several genera within the order Sphingomonadales, and to identify novel genes that may have a role in carotenoid biosynthesis or modification among these bacteria.
Methods
Selection of bacterial strains
Bacterial strains included in this study were carefully selected so that type strains representing the major genera and species of the two families of the order Sphingomonadales made up at least 50 % of the pigmented cohort. This selection ensured that an authentic publication/reference for the colour/phenotype would be available. Other strains were selected based on the availability of their genome sequences in GenBank. Among these ‘other strains’, the majority had information about their colour/phenotype (either in the form of a publication or through personal communication). The final list included 41 strains, of which 34 had their genomes completely sequenced (Table 1). These strains represented the genera Sphingomonas (12), Sphingobium (8), Sphingopyxis (6), Altererythrobacter (6), Novosphingobium (3), Erythrobacter (2), Citromicrobium (1), Croceicoccus (1), Porphyrobacter (1) and Sphingorhabdus (1). Two of the strains (JL477 and DSM 9434) had already been confirmed to contain PGCs [15, 24].
Genome annotation and identification of orthologues
The genome sequences of the 41 strains were retrieved from GenBank and annotated using the rast (rapid annotation using subsystem technology) server (http://rast.nmpdr.org/). The crt ORFs within these annotated genomes were identified by blastp analyses using the protein sequences from Sphingomonas elodea strain ATCC 31461 [21] as queries. The PGCs within these genomes were identified using protein sequences from the PGC of P. neustonensis DSM 9434 [15] as queries. The ORFs were deemed orthologous if the putative proteins encoded by them had at least 30 % identity and less than 20 % difference in length (with a bit score of at least 50 and an E value <10−10) during blastp analyses. The order and orientation of the crt ORFs in complete genomes were checked using the sequence-based comparison tool available within the SEED Viewer. The synteny of ORFs (i.e. the occurrence of ORFs in the same order and orientation within a locus) was also an important criterion in the identification of orthologues. In strains/genomes where more than one potential orthologue was identified for a given crt ORF, the orthologue with the highest percent identity was chosen. The locations of the crt loci/ORFs within the respective chromosomes were assessed using the ‘Feature Table’ option of the SEED Viewer. Overlapping ORFs were checked and confirmed by manual curation of their sequences. Raw maps of the crt loci were copied from SEED Viewer and edited/refined using Microsoft Paint.
Analyses of protein sequences
Almost all protein sequences were retrieved from GenBank and identified using their standard locus tags. For some strains, the sequences had to be derived from their genome annotations within rast and assigned convenient locus tags. A preliminary alignment of the orthologous protein sequences was generated using Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/) with default parameters. Orthologues showing misalignment were excluded from further analyses. Clustal Omega alignment was also crucial to predict the length of the proteins and assign start codons (ATG, GTG, TTG or CTG in that preferential order). Manual trimming (or ‘extension’) of ORFs that appeared to be incorrectly annotated in GenBank or rast was performed based on this alignment. ClustalW (http://embnet.vital-it.ch/software/ClustalW.html) was the tool of choice to obtain a ‘pir’ output of the protein sequences. The boxshade server (http://www.ch. embnet.org/software/BOX_form.html) was used to further align the ‘pir’ output from ClustalW and visualise the conserved positions within the protein sequences.
Protein secondary structure prediction and phylogenetic analyses
Homologous protein sequences were identified using blastp analysis and acquired from GenBank, rast or the Protein Data Bank (PDB). Protein homologues with known crystal structures were used for predicting the secondary structure of the query. Multiple sequence alignment of the protein homologues was performed using Clustal Omega. The aligned sequences were analysed using ESPript (http://espript.ibcp.fr/ESPript/ESPript/) to depict similarities and secondary structure information. For phylogenetic analyses, relevant homologues (showing at least 40 % identity) from bacteria outside the order Sphingomonadales were included in the dataset when necessary. One of the datasets contained 31 sequences, whereas the other had just 7. Sequences were aligned using ClustalW and evolutionary distances were computed by analyses using the Molecular Evolutionary Genetics Analysis (mega 7.0) software package. Phylogeny was reconstructed using the maximum-likelihood method with the JTT or LG+F substitution model. Reliability of the trees was estimated using bootstrap methods (minimum of 500 replicates).
Results and Discussion
Clustering patterns of crt ORFs in strains of Sphingomonadales are variable
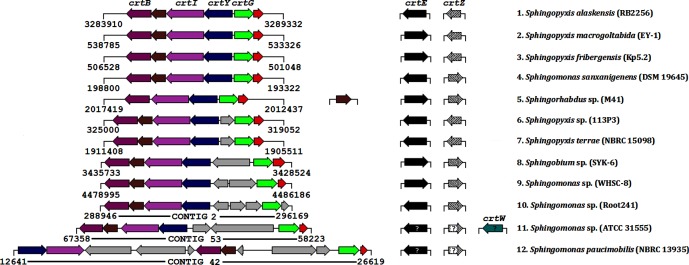
A gene cluster is defined as a ‘set of functionally related genes located in close physical proximity in a genome’ [25]. Previous studies have shown that ORFs encoding proteins involved in C40/C50 carotenoid biosynthesis form a cluster in the chromosomes of some bacteria [26–29]. In Sm. elodea strain ATCC 31461, which is yellow pigmented and shown to produce nostoxanthin, a carotenoid biosynthesis (crt) locus containing seven ORFs was identified [21]. Only four of these ORFs were assigned functions based on genetic analyses and were designated crtB, crtI, crtY and crtG [21]. blastp analyses of the genomes of 41 strains (Table 1) of Sphingomonadales indicated that the crt ORFs were adjacent to each other only in five of them (Fig. 1, serial numbers 1–5). The crt loci within these genomes contained a set of 5–6 ORFs, wherein the crtG ORF appeared to be located on the ‘opposite’ strand (Fig. 1).
Fig. 1.
Comparison of crt ORFs/loci in 12 strains of Sphingomonadales. Serial numbers of the bacteria are the same as in Table 1. The crt ORF that each colour-coded or shaded arrow represents is indicated on the top. The brown and red arrows represent ORFs encoding the putative LOG and DUF2141 proteins, respectively. The grey arrows represent the separating ORFs that are unlikely to be involved in carotenoid biosynthesis. Numbers at the beginning and end of gene clusters indicate the coordinates within each genome or contig. In the case of crtE and crtZ, arrows pointing to the right indicate that they were located on the plus strand, while those pointing to the left indicate that they were located on the minus strand. The uncertainty of the location of these ORFs on the plus or minus strands in the draft genomes of strains ATCC 31555 and NBRC 13935 is denoted by a ‘?’ within the respective arrows.
Although the arrangement of the crt ORFs was similar in six other genomes (Fig. 1, serial numbers 6–11), crtG in these loci was separated from the crtY, crtI and crtB ORFs by the insertion of 1–3 ORFs. The draft genome of the yellow-pigmented Sphingomonas paucimobilis NBRC 13935 appeared to contain an atypical crt locus, wherein crtG, crtB and crtI were set apart from each other (Fig. 1). Furthermore, crtY and crtI were translocated upstream of crtB, and were present on the same strand as crtG in this genome (Fig. 1). All 12 genomes compared in Fig. 1 contained crtE and crtZ [encoding a putative geranylgeranyl pyrophosphate (GGPP) synthase and a β-carotene hydroxylase (CrtZ), respectively]. In each strain, these two ORFs were present in different regions of the respective chromosomes and were not part of the crt locus. The genome of Sphingomonas sp. ATCC 31555 contained crtW (encoding a putative β-carotene ketolase) that was also not part of the crt locus (Fig. 1).
The separating ORFs in most of the crt loci shown in Fig. 1 were unrelated to each other, and their roles in carotenoid biosynthesis remain to be investigated. Sm. paucimobilis NBRC 13935 appeared to be a prototype for the separation of the crt ORFs in species and strains of Sphingomonadales that were capable of pigment biosynthesis, because two distinct patterns of separation were observed among 23 other genomes. In 21 genomes, crtY, crtI and crtB occurred together (all oriented in the same direction), and were set apart from crtG (Fig. S1, available in the online Supplementary Material, serial numbers 14–34). In two genomes, crtY, crtI and crtG occurred together, and were set apart from crtB (Fig. S1, serial numbers 35 and 36). Notably, only seven genomes (serial numbers 13, 21, 24, 25, 30, 35 and 36) contained crtW. Based on these patterns, it appears that in vast majority of species and strains, crtY, crtI and crtB occur together, and that each of the other four ORFs (crtE, crtG, crtZ and crtW) is located elsewhere. It also appears that in at least 50 % of the genomes, crtB and crtG are oriented opposite to each other, irrespective of whether they are located close to each other or are set apart. Although crtY, crtI, crtB and crtG are located together in a single locus in a few species and strains (Fig. 1), it appears that their clustering is not required for pigmentation (e.g. in the orange- pigmented Sphingomonas taxi ATCC 55669, crtB is set apart, and in the yellow-pigmented Sphingobium japonicum UT26S, crtB and crtG are set apart; Fig. S1, serial numbers 35 and 37). Genome comparisons revealed that at least five ORFs (crtE, crtB, crtI, crtY and crtZ) are commonly found in bacteria producing carotenoid pigments. Furthermore, Sphingomonas sp. MM-1, Sphingobium baderi DE-13, Sphingomonas wittichii RW1 and Sphingopyxis granuli TFA were found to be lacking one or more of these five crt ORFs (Table S1), and some of them have been reported to be non-pigmented (Table 1).
Co-occurrence of crtY and crtI is an evolutionarily conserved feature
Operonic organization and overlapping of ORFs are the hallmark of prokaryotic genomes, and these features have functional consequences [30, 31]. When the sequences of carotenoid biosynthesis genes from the purple non-sulfur photosynthetic bacterium Rhodobacter capsulatus were described, it was noted that the start codon (ATG) of crtB overlapped with the stop codon (TGA) of crtI [32]. Similar physical linkages of the crt ORFs have been observed in Pantoea ananatis (previously Erwinia uredovora) [26], Paracoccus sp. strain N81106 (previously Agrobacterium aurantiacum) [33], Bradyrhizobium sp. ORS278 [34] and Sphingobium yanoikuyae XLDN2-5 [22]. Overlapping of crt ORFs is likely to facilitate coordinated gene expression, and it has been shown that the preceding ORFs (e.g. crtY and crtI) contain the ribosome binding site (the Shine–Dalgarno sequence) for the subsequent ORFs (e.g. crtI and crtB, respectively) for translational coupling [33]. Genome comparisons showed that crtY and crtI occur together and have the same orientation in 38 strains of Sphingomonadales (Figs 1 and S1). In 28 of these strains, the reading frames of crtY and crtI overlapped by 1–8 bp, with 4 bp being the most common overlap (Table S2). In 10 strains where crtY and crtI occur together without an overlap of the ORFs, their separation range was only 6–25 bp (Table S2). Thus, it appears that the co-occurrence of crtY and crtI is an evolutionarily conserved feature in carotenogenic bacteria.
The N-terminal regions of bacterial CrtY (lycopene cyclase) and CrtI (phytoene desaturase/dehydrogenase) contain characteristic flavin adenine dinucleotide (FAD)-binding motifs, and the respective enzymes from P. ananatis have been shown to require FAD for their catalytic activity [35–37]. Sequence analyses showed that the N-terminal regions of most of the CrtY and CrtI orthologues from Sphingomonadales contain ‘GxGxxG(x)19E’ and ‘GxGxxG(x)17E’ motifs, respectively (data not shown). Although crtI in P. ananatis is located on a plasmid, the identity between the protein it encodes (GenBank accession no. ADD79328) and the CrtI orthologues of Sphingomonadales was high (50–59 %). In this context, the structural and functional similarities between different CrtI homologues need to be elucidated.
crt loci in many strains contain an ORF encoding a putative homologue of LONELY GUY (LOG: cytokinin phosphoribohydrolase)
In bacteria that contain crt loci, crtB usually occurs immediately downstream of crtI, and in some cases their ORFs overlap [26–28, 33, 34]. Such occurrence appears to be rare among Sphingomonadales, because it was found only in three strains: Sphingorhabdus sp. M41, Sphingomonas astaxanthinifaciens DSM 22298 and Croceicoccus naphthovorans PQ-2 (Figs 1 and S1). Surprisingly, in the genomes of 24 strains of Sphingomonadales, an unusual ORF was present between crtI and crtB (Figs 1 and S1). This ORF encodes a putative protein that appeared to be a homologue of LOG involved in the direct pathway of biosynthesis of cytokinins in plants and bacteria [38–40]. Furthermore, most strains of Sphingomonadales had a homologue of miaA [which encodes a putative tRNA delta(2)-isopentenylpyrophosphate transferase (dimethylallyl diphosphate:tRNA dimethylallyltransferase)] (data not shown). In bacteria, MiaA prenylates tRNA and generates tRNA i6A37, which is a precursor for cytokinin biosynthesis [40, 41].
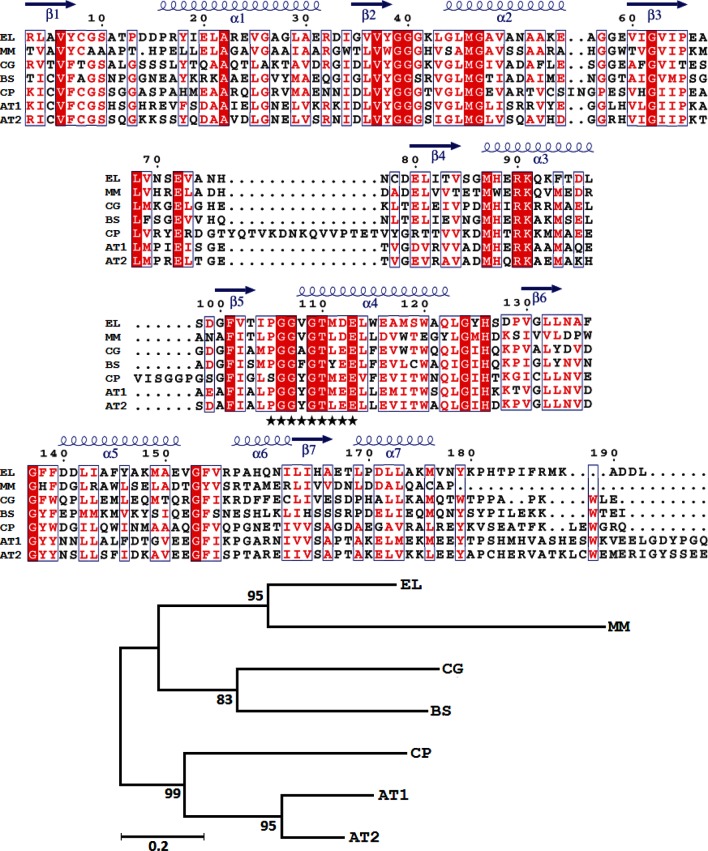
A search of the PDB using the putative LOG of Erythrobacter litoralis HTCC2594 (ELI_09890, 193 aa) showed that it was related to the predicted LOG, nucleotide-binding or molybdenum cofactor carrier proteins from other bacteria and eukaryotes. Although phylogenetic analysis showed that the LOG homologues from bacteria and eukaryotes cluster on different branches, the secondary structural features of these proteins were found to be conserved (Fig. 2). Furthermore, the characteristic PGGxGTxxE motif previously identified in LOG homologues [38, 40] was conspicuous in all of them (Fig. 2). The secondary structure comparison further supports the annotation of ELI_09890 as LOG, and a model of the 3D structure (data not shown) indicates that it may be a nucleotide-binding protein.
Fig. 2.
Comparison of LOG homologues. Sequences of the putative LOG (ELI_09890, 193 aa) of Erythrobacter litoralis (EL) and LOG of Mycobacterium marinum (MM: PDB code 3SBX), Corynebacterium glutamicum (CG: 5ITS), Bacillus subtilis (BS: 1T35), Claviceps purpurea (CP: 5AJT) and Arabidopsis thaliana (AT1 : 2A33, AT2 : 1YDH) were aligned. The secondary structure of ELI_09890 was inferred from the homology model constructed using 3SBX as the template (data not shown). Coils (α1–α7) and arrows (β1–β7) represent the predicted helices and strands, respectively, within ELI_09890. Residues identical in all seven homologues are shaded red, and sites with ≥70 % similarity are in red font. Blocks of conserved sites are boxed. Asterisks denote the conserved PGGxGTxxE motif. The numbers above the sequence indicate the aa positions within ELI_09890. The tree (derived using the maximum-likelihood method in mega 7.0 with the JTT substitution model) shows the phylogenetic relationship among the seven LOG homologues. All positions with less than 95 % site coverage were eliminated, and the final dataset contained 175 positions. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (500 replicates) are shown next to the branches. The tree is drawn to scale. Bar, the number of amino acid substitutions per site.
It appears that LOG is not essential for bacterial survival [42], and has no direct role in carotenogenesis since deletion of the ORF encoding the putative LOG in Sm. elodea ATCC 31461 did not affect the pigment profile [21]. However, previously it has been shown that the cytokinin 6-benzylaminopurine enhances carotenoid accumulation in radish seedlings [43], Ricinus cell cultures [44] and the green alga Chlorella pyrenoidosa [45]. Furthermore, it has been reported that the deletion of crtR, which encodes a MarR-type repressor of the crt operon, enhances carotenoid accumulation in Corynebacterium glutamicum ATCC 13032, and that GGPP and IPP (isopentenyl diphosphate) prevent the binding of CrtR to its target DNA [29]. In view of these observations, the role of cytokinins and their precursors in bacterial carotenogenesis remains to be investigated. While cytokinin biosynthesis and the function of LOG homologues have been reported in bacteria [40, 42, 46, 47], this is the first time such homologues have been discovered within crt loci.
crtG is co-located with a hitherto unidentified ORF in many strains
Carotenoid 2,2′-β-hydroxylase (CrtG) was first identified in Brevundimonas sp. strain SD212 [28]. This is a key enzyme that can convert zeaxanthin to 2, 2′-dihydroxyzeaxanthin, and astaxanthin to 2-hydroxyastaxanthin [28]. Homologues of crtG appear to be present in many branches of the bacterial domain, including the cyanobacterium Thermosynechococcus elongatus strain BP-1 [48]. The identity between CrtG of strain SD212 (GenBank accession no. BAD99415) and the CrtG orthologues from Sphingomonadales was rather low (40–48 %). The most conspicuous feature of crtG in many strains of Sphingomonadales was its co-location with another ORF (Figs 1 and S1). The putative proteins encoded by this ORF in different strains varied in their lengths (data not shown). blastp analyses showed that these orthologues belong to the DUF2141 superfamily of proteins that have no known function. Interestingly, members of this superfamily are found only among bacteria and have received very little attention most likely because they are not essential for cell survival. Further analyses are required to establish the role, if any, of proteins of the DUF2141 superfamily in carotenoid biosynthesis or modification.
crtZ is co-located with crtW in some strains
The ORF encoding β-carotene hydroxylase (also referred to as carotenoid 3,3′-β-hydroxylase), which catalyses the hydroxylation of β-carotene to zeaxanthin, was identified in P. ananatis and was designated crtZ [26]. Homologues of crtZ have been found in numerous pigmented members of the class Alphaproteobacteria, and a crtZ deletion mutant of Sm. elodea ATCC 31461 has been shown to lack 3-hydroxy carotenoids [21]. Furthermore, crtZ of Sphingomonas lacus strain PB304 has been shown to convert β-carotene to zeaxanthin in an Escherichia coli strain that expresses CrtE (GGPP synthase), CrtB, CrtI and CrtY homologues from Pantoea agglomerans [23]. Multiple sequence alignment revealed the presence of two His-rich motifs (HxxHH near the N-terminal and HxLHH near the C-terminal) among the CrtZ orthologues of 37 strains of Sphingomonadales (data not shown). Although crtZ was located away from crtY, crtI, crtB and crtG in 36 strains, it was located adjacent to crtB, but in the opposite orientation, in Sm. astaxanthinifaciens DSM 22298 (Fig. S1). A similar occurrence has been reported in the crt locus of the related strain PB304 [23].
The ORF encoding β-carotene ketolase (also referred to as carotenoid 4,4′-β-oxygenase or carotenoid 4,4′-β-ketolase), which catalyses the conversion of β-carotene to canthaxanthin via echinenone, was discovered in Haematococcus pluvialis [49, 50]. A homologue of this gene was identified in two marine bacteria, and was designated crtW [33, 51]. Furthermore, crtW was located adjacent to crtZ and crtB in Paracoccus sp. strain N81106 and Bradyrhizobium sp. strain ORS278, respectively [33, 34]. In Brevundimonas sp. strain SD212 (where crtW was located adjacent to crtY, but in the opposite orientation) the presence of crtZ, crtW and crtG was predicted to bestow the ability to synthesize adonixanthin, astaxanthin, canthaxanthin and erythroxanthin [28]. Genomic analyses indicated that of the 37 strains of Sphingomonadales that contained crtZ, only 8 (~22 %) contained crtW (Figs 1 and S1). In four strains (JCM 16345, NBRC 107699, HTCC2594 and DSM 9434, which are members of Erythrobacteraceae), crtW was located immediately upstream of crtZ (Fig. S1). Conversely, in two strains (ATCC 55669 and RIT328, which are members of Sphingomonas), crtW was located immediately downstream of crtZ (Fig. S1). Interestingly, in two other strains (ATCC 31555 and DSM 22298, which are also members of Sphingomonas), crtW was located away from other crt ORFs (Figs 1 and S1). Three His-rich motifs (HDxxH, HxxHH and HxxHH) have been shown to be essential for β-carotene ketolase activity in Paracoccus sp. strain N81106 [52], and all three of them were conserved in the diverse set of CrtW (β-carotene ketolase) proteins from Sphingomonadales and other bacteria (data not shown).
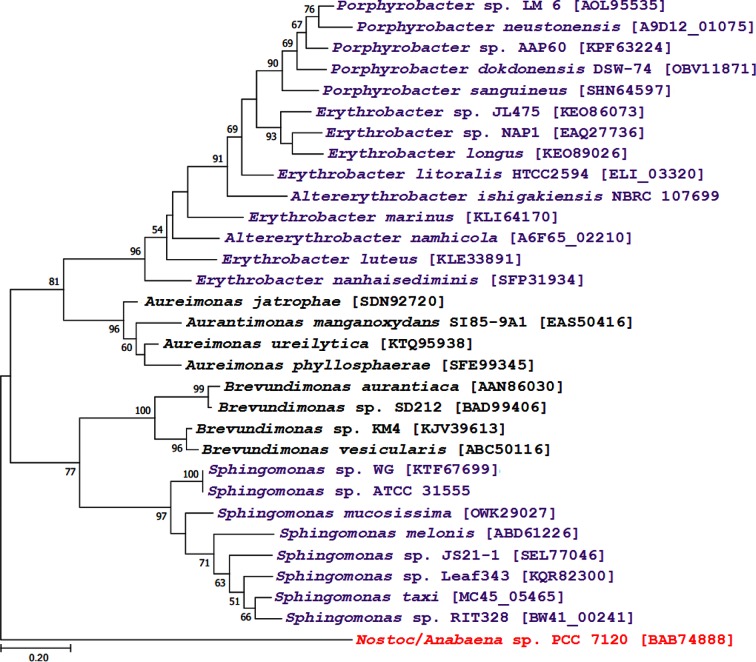
blastp and psi-blast analyses showed that the top hits of CrtW of strains ATCC 55669 and RIT328 were from strains of the genus Sphingomonas and other bacteria (e.g. Brevundimonas spp.), but not from strains of Erythrobacteraceae. Similarly, the top hits of CrtW of strains JCM 16345, NBRC 107699, HTCC2594 and DSM 9434 were from strains of Erythrobacteraceae and other bacteria (e.g. Aureimonas spp.), but not from strains of the genus Sphingomonas. Phylogenetic analysis confirmed these results: CrtW of strains of Sphingomonas spp. clustered on a main branch that contained CrtW of Brevundimonas spp., whereas CrtW of strains of Erythrobacteraceae clustered on yet another main branch that contained CrtW of Aureimonas spp. (Fig. 3). Further analyses are required to establish whether these outcomes, and the rarity of CrtW among strains of Sphingomonadales, are due to preferential acquisition (through horizontal gene transfer) or differential gene loss. Similar interpretations have been made previously based on phylogenetic analysis of carotenoid hydroxylases and ketolases from various bacteria [53]. Despite their importance in carotenoid biosynthesis, many aspects of the catalytic activity of CrtZ, CrtW and CrtG remain unknown because no crystal structures are available for these proteins, and attempts to purify them for in vitro enzymatic characterisation were reportedly futile [54].
Fig. 3.
Phylogenetic tree based on CrtW homologues. The analysis involved 31 sequences and the tree was constructed using the maximum-likelihood method (with the LG+F substitution model) in mega 7.0. Bootstrap values of 1000 replicates are indicated as numbers out of 100 at the nodes (only values >50 are shown). All positions containing gaps and missing data were eliminated, and the final dataset contained 214 positions. Purple text indicates CrtW from strains of Sphingomonadales. Black text indicates CrtW from taxa distantly related to Sphingomonadales. Red text indicates the outgroup CrtW sequence. GenBank accession numbers are provided in square brackets. The tree is drawn to scale. Bar, the number of amino acid substitutions per site.
crtX appears to be very rare among strains of Sphingomonadales
An ORF encoding a zeaxanthin glycosyltransferase was identified in the crt locus of P. ananatis and was designated crtX [26]. Homologues of this ORF have been characterised only in a few bacteria, including P. agglomerans [55], Xanthobacter autotrophicus [56] and Paracoccus haeundaensis [57]. An ORF encoding a putative carotenoid glycosyltransferase (CrtX, 407 aa) was identified immediately downstream of crtW in Sm. lacus PB304 [23]. A homologue of this ORF was found in Sm. astaxanthinifaciens DSM 22298 (68 % identity across 392 aa). The arrangement of crtW and crtX was similar in strains PB304 and DSM 22298, which were isolated from aquatic sources in Japan and Korea, respectively [58, 59]. Furthermore, the closest homologues of CrtX of these strains were found in Aureimonas and Aurantimonas (data not shown). The function of crtX in strains PB304 and DSM 22298, which have been reported to produce astaxanthin dideoxyglycosides [23, 60], is unknown.
crtE occurs even in non-carotenogenic strains of Sphingomonadales
Although the condensation of two molecules of GGPP by phytoene synthase (CrtB) to produce phytoene, which is a C40 hydrocarbon and a colourless carotene, is the first committed step in carotenoid biosynthesis, there is reason to believe that the pathway in many organisms actually starts with the production of GGPP. This belief is supported by the fact that crtE, which encodes GGPP synthase, is part of the crt locus in some bacteria [28]. However, crtE was not a part of the crt locus either in Sm. elodea ATCC 31461 or in Sm. lacus PB304 [21, 23]. The same appeared to be true in the current comparisons, because crtE never co-occurred with any other crt ORF (Figs 1 and S1). Notably, most strains appeared to contain a single homologue of crtE, and strains that were non-carotenogenic also contained crtE. Furthermore, the CrtE orthologues of most strains of Sphingomonadales contained ~300 aa (Table S1), and multiple sequence alignment revealed the presence of five motifs (GKxxR, DDxxxxD, GQxxD, KT and DDxxx; data not shown) that have been shown to be conserved in bacterial, archaeal, yeast, plant and human GGPP synthases [61]. Isoprenoids and isoprenyl diphosphates are substrates for the synthesis of a variety of biologically important compounds, and E. coli reportedly contains three isoprenyl diphosphate synthases, but lacks a GGPP synthase [62]. In this context, the function of crtE in non-carotenogenic strains of Sphingomonadales requires further characterisation.
PGCs are not pervasive among strains of Sphingomonadales
As noted previously, only a few Bchl a-containing species/strains of Sphingomonadales have been described, and PGCs were identified in the genomes of only four strains [14, 15, 24]. Comparative analysis revealed that similar PGCs also occur in the genomes of Sphingomonas sanxanigenens DSM 19645, Sphingomonas hengshuiensis WHSC-8, Sm. astaxanthinifaciens DSM 22298 and Altererythrobacter ishigakiensis NBRC 107699 (Table 1). However, the latter strain was reported to lack Bchl a [63], and no information is available about the presence or absence of Bchl a in the descriptions of the type strains DSM 22298, DSM 19645 and WHSC-8 [57, 64, 65]. Nevertheless, it is evident from Table 1 that PGCs are generally found in strains that are coloured, and strains that are non-carotenogenic due to the absence of one or more crt ORFs lack PGCs. Whether β-carotene and its derivates have any role in anoxygenic phototrophy in Sphingomonadales remains to be investigated.
Conclusions
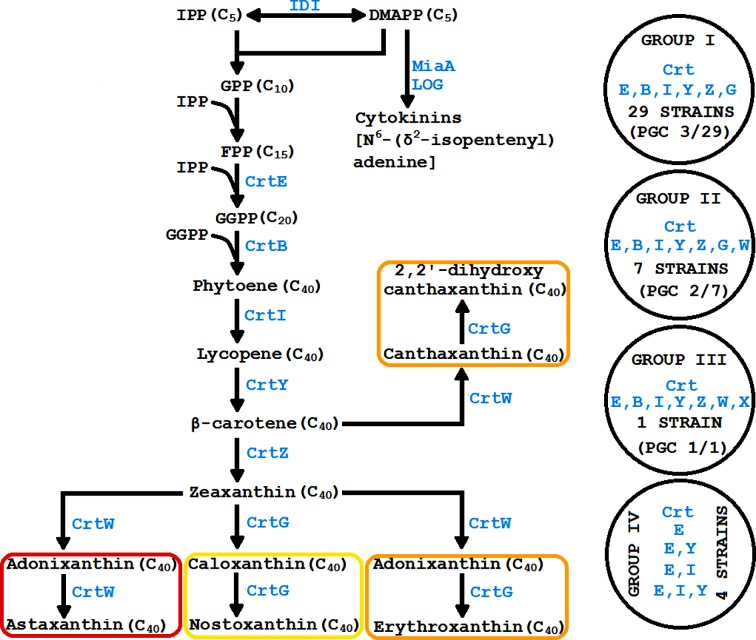
This study sought to understand the genetic basis for the vibrant colours of members of the Sphingomonadales through comparative genomic analysis. It was found that the crt ORFs were scattered across the chromosome in many strains, indicating that clustering of crt ORFs is not necessary for the coloured phenotype. Based on the current analyses, strains of Sphingomonadales could broadly be categorised into those that are coloured (with subcategories of yellow, orange and red) and colourless. Yellow coloured strains most likely accumulate nostoxanthin, and contain six ORFs (group I: crtE, crtB, crtI, crtY, crtZ, crtG; Table 1, Fig. 4). However, orange coloured strains (group II: crtE, crtB, crtI, crtY, crtZ, crtG, crtW; Table 1, Fig. 4) could contain some of the carotenoids predicted to be produced by Brevundimonas sp. strain SD212 [28], in addition to echinenone, zeaxanthin, caloxanthin and/or nostoxanthin. Red coloured strains may contain six (crtE, crtB, crtI, crtY, crtZ, crtW) or seven (crtE, crtB, crtI, crtY, crtZ, crtW, crtX) ORFs (group III; Table 1, Fig. 4) that enable the production of astaxanthin and its derivatives. Non-pigmented strains may contain a smaller subset of crt ORFs, and are thus unable to produce any carotenoids (group IV; Table 1, Fig. 4). The four groups appear to transcend phylogenetic relationships within the order Sphingomonadales.
Fig. 4.
Presumptive pathways of carotenoid biosynthesis in strains of Sphingomonadales. Shorthand notations of enzymes are indicated in blue (e.g. IDI – isopentenyl diphosphate isomerase). Zeaxanthin (whose biosynthesis is enabled by the presence of CrtE, CrtB, CrtI, CrtY and CrtZ) appears to be the branchpoint for three different pathways that produce astaxanthin (using CrtW), nostaxanthin (using CrtG) and erythroxanthin (using CrtW and CrtG). Additionally, β-carotene is the branchpoint for the pathway that produces 2,2′-dihydroxycanthaxanthin (using CrtW and CrtG). The precursor for cytokinin production (using MiaA and LOG) and carotenoid biosynthesis is DMAPP. Circles on the right represent the grouping of bacterial strains based on their crt genotype as shown in Table 1.
It is important to note that the relative quantities of the different carotenoids (and their isomers) could determine the type and intensity of colour. This notion is supported by the observation that cells containing lower levels of lycopene are pink, whereas those accumulating higher levels can be red [21, 33, 37, 66–69]. Similarly, cells containing low/moderate levels of β-carotene are yellow, whereas those accumulating higher levels can be orange [23, 26, 33, 35, 36, 67, 70–72]. Zeaxanthin producing cells are no exception in this context; they can be yellow, orange or yellow–orange [23, 26, 65, 73–76]. Furthermore, the absorption spectra of β-carotene, zeaxanthin, caloxanthin and nostoxanthin are apparently similar. Thus, the crt genotype identified in strains of Sphingomonadales will have to be reconciled with the ‘actual’ colour phenotype and vice-versa. Occasionally, some strains of this order could contain the minimal crt genotype to be coloured, but appear colourless (e.g. Sm. sanxanigenens DSM 19645 and Sphingorhabdus sp. M41). Such strains could accumulate GGPP or phytoene, which are colourless compounds, due to dysfunctional crtB or crtI, respectively. They could also be colourless due to repression of crtB and/or crtI by as yet unknown mechanisms.
Data bibliography
Lauro FM et al. GenBank accession no. CP000356 (2006).
Ohtsubo Y et al. GenBank accession no. CP012700 (2015).
Oelschlagel M et al. GenBank accession no. CP009122 (2014).
Ma T et al. GenBank accession no. CP006644 (2013).
Jeong H et al. GenBank accession no. CP014545 (2015).
Ohtsubo Y et al. GenBank accession no. CP009452 (2015).
Ohtsubo Y et al. GenBank accession no. CP013342 (2015).
Masai E et al. NCBI RefSeq accession no. NC_015976 (2011).
Wei S et al. GenBank accession no. CP010836 (2015).
Bai Y et al. NCBI RefSeq accession no. NZ_LMIV01000000 (2015).
Wang X et al. NCBI RefSeq accession no. NZ_ALBQ00000000 (2012).
Hosoyama A et al. NCBI RefSeq accession no. NZ_BBJS01000000 (2014).
Kyrpides N et al. NCBI RefSeq accession no. NZ_JONN00000000 (2014).
Lucas S et al. GenBank accession no. CP002798 (2011).
Tabata M et al. GenBank accession no. CP005188 (2013).
Lee C-M et al. GenBank accession no. CP015986 (2016).
Yan X. GenBank accession no. CP010954 (2015).
Copeland A et al. GenBank accession no. CP000248 (2006).
Zheng Q et al. GenBank accession no. CP011344 (2015).
Cheng H et al. GenBank accession no. CP016591 (2016).
Cheng H et al. GenBank accession no. CP016545 (2016).
Li Z et al. GenBank accession no. CP012669 (2015).
Wu YH et al. GenBank accession no. CP011452 (2015).
Wu YH et al. GenBank accession no. CP015963 (2016).
Giovannoni SJ et al. GenBank accession no. CP000157 (2005).
Zhuang L et al. GenBank accession no. CP011310 (2015).
Earl A et al. NCBI RefSeq accession no. NZ_AGZU00000000 (2012).
Petrillo M. GenBank accession no. FR856862 (2011).
Choi DH et al. GenBank accession no. CP009291 (2014).
Shi XL et al. GenBank accession no. CP016033 (2016).
Zhu X et al. GenBank accession no. CP015521 (2016).
Wang H, Zhu P. NCBI RefSeq NZ_LQCK00000000 (2015).
Kim KM. GenBank accession no. CP011805 (2015).
Zeng Y, Huang Y. GenBank accession no. CP011770 (2015).
Zhou Y et al. GenBank accession no. CP009571 (2014).
Gan HY et al. NCBI RefSeq accession no. NZ_JFYV01000000 (2014).
Nagata Y et al. NCBI RefSeq accession no. NC_014006 (2008).
Tabata M et al. GenBank accession no. CP004036 (2013).
Cheng M et al. GenBank accession no. CP013264 (2015).
Copeland A et al. GenBank accession no. CP000699 (2007).
Garcia-Romero I et al. GenBank accession no. CP012199 (2015).
Funding information
Research at the Institute of Bioinformatics and Applied Biotechnology (IBAB) is continuously supported by the Department of Innovation and Technology of the Government of Karnataka, India. Research infrastructure at IBAB is made possible by the Department of Science and Technology of the Government of India through a program called ‘Fund for Improvement of S&T Infrastructure in Universities and Higher Educational Institutions’ (DST-FIST: level 0 from 2006 to 2011, and level 1 from 2012 to 2017). The Department of Science and Technology of the Government of India generously supported the academic endeavours of V. V. through an INSPIRE fellowship (sanction number IF120372). The Department of Science and Technology of the Government of India also supported the work of S.T.
Acknowledgements
The authors are grateful to their colleagues worldwide for freely sharing the genome sequences of the strains of Sphingomonadales in the public databases. Without such a voluntary ‘open data’ policy, work of the kind described here would be unfeasible. The authors thank the three anonymous reviewers whose comments immensely improved the manuscript.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Ethical statement
No human/animal experiments were involved in this study.
Supplementary Data
Footnotes
Abbreviations: AAP, aerobic anoxygenic phototroph; Bchl a, bacteriochlorophyll a; CrtB, phytoene synthase; CrtE, GGPP synthase; CrtG, carotenoid 2,2′-β-hydroxylase; CrtI, phytoene desaturase/dehydrogenase; CrtW, β-carotene ketolase; CrtX, carotenoid glycosyltransferase; CrtY, lycopene cyclase; CrtZ, β-carotene hydroxylase; DMAPP, dimethylallyl pyrophosphate; DUF, domain of unknown function; FAD, flavin adenine dinucleotide; FPP, farnesyl diphosphate; GGPP, geranylgeranyl pyrophosphate; GPP, geranyl diphosphate; IPP, isopentenyl diphosphate; LOG, lonely guy (cytokinin phosphoribohydrolase); MiaA, tRNA delta(2)-isopentenylpyrophosphate transferase (dimethylallyl diphosphate:tRNA dimethylallyltransferase); PDB, Protein Data Bank; PGC, photosynthesis gene cluster; RAST, rapid annotation using subsystem technology.
All supporting data, code and protocols have been provided within the article or through supplementary data files. One supplementary figure and two supplementary tables are available with the online Supplementary Material.
References
- 1.Garrity GM, Bell JA, Lilburn T. Phylum XIV. Proteobacteria phyl. nov. In: Brenner D, Krieg NR, Staley JT, Garrity GM, editors. Bergey’s Manual of Systematic Bacteriology (The Proteobacteria), part B (The Gammaproteobacteria) 2nd ed. vol. 2. New York:: Springer; 2005. p. 1. (editors) [Google Scholar]
- 2.Garrity GM, Bell JA, Lilburn T. Class I. Alphaproteobacteria class. nov. In: Brenner DJ, Krieg NR, Staley JT, Garrity GM, editors. Bergey’s Manual of Systematic Bacteriology (The Proteobacteria), part C (The Alpha-, Beta-, Delta-, and Epsilonproteobacteria. 2nd ed. vol. 2. New York:: Springer; 2005. p. 1. (editors) [Google Scholar]
- 3.Yabuuchi E, Kosako Y. Order IV. Sphingomonadales ord. nov. In: Brenner DJ, Krieg NR, Staley JT, Garrity GM, editors. Bergey’s Manual of Systematic Bacteriology (The Proteobacteria), part C (The Alpha-, Beta-, Delta-, and Epsilonproteobacteria) 2nd ed. vol. 2. New York:: Springer; 2005. pp. 230–286. (editors) [Google Scholar]
- 4.Lee KB, Liu CT, Anzai Y, Kim H, Aono T, et al. The hierarchical system of the 'Alphaproteobacteria': description of Hyphomonadaceae fam. nov., Xanthobacteraceae fam. nov. and Erythrobacteraceae fam. nov. Int J Syst Evol Microbiol. 2005;55:1907–1919. doi: 10.1099/ijs.0.63663-0. [DOI] [PubMed] [Google Scholar]
- 5.Swingley WD, Blankenship RE, Raymond J. Evolutionary relationships among purple photosynthetic bacteria and the origin of proteobacterial photosynthetic systems. In: Hunter CN, Daldal F, Thurnauer MC, Beatty JT, editors. The Purple Phototrophic Bacteria. New York:: Springer; 2009. pp. 17–29. (editors) [Google Scholar]
- 6.Yurkov VV, Beatty JT. Aerobic anoxygenic phototrophic bacteria. Microbiol Mol Biol Rev. 1998;62:695–724. doi: 10.1128/mmbr.62.3.695-724.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yurkov V, Csotonyi JT. New light on aerobic anoxygenic phototrophs. In: Hunter CN, Daldal F, Thurnauer MC, Beatty JT, editors. The Purple Phototrophic Bacteria. New York:: Springer; 2009. pp. 31–55. (editors) [Google Scholar]
- 8.Shiba T, Simidu U. Erythrobacter longus gen. nov., sp. nov., an aerobic bacterium which contains bacteriochlorophyll a. Int J Syst Bacteriol. 1982;32:211–217. doi: 10.1099/00207713-32-2-211. [DOI] [Google Scholar]
- 9.Koblížek M. Ecology of aerobic anoxygenic phototrophs in aquatic environments. FEMS Microbiol Rev. 2015;39:854–870. doi: 10.1093/femsre/fuv032. [DOI] [PubMed] [Google Scholar]
- 10.Fuerst JA, Hawkins JA, Holmes A, Sly LI, Moore CJ, et al. Porphyrobacter neustonensis gen. nov., sp. nov., an aerobic bacteriochlorophyll-synthesizing budding bacterium from fresh water. Int J Syst Bacteriol. 1993;43:125–134. doi: 10.1099/00207713-43-1-125. [DOI] [PubMed] [Google Scholar]
- 11.Kosako Y, Yabuuchi E, Naka T, Fujiwara N, Kobayashi K. Proposal of Sphingomonadaceae fam. nov., consisting of Sphingomonas Yabuuchi et al. 1990, Erythrobacter Shiba and Shimidu 1982, Erythromicrobium Yurkov et al. 1994, Porphyrobacter Fuerst et al. 1993, Zymomonas Kluyver and van Niel 1936, and Sandaracinobacter Yurkov et al. 1997, with the type genus Sphingomonas Yabuuchi et al. 1990. Microbiol Immunol. 2000;44:563–575. doi: 10.1111/j.1348-0421.2000.tb02535.x. [DOI] [PubMed] [Google Scholar]
- 12.Yabuuchi E, Kosako Y, Fujiwara N, Naka T, Matsunaga I, et al. Emendation of the genus Sphingomonas Yabuuchi et al. 1990 and junior objective synonymy of the species of three genera, Sphingobium, Novosphingobium and Sphingopyxis, in conjunction with Blastomonas ursincola. Int J Syst Evol Microbiol. 2002;52:1485–1496. doi: 10.1099/00207713-52-5-1485. [DOI] [PubMed] [Google Scholar]
- 13.Hiraishi A, Yonemitsu Y, Matsushita M, Shin Y, Kuraishi H, et al. Characterization of Porphyrobacter sanguineus sp. nov., an aerobic bacteriochlorophyll-containing bacterium capable of degrading biphenyl and dibenzofuran. Arch Microbiol. 2002;178:45–52. doi: 10.1007/s00203-002-0423-5. [DOI] [PubMed] [Google Scholar]
- 14.Zheng Q, Zhang R, Koblížek M, Boldareva EN, Yurkov V, et al. Diverse arrangement of photosynthetic gene clusters in aerobic anoxygenic phototrophic bacteria. PLoS One. 2011;6:e25050. doi: 10.1371/journal.pone.0025050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Q, Wu YH, Cheng H, Xu L, Wang CS, et al. Complete genome sequence of bacteriochlorophyll-synthesizing bacterium Porphyrobacter neustonensis DSM 9434. Stand Genomic Sci. 2017;12:32. doi: 10.1186/s40793-017-0243-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Glaeser SP, Kämpfer P. The family Sphingomonadaceae. In: Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F, et al., editors. The Prokaryotes (Alphaproteobacteria and Betaproteobacteria) 4th ed. New York:: Springer; 2014. pp. 641–707. (editors) [Google Scholar]
- 17.Tonon LAC, Moreira APB, Thompson F. The family Erythrobacteraceae. In: Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F, et al., editors. The Prokaryotes (Alphaproteobacteria and Betaproteobacteria) 4th ed. New York:: Springer; 2014. pp. 213–235. (editors) [Google Scholar]
- 18.Nupur LN, Vats A, Dhanda SK, Raghava GP, Pinnaka AK, et al. ProCarDB: a database of bacterial carotenoids. BMC Microbiol. 2016;16:96. doi: 10.1186/s12866-016-0715-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yabuzaki J. Carotenoids database: structures, chemical fingerprints and distribution among organisms. Database. 2017;2017::bax004. doi: 10.1093/database/bax004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takaichi S. Distribution and biosynthesis of carotenoids. In: Hunter CN, Daldal F, Thurnauer MC, Beatty JT, editors. The Purple Phototrophic Bacteria. New York:: Springer; 2009. pp. 97–117. (editors) [Google Scholar]
- 21.Zhu L, Wu X, Li O, Qian C, Gao H. Cloning and characterization of genes involved in nostoxanthin biosynthesis of Sphingomonas elodea ATCC 31461. PLoS One. 2012;7:e35099. doi: 10.1371/journal.pone.0035099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu X, Gai Z, Tao F, Tang H, Xu P. Carotenoids play a positive role in the degradation of heterocycles by Sphingobium yanoikuyae. PLoS One. 2012;7:e39522. doi: 10.1371/journal.pone.0039522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim SH, Kim JH, Lee BY, Lee PC. The astaxanthin dideoxyglycoside biosynthesis pathway in Sphingomonas sp. PB304. Appl Microbiol Biotechnol. 2014;98:9993–10003. doi: 10.1007/s00253-014-6050-7. [DOI] [PubMed] [Google Scholar]
- 24.Zheng Q, Liu Y, Jeanthon C, Zhang R, Lin W, et al. Geographic impact on genomic divergence as revealed by comparison of nine citromicrobial genomes. Appl Environ Microbiol. 2016;82:7205–7216. doi: 10.1128/AEM.02495-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu M, Huang H, Li G, Ren Y, Shi Z, et al. The evolutionary life cycle of the polysaccharide biosynthetic gene cluster based on the Sphingomonadaceae. Sci Rep. 2017;7:46484. doi: 10.1038/srep46484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Misawa N, Nakagawa M, Kobayashi K, Yamano S, Izawa Y, et al. Elucidation of the Erwinia uredovora carotenoid biosynthetic pathway by functional analysis of gene products expressed in Escherichia coli. J Bacteriol. 1990;172:6704–6712. doi: 10.1128/jb.172.12.6704-6712.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sedkova N, Tao L, Rouvière PE, Cheng Q. Diversity of carotenoid synthesis gene clusters from environmental Enterobacteriaceae strains. Appl Environ Microbiol. 2005;71:8141–8146. doi: 10.1128/AEM.71.12.8141-8146.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nishida Y, Adachi K, Kasai H, Shizuri Y, Shindo K, et al. Elucidation of a carotenoid biosynthesis gene cluster encoding a novel enzyme, 2,2'-beta-hydroxylase, from Brevundimonas sp. strain SD212 and combinatorial biosynthesis of new or rare xanthophylls. Appl Environ Microbiol. 2005;71:4286–4296. doi: 10.1128/AEM.71.8.4286-4296.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Henke NA, Heider SAE, Hannibal S, Wendisch VF, Peters-Wendisch P. Isoprenoid pyrophosphate-dependent transcriptional regulation of carotenogenesis in Corynebacterium glutamicum. Front Microbiol. 2017;8:633. doi: 10.3389/fmicb.2017.00633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sakharkar KR, Sakharkar MK, Verma C, Chow VTK. Comparative study of overlapping genes in bacteria, with special reference to Rickettsia prowazekii and Rickettsia conorii. Int J Syst Evol Microbiol. 2005;55:1205–1209. doi: 10.1099/ijs.0.63446-0. [DOI] [PubMed] [Google Scholar]
- 31.Mir K, Neuhaus K, Scherer S, Bossert M, Schober S. Predicting statistical properties of open reading frames in bacterial genomes. PLoS One. 2012;7:e45103. doi: 10.1371/journal.pone.0045103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Armstrong GA, Alberti M, Leach F, Hearst JE. Nucleotide sequence, organization, and nature of the protein products of the carotenoid biosynthesis gene cluster of Rhodobacter capsulatus. Mol Gen Genet. 1989;216:254–268. doi: 10.1007/BF00334364. [DOI] [PubMed] [Google Scholar]
- 33.Misawa N, Satomi Y, Kondo K, Yokoyama A, Kajiwara S, et al. Structure and functional analysis of a marine bacterial carotenoid biosynthesis gene cluster and astaxanthin biosynthetic pathway proposed at the gene level. J Bacteriol. 1995;177:6575–6584. doi: 10.1128/jb.177.22.6575-6584.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hannibal L, Lorquin J, D'Ortoli NA, Garcia N, Chaintreuil C, et al. Isolation and characterization of canthaxanthin biosynthesis genes from the photosynthetic bacterium Bradyrhizobium sp. strain ORS278. J Bacteriol. 2000;182:3850–3853. doi: 10.1128/JB.182.13.3850-3853.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cunningham FX, Sun Z, Chamovitz D, Hirschberg J, Gantt E. Molecular structure and enzymatic function of lycopene cyclase from the cyanobacterium Synechococcus sp strain PCC7942. Plant Cell. 1994;6:1107–1121. doi: 10.1105/tpc.6.8.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu Q, Schaub P, Ghisla S, Al-Babili S, Krieger-Liszkay A, et al. The lycopene cyclase CrtY from Pantoea ananatis (formerly Erwinia uredovora) catalyzes an FADred-dependent non-redox reaction. J Biol Chem. 2010;285:12109–12120. doi: 10.1074/jbc.M109.091843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schaub P, Yu Q, Gemmecker S, Poussin-Courmontagne P, Mailliot J, et al. On the structure and function of the phytoene desaturase CRTI from Pantoea ananatis, a membrane-peripheral and FAD-dependent oxidase/isomerase. PLoS One. 2012;7:e39550. doi: 10.1371/journal.pone.0039550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Frebort I, Kowalska M, Hluska T, Frebortova J, Galuszka P. Evolution of cytokinin biosynthesis and degradation. J Exp Bot. 2011;62:2431–2452. doi: 10.1093/jxb/err004. [DOI] [PubMed] [Google Scholar]
- 39.Samanovic MI, Darwin KH. Cytokinins beyond plants: synthesis by Mycobacterium tuberculosis. Microb Cell. 2015;2:168–170. doi: 10.15698/mic2015.05.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seo H, Kim S, Sagong HY, Son HF, Jin KS, et al. Structural basis for cytokinin production by LOG from Corynebacterium glutamicum. Sci Rep. 2016;6:31390. doi: 10.1038/srep31390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Taller BJ. Distribution, biosynthesis, and function of cytokinins in tRNA. In: DWS Mok, Mok MC., editors. Cytokinins: Chemistry, Activity and Function. Boca Raton, FL:: CRC Press; 1994. pp. 101–112. (editors) [Google Scholar]
- 42.Samanovic MI, Tu S, Novák O, Iyer LM, McAllister FE, et al. Proteasomal control of cytokinin synthesis protects Mycobacterium tuberculosis against nitric oxide. Mol Cell. 2015;57:984–994. doi: 10.1016/j.molcel.2015.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Buschmann C, Lichtenthaler HK. The effect of cytokinins on growth and pigment accumulation of radish seedlings (Raphanus sativus L.) grown in the dark and at different light quanta fluence rates. Photochem Photobiol. 1982;35:217–221. doi: 10.1111/j.1751-1097.1982.tb03835.x. [DOI] [Google Scholar]
- 44.Gemmrich A, Kayser H. Hormone induced changes in carotenoid composition in Ricinus cell cultures. II. Accumulation of rhodoxanthin during auxin-controlled chromoplast differentiation. Z Naturforsch. 1984;39c:753–757. [Google Scholar]
- 45.Czerpak R, Bajguz A. Stimulatory effect of auxins and cytokinins on carotenes, with differential effects on xanthophylls in the green alga Chlorella pyrenoidosa chick. Acta Societatis Botanicorum Poloniae. 1997;66:41–46. doi: 10.5586/asbp.1997.006. [DOI] [Google Scholar]
- 46.Morris RO, Powell GK. Genes specifying cytokinin biosynthesis in prokaryotes. Bioessays. 1987;6:23–28. doi: 10.1002/bies.950060107. [DOI] [Google Scholar]
- 47.Akiyoshi DE, Regier DA, Gordon MP. Cytokinin production by Agrobacterium and Pseudomonas spp. J Bacteriol. 1987;169:4242–4248. doi: 10.1128/jb.169.9.4242-4248.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Iwai M, Maoka T, Ikeuchi M, Takaichi S. 2,2'-Beta-hydroxylase (CrtG) is involved in carotenogenesis of both nostoxanthin and 2-hydroxymyxol 2'-fucoside in Thermosynechococcus elongatus strain BP-1. Plant Cell Physiol. 2008;49:1678–1687. doi: 10.1093/pcp/pcn142. [DOI] [PubMed] [Google Scholar]
- 49.Lotan T, Hirschberg J. Cloning and expression in Escherichia coli of the gene encoding β-C-4-oxygenase, that converts β-carotene to the ketocarotenoid canthaxanthin in Haematococcus pluvialis. FEBS Lett. 1995;364:125–128. doi: 10.1016/0014-5793(95)00368-j. [DOI] [PubMed] [Google Scholar]
- 50.Kajiwara S, Kakizono T, Saito T, Kondo K, Ohtani T, et al. Isolation and functional identification of a novel cDNA for astaxanthin biosynthesis from Haematococcus pluvialis, and astaxanthin synthesis in Escherichia coli. Plant Mol Biol. 1995;29:343–352. doi: 10.1007/BF00043657. [DOI] [PubMed] [Google Scholar]
- 51.Misawa N, Kajiwara S, Kondo K, Yokoyama A, Satomi Y, et al. Canthaxanthin biosynthesis by the conversion of methylene to keto groups in a hydrocarbon β-carotene by a single gene. Biochem Biophys Res Commun. 1995;209:867–876. doi: 10.1006/bbrc.1995.1579. [DOI] [PubMed] [Google Scholar]
- 52.Rw Y, Stead KJ, Yao H, He H. Mutational and functional analysis of the β-carotene ketolase involved in the production of canthaxanthin and astaxanthin. Appl Environ Microbiol. 2006;72:5829–5837. doi: 10.1128/AEM.00918-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Klassen JL. Phylogenetic and evolutionary patterns in microbial carotenoid biosynthesis are revealed by comparative genomics. PLoS One. 2010;5:e11257. doi: 10.1371/journal.pone.0011257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Misawa N. Carotenoid β-ring hydroxylase and ketolase from marine bacteria-promiscuous enzymes for synthesizing functional xanthophylls. Mar Drugs. 2011;9:757–771. doi: 10.3390/md9050757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hundle B, Alberti M, Nievelstein V, Beyer P, Kleinig H, et al. Functional assignment of Erwinia herbicola Eho10 carotenoid genes expressed in Escherichia coli. Mol Gen Genet. 1994;245:406–416. doi: 10.1007/BF00302252. [DOI] [PubMed] [Google Scholar]
- 56.Larsen R, Wilson M, Guss A, Metcalf W. Genetic analysis of pigment biosynthesis in Xanthobacter autotrophicus Py2 using a new, highly efficient transposon mutagenesis system that is functional in a wide variety of bacteria. Arch Microbiol. 2002;178:193–201. doi: 10.1007/s00203-002-0442-2. [DOI] [PubMed] [Google Scholar]
- 57.Seo YB, Choi SS, Nam SW, Lee JH, Kim YT. Cloning and characterization of the zeaxanthin glucosyltransferase gene (crtX) from the astaxanthin-producing marine bacterium, Paracoccus haeundaensis. J Microbiol Biotechnol. 2009;19:1542–1546. doi: 10.4014/jmb.0904.04033. [DOI] [PubMed] [Google Scholar]
- 58.Asker D, Beppu T, Ueda K. Sphingomonas astaxanthinifaciens sp. nov., a novel astaxanthin-producing bacterium of the family Sphingomonadaceae isolated from Misasa, Tottori, Japan. FEMS Microbiol Lett. 2007;273:140–148. doi: 10.1111/j.1574-6968.2007.00760.x. [DOI] [PubMed] [Google Scholar]
- 59.Kim JH, Kim SH, Kim KH, Lee PC Sphingomonas lacus sp. nov., an astaxanthin-dideoxyglycoside-producing species isolated from soil near a pond. Int J Syst Evol Microbiol. 2015;65:2824–2830. doi: 10.1099/ijs.0.000337. [DOI] [PubMed] [Google Scholar]
- 60.Asker D, Amano S, Morita K, Tamura K, Sakuda S, et al. Astaxanthin dirhamnoside, a new astaxanthin derivative produced by a radio-tolerant bacterium, Sphingomonas astaxanthinifaciens. J Antibiot. 2009;62:397–399. doi: 10.1038/ja.2009.50. [DOI] [PubMed] [Google Scholar]
- 61.Kavanagh KL, Dunford JE, Bunkoczi G, Russell RG, Oppermann U. The crystal structure of human geranylgeranyl pyrophosphate synthase reveals a novel hexameric arrangement and inhibitory product binding. J Biol Chem. 2006;281:22004–22012. doi: 10.1074/jbc.M602603200. [DOI] [PubMed] [Google Scholar]
- 62.Umeno D, Tobias AV, Arnold FH. Diversifying carotenoid biosynthetic pathways by directed evolution. Microbiol Mol Biol Rev. 2005;69:51–78. doi: 10.1128/MMBR.69.1.51-78.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Matsumoto M, Iwama D, Arakaki A, Tanaka A, Tanaka T, et al. Altererythrobacter ishigakiensis sp. nov., an astaxanthin-producing bacterium isolated from a marine sediment. Int J Syst Evol Microbiol. 2011;61:2956–2961. doi: 10.1099/ijs.0.024729-0. [DOI] [PubMed] [Google Scholar]
- 64.Huang HD, Wang W, Ma T, Li GQ, Liang FL, et al. Sphingomonas sanxanigenens sp. nov., isolated from soil. Int J Syst Evol Microbiol. 2009;59:719–723. doi: 10.1099/ijs.0.000257-0. [DOI] [PubMed] [Google Scholar]
- 65.Wei S, Wang T, Liu H, Zhang C, Guo J, et al. Sphingomonas hengshuiensis sp. nov., isolated from lake wetland. Int J Syst Evol Microbiol. 2015;65:4644–4649. doi: 10.1099/ijsem.0.000626. [DOI] [PubMed] [Google Scholar]
- 66.Schmidt-Dannert C, Umeno D, Arnold FH. Molecular breeding of carotenoid biosynthetic pathways. Nat Biotechnol. 2000;18:750–753. doi: 10.1038/77319. [DOI] [PubMed] [Google Scholar]
- 67.Arrach N, Fernández-Martín R, Cerdá-Olmedo E, Avalos J. A single gene for lycopene cyclase, phytoene synthase, and regulation of carotene biosynthesis in Phycomyces. Proc Natl Acad Sci USA. 2001;98:1687–1692. doi: 10.1073/pnas.98.4.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Maresca JA, Graham JE, Wu M, Eisen JA, Bryant DA. Identification of a fourth family of lycopene cyclases in photosynthetic bacteria. Proc Natl Acad Sci USA. 2007;104:11784–11789. doi: 10.1073/pnas.0702984104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xu Z, Tian B, Sun Z, Lin J, Hua Y. Identification and functional analysis of a phytoene desaturase gene from the extremely radioresistant bacterium Deinococcus radiodurans. Microbiology. 2007;153:1642–1652. doi: 10.1099/mic.0.2006/002857-0. [DOI] [PubMed] [Google Scholar]
- 70.Misawa N, Yamano S, Ikenaga H. Production of β-carotene in Zymomonas mobilis and Agrobacterium tumefaciens by introduction of the biosynthesis genes from Erwinia uredovora. Appl Environ Microbiol. 1991;57:1847–1849. doi: 10.1128/aem.57.6.1847-1849.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Verwaal R, Wang J, Meijnen JP, Visser H, Sandmann G, et al. High-level production of beta-carotene in Saccharomyces cerevisiae by successive transformation with carotenogenic genes from Xanthophyllomyces dendrorhous. Appl Environ Microbiol. 2007;73:4342–4350. doi: 10.1128/AEM.02759-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wassef L, Wirawan R, Chikindas M, Breslin PA, Hoffman DJ, et al. β-Carotene-producing bacteria residing in the intestine provide vitamin A to mouse tissues in vivo. J Nutr. 2014;144:608–613. doi: 10.3945/jn.113.188391. [DOI] [PubMed] [Google Scholar]
- 73.Berry A, Janssens D, Hümbelin M, Jore JP, Hoste B, et al. Paracoccus zeaxanthinifaciens sp. nov., a zeaxanthin-producing bacterium. Int J Syst Evol Microbiol. 2003;53:231–238. doi: 10.1099/ijs.0.02368-0. [DOI] [PubMed] [Google Scholar]
- 74.Hameed A, Shahina M, Lin SY, Lai WA, Hsu YH, et al. Aquibacter zeaxanthinifaciens gen. nov., sp. nov., a zeaxanthin-producing bacterium of the family Flavobacteriaceae isolated from surface seawater, and emended descriptions of the genera Aestuariibaculum and Gaetbulibacter. Int J Syst Evol Microbiol. 2014;64:138–145. doi: 10.1099/ijs.0.052621-0. [DOI] [PubMed] [Google Scholar]
- 75.Lee JH, Hwang YM, Baik KS, Choi KS, Ka JO, et al. Mesoflavibacter aestuarii sp. nov., a zeaxanthin-producing marine bacterium isolated from seawater. Int J Syst Evol Microbiol. 2014;64:1932–1937. doi: 10.1099/ijs.0.061085-0. [DOI] [PubMed] [Google Scholar]
- 76.Zhang W, Hu X, Wang L, Wang X. Reconstruction of the carotenoid biosynthetic pathway of Cronobacter sakazakii BAA894 in Escherichia coli. PLoS One. 2014;9:e86739. doi: 10.1371/journal.pone.0086739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Vancanneyt M, Schut F, Snauwaert C, Goris J, Swings J, et al. Sphingomonas alaskensis sp. nov., a dominant bacterium from a marine oligotrophic environment. Int J Syst Evol Microbiol. 2001;51:73–79. doi: 10.1099/00207713-51-1-73. [DOI] [PubMed] [Google Scholar]
- 78.Oelschlägel M, Rückert C, Kalinowski J, Schmidt G, Schlömann M, et al. Sphingopyxis fribergensis sp. nov., a soil bacterium with the ability to degrade styrene and phenylacetic acid. Int J Syst Evol Microbiol. 2015;65:3008–3015. doi: 10.1099/ijs.0.000371. [DOI] [PubMed] [Google Scholar]
- 79.Hatanaka T, Asahi N, Tsuji M. Purification and characterization of poly(vinyl alcohol) dehydrogenase from Pseudomonas sp. 113P3. Biosci Biotechnol Biochem. 1995;59:1813–1816. doi: 10.1271/bbb.59.1813. [DOI] [Google Scholar]
- 80.Takeuchi M, Kawai F, Shimada Y, Yokota A. Taxonomic study of polyethylene glycol-utilizing bacteria: emended description of the genus Sphingomonas and new descriptions of Sphingomonas macrogoltabidus sp. nov., Sphingomonas sanguis sp. nov. and Sphingomonas terrae sp. nov. Syst Appl Microbiol. 1993;16:227–238. doi: 10.1016/S0723-2020(11)80473-X. [DOI] [Google Scholar]
- 81.Pollock TJ. Gellan-related polysaccharides and the genus Sphingomonas. J Gen Microbiol. 1993;139:1939–1945. doi: 10.1099/00221287-139-8-1939. [DOI] [Google Scholar]
- 82.Yabuuchi E, Yano I, Oyaizu H, Hashimoto Y, Ezaki T, et al. Proposals of Sphingomonas paucimobilis gen. nov. and comb. nov., Sphingomonas parapaucimobilis sp. nov., Sphingomonas yanoikuyae sp. nov., Sphingomonas adhaesiva sp. nov., Sphingomonas capsulata comb. nov., and two genospecies of the genus Sphingomonas. Microbiol Immunol. 1990;34:99–119. doi: 10.1111/j.1348-0421.1990.tb00996.x. [DOI] [PubMed] [Google Scholar]
- 83.Nohynek LJ, Suhonen EL, Nurmiaho-Lassila E-L, Hantula J, Salkinoja-Salonen M. Description of four pentachlorophenol-degrading bacterial strains as Sphingomonas chlorophenolica sp. nov. Syst Appl Microbiol. 1995;18:527–538. doi: 10.1016/S0723-2020(11)80413-3. [DOI] [Google Scholar]
- 84.Ito M, Prokop Z, Klvana M, Otsubo Y, Tsuda M, et al. Degradation of β-hexachlorocyclohexane by haloalkane dehalogenase LinB from γ-hexachlorocyclohexane-utilizing bacterium Sphingobium sp. MI1205. Arch Microbiol. 2007;188:313–325. doi: 10.1007/s00203-007-0251-8. [DOI] [PubMed] [Google Scholar]
- 85.Sun J-Q, Huang X, Chen Q-L, Liang B, Qiu J-G, et al. Isolation and characterization of three Sphingobium sp. strains capable of degrading isoproturon and cloning of the catechol 1,2-dioxygenase gene from these strains. World J Microbiol Biotechnol. 2009;25:259–268. doi: 10.1007/s11274-008-9888-y. [DOI] [Google Scholar]
- 86.Balkwill DL, Drake GR, Reeves RH, Fredrickson JK, White DC, et al. Taxonomic study of aromatic-degrading bacteria from deep-terrestrial-subsurface sediments and description of Sphingomonas aromaticivorans sp. nov., Sphingomonas subterranea sp. nov., and Sphingomonas stygia sp. nov. Int J Syst Bacteriol. 1997;47:191–201. doi: 10.1099/00207713-47-1-191. [DOI] [PubMed] [Google Scholar]
- 87.Fan ZY, Xiao YP, Hui W, Tian GR, Lee JS, et al. Altererythrobacter dongtanensis sp. nov., isolated from a tidal flat. Int J Syst Evol Microbiol. 2011;61:2035–2039. doi: 10.1099/ijs.0.024380-0. [DOI] [PubMed] [Google Scholar]
- 88.Park SC, Baik KS, Choe HN, Lim CH, Kim HJ, et al. Altererythrobacter namhicola sp. nov. and Altererythrobacter aestuarii sp. nov., isolated from seawater. Int J Syst Evol Microbiol. 2011;61:709–715. doi: 10.1099/ijs.0.021196-0. [DOI] [PubMed] [Google Scholar]
- 89.Li ZY, Wu YH, Huo YY, Cheng H, Wang CS, et al. Complete genome sequence of a benzo[a]pyrene-degrading bacterium Altererythrobacter epoxidivorans CGMCC 1.7731T. Mar Genomics. 2016;25:39–41. doi: 10.1016/j.margen.2015.11.009. [DOI] [PubMed] [Google Scholar]
- 90.Wu YH, Xu L, Meng FX, Zhang DS, Wang CS, et al. Altererythrobacter atlanticus sp. nov., isolated from deep-sea sediment. Int J Syst Evol Microbiol. 2014;64:116–121. doi: 10.1099/ijs.0.052951-0. [DOI] [PubMed] [Google Scholar]
- 91.Oh HM, Giovannoni SJ, Ferriera S, Johnson J, Cho JC. Complete genome sequence of Erythrobacter litoralis HTCC2594. J Bacteriol. 2009;191:2419–2420. doi: 10.1128/JB.00026-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhuang L, Wang W, Shao Z, Liu Y, Wang L. Erythrobacter atlanticus sp. nov., a bacterium from ocean sediment able to degrade polycyclic aromatic hydrocarbons. Int J Syst Evol Microbiol. 2015;65:3714–3719. doi: 10.1099/ijsem.0.000481. [DOI] [PubMed] [Google Scholar]
- 93.Notomista E, Pennacchio F, Cafaro V, Smaldone G, Izzo V, et al. The marine isolate Novosphingobium sp. PP1Y shows specific adaptation to use the aromatic fraction of fuels as the sole carbon and energy source. Microb Ecol. 2011;61:582–594. doi: 10.1007/s00248-010-9786-3. [DOI] [PubMed] [Google Scholar]
- 94.Sohn JH, Kwon KK, Kang JH, Jung HB, Kim SJ. Novosphingobium pentaromativorans sp. nov., a high-molecular-mass polycyclic aromatic hydrocarbon-degrading bacterium isolated from estuarine sediment. Int J Syst Evol Microbiol. 2004;54:1483–1487. doi: 10.1099/ijs.0.02945-0. [DOI] [PubMed] [Google Scholar]
- 95.Wang H, Xie C, Zhu P, Zhou NY, Lu Z. Two novel sets of genes essential for nicotine degradation bySphingomonas melonis TY. Front Microbiol. 2016;7:2060. doi: 10.3389/fmicb.2016.02060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Seo SH, Lee SD. Altererythrobacter marensis sp. nov., isolated from seawater. Int J Syst Evol Microbiol. 2010;60:307–311. doi: 10.1099/ijs.0.011031-0. [DOI] [PubMed] [Google Scholar]
- 97.Huang Y, Zeng Y, Feng H, Wu Y, Xu X. Croceicoccus naphthovorans sp. nov., a polycyclic aromatic hydrocarbons-degrading and acylhomoserine-lactone-producing bacterium isolated from marine biofilm, and emended description of the genus Croceicoccus. Int J Syst Evol Microbiol. 2015;65:1531–1536. doi: 10.1099/ijs.0.000132. [DOI] [PubMed] [Google Scholar]
- 98.Page M, Landry N. Bacterial mass production of taxanes with Erwinia. US Patent. 1996:5561055 [Google Scholar]
- 99.Pal R, Bala S, Dadhwal M, Kumar M, Dhingra G, et al. Hexachlorocyclohexane-degrading bacterial strains Sphingomonas paucimobilis B90A, UT26 and Sp+, having similar lin genes, represent three distinct species, Sphingobium indicum sp. nov., Sphingobium japonicum sp. nov. and Sphingobium francense sp. nov., and reclassification of [Sphingomonas] chungbukensis as Sphingobium chungbukense comb. nov. Int J Syst Evol Microbiol. 2005;55:1965–1972. doi: 10.1099/ijs.0.63201-0. [DOI] [PubMed] [Google Scholar]
- 100.Li Y, Chen Q, Wang CH, Cai S, He J, et al. Degradation of acetochlor by consortium of two bacterial strains and cloning of a novel amidase gene involved in acetochlor-degrading pathway. Bioresour Technol. 2013;148:628–631. doi: 10.1016/j.biortech.2013.09.038. [DOI] [PubMed] [Google Scholar]
- 101.Yabuuchi E, Yamamoto H, Terakubo S, Okamura N, Naka T, et al. Proposal of Sphingomonas wittichii sp. nov. for strain RW1T, known as a dibenzo-p-dioxin metabolizer. Int J Syst Evol Microbiol. 2001;51:281–292. doi: 10.1099/00207713-51-2-281. [DOI] [PubMed] [Google Scholar]
- 102.García-Romero I, Pérez-Pulido AJ, González-Flores YE, Reyes-Ramírez F, Santero E, et al. Genomic analysis of the nitrate-respiring Sphingopyxis granuli (formerly Sphingomonas macrogoltabida) strain TFA. BMC Genomics. 2016;17:93. doi: 10.1186/s12864-016-2411-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.