Graphical abstract
Our study aims to exploit the denitrifying bacteria MMT and its NAP enzyme in dual missions, nitrate removal from wastewater and nanoparticles synthesis. Therefore, the crud NAP enzyme was characterized and immobilized by entrapment technique for aforementioned application. To the best of author knowledge, the immobilization for denitrifying bacteria and NAP enzyme for simultaneous bioremediation and bionanoparticles synthesis was not studied previously.
Keywords: Immobilization, Nitrate reductase, Nanoparticles, Denitrification, Bioremediation
Highlights
-
•
This study employed the immobilized strain MMT and its NAP enzyme in concurrent denitrification and NPs synthesis.
-
•
The properties of crude NAP enzyme were assessed at different ranges of pH and temperatures and also its stability at 4 °C and 30 °C was studied.
-
•
The effect of several types of additives were evaluated at concentration rang (1 mM, 2.5 mM, 5 mM and 10 mM).
-
•
Concurrently, the immobilized MMT cells completely removed NO3− upon 8th day with AgNPs synthesis ranging from 23.26 to 58.14.
-
•
Immobilized NAP exhibited lower efficiency with 28.6% of NO3 elimination and large aggregated AgNPs ranging from 94.44 nm to 172.22 nm.
Abstract
The periplasmic nitrate reductase enzyme (NAP) has become attractive catalyst, whose exploitation has emerged as one of the indispensable strategies toward environmentally benign applications. To achieve them efficiently and overcome the sensitivity of NAP in harsh environmental circumstances, the immobilization for denitrifying bacteria and NAP enzyme for simultaneous bioremediation and bionanoparticles synthesis was studied. NAP catalyzed NO3− reduction at Vmax of 0.811 μM/min and Km of 14.02 mM. Concurrently, the immobilized MMT cells completely removed NO3- upon 192 h with AgNPs synthesis ranging from 23.26 to 58.14 nm as indicated by SEM. Wherase, immobilized NAP exhibited lower efficiency with 28.6% of NO3− elimination within 288 h and large aggregated AgNPs ranging from 94.44 nm to 172.22 nm. To the best of author knowledge, the immobilization for denitrifying bacteria and NAP enzyme for simultaneous bioremediation and bionanoparticles synthesis was not studied before.
1. Introduction
Currently, due to the water scarcity problem, especially with rapidly increasing in population and consumption, governments directed toward the utilization of non-conventional water resources such as recycled drainage water and treated Sewage water. It is plausible that such types of resources filled with heavy metals, organic and inorganic contaminants from varied sources such as agricultural fertilizers residues, pesticides, mining and smelting manufacturing. Silver and nitrate are among the pollutants that usually present in porcelain, ink, photographic, electroplating and manufacturing industries. Many physical and chemical methods were assigned for their removal from wastewater as reported by [1]. However these approaches restricted their utility due to expensiveness and waste brine accumulation. Hence, it is inevitable to utilize ecofriendly methods to remove nitrate and silver from effluents. The exploitation of biological catalysts (microorganisms and their enzymes) impregnated with new immobilization techniques would be resulted in effective nitrate and silver removal through denitrification and AgNPs synthesis simultaneously. Therefore, the main target of technology will be achieved in a cost-effective manner with low time consuming.
It is noteworthy that nitrate reductases (NRs) enzymes catalyze nitrate reduction and have vital environmental, agricultural and public health implications [2]. In prokaryotes, three classes of NRs have been identified : assimilatory nitrate reductases (NAS) and two dissimilatory classes of (respiratory membrane bound nitrate reductases (NAR) and periplasmic nitrate reductases (NAP). In spite of the disparity in physiological function, structure, level of cellular localization, biochemical charactristics, organization and regulation genes, but all of them are molybdenum dependent enzymes, where the molybdenum (Mo) and molybdenum cofactor (Mo-co) present in the enzyme active center as the molybdopterin guanine dinucleotide (MGD) [3].
NAP was detected in phototrophic, denitrifying bacteria and widely spread among gram-negative bacteria, including Alcaligenes eutrophus (Ralstonia eutropha), Paracoccus pantotrophus, Wolinella succinogenes, P. denitrificans, E. coli, and Rhodobacter species [2]. Different physiological functions were exhibited by NAP in different organisms or even in the same bacterium under different metabolic conditions. NAP enzyme considers being responsible for aerobic denitrification, adaptation to anaerobic metabolism after transition from aerobic conditions and microaerobically growth. Besides, maintenance of the redox balance, oxidative metabolism of highly reduced carbon substrates in aerobic heterotrophs and anaerobic photoheterotrophic growth of photosynthetic bacteria [[4], [5], [6]].
Generally, NRs are the building block in the denitrification pathway in conjugation with series of different enzymes such as nitrite reductases (Nir), nitric oxide reductase (Nor) and nitrous oxide reductase (Nos) [7]. In the recent decade, bionanotechnology has emerged as integration among biotechnology and nanotechnology for developing environmentally benign technology [8]. Interestingly, oxidoreductases, particularly NRs are among several biomolecules that contribute mainly in biosynthesis of various nanoparticles (NPs) [[9], [10], [11]].
The high cost of enzymes isolation and purification, besides their fragility and sensitivity to adverse environmental conditions leads to limiting its operational lifetime. The immobilization technique overcomes most of the constraints that limit application of living cell or their biomolecules especially in bioremediation or wastewater treatment [12]. Th microencapsulation on suitable materials is one of the modern and vital methods in bioprocess biotechnology and subsequent wide applications. The maintenance of high cell viability and the catalytic activity of enzymes at a maximum level were the main goals of immobilization [13]. The encapsulation approach involves bidirectional diffusion of molecules such as the influx of oxygen and nutrients that are essential for cell metabolism, concomitantly, the outward diffusion of waste products and secondary metabolites [13]. Entrapment is a simple, physically stable, easy production, cost effective technique and efficient removal from the reaction medium for reuse [14]. The nontoxic nature of alginate polymer prompts its application in heavy metals remediation
In the light of that mentioned previously, our study aims to exploit the denitrifying bacteria Achromobacter sp. MMT and its NAP enzyme in dual mission, nitrate removal from wastewater and NPs synthesis. Therefore, the crud NAP enzyme was characterized and immobilized by entrapment technique for aformentioned application. To the best of author knowledge, the immobilization for denitrifying bacteria Achromobacter sp. and NAP enzyme for simultaneous bioremediation and bionanoparticles synthesis was not studied previously.
2. Material and methods
2.1. Medium composition and growth conditions
The bacterial strain Achromobacter sp. MMT was isolated from Mariot Lake basin 3, Alexandria, Egypt. The strain MMT was one of the previously screened isolates exhibiting denitrification in addition to bionanaoparticle synthesis capability [15]. The maximum NAP activity was obtained by inoculation of 0.5 McFarland (≈108 CFU/ml) of MMT on optimized media reported by [16] at 30 °C in an orbital shaker (STUART SI500) at 150 rpm.
2.2. Crud enzyme preparation
The exponentially growing cells were harvested by centrifugation (10,000g, 20 min). The cells were washed three times in a potassium phosphate buffer 80 mM (pH = 7) and disrupted by mild osmotic shock through suspending the bacterial pellets in ice cold lysis buffer containing (0.1 M Tris−HCl buffer, pH 8.0, 5mMEDTA and 0.5 M sucrose) using 1 ml of the solution for each 0.1 g m of pellet and the mixture were mixed thoroughly and softly for 20 min at 30 °C. Then, Lysozyme (20 mg/ml) was added and the cells were stirred vigorously by vortex for further 20 min at room temperature. Additionally, about 50 μg/ml of DNAse was added to reduce the viscosity caused by DNA release. The resulting suspension was centrifuged for 10 min at 5000 g to remove cell debris and unbroken cells. Spheroplast (supernatant fraction) obtained was subjected for centrifugation at 100,000 g 4 °C for 60 min. The supernatant was saved as the crude soluble periplasmic fraction [17,18,19].
2.3. Enzyme assay
NAP activity was determined based on diazotization of nitrite. Nitrite liberated by reduction of nitrate by NAP using dithionite reduced benzyle viologen as an artificial electron donor. The reaction mixture and incubation conditioned was described in details previously by [20]. The nitrite concentration in the mixture was measured spectrophotometrically at 540 nm (T60 UV/VIS spectrophotometer). One unit of NAP activity corresponds to the amount of enzyme that catalyzes the formation of 1 μmol of nitrite per minute or 1 μmol of nitrate reduced per minute under standard assay conditions [21].
2.4. Determination of specific activity
Protein concentration was determined according to the original method of Lowry et al. [22]. The bovine serum albumin as a standard was routinely used. Crude NR was also measured by the same method. Specific activity of the NR was calculated by the activity of NR per milligram of protein per minute [23] according to the following equation:
| Specific activity = NR activity * (Protein concentration)−1 * (min)−1 |
2.5. Determination of enzyme properties
2.5.1. Activity profiles at different temperatures and pH
The assays were performed as described above. Temperature activity profile was determined by incubating the reaction mixture in a water bath at 4, 10, 20, 30, 40, 50, 60 and 70 °C for 30 min. For pH activity profile, the reaction mixtures were examined using Citrate-phosphate (pH 2–6), sodium phosphate, pH, 7.0, Trise−HCl pH 8 and glycine NaOH (pH 9–13). Enzymatic activity was measured and plotted against the temperature and pH [24,25].
2.5.2. Determination of NAP stability at 4 °C and 30 °C
By incubating the crude enzyme in phosphate buffer pH 7 at 30 °C and 4 °C for 240 h, the activity was measured each 12 h time interval under the standard assay. The relative activity was determined (The residual or relative activity was measured as a percentage (%) by comparing it with the control at optimum conditions). The relative activity was plotted against each time interval [25,26].
2.5.3. Effect of different additives, metals, anions, cations and detergents on NAP activity
The effects of inhibitors (sodium azide, potassium cyanide, sodium thiocyanate, DTT (Dithiothreitol), PMSF (phenylinethylsulfonylfluoride), ß-mercaptoethanol, H2O2, phenol and urea), metal ions (Ag 1+, Cu 2+, Co 2+, Fe 3+, Fe 2+, Mn 2+, Zn 2+, Cr +3, Cd +2, Hg +2, Pb +2 and Ni +2), cations (Na+, K+, NH4+, Mg2+, Ca2+ and Ba2+) and anions (Cl−, I−, SO42- and CO32-) and chelating agents (EDTA and 8-Hydroxyquinolon (8HQ)) were examined in concentrations of (1, 2.5, 5 and 10 mM), using the enzyme assay procedure. Additionally, detergents (SLS, Tween-20 and Triton-X-100) and organic solvent such as DMSO were examined in concentrations (0.5, 1, 3 and 5%). The relative enzyme activities were expressed in percentages by comparing with the control assay mixture, without any additions [26]. Activity inhibition percentage or stimulation % of the enzyme activities were calculated relative to the controls using the following formula [27]:
| Inhibition (%) = [(EC-ET) /EC]*100 |
Where, EC: Enzyme activity of the control, ET: Enzyme activity of the treatment.
2.5.4. Preservation of NAP in −20 °C
Long storage of crude enzyme was performed by incubating it in a phosphate buffer (pH 7) at −20 °C in the absence and presence of preservation buffer (20% glycerol and 0.4 mM EDTA) for 3 months. The activity was measured weekly as time interval under the standard assay. The relative activity was determined as previously. The relative activity was plotted against each time interval [28].
2.5.5. Determination of kinetic parameters (Km and Vmax)
Effect of substrate concentration was determined using different concentrations of nitrate in the range of 10 mM to 100 mM under optimum incubation conditions. The kinetic parameters Km & Vmax were calculated from Lineweaver Burk plot. A Lineweaver Burk plot was made between reciprocal of nitrate concentration (I/[S]) vs reciprocal of enzyme activity (1/V). Apparent Km & Vmax were calculated from the plot or from Lineweaver-Burk equation: [29].
| 1/ V = (Km/Vmax) (1/ S) + 1/Vmax |
2.6. Immobilization of MMT-cells and NAP-enzyme
The cells of 6 × 108 CFU/ml of strain MMT and its corresponding NAP crud extract (100 mg protein with 900 U/mg-NAP activity) were immobilized using the entrapment approach in calcium alginate beads. Slurry of 4% solutions of sodium alginate were prepared and mixed with equal volume of MMT-cells or crude extract of NAP (25 ml for each) to get the final concentration of sodium alginate 2%. The entrapped cells or enzyme in calcium alginate gel was poured drop wise through a sterile syringe into a beaker containing 0.5 M chilled sterile CaCl2 with gentle stirring on a magnetic stirrer [30]. Hardening of beads was occurring through 15–30 min at room temperature. The hardened beads washed and preserved in sterile distilled water [14,31]. Benzyle viologen was immobilized with NR as the electron donor in alginate beads.
2.6.1. Concurrent denitrification and AgNPs synthesis via immobilized MMT-cells and NAP-enzyme
The beads entrapped with MTT-cells / NAP-enzyme were applied in sterile distilled water artificially inoculated with 3 mM of AgNO3 for 288 h (12 days). The concentrations of NO3− and NO2− were determined according to [32] every 12 h interval [33,34]. For assuring AgNPs synthesis by both systems, the immobilized beads were examined SEM, EDX and XRD at the end of the experiment.
2.6.1.1. Scanning electron microscopy (SEM)
The MTT-cells and NAP-enzyme immobilized beads were fixed overnight with 2.5% glutaraldehyde, then dehydrated in a series of graded ethanol, finally with hexamethyldisilazane and drop coated on silicon wafers. Air dried samples were sputter coated with gold and observed under SEM (JEOL JEM-1230, Japan).
2.6.1.2. Dispersive X-ray spectra (EDX)
The elemental composition of both immobilized systems was examined using EDX (JEOL JSM 6360 L A, Japan – Electron Microscope Unit, Faculty of Science- Alexandria University) scanning electron microscope equipped with EDX controlled system.
2.6.1.3. X-ray diffraction analysis (XRD)
The crystallographic identity, crystalline state and purity of both examined immobilized systems were determined by X-ray diffraction. The samples were exposed to Cu Kα radiation (λ = 1.504 A°) in X-ray diffractometer (Schimadzu-7000, USA). The voltage and current of the X-ray source were 30 kV and 30 mA; respectively. The instrument was operated in continuous mode in increments of 4°/min and scanned over wide range of Bragg angles 10°≤ 2θ ≤ 120.
3. Results and discussion
3.1. Determination of enzyme properties
3.1.1. Activity profiles at different temperatures and pH
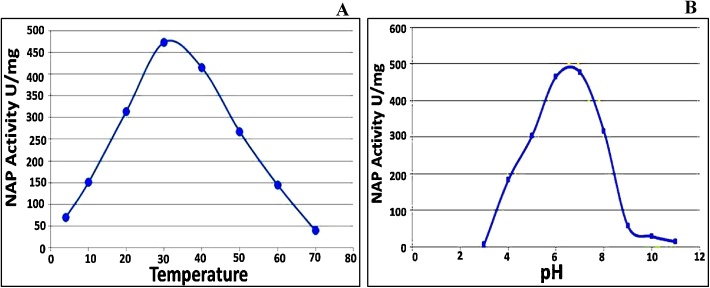
NAP enzyme catalyzed NO3− reduction in the temperature range of 4–70 °C and optimally at 30 °C–40 °C with maximum activity 487.43 and 414.64 U/mg, respectively. On the other hands, the NAP activity in below/above optimum temperature was detectable but in a lower value as illustrated in Fig. 1A. Clearly, at lower temperature (4 °C and 10 °C), low kinetic energy of nitrate molecules is displayed and subsequently move into the NAP active site more slowly. While, increasing temperature elevates kinetic energy of nitrate molecules which causes more random collisions between nitrate molecules per unit time. This leads to increase the rate of reaction and formation of more product (nitrite), which was expressed at (30 °C and 40 °C). Upon temperature elevation, the cleavage in weaker bonds, especially the hydrogen and ionic bonds within NAP enzyme occurred which thereafter led to changing in conformational secondary and tertiary structures of enzyme active site and eventually enzyme denaturation as documented by [35].
Fig. 1.
The activity profile of NAP enzyme of strain MMT under different ranges of temperature (A) and pH (B).
Additionally, NAP activity was optimally active at pH 6–7 with 477 and 465 U/mg, respectively (Fig. 1B). The data indicated that NAP enzyme was still active, but with gradual decrease at both acidic and alkaline conditions (pH 4–8). However, the absence of NAP activity at pH (3 & 11) could be attributed to the changes in enzyme configuration due to alteration in the active sites [36]. Besides, adversely influence of the reducibility of benzyle viologen at extreme pH levels as reported by [37].
3.1.2. Determination of NAP stability at 4 °C and 30 °C
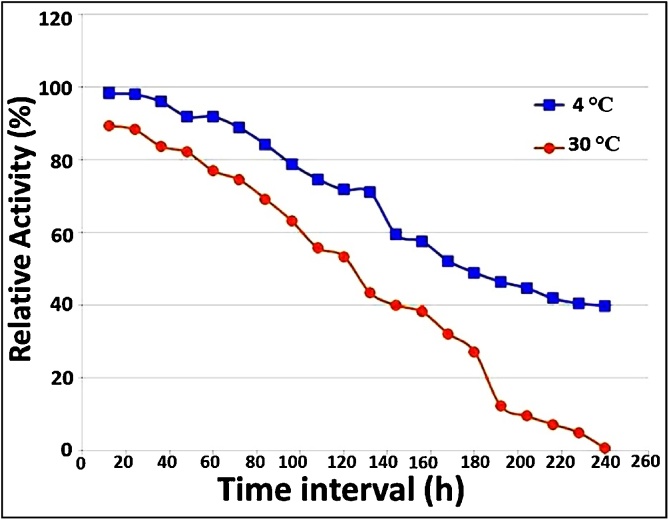
The stabilization of NAP enzyme at different temperature enables its subsequent employment at various seasons that accompanying with temperature fluctuations. As demonstrated in Fig. 2, the NAP activity was more stable at 4 °C than at 30 °C. In addition, NAP enzyme was able to retain more than 91% and 77% of residual activity up to 60 h at 4 °C and 30 °C, respectively. In accordance with our finding, [38] reported that NAR derived from Escherichia coli (E.C. 1.9.6.1) showed higher stablity at 6 °C than at 23 °C; while, lost complete activity after 9 days of incubation.
Fig. 2.
Relative activity (%) at 4 °C and 30 °C for 240 h.
3.1.3. Effect of different additives, metals, anions, cations and detergents on NAP activity
The influence of several categories of inhibitors on the activity of NAP was indicated in Table 1. In coincidence with our results, [36] documented the inactivation of partially purified NAP of Pseudomonas sp. SH7 by applying 500 μM of cyanide, while the exact concentration of azide caused a 76% reduction in NAP activity.
Table 1.
Inhibitors effect on NR activity.
| Inhibitor | Concentration | Relative activity (%) |
|---|---|---|
| Sodium Azid | 1.0 mM | 50.67 |
| 2.5 mM | 22.72 | |
| 5 mM | 8.81 | |
| 10 mM | 0 | |
| Potassium Cyanid | 1.0 mM | 39.74 |
| 2.5 mM | 13.61 | |
| 5 mM | 3.64 | |
| 10 mM | 0 | |
| Sodium Thiocyanate | 1.0 mM | 62.02 |
| 2.5 mM | 30.8 | |
| 5 mM | 11.5 | |
| 10 mM | 7.14 | |
| DTT | 1.0 mM | 43.68 |
| 2.5 mM | 9.1 | |
| 5 mM | 2.77 | |
| 10 mM | 0 | |
| PMSF | 1.0 mM | 36.84 |
| 2.5 mM | 6.92 | |
| 5 mM | 1.46 | |
| 10 mM | 0 | |
| H2O2 | 1.0 mM | 37.27 |
| 2.5 mM | 15.29 | |
| 5 mM | 5.61 | |
| 10 mM | 1.09 | |
| ß-mercaptoethanol | 1.0 mM | 20.02 |
| 2.5 mM | 0 | |
| 5 mM | 0 | |
| 10 mM | 0 | |
| Urea | 1 mM | 99.9 |
| 2.5 mM | 98.17 | |
| 5 mM | 62.18 | |
| 10 mM | 53.44 | |
| Phenol | 1 mM | 99.1 |
| 2.5 mM | 93.85 | |
| 5 mM | 46.74 | |
| 10 mM | 42.08 | |
Whereas, thiocyanate was un-competitive type of respiratory inhibitors in agreement with [39].
On the other hand, potent inhibition was noticed by serine protease inhibitor as PMSF, thiol inhibitors (reducing agents) as ß-mercaptoethanol and oxidizing agents represented by H2O2 by 63.16%, 79.98% and 63.72%, respectively at the lowest concentration examined. H2O2 caused inactivation by oxidation of sulfhydryl groups of amino acid residues close to metal-binding sites [40]. Consistent with our results, G. alkanivorans S7 NR was inactivated by thiol inhibitors [41]. Regarding to protein denaturant (urea) and phenolic compounds as phenol, mild or moderate inhibition was observed by 46.56% and 57.92% inhibition at the highest concentration applied.
Interestingly, NAP activity was slightly stimulated by 1 mM of Fe3+, Fe2+ and Cu+2 as highlighted in Table 2. Remarkably, oxidoreductases among enzymes that require some metals for stabilizing their protein structure, or act as an activator [42,43] such as Mo6+, Fe2+, Fe3+, Zn 2+, Mn2+, Co2+ and Cu2+. The presence of [4Fe-4S] cluster incorporated at a NAP active site in the catalytic subunit reflects enhancement role of iron ions in enzyme activity [5,44]. And conversely, the lethal effect of Cd2+, Ag+ and Hg2+ on NAP activity could be attributed to either their binding to sulfhydryl groups of the cysteine active site and forming of mercurial derivatives" mercaptide" that disrupts the surface configuration, or eventual substitution for the essential metal in the metalloprotein and so induce destabilization [[45], [46], [47]] reported that the high affinity of Hg2+ and Cd2+ to thiol group of NAP active site resulted in complete inactivation of NAP in strains E. coli and Pseudomonase sp. in concentration range from 0.2 to 1 mM.
Table 2.
Heavy Metals effect on NR activity.
| Heavy metal | concentration | Relative activity (%) |
|---|---|---|
| Fe 3+ | 1 mM | 101.09 |
| 2.5 mM | 92.73 | |
| 5 mM | 72.07 | |
| 10 mM | 53.16 | |
| Fe 2+ | 1 mM | 100.57 |
| 2.5 mM | 94.55 | |
| 5 mM | 93.09 | |
| 10 mM | 56.95 | |
| Ag + | 1 mM | 10.18 |
| 2.5 mM | 4 | |
| 5 mM | 0 | |
| 10 mM | 0 | |
| Mn 2+ | 1 mM | 72.72 |
| 2.5 mM | 70.76 | |
| 5 mM | 55.93 | |
| 10 mM | 37.31 | |
| Hg 2+ | 1 mM | 12.43 |
| 2.5 mM | 4.8 | |
| 5 mM | 1.45 | |
| 10 mM | 0 | |
| Zn 2+ | 1 mM | 98.88 |
| 2.5 mM | 72.73 | |
| 5 mM | 59.56 | |
| 10 mM | 45.24 | |
| Co 2+ | 1 mM | 66.25 |
| 2.5 mM | 58.98 | |
| 5 mM | 51.13 | |
| 10 mM | 37.67 | |
| Ni 2+ | 1 mM | 64.68 |
| 2.5 mM | 52.92 | |
| 5 mM | 49.11 | |
| 10 mM | 32.92 | |
| Cr 3+ | 1 mM | 45.52 |
| 2.5 mM | 40.73 | |
| 5 mM | 22.69 | |
| 10 mM | 7.93 | |
| Cu 2+ | 1 mM | 100.4 |
| 2.5 mM | 93.82 | |
| 5 mM | 78.55 | |
| 10 mM | 49.67 | |
| Cd 2+ | 1 mM | 9.96 |
| 2.5 mM | 3.27 | |
| 5 mM | 0 | |
| 10 mM | 0 | |
| Pb 2+ | 1 mM | 53.09 |
| 2.5 mM | 48.22 | |
| 5 mM | 35.42 | |
| 10 mM | 13.24 | |
The effects of anions and cations were presented in Table 3. Generally, at low concentrations of both, NAP activity appeared to be stabilized through nonspecific electrostatic interaction with the enzyme [48]. However, divalent cations reduced NAP activity by 40% via turning the system thermodynamically less stable. Hence induce changes in the nature, polarity of NAP surface, hydration and charged state which resuledt in the molecular configuration of the active site [48].
Table 3.
Cations, anions and chelating agents effect on NAP activity.
| Cations | Concentration | Relative activity (%) | Anions & chelating agents | Relative activity (%) |
|---|---|---|---|---|
| Na+ | 1 mM | 99.57 | SO42− | 99.57 |
| 2.5 mM | 90.04 | 70.56 | ||
| 5 mM | 89.61 | 69.99 | ||
| 10 mM | 89.26 | 69.84 | ||
| K+ | 1 mM | 99.71 | CO32− | 58.38 |
| 2.5 mM | 89.11 | 51.00 | ||
| 5 mM | 88.18 | 44.77 | ||
| 10 mM | 85.82 | 38.47 | ||
| NH4+ | 1 mM | 99.93 | Cl− | 99.57 |
| 2.5 mM | 92.34 | 90.04 | ||
| 5 mM | 91.05 | 89.61 | ||
| 10 mM | 88.11 | 89.26 | ||
| Mg2+ | 1 mM | 93.12 | I − | 77.01 |
| 2.5 mM | 65.26 | 58.17 | ||
| 5 mM | 62.68 | 56.23 | ||
| 10 mM | 62.20 | 43.91 | ||
| Ca2+ | 1 mM | 92.55 | EDTA | 89.74 |
| 2.5 mM | 66.40 | 62.26 | ||
| 5 mM | 61.46 | 54.85 | ||
| 10 mM | 61.39 | 32.01 | ||
| Ba 2+ | 1 mM | 92.34 | 8-Hydroxyquinolon (8HQ) |
38.47 |
| 2.5 mM | 64.61 | 22.83 | ||
| 5 mM | 58.24 | 7.34 | ||
| 10 mM | 56.52 | 1.03 | ||
Moreover, Table 3 results follow "Hofmeister series" which was ordered as follows: NH4+ < K+ < Na+ < Cs+ < Li+ < Mg2+ < Ca2+ < Ba2+ for cations and F− < SO42- < Cl− < Br − < I− < SCN− for anions. This series is an empirical ranking of how various ions influence protein solubility/aggregation, which in turn is related to the thermodynamic stability of the native state [49,50]. Hofmeister ions present on the left of the order called kosmotrope consider being more efficient in maintaining the protein stability than that present in the right order which calling chaotrope. So NAP exhibited stability at SO42- and Cl− than in presence of I− especially at higher concentrations [51]. It is noteworthy that CO32- ions inactivated NAP even at the lowest concentration. That could be attributed to an alternation of pH to alkalinity which led to change the chemical equilibrium of enzymatic reactions [52].
Regarding to metal chleating agents, 8HQ was more inhibitory than EDTA as observed in Table 3. 8HQ is the only one, among seven isomeric monohydroxyquinolines, capable of forming complexes with divalent metal ions as Fe 2+, Zn 2+, Cu 2+ and Mo 2+ through chelation, which subsequently causes inactivation of chelated complex biomolecule [53]. NAP enzyme is considered to be metalloenzymes in which the enzyme activity is dependent on some kinds of metal ions such as molybdenum and iron. Therefore, addition of chelating agents to the reaction mixture results in the formation of complexes with the ions in the active site, which cause inhibition of enzyme activity [54].
Various types of detergents were examined varied from nonionic as Triton X-100 and Tween- 20 to anionic as SLS as indicated in Table 4. The progressive reduction of activity was noticed with increasing the detergents concentration could be attributed to their denaturation potency by disruption of hydrogen bond and binding to hydrophobic regions of the polypeptide chain as a necklace like shape [55]. Whereas, DMSO (up to1%) stabilized NAP activity. [56] recorded the enhancement effect of DMSO on activity of esterase enzyme of Talaromyces stipitatus.
Table 4.
Influence of various detergents and organic solvent on NAP activity.
| Agent | Concentration | Relative activity (%) |
|---|---|---|
| DMSO | 0.50% | 95.45 |
| 1% | 90.09 | |
| 3% | 59.62 | |
| 5% | 20.78 | |
| SLS | 0.50% | 21.83 |
| 1% | 18.11 | |
| 3% | 8.29 | |
| 5% | 4.24 | |
| Triton- X-100 | 0.50% | 31.73 |
| 1% | 21.74 | |
| 3% | 12.87 | |
| 5% | 7.24 | |
| Tween-20 | 0.50% | 40.88 |
| 1% | 28.40 | |
| 3% | 19.38 | |
| 5% | 10.30 | |
3.1.4. Preservation of NAP at −20 °C
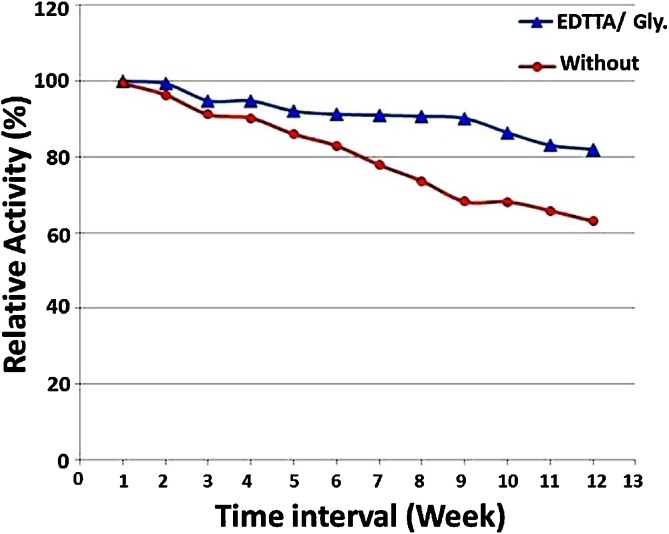
The adequate storage conditions are one of the most critical parameters in enzyme preservation for subsequent application [57]. As shown in Fig. 3, The preservation buffer was able to save about 20% of the activity with 82% residual activity retained in its presence compared to 63% in its absence after 3 months of experimenting. The glycerol in the preservation buffer increased the longevity of the enzyme’s activity [27] documented that mixture of phosphate buffer containing glycerol 50% and EDTA 0.4 mM is effective for D. desulfuricans ATCC 27,774 purified NR preservation.
Fig. 3.
Preservation of NAP at −20 °C in the presence/absence of preservation buffer.
Generally, the presence of NAP in crude form accompanying with other proteins could support its stability and provide protection against harsh circumstances, which enables the enzyme for posterior application in a wide range of environmental conditions particularly in waste treatment. This point of view was supported previously by [57], who mentioned that α- amylase of Bacillus licheniformis ATCC 6346 retained the unique features in crude compared to the pure enzyme.
3.1.5. Kinetic parameters for NR (Km and Vmax)
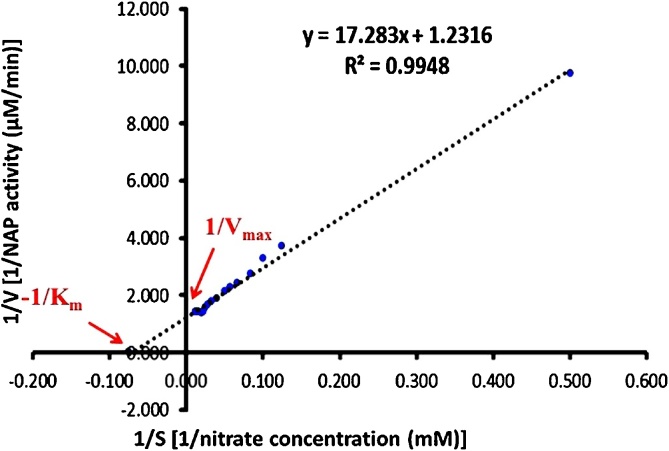
The quality of any enzymatic reaction was determined by Michaelis menten constant (Km) and Vmax [58]. The enzyme followed the Michealis Menten kinetics of catalysis. The use of the double reciprocal or Lineweaver–Burk plot yields much more accurate numerical values for Km and Vmax than the Michaelis-Menten curve as depicted in Fig. 4. Where, NAP catalyzed NO3− reduction at Vmax of 0.811 μM/min and Km of 14.02 mM.
Fig. 4.
Lineweaver-Burk plot of velocity versus substrate concentration.
3.2. Concurrent denitrification and AgNPs synthesis via immobilized MMT and NAP
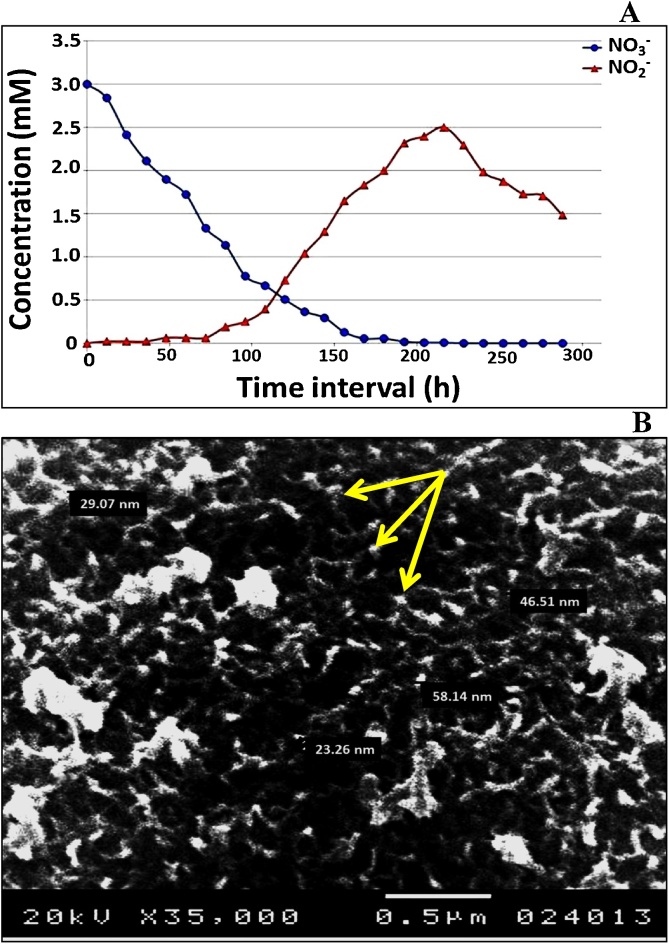
The immobilized MMT-cells and NAP-enzyme were applied in artificial wastewater containing 3 mM of AgNO3. The removal rate of NO3− and NO2− was measured for 288 h As displayed in Fig. 5A, 90% of NO3− was removed within 144 h incubation and complete removal was achieved upon 204 h by the action of immobilized cells. The concentration of NO2- increased until complete NO3- removal then decrease gradually by cell exhaustion. During nitrate bioremediation process, the immobilized cells transform Ag+ to AgNPs indicated by precipitation of tiny dark particles on alginate beads ranging from 23.26 nm to 58.14 nm as illustrated in (Fig. 5B). it is noteworthy to clarify that nitrate reductase enzyme together with photosensitive electron shuttling compounds initiate the nitrate reduction and during the metabolic process, the enzymes may shuttle electrons to the metal ions that capable of undergoing redox reaction; which finally leads to NPs formation [59]. As a consequence, nitrate removal and NPs synthesis come in parallel to each other. Remarkably, the denitrification process was suggested to be accomplished indicated by absence of nitrate and nitrite within 288 h which implies that the emission of gas as final product of this process (not detected).
Fig. 5.
Application of immobilized MMT cells in A) removal of NO3− and NO3− from artificial wastewater, B) Biosynthesized AgNPs. Arrows pointing to tiny AgNPs.
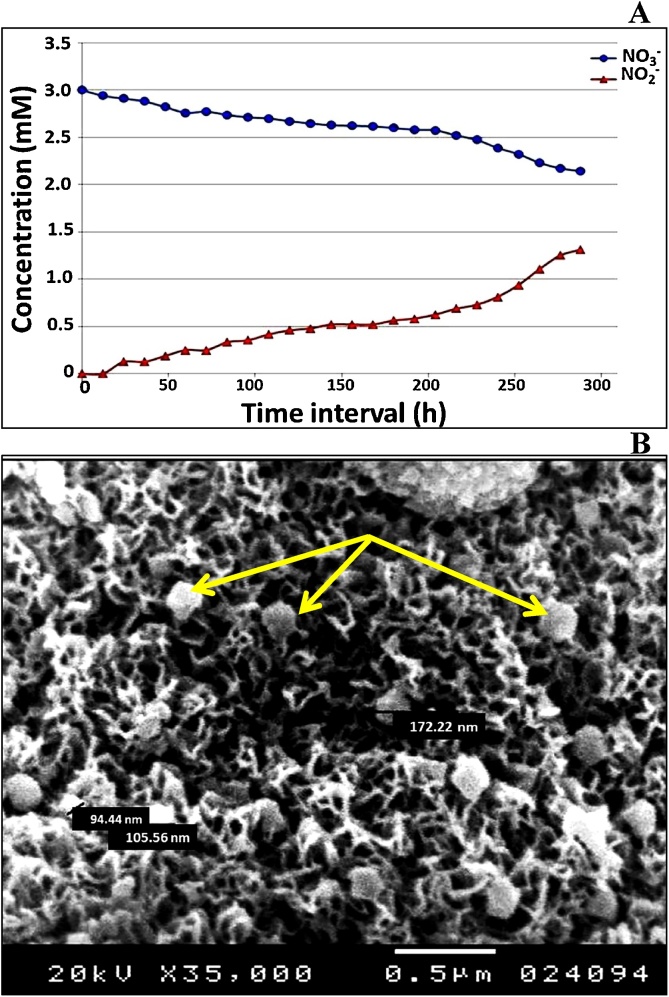
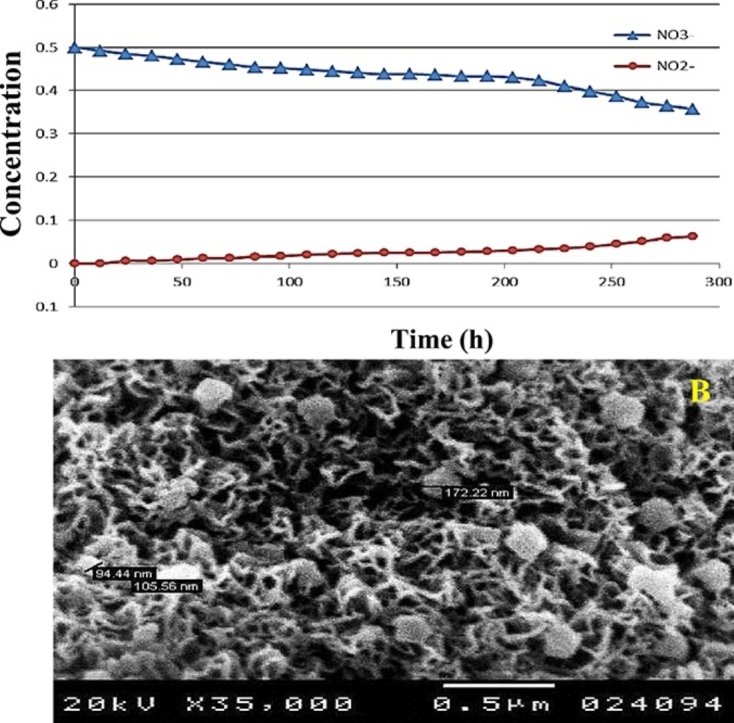
On the other hand, the immobilized NAP-enzyme exhibited lower efficiency, whether in NO3− removal or AgNPs formation. As observed in Fig. 6A, 28.6% of NO3− was eliminated within 288 h of incubation. Besides, very low concentration of AgNPs was synthesized within the exact time (Fig. 6B). Large particles ranging from 94.44 nm to 172.22 nm were deposited on the humps of alginate surface. This suggested being tiny AgNPs agglomerated together in aggregates.
Fig. 6.
Application of immobilized NAP-enzyme in: A) removal of NO3− from artificial wastewater, B) Biosynthesized AgNPs, Arrows pointing to AgNPs aggregates.
Generally, enzyme immobilization suffers from several disadvantages as the limitation in mass transfer and inactivation of enzyme during the immobilization process. That disadvantages appeared clearly in our results with lower performance of immobilized crude NAP in NO3− removal and AgNPs synthesis. The lower activity of immobilized NAP could be attributed to the change in the three dimensional conformation of the protein, particularly in the catalytic active sites when linked with the support matrix which led to change in the enzyme properties. Further [60], suggested that the physical, chemical characteristics and polyanionic nature of the matrices are responsible for a particular physiology and performance of immobilized material (cells or proteins) with the space available for metabolic activity and ion exchange between the immobilized material and environment [61] also suggested that Ca-induced supramolecular conformation of alginate which influence on proteins behavior. In the light of such suggestion, the entrapment in alginate polymer seemed inadequate matrix or method for crude NAP-enzyme immobilization. That’s leads to propose applying alternative matrices as agarose, polyacrylamide and silica or other methods as adsorption and cross linking in further investigations.
Remarkably, NAS enzyme was involved in enzymatic electrodes or amperometric biosensors for nitrate detection/removal as green chemistry solution. NAS enzyme derived from Arabidopsis thaliana has been successfully expressed in methylotrophic yeast Pichia pastoris and employed for nitrate analysis in real water samples [62]. Also [63], constructed electro-bioreactor composed of immobilized NAS from Zea mays in combination with crude-periplasmic nitrite reductase and nitrous oxide reductase of Rhodopseudomonas spheroids. Recently, nitrate reductase-(NAD(P)H) from Aspergillus niger was immobilized on surface functionalized nanoscale iron oxide and zinc oxide particles in an attempt to enhance stability and kinetic properties of NR in water and soil [64]. However, the NADH-dependent nitrate reductase from Fusarium oxysporum was directly investigated in AgNPs formation by silver nitrate reduction either in free form [65] or immobilized as cross linked enzyme aggregates (CLEAs) [66]. It is worthy noting that there is no study reported formerly immobilization of NAP-enzyme of Achromobacter sp. As a consequence, the current study deemed being the first attempt in concurrent nitrate removal and AgNPs sythesise which would be improved and harnessed in vital environmental processes.
3.2.1. EDX
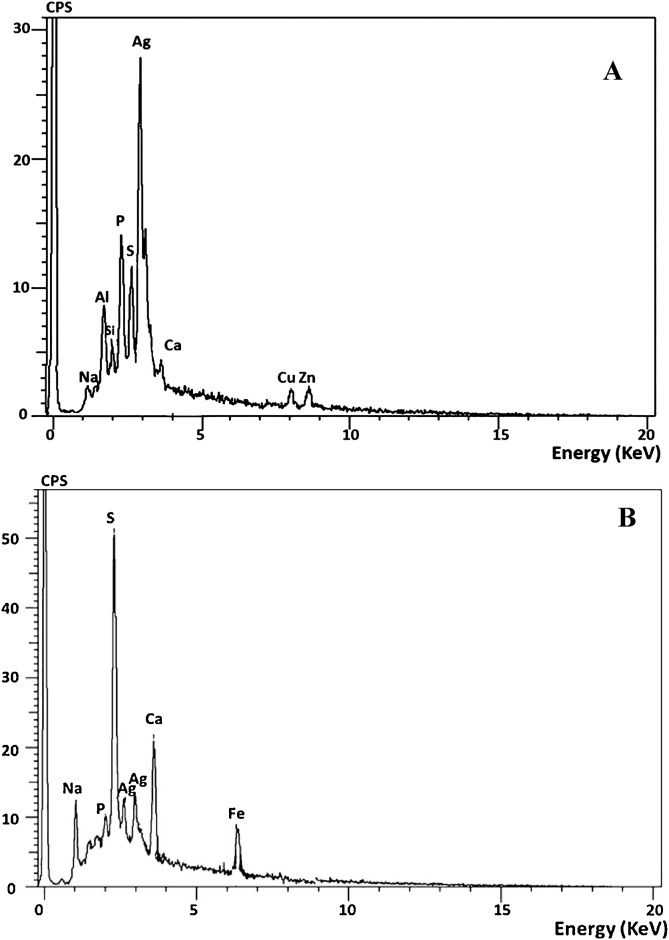
It is main compositional analysis technique that identifies elements qualitatively as well as quantitatively in examined sample. The elemental profile of both immobilized systems assured that silver is the major constituent element by 53% and 23% for immobilized MMT-cells and NAP-enzyme, respectively (Fig. 7A and B). The biosynthesized AgNPs displayed typical peak approximately at 3 keV due to surface plasmon resonance which confirms formation of Ag nano-crystals. These obtained elemental patterns are coincident with several earlier reports of AgNPs synthesis by green methods [67,68]. Moreover, signals for other elements were detected such as for P, S, Na, Ca and Fe. Presence of P and S peaks could be attributed to the association of AgNPs with bacterial biomolecules such as DNA, RNA and amino acids. On the other hand, Na and Ca are the constituents of sodium alginate and calcium chloride which embedded in the microspheres. Presence of Zn and Cu in EDX pattern of immobilized MMT-cells could be attributed to cell constituents in the functional group of proteinogenic amino acids. Our results are harmonized with [69]. Obviously, the presence of Fe peak in EDX spectrum of immobilized NAP-enzyme was suggested to be due to its incorporation into the enzyme active center in catalytic subunit as referred by [70,71].
Fig. 7.
EDX pattern of immobilized MMT-cells (A) and immobilized NAP-enzyme (B) beads at the end of incubation period.
3.2.2. XRD
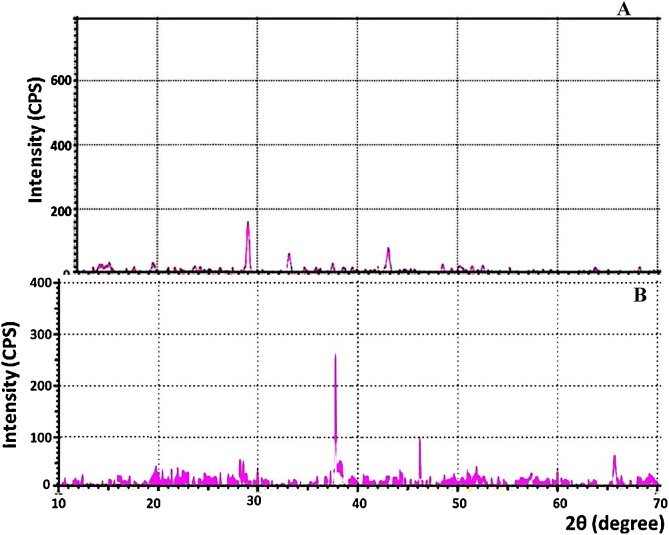
It is the fundamental crystallographic characterization approach for bulk, nano and thin film materials. XRD pattern is characterized by the interplanar d-spacing/2θ degree and the relative intensities (I/I0) of the strongest peaks. The X-ray difractograms of immobilized MMT-cells and NAP-enzyme beads were illustrated in (Fig. 8A and B). XRD patterns revealed three well resolved diffraction peaks at 2 theta (degree) as 38.3°, 46.3° and 67.4°, which could be indexed to (200, 211and 222) planes of face centered cubic silver, respectively. These peaks are matched with the standard powder diffraction card of Joint Committee on Powder Diffraction Standards (JCPDS), silver file No. 76‐1393. The positions and relative intensities of the reflection peak indicated that both immobilization systems fabricated AgNPs with crystalline nature. In addition, the results proposed that the biosynthesized AgNPs are single phase and no diffraction peaks related to other phases were detected which indicating the phase purity [72]. These results came in consistent with [15].
Fig. 8.
XRD crystallographic pattern of immobilized MMT-cells (A) and immobilized NAP-enzyme (B) beads at the end of incubation period.
4. Conclusion
In summery, this study aims to employ the immobilized denitrifying strain MMT and its NAP enzyme in concurrent denitrification and NPs synthesis, for the first time to study. The properties of crude NAP enzyme were assessed indicating a higher activity at pH range (6–7) and temperature range (30–40 °C). NAP enzyme exhibited higher stability at 4 °C more than 30 °C with 91% and 77% of residual activity, respectively up to 60 °C. The effect of several types of additives (respiratory inhibitor, serine protease inhibitor, thiol-inhibitors, oxidizing agent, denaturants, heavy metals, anions, cations, chelating agents, detergents and organic solvents) were evaluated at different concentrations (1 mM 2.5 mM, 5 mM and 10 mM). The kinetic parameters of NAP enzyme were 14.02 mM and 0.811 μM/min for Km and Vmax, respectively. Finally, MMT-cells and NAP-enzyme were immobilized with entrapement in alginate beads and applied in artificial wastewater with 3 mM AgNO3. Both immobilization systems removed nitrate and synthesized AgNPs with different effeciencies. SEM, EDX and XRD approaches were employed to affirm AgNPs biofabrication by both immobilization systems.
Acknowledgment
This work was supported by PhD grant from the Academy of Scientific Research and Technology (Egypt) (C-15).
Contributor Information
Marwa Eltarahony, Email: meltarahony@srtacity.sci.eg.
Sahar Zaki, Email: szaki@srtacity.sci.eg.
Zeinab Kheiralla, Email: kheiralla@hotmail.com.
Desouky Abd-El-haleem, Email: dabdelhaleem@srtacity.sci.eg.
References
- 1.Shrimali M., Singh K. New methods of nitrate removal from water$ Environ. Pollut. 2001;112:351–359. doi: 10.1016/s0269-7491(00)00147-0. [DOI] [PubMed] [Google Scholar]
- 2.Okolo J., Nweke C., Nwabueze R., Dike C., Nwanyanwu C. Toxicity of phenolic compounds to oxidoreductases of Acinetobacter species isolated from a tropical soil. Sci. Res. Essays. 2007;2:244–250. [Google Scholar]
- 3.Sparacino-Watkins C., Stolz J., Basua P. Nitrate and periplasmic nitrate reductases. Chem. Soc. Rev. 2014;43(2):676–706. doi: 10.1039/c3cs60249d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moreno-Vivia´ N., Cabello P., Marti´nez-Luque M., Blasco R., Castillo F. Prokaryotic nitrate reduction: molecular properties and functional distinction among bacterial nitrate reductases, mini-review. J. Bacteriol. 1999;181(21):6573–6584. doi: 10.1128/jb.181.21.6573-6584.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morozkina E., Zvyagilskaya R. Nitrate reductases: structure, functions, and effect of stress factors. Biochemistry (Mosc.) 2007;72(10):1151–1160. doi: 10.1134/s0006297907100124. [DOI] [PubMed] [Google Scholar]
- 6.Sparacino-Watkins C., Stolz J., Basua P. Nitrate and periplasmic nitrate reductases. Chem. Soc. Rev. 2014;43(2):676–706. doi: 10.1039/c3cs60249d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jones C. Department of Microbiology- Faculty of Natural Resources and Agricultural Sciences- Swedish University of Agricultural Sciences; 2010. Denitrification: From Genes to Ecosystems. PHD in Microbiology. [Google Scholar]
- 8.Behera S., Ak O., Rout J., Nayak P. Plant mediated synthesis of silver nanoparticles: opportunity and challenges. Int. J. Biol. Pharm. Allied Sci. 2012;1(11):1637–1658. [Google Scholar]
- 9.F. Ramezani, M. Ramezanib, S. Talebi, Mechanistic aspects of biosynthesis of nanoparticles by several microbes, Nano Con. 12 (2010) 14, Olomouc, Czech Republic, EU.
- 10.Zaki S., Eltarahony M., Elkady M., Abd-El-Haleem D. The use of bioflocculant and bioflocculant-producing bacillus mojavensis strain 32A to synthesize silver nanoparticles. J. Nanomater. 2014:1–7. [Google Scholar]
- 11.Eltarahony M., Zaki S., Kheiralla Z., Abd-El-haleem D. Biogenic synthesis of iron oxide nanoparticles via optimization of nitrate reductase enzyme using statistical experimental design. J. Adv. Biotechnol. 2016;5:667–684. [Google Scholar]
- 12.Prakash O., Jaiswal N. Immobilization of a thermostable –amylase on agarose and agar matrices and its application in starch stain removal. World Appl. Sci. J. 2011;13(3):572–577. [Google Scholar]
- 13.Badalova M., Evstatieva Y., Mushekova L., Licheva T., Nikolova D., Savov V. Natural and modified zeolites as matrices for the immobilization of trichoderma viride SL-45. Bulg. J. Agric. Sci. 2014:93–96. [Google Scholar]
- 14.Tee B., Kaletun G. Immobilization of a thermostable a-amylase by covalent binding to an alginate matrix increases high temperature usability. Biotechnol. Prog. 2009;25(2):436–445. doi: 10.1002/btpr.117. [DOI] [PubMed] [Google Scholar]
- 15.Eltarahony M., Zaki S., Kheiralla Z., Abd-El-haleem D. Isolation, characterization and identification of nitrate reductase producing bacteria. Int. J. Recent Sci. Res. 2015;6:7225–7233. [Google Scholar]
- 16.Eltarahony M., Zaki S., Kheiralla Z., Abd-El-haleem D. Bioconversion of silver and nickel ions into nanoparticles by Achromobacter sp. strain MMT and their application in wastewater disinfection. Int. J. Recent Sci. Res. 2016;7:8838–8847. [Google Scholar]
- 17.Giordani R., Buc J., Cornish-Bowden A., Rdenas M. Kinetics of membrane-bound nitrate reductase a from Escherichia coli with analogues of physiological electron donors different reaction sites for menadiol and duroquinol. Eur. J. Biochem. 1997;250:567–577. doi: 10.1111/j.1432-1033.1997.0567a.x. [DOI] [PubMed] [Google Scholar]
- 18.Frankenberger W. Technical Completion Report (University of California Water Resources Center); 2003. Perchlorate Removal in Groundwater by Perchlorate Reductases from the Perchlorate Respiring Bacterium, perc1ace. Project Number W-950. [Google Scholar]
- 19.Filimonenkov A., Zvyagilskaya R., Tikhonova T., Popov V. Isolation and characterization of nitrate reductase from the halophilic sulfur_oxidizing bacterium Thioalkalivibrio nitratireducens. Biochemistry (Mosc.) 2010;75(6):744–751. doi: 10.1134/s000629791006009x. [DOI] [PubMed] [Google Scholar]
- 20.Eltarahony M., Zaki S., Kheiralla Z., Abd-El-haleem D. Isolation, characterization and identification of nitrate reductase producing bacteria. Int. J. Recent Sci. Res. 2015;6:7225–7233. [Google Scholar]
- 21.Aoki K., Shinke R., Nishira H. Isolation and identification of respiratory nitrate reductase-producing bacteria from soli and production of enzyme. Agric. Biol. Chem. 1981;45(4):817–822. [Google Scholar]
- 22.Lowry O., Rosebrough H., Farr A., Randall R. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 23.Akhtaruzzama M., Mozumder N., Jamal R., Rahman A., Rahman T. Isolation and characterization protease enzyme from leguminous seeds. Agric. Sci. Res. J. 2012;2(8):434–440. [Google Scholar]
- 24.Karn N., Karn S. Evaluation and characterization of protease production by Bacillus sp. induced By UV-mutagenesis. Enzyme Eng. 2014;3:1. [Google Scholar]
- 25.Kumar D., Rakshitha R., Vidhya M., Jennifer Production, optimization and characterization of fibrinolytic enzyme by Bacillus subtilis RJAS 19. Pak. J. Biol. Sci. 2014;17(4):529–534. doi: 10.3923/pjbs.2014.529.534. [DOI] [PubMed] [Google Scholar]
- 26.Ulker S., Ozel A., Colak A., Karaoglu Ş. Isolation, production, and characterization of an extracellular lipase from Trichoderma harzianum isolated from soil. Turk. J. Biol. 2010;35:543–550. [Google Scholar]
- 27.Orji J., Ogwude D. Effects of phenolic compounds on periplasmic nitrate reductase and dehydrogenase enzymes of Escherichia sp. Niger. J. Biochem. Mol. Biol. 2009;24(2):11–16. [Google Scholar]
- 28.Bursakov S., Carneiro C., Almendra M., Duarte R., Caldeira J., Moura I., Moura J. Enzymatic properties and effect of ionic strength on periplasmic nitrate reductase (NAP) from Desulfovibrio desulfuricans ATCC 27774. Biochem. Biophys. Res. 1997;239:816–822. doi: 10.1006/bbrc.1997.7560. [DOI] [PubMed] [Google Scholar]
- 29.UL-Haq I., Javed M., Hameed U., Adnan F. Kinitics and thermodynamics of alpha amylase from Bacillus lichneformis mutant. Pak. J. Biotechnol. 2010;42(5):3507–3516. [Google Scholar]
- 30.Mahajan R., Gupta V., Sharma J. Comparison and suitability of gel matrix for entrapping higher content of enzymes for commercial applications. Ind. J. Pharm. Sci. 2010;72(2):223–228. doi: 10.4103/0250-474X.65010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuldeep K., Sandeep P., Jagjit K., Teena P. Development of plant asparagine biosensor for detection of leukemia. J. Pharm. Biomed. Sci. 2013;35(35):1796–1801. [Google Scholar]
- 32.American Public Health Association (APHA, 1999): American Water Works Association- Water Environment Federation; 1999. Standard Methods for the Examination of Water and Wastewater. [Google Scholar]
- 33.Wu Z., Huang S., Yang Y., Xu F., Zhang Y., Jiang R. Isolation of an aerobic denitrifying bacterial strain from a biofilter for removal of nitrogen oxide. Aerosol Air Qual. Res. 2013;13:1126–1132. [Google Scholar]
- 34.Joshi K., Joseph J., Srinikethan G., Saidutta M. Isolation and characterization of Pseudomonas syringae for nitrate removal under aerobic conditions. J. Biochem. Tech. 2014;5(2):693–697. [Google Scholar]
- 35.Mahesh M., Neha G., Rajesh T., Somashekhar R., Puttaiah E. Isolation and characterization of extracellulare thermostable alkaline phosphatase enzyme from Bacillus spp. Int. J. Appl. Biol. Pharm. Technol. I. 2010;1:21–33. [Google Scholar]
- 36.Al-Rajhi S., Al-Gelawi M., Aziz G. Purification and characterization of nitrate reductase (NAR) from Pseudomonas sp. SH7 isolate. J. Biotechnol. Res. Cent. 2010;4:23–36. [Google Scholar]
- 37.Kim S., Song Y., Yoo Characterization of membrane bound nitrate reductase from denitrifying bacteria Ochrobactrum anthropii SY509. Biotechnol. Bioprocess Eng. 2006;11:32–37. [Google Scholar]
- 38.Schiller J., Liu C. Immobilization of nitrate reductase within polyacrylamide gels. Biotechnol. Bioeng. 1976;18:1643–1645.S. doi: 10.1002/bit.260181115. [DOI] [PubMed] [Google Scholar]
- 39.Khan A. NarGHJI) from M. tuberculosis H37Ra.PhD - University of Pune; 2008. Isolation, Purification and Partial Characterization of Respiratory Nitrate Reductase. [Google Scholar]
- 40.Imlay J. Transcription factors that defend bacteria against reactive oxygen species. Ann. Rev. Microbiol. 2015;69:93–108. doi: 10.1146/annurev-micro-091014-104322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Romanowska I., Kwapisz E., Mitka M., Bielecki S. Isolation and preliminary characterization of a respiratory nitrate reductase from hydrocarbon-degrading bacterium Gordonia alkanivorans S7. J. Ind. Microbiol. Biotechnol. 2010;7:625–629. doi: 10.1007/s10295-010-0717-6. [DOI] [PubMed] [Google Scholar]
- 42.Wyszkowska J., Borowik A., Kucharski M., Kucharski J. Effect of cadmium, copper and zinc on plants, soil microorganisms and soil enzymes. J. Elem. 2013:769–796. [Google Scholar]
- 43.Hussein K., Joo J. Heavy metal resistance of bacteria and its impact on the production of antioxidant enzymes. Afr. J. Microbiol. Res. 2013;7(20):2288–2296. [Google Scholar]
- 44.Marangon J., de Sousa P., Moura I., Brondino C., Moura J., González P. Substrate-dependent modulation of the enzymatic catalytic activity: reduction of nitrate, chlorate and perchlorate by respiratory nitrate reductase from Marinobacter hydrocarbonoclasticus 617. Biochim. Biophys. Acta. 2012;1817:1072–1082. doi: 10.1016/j.bbabio.2012.04.011. [DOI] [PubMed] [Google Scholar]
- 45.Aiken A. Department of chemical engineering-Washington State University; 2002. Heavy Metal Induced Inhibition of Aspergillus niger Nitrate Reductase: Application for Rapid Contaminant Detection in Aqueous Samples. M.Sc. in Chemical Engineering. [Google Scholar]
- 46.Sharma J., Subhadra V. The effect of mercury on nitrate reductase activity in bean leaf segments (Phaseolus vulgaris) and its chelation by phytochelatin synthesis. Life Sci. Med. Res. 2010;13 [Google Scholar]
- 47.Orji J., Nweke O., Nwabueze R., Anyaegbu B., Chukwu J., Chukueke P., Nwanyanwu C. Impact of some cations on periplasmic nitrate reductase and dehydrogenase enzymes of E. coli, Pseudomonas and Acinetobacter species. An Interdis. J. Appl. Sci. 2008;3(2):5–18. [Google Scholar]
- 48.Hamada H., Arakawa T., Shiraki K. Effect of additives on protein aggregation. Curr. Pharm. Biotechnol. 2009;10:400–407. doi: 10.2174/138920109788488941. [DOI] [PubMed] [Google Scholar]
- 49.Schneider C., Shukla D., Trout B. Arginine and the Hofmeister series: the role of ion–ion interactions in protein aggregation suppression. J. Phys. Chem. B. 2011;115(22):7447–7458. doi: 10.1021/jp111920y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Salis A., Ninham B. Models and mechanisms of Hofmeister effects in electrolyte solutions, and colloid and protein systems revisited. Chem. Soc. Rev. 2014;43:7358–7377. doi: 10.1039/c4cs00144c. [DOI] [PubMed] [Google Scholar]
- 51.Paggir R., Casrton O., Kerber O., Garci A. Effect of salts on the activity of nitrate reductase from the photosynthetic bacterium Rhodobacter sphaeroides SW. J. Basic Microbiol. 1995;35(1):47–52. [Google Scholar]
- 52.Sharma R. Enzyme inhibition: mechanisms and scope. Enzyme Inhib. Bioappl. 2012:1–35. [Google Scholar]
- 53.Garrett R., Nason A. Furthur purification and properties of Neurospora nitrate reductase. J. Boil. Chem. 1969;244(11):2870–2882. [PubMed] [Google Scholar]
- 54.Martnez-Espinosa R., Marhuenda-Egea F., Bonete M. Assimilatory nitrate reductase from the haloarchaeon Haloferax mediterranei: puri¢cation and characterization. FEMS Microbiol. Lett. 2001;204:381–385. doi: 10.1016/s0378-1097(01)00431-1. [DOI] [PubMed] [Google Scholar]
- 55.Stumpe M., Grubmuller H. Interaction of urea with amino acids: implications for urea-induced protein denaturation. J. Am. Chem. Soc. 2007;129:16126–16131. doi: 10.1021/ja076216j. [DOI] [PubMed] [Google Scholar]
- 56.Faulds C., Pérez-Boada M., Martinez A. Influence of organic co-solvents on the activity and substrate specificity of feruloyl esterases. Bioresour. Technol. 2011;102:4962–4967. doi: 10.1016/j.biortech.2011.01.088. [DOI] [PubMed] [Google Scholar]
- 57.Vengadaramana A., Balakumar S., Arasaratnam V. Characteristic analysis of crude and purified α-amylase from Bacillus licheniformis ATCC 6346 and comparison with commercial enzyme. Schol. Acad. J. Pharm. (SAJP) 2013;2(2):31–35. [Google Scholar]
- 58.Bobade S., Khyade V. Influence of inorganic nutrients on the activity of enzyme, nitrate reductase in the leaves of Mulberry, Morus alba (L) (M-5 variety) Res. J. Rec. Sci. 2012;1(5):14–21. [Google Scholar]
- 59.Lin I., Lok C., Che C. Biosynthesis of silver nanoparticles from silver (I) reduction by the periplasmic nitrate reductase ctype cytochrome subunit NapC in a silver-resistant E. coli. Chem. Sci. 2014;5:3144–3150. [Google Scholar]
- 60.Hahn-Hagerdal B. Physiological aspects of immobilized cells: a general overview. Proceedings of an International Symposium; Wageningen, The Netherlands; 1989. Elsevier Science, Amsterdam. [Google Scholar]
- 61.Roger D., David A., David H. Immobilization of flax protoplasts in agarose and alginate beads. Plant Physiol. 1996;112:1191–1199. doi: 10.1104/pp.112.3.1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Campbell W., Song P., Barbier G. Nitrate reductase for nitrate analysis in water. Environ. Chem. Lett. 2006;4:69–73. [Google Scholar]
- 63.Mellor R., Ronnenberg J., Campbell W., Diekmann S. Reduction of nitrate and nitrite in water by immobilized enzymes. Lett. Nat. 1992;355:717–719. [Google Scholar]
- 64.Sachdeva V., Hooda V. Effect of changing the nanoscale environment on activity and stability of nitrate reductase. Enzyme Microb. Technol. 2016;89:52–62. doi: 10.1016/j.enzmictec.2016.03.007. [DOI] [PubMed] [Google Scholar]
- 65.Kumar S., Abyaneh M., Gosavi S., Kulkarni S., Pasricha R., Ahmad A., Khan M. Nitrate reductase-mediated synthesis of silver nanoparticles from AgNO3. Biotechnol. Lett. 2007;29:439–445. doi: 10.1007/s10529-006-9256-7. [DOI] [PubMed] [Google Scholar]
- 66.Talekar S., Joshi G., Chougle R., Nainegali B., Desai S., Joshi A., Kambale S., Kamat P., Haripurkar R., Jadhav S., Nadar S. Preparation of stable cross-linked enzyme aggregates (CLEAs) of NADHdependent nitrate reductase and its use for silver nanoparticle synthesis from silver nitrate. Catal. Commun. 2014;53:62–66. [Google Scholar]
- 67.Khan M., Khan M., Adil S., Tahir M., Tremel W., Alkhathlan H., Al-Warthan A., Rafiq M., Siddiqui H. Green synthesis of silver nanoparticles mediated by Pulicaria glutinosa extract. Int. J. Nanomed. 2013;8:1507–1516. doi: 10.2147/IJN.S43309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ibraheem H. Green synthesis and characterization of silver nanoparticles using banana peel extract and their antimicrobial activity against representative microorganisms. J. Radiat. Res. Appl. Sci. 2015;8(3):265–275. [Google Scholar]
- 69.ZhaoQing L., Li L. Preparation and characterization of radiopaque calcium alginate microspheres embedded silver nanoparticles. Adv. Mater. Res. 2015;1120(1121):867–872. [Google Scholar]
- 70.Morozkina E., Zvyagilskaya R. Nitrate reductases: structure, functions, and effect of stress factors. Biochemistry (Mosc.) 2007;72(10):1151–1160. doi: 10.1134/s0006297907100124. [DOI] [PubMed] [Google Scholar]
- 71.Marangon J., de Sousa P., Moura I., Brondino C., Moura J., González P. Substrate-dependent modulation of the enzymatic catalytic activity: reduction of nitrate, chlorate and perchlorate by respiratory nitrate reductase from Marinobacter hydrocarbonoclasticus 617. Biochim. Biophys. Acta. 2012;1817:1072–1082. doi: 10.1016/j.bbabio.2012.04.011. [DOI] [PubMed] [Google Scholar]
- 72.Mohameed Q., Hattab F., Fakhry M. Effect of substrate temperature on structural characteristics of nano silver oxide prepared by pulsed-laser deposition. Iraqi J. Appl. Phys. 2015;11(2):33–36. [Google Scholar]