Summary
Human neural development begins at embryonic day 19 and marks the beginning of organogenesis. Neural stem cells in the neural tube undergo profound functional, morphological, and metabolic changes during neural specification, coordinated by a combination of exogenous and endogenous cues. The temporal cell signaling activities that mediate this process, during development and in the postnatal brain, are incompletely understood. We have applied gene expression studies and transcription factor-activated reporter lentiviruses during in vitro neural specification of human pluripotent stem cells. We show that nuclear factor κB orchestrates a multi-faceted metabolic program necessary for the maturation of neural progenitor cells during neurogenesis.
Keywords: NF-κB signaling, neurogenesis, human embryonic development, metabolism, autophagy, cell cycle
Highlights
-
•
NF-κB activation is required during normal human neurogenesis
-
•
NPCs that have the capacity to mature into neurons have high NF-κB signaling
-
•
NPC maturation involves changes in metabolism, autophagy, and cell-cycle regulation
-
•
NF-κB activation inhibits the expression of neural stem cell markers
In this research article, FitzPatrick and colleagues have highlighted the requirement for NF-κB signaling in neural specification of human embryonic stem cells. They demonstrate that its activity orchestrates a metabolic shift toward oxidative phosphorylation in committing neural progenitor cells. Moreover, they demonstrate that progenitor cells with increased endogenous NF-κB activity have a higher propensity for maturation.
Introduction
Multipotent neural stem cells (NSCs) of the neural tube differentiate and populate the entire CNS. A small population of NSCs resides in a niche in the subventricular zone of the adult brain, for neurogenesis in response to injury. They are epithelial-like in morphology and undergo epithelial-to-mesenchymal transition when migrating in response to developmental or regenerative cues. The resultant self-renewing neural progenitor cells (NPCs), with features of transit amplifying cells, are capable of long-distance migration and terminal differentiation to mature neural cell types (Worlitzer and Schwamborn, 2014). NSCs primarily rely on glycolysis for energy production, but mature neurons require mitochondrial oxidative phosphorylation (OXPHOS). At some point during neural commitment, there is a metabolic switch in glucose utilization that has not been clearly defined. Furthermore, macroautophagy (herein referred to as autophagy)—the intracellular degradation system—is important for maintaining nutrient homeostasis during neurogenesis; impairment in autophagy results in neural tube defects in mice (Fimia et al., 2007).
It is clear that nuclear factor κB (NF-κB) activity plays a role in neural differentiation during perinatal development (Nolan et al., 2014, Yamanishi et al., 2015) and throughout adulthood (Denis-Donini et al., 2005). However, there is ambiguity in terms of its longitudinal molecular function. Recent studies in murine pathogenic models have implicated NF-κB activation in NSC commitment (Denis-Donini et al., 2008, Li et al., 2012, Rolls et al., 2007, Zhang et al., 2013b). NF-κB has previously been associated with metabolic reprogramming in stem cells and cancer (Iliopoulos et al., 2009, Kawauchi et al., 2008, Mauro et al., 2011), but its involvement in metabolic and autophagic reprogramming in human embryonic neurogenesis has not yet been elucidated. Here, we use a five-stage neural differentiation protocol using human pluripotent stem cells to interrogate NF-κB activity and function during neurogenesis. We find that NF-κB orchestrates a maturation program involving alterations in metabolism, autophagy, and cell cycle.
Results
Validation of Our Stage-Wise In Vitro Neural Differentiation Protocol
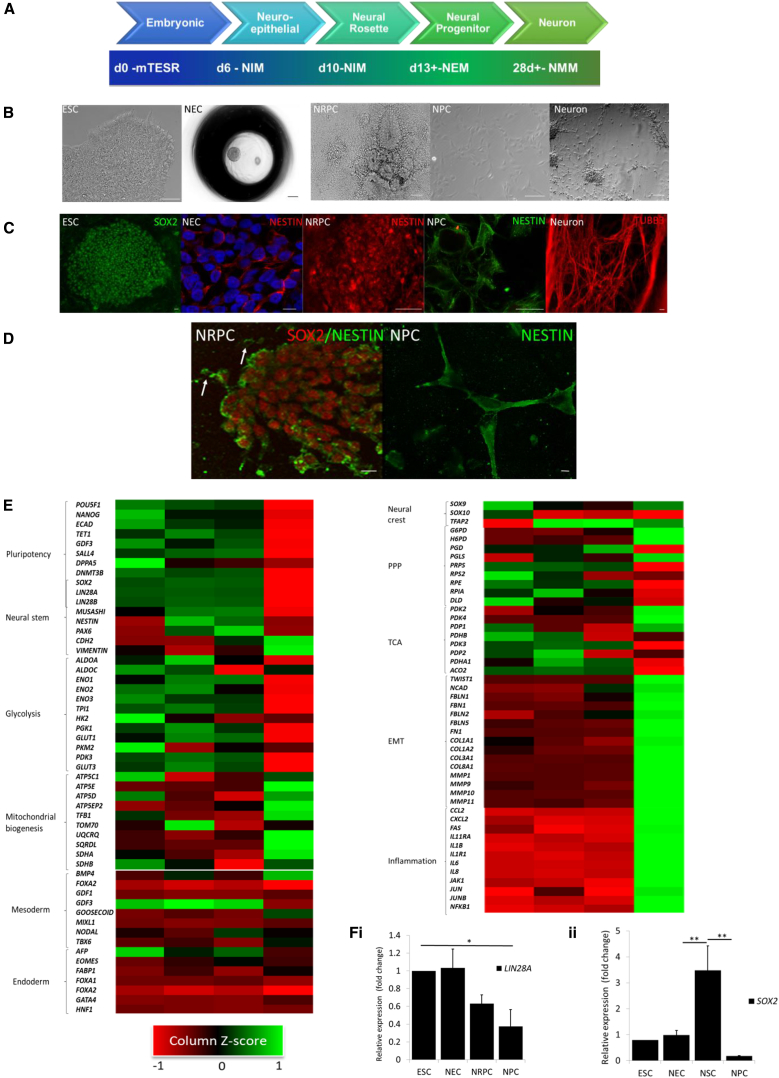
We first designed a five-stage neural differentiation protocol from pluripotency based largely on self-organization with minimal growth factor application to avoid exogenous stimulation (summarized in Figure 1A) (Burridge et al., 2011, Dottori and Pera, 2008, Zhang and Zhang, 2010). The human embryonic stem cell (hESC) line Shef3 (obtained from the UK Stem Cell Bank under the project SCSC10-48) was progressively differentiated through stages aligning to neuroepithelial clusters (NECs), neural rosette-forming progenitor cells (NRPCs), committed NPCs, and mature neurons (Figure 1B). When neural rosettes were mechanically isolated and replated, migrating cells with a mesenchymal morphology rapidly assumed a SOX2−/Nestin+ phenotype (Figures 1C and D). As neural differentiation progressed, pluripotency markers such as OCT4 and Tra-1-81 were no longer apparent, and SOX2 and Nestin expression also decreased in terminally differentiated cells (Figures S1A and S6).
Figure 1.
Evaluation of Stage-Wise Targeted Differentiation of hESCs to Mature Neurons
(A) Schematic representation of our five-stage differentiation protocol.
(B and C) Morphological evaluation (B) and immunocytochemical validation (C) of (i) embryonic stem cells (SOX2), (ii) neuroepithelial cells, (iii) neural rosette-forming progenitor cells, (iv) neural progenitor cells (all Nestin), and (v) neuronal cell cultures (β-III-tubulin). Scale bar, 100 μm.
(D) Co-immunocytochemistry shows SOX2+/Nestin+ neural rosette structures and adjacent SOX2−/Nestin+ committed migratory cells (arrows). Scale bars, 100 μm.
(E) Transcriptomic analysis of our stage-wise neural differentiation.
(F) qRT-PCR validation of transcriptional expression of the neural stem cell markers (i) LIN28A and (ii) SOX2 gene expression during neural specification (n = 3 independent biological repeats; ∗p ≤ 0.05, ∗∗p ≤ 0.01; error bars, ±SEM).
Transcriptomic Evaluation of hESC Neural Differentiation
Global gene expression was compared using Illumina microarray across our neural differentiation protocol. Hierarchical clustering of biological repeats demonstrated that cells at the NEC and NRPC stages were most similar and had a transcriptome more similar to ESCs than NPCs (Figure S1B). As we would predict, the pluripotency-associated transcripts for OCT4 and Nanog were downregulated gradually over differentiation and became undetectable by the NPC stage. SOX2 and LIN28 expression are associated with both pluripotency and NSC maintenance. Transcriptomics and qRT-PCR confirmed expression of both was maintained in NRPCs before dropping to undetectable levels in NPCs (Figures 1E, 1Fi, and 1Fii). The highest expression of Nestin and PAX6 transcripts was at the NRPC stage whereas markers of a more committed neural phenotype; Vimentin and CDH2, were highest at the NPC stage. Markers of endodermal (such as SOX17, GATA4, and HNF1) and mesodermal specification (Goosecoid, MIXL1, and Eomes) were unchanged during our neural specification protocol. These data validated our neurogenesis model. Analysis of regionalization suggested that NPCs are hindbrain precursors, most likely due to continued culture in fibroblast growth factor (FGF) throughout expansion (Figure S1C).
Metabolic reprogramming occurs during the differentiation of pluripotent stem cells to neurons, so we interrogated our transcriptomic dataset for indicators of this process. Our dataset showed that transcripts encoding proteins involved in glycolysis were strongly represented up to and including NRPCs but were reduced in committed NPCs. We saw an inverse correlation of transcripts associated with mitochondrial biogenesis. Interestingly, transcripts encoding proteins involved in the tricarboxylic acid (TCA) cycle, responsible for mitochondrial respiration, were not universally upregulated between NRPCs and NPCs. We also evaluated the anabolic utilization of glucose through the pentose phosphate pathway (PPP). Although there was not unilateral upregulation of PPP transcripts, the rate-limiting enzyme glucose-6-phosphate dehydrogenase (G-6-PD) was increased between NRPC and NPC, suggesting that there could be an increase in PPP activity as cells commit to the NPC phenotype (Figure 1E).
Gene Ontogeny of NF-κB Activity in Differentiating Cultures
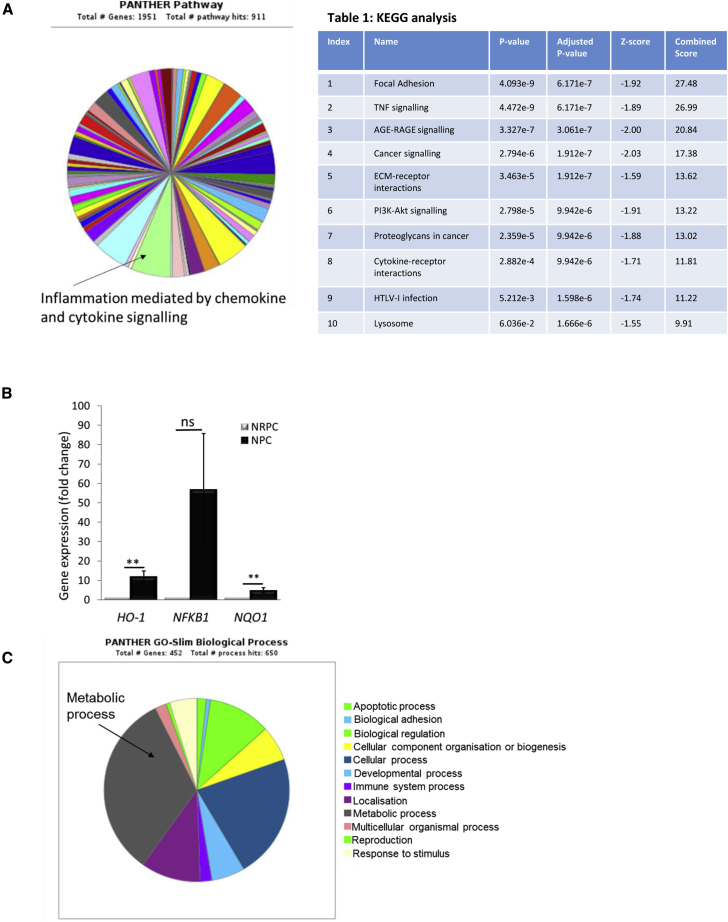
Gene ontogeny analysis of our transcriptomic dataset (KEGG and PANTHER) showed that “TNFα signaling,” “inflammation,” “cytokine signaling,” and more specifically “NF-κB targets” were collectively the most over-represented cellular processes during NPC commitment from NRPC (Figures 2A and 1E; Table S2). Normalized average array signals for classical canonical NF-κB signal transducers and transcriptional targets (IL6/8, MMP9, CCL2) were significantly increased in NPCs compared with NRPCs. Signal transduction of canonical NF-κB concludes with nuclear translocation of the p50/p65 heterodimer, which binds to DNA consensus sites at gene regulatory sites and activates transcription. Upregulation of the p50 precursor NFKB1, and metabolism associated NF-κB transcriptional target NQO1, in NPCs compared with NRPCs, were confirmed by qRT-PCR (Figure 2B). Similarly, we developed a script for meta-analysis (R studio and Linux) to isolate correlation coefficients with p65 in open-access mouse neural differentiation arrays. Analysis of the top 500 genes associated with p65 (correlation coefficients ranged from 0.82 to 0.53) using EnrichR (KEGG2015) resulted in the “oxidative phosphorylation” category obtaining the highest combined score (p = 0.0002). In the context of neural differentiation, genes such as ATP synthases and ubiquinol-cytochrome c reductases were found to show high correlation with p65 expression as well as a number of NADH dehydrogenases. PANTHER analysis of the largest group (213 of the 452 genes identified) represented genes contributing to metabolic processes (Figure 2C).
Figure 2.
Gene Ontology Analysis of Illumina HT-12 Microarray and Publicly Available Datasets
(A) PANTHER and KEGG pathway analysis of our transcriptomic dataset.
(B) qRT-PCR validation of transcriptional expression of the NF-κB targets NFKB1 and NQO1 during neural specification (n = 3 independent biological repeats; ∗∗p ≤ 0.01; ns, not significant; error bars, ±SEM).
(C) PANTHER meta-analysis of genes correlating with RELA expression in open-access mouse neural differentiation databases (GPL1261 platform).
NF-κB Activity Is Increased during NPC Maturation
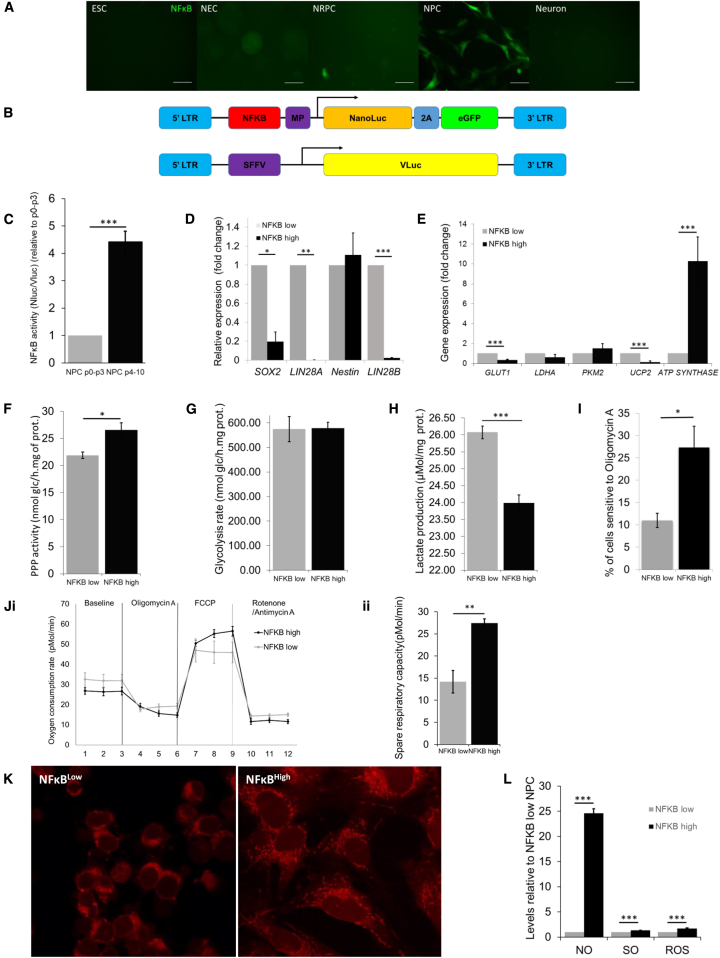
To further interrogate the role of NF-κB during neural differentiation, we employed a lentiviral NF-κB-activated firefly luciferase (FLuc)-2A-eGFP expressing reporter vector (LNT-NFκB-FLuc/EGFP) to assess NF-κB activity in living, differentiating cultures. Feeder-free hESCs were transduced with LNT-NFκB/FLuc-eGFP. hESCs containing a single genomic integration of the NFκB-eGFP expression cassette were subjected to our neural differentiation protocol and GFP+ cells were observed only at the NPC stage (Figure 3A). In future experiments, we employed a further iteration of the NF-κB reporter cassette containing a secreted luciferase variant; NanoLuc, to measure real-time NF-κB activity in living differentiating NPC cultures (LNT-NFKB-NanoLuc/EGFP, Figure 3B). Interestingly, GFP amplification was only observed after extended passage of NPC, implying a maturation process (Figures 3B and S1D). By quantifying NFκB-NanoLuc activity we were able to separate early-passage (P2) NFκBlow and later-passage (P9) NFκBhigh NPC populations (Figure 3C) for phenotypic comparison. Although morphologically similar, NFκBlow NPC were broadly SOX2+/LIN28+/Nestin+ whereas NFκBhigh NPC were SOX2−/LIN28−/Nestin+ (Figure 3D). Fluorescence-activated cell sorting (FACS) analysis to elucidate lineage and commitment of NFκBhigh and NFκBlow NPCs demonstrated that in NFκBhigh NPCs, a large portion of the cells (73%) stained positively for Nestin but the neuronal marker, β-III-tubulin, was not expressed in a large fraction of the population (2.9%) (Figure S2A). Moreover, only a small proportion of the NF-κB positive cells expressed the neural crest marker, SOX10 (6.8%). Similarly, in NF-κB-negative NPCs, Nestin was present in 69% of cells and β-III-tubulin and SOX10 were expressed in only a small fraction of cells (9.4% and 0.02%, respectively) (Figure S2B). We demonstrated that this observation is not specific to the Shef3 hESC line. Shef6 hESCs behaved in a similar manner; when subjected to neural induction media, rosettes developed and early-passage NPCs stained positively for SOX2 and Nestin (Figure S3A). Moreover, we observed a significant upregulation of NF-κB signaling in NPC by P6, using the LNT- NFκB-NanoLuc reporter (Figure S3B).
Figure 3.
NF-κB Activity Modulates Metabolic Activity during Neural Specification
(A) Schematic representation of the lentiviral reporter system used to quantify NF-κB activity during NPC passage. Scale bars, 100 μm.
(B) Lentiviral NFκB-NanoLuc/EGFP reporter assay during neural specification shows NF-κB activation at the NPC stage.
(C and D) Lentiviral NFκB-NLuc quantitative assay on living, differentiating cultures during serial passage of NPCs (n = 3 independent biological repeats; ∗∗∗p ≤ 0.001; error bars, ±SEM) (C). NFκBlow-NPC were characterized between P1 and P3 and NFκBhigh-NPC from P5 to P8. (D) Comparison of NFκBlow- and NFκBhigh-NPC by qRT-PCR of the neural stem cell markers SOX2 and LIN28A and B, and Nestin (n = 3 independent biological repeats; ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.01; error bars, ±SEM).
(E–H) qRT-PCR for GLUT1, LDHA, PKM2, UCP2, and ATP Synthase expression (n = 3 independent biological repeats; error bars, ±SEM), glycolysis as measured by peak medium 3H2O in NPCs loaded with radiolabeled [5-3H]glucose (F), medium lactate (G), and PPP (H) as assessed by quantifying the ratio of [1-14C]glucose conversion to 14CO2 by decarboxylation through 6-phosphogluconate dehydrogenase compared with [6-14C]glucose decarboxylation through the TCA cycle (n = 3 independent biological repeats; ∗p ≤ 0.05, ∗∗∗p ≤ 0.001; error bars, ±SEM).
(I–L) OXPHOS as measured by the increased percentage of cells sensitive to oligomycin (I) (n = 3 independent biological repeats), Seahorse Bioanalyzer Mitostress test (Ji and Jii), MitoTracker staining (K), and ROS/SO/NO analysis (L) using a quantifiable fluorescent assay (n = 3 independent biological repeats; ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001; error bars, ±SEM).
Metabolic Reprogramming Occurs as NPCs Assume a Maturation Phenotype
We endeavored to resolve NFκBlow and NFκBhigh-NPC phenotypes at the metabolic level. The glucose transporter GLUT1 and the mitochondrial uncoupling protein UCP2 were both transcriptionally downregulated whereas mitochondrial ATP synthase showed a >10-fold increase indicative of a molecular switch to OXPHOS (Figure 3E). We assessed glucose distribution during NPC maturation from NFκBlow- to NFκBhigh-NPC. Glycolysis was compared specifically by measuring peak medium 3H2O in NPCs loaded with radiolabeled [5-3H]glucose and was found to be unchanged (Figure 3F). A single hydrogen atom of H2O is interchanged with [3H] in [3-3H]glucose at the triose-phosphate isomerase-catalyzed reaction in the glycolytic process. This results in pyruvate production, which is then either converted to lactate or fed into the mitochondrial TCA cycle. Medium lactate was measured and found to be significantly decreased in NFκBhigh compared with NFκBlow-NPC (Figure 3G), suggesting that pyruvate is being preferentially distributed to the TCA cycle. We further assessed anabolic utilization of glucose via the PPP in NPCs by quantifying the ratio of [1-14C]glucose conversion to 14CO2 by decarboxylation through 6-phosphogluconate dehydrogenase compared with [6-14C]glucose decarboxylation through the TCA cycle. Interestingly, the PPP was significantly increased in NFκBhigh-NPC, suggesting that glucose is also redistributed to the PPP during NPC metabolic reprogramming (Figure 3H).
We also assessed mitochondrial activity in NPCs by direct inhibition of ATP synthase using oligomycin A and measuring ATP production (Figure 3I), as well as carrying out a “MitoStress test” using the Seahorse Bioanalyzer (Figures 3Ji and 3Jii). In both experiments, we observed a significant increase in the contribution of OXPHOS to ATP production between NFκBlow- and NFκBhigh-NPC. We saw similar metabolic function in Shef6-derived NPCs, where NFκBhigh-NPC had a significantly higher spare mitochondrial capacity when compared with NFκBlow-NPC (Figure S3C).
Furthermore, MitoTracker staining in NPCs showed evidence of a greater mitochondrial load in NFκBhigh-NPC (Figure 3K). We compared reactive oxygen species (ROS), superoxide (SO), and nitric oxide (NO) production in NFκBlow- and NFκBhigh-NPC and found that all three species were significantly upregulated (Figure 3L), further corroborating the increase in mitochondrial activity that we observed in NFκBhigh-NPC. NF-κB also induces the upregulation of NO via NO synthases in hepatocytes (Hatano et al., 2001) and promotes glucose 6-phosphate dehydrogenase induction in astrocytes (García-Nogales et al., 1999). Our data in NPCs are consistent with these previous observations and suggest that a similar mechanism is active in committed NPCs.
Autophagic Flux Is Reduced during NPC Maturation
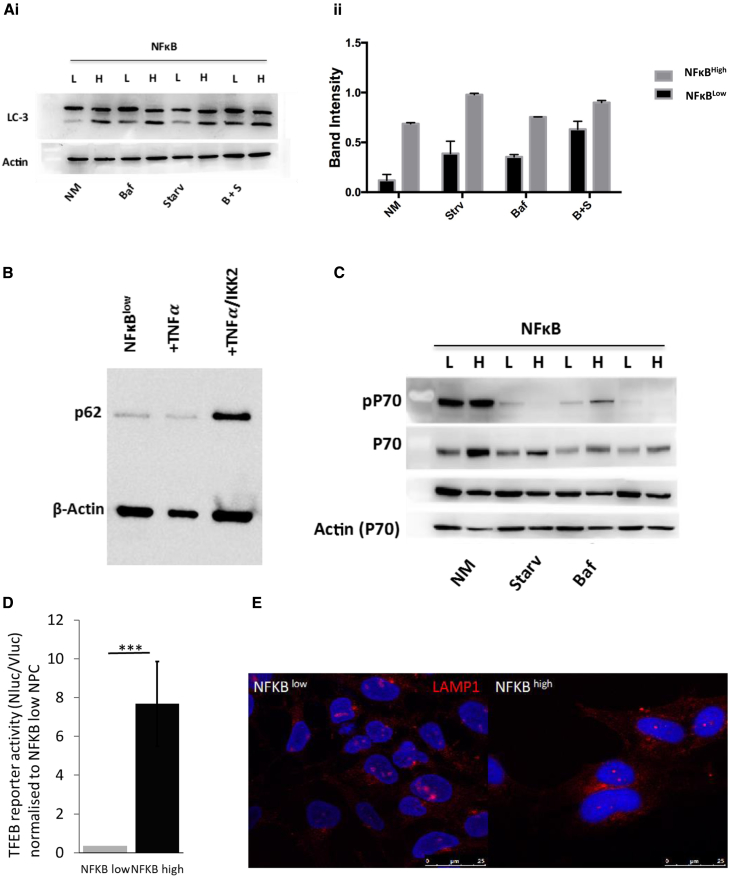
We interrogated alterations in degradative capacity between NFκBlow- and NFκBhigh-NPC by investigating autophagy under different nutrient conditions. As one might expect, starvation increased autophagic flux and, subsequently, the ratio of LC3-II to LC3-I in NFκBlow-NPC. Moreover, blocking autophagosome-lysosome fusion and degradation with bafilomycin A (Baf) treatment shunted autophagic flux, causing further accumulation of LC3-II (Figures 4Ai, 4Aii, and S5). However, in NFκBhigh-NPC under normal and serum-starvation conditions we saw increased LC3-II levels, but blocking autophagy with Baf had little effect alone or an additive effect to serum starvation. This would imply that autophagic flux is lower in NFκBhigh-NPC compared with NFκBlow-NPC. Critically, induction of NF-κB by tumor necrosis factor α (TNFα)/IKK-2 induction in NFκBlow-NPC resulted in an accumulation of the p62 protein, an indication of autophagic block (Figure 4B).
Figure 4.
NF-κB Activity Modulates Autophagy during Neural Specification
(Ai and Aii) NFκBHigh- and NFκBlow-NPC populations were compared for LC-3 activation status by western blot in normal media conditions, after treatment with bafilomycin A, under starvation conditions, or with both starvation and bafilomycin A treatment (n = 3 independent biological repeats ± SEM).
(B and C) p62 accumulation in NFκBhigh- and NFκBlow-NPCs stimulated with IKK2 and TNFα (B) and P70S6K (P70) phosphorylation (C) was assessed by western blot under the same conditions as described in (A).
(D) TFEB-NanoLuc/EGFP reporter activity assay (n = 3 independent biological repeats; ∗∗∗p ≤ 0.001; error bars, ±SEM).
(E) LAMP1 staining.
We also evaluated the activity of mammalian target of rapamycin complex 1 (mTORC1) by comparing phosphorylation of its target P70S6K in NPCs. In normal culture conditions, there appeared to be similar levels of P70S6K phosphorylation that was abolished by upregulation of autophagy after serum starvation, and partially restored by blocking autophagic flux with Baf treatment (Figure 4C). A decrease in autophagic flux would be consistent with increased mTORC1 activity, and we indeed observed an increase in phosphorylated P70S6K under normal conditions after Baf treatment. Lysosomal biogenesis is controlled by transcription factor E box (TFEB) where nuclear translocation is itself inhibited by mTORC1-mediated phosphorylation. We used a TFEB-activated transcription factor-activated reporter (TFAR) lentiviral expression cassette to assay for TFEB nuclear translocation in NPCs and surprisingly saw that it was >7-fold higher in NFκBhigh-NPC compared with NFκBlow-NPC (Figure 4D). LAMP1 immunostaining confirmed a higher density of lysosomes in NFκBhigh-NPC (Figure 4E).
Cell-Cycle Arrest Occurs as NPCs Assume a Maturation Phenotype
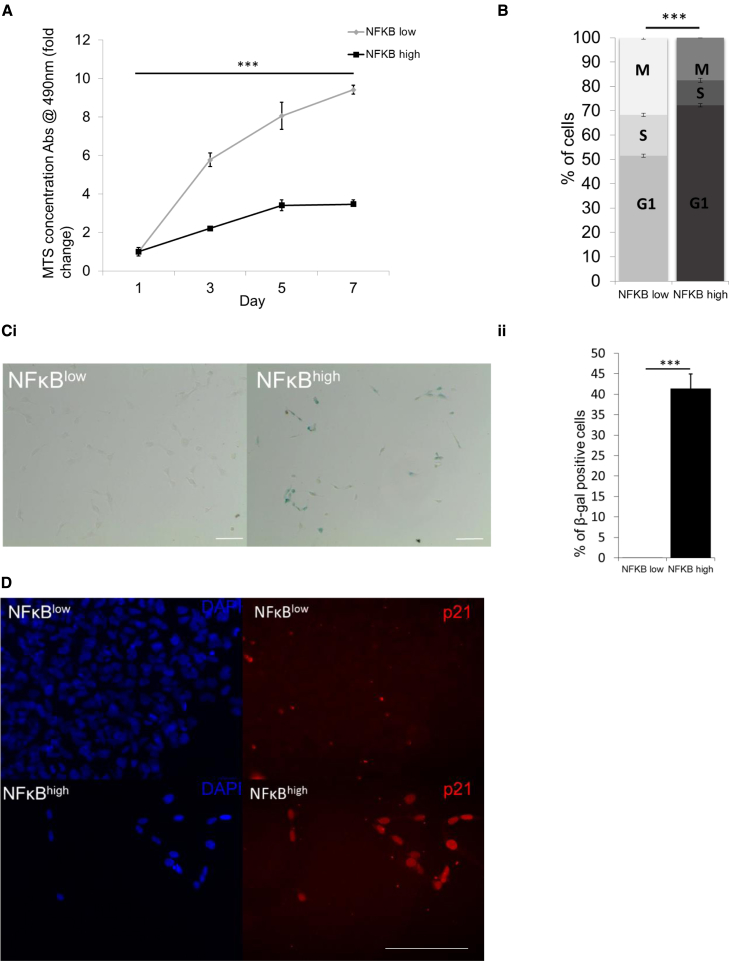
Terminally differentiated mature neurons are post-mitotic and so cell-cycle arrest is another key component of the development process. Although NFκBlow- and NFκBhigh-NPC continued to express equivalent levels of Nestin and there was no evidence of spontaneously differentiated neurons in either cultures, NFκBhigh-NPC had a lower proliferation index (Figure 5A) and an increased proportion of cells in G0/G1 compared with NFκBlow-NPC (Figure 5B). Consistently, NFκBhigh-NPC presented with significantly increased senescence-associated β-galactosidase (SA-β-Gal; Figures 5Ci and 5Cii) activity and increased p21 expression (Figure 5D) without exhibiting an overt change in cell morphology.
Figure 5.
NF-κB Activity Modulates Cell Cycle and p21 during Neural Specification
Cell proliferation by serial cell counts (A) (n = 3 independent biological repeats ± SEM; ∗∗∗p ≤ 0.001), PI incorporation (B) (n = 3 independent biological repeats ± SEM, ∗∗∗p ≤ 0.001), senescence-associated β-galactosidase assay (Ci and Cii) (n = 3 independent biological repeats [200 counts/sample]; ∗∗∗p ≤ 0.001; error bars, ±SEM), and p21 immunocytochemistry (D). Scale bars, 100 μm.
NF-κB Modulation Induces a Neural Maturation Phenotype in NPCs
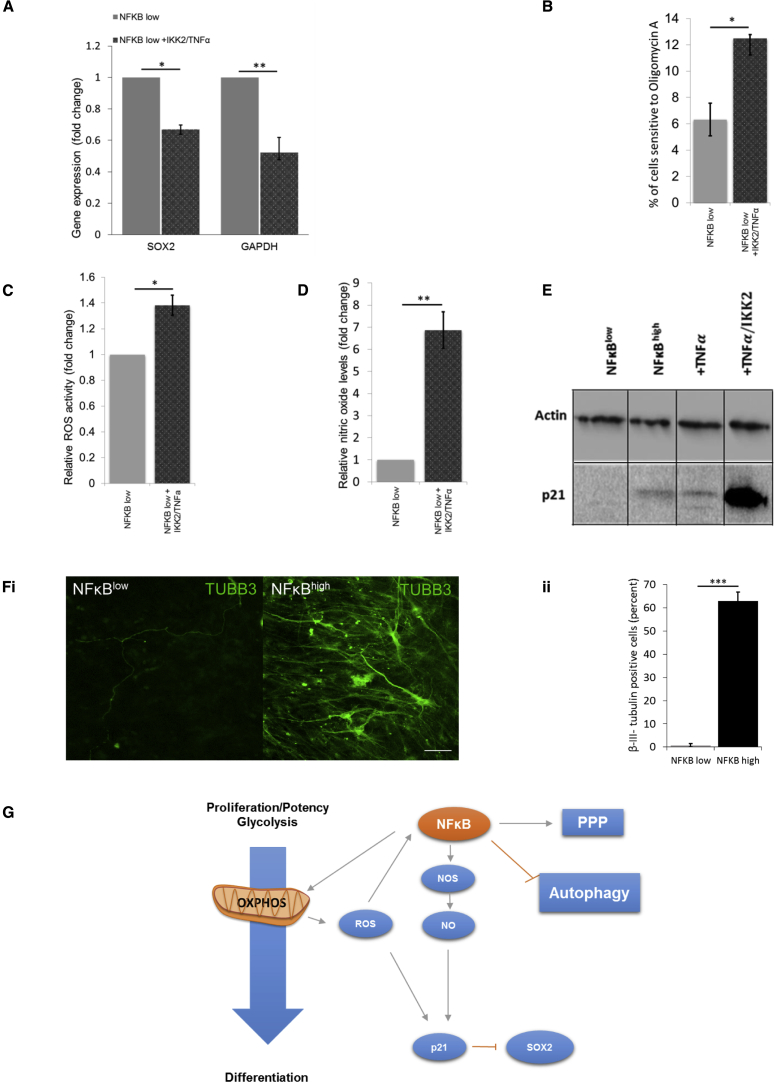
To mechanistically link NF-κB activity to neural maturation, metabolic reprogramming, and cell-cycle arrest in NPCs, we again used a combination of IKK-2 transgene expression and exogenous TNFα application to stimulate NF-κB in early-passage NFκBlow-NPC. Cells transduced with the LNT-NFκB-NanoLuc/EGFP reporter were responsive to both TNFα and IKK-2 induction (Figures S4A and S4B). NFκBlow-NPC subjected to IKK-2/TNFα treatment significantly decreased SOX2 and the glycolytic transcript GAPDH (Figure 6A) under maturation conditions. The proportion of ATP production susceptible to oligomycin A inhibition (OXPHOS, Figure 6B), ROS, and NO accumulation were all increased (Figures 6C and 6D). TNFα induced p21 in early-passage NPCs to broadly equivalent amounts but IKK2/TNFα treatments appeared to act synergistically to hyperactivate p21 (Figure 6E). In conclusion, NF-κB activation induces the neural maturation program in NPCs by redirecting glucose metabolism toward mitochondrial respiration for ATP production and the PPP for nucleotide production while slowing the cell cycle prior to any loss of Nestin expression or morphological changes indicative of neural maturation. Remarkably, we consistently observed that NPC would only undergo terminal differentiation to mature neurons after endogenous activation of NF-κB (Figure 6F). We observed ∼63% of generated β-III-tubulin-positive neurons compared with <1% in NFκBlow-NPC (Figure 6Fii). Furthermore, very few GFAP+ cells were present in terminally matured NFκBhigh neuronal cultures (Figure S6).
Figure 6.
NF-κB Agonists Induce the NFκBhigh-NPC Phenotype in Early-Passage NFκBlow-NPC
(A) SOX2 and GAPDH transcripts in NF-κB modulated NPCs (n = 3 independent biological repeats; ∗p ≤ 0.05, ∗∗p ≤ 0.01; error bars, ±SEM).
(B–D) Percentage of cells sensitive to oligomycin A (OXPHOS) (B), intracellular NO (C), and ROS activity (D) (all n = 3 independent biological repeats; ∗p ≤ 0.05, ∗∗p ≤ 0.01; error bars, ±SEM).
(E) p21 protein as assessed by western blot.
(F) (Fi) β-III-Tubulin immunocytochemistry of NFκBlow- and NFκBhigh-NPC subjected to our standard neural maturation protocol (representative images, n = 3). Scale bars, 100 μm. (Fii) proportion of β-III-tubulin-positive cells in each population (n = 3 independent biological repeats; ∗∗∗p ≤ 0.001; error bars, ±SEM).
(G) Model of NF-κB activity during neural specification. Upregulation of NF-κB, potentially due to an accumulation of ROS, initiates a feedforward loop increasing mitochondrial biogenesis and respiration, and increased NO production resulting in upregulation of p21, which slows the cell cycle and represses SOX2. NF-κB also inhibits autophagy and increases anabolic PPP to initiate a maturation program capable of efficient differentiation to mature neurons.
Discussion
In our study, we propose a model of neurogenesis whereby endogenous NF-κB activation in committed NPCs induced a maturation phenotype by orchestrating metabolic reprogramming (Figure 6G). Two recent studies showed that NF-κB signaling promotes mouse hippocampal NSC differentiation when extrinsically activated by inflammation (Zhang et al., 2013a) (Li et al., 2012). However, neither study explored the molecular mechanisms underlying NFκB-mediated differentiation.
We were able to discriminate between two discrete populations of proliferating NPC, NFκBhigh- and NFκBlow-NPC. Although maintained in medium containing heparin and FGF2, NFκBhigh-NPC expressed lower levels of multipotency markers, demonstrated an elongated cell cycle, and more efficiently generated neurons compared with their NFκBlow counterparts. Simultaneous loss of multipotency and slowing of the cell cycle toward terminal maturation has only previously been observed after induction with exogenous molecules such as dbCAMP or retinoic acid (Hansen et al., 2000, Tojima et al., 2003, López-Carballo et al., 2002, Qiao et al., 2012, Cañón et al., 2004).
The metabolic profile of our two NPC populations also differed significantly; the mitochondrial function of NFκBhigh-NPC was characteristically similar to that of neurons. NFκBhigh-NPC had elongated mitochondria and increased mitochondrial membrane potential, and were significantly more dependent on OXPHOS for energy production compared with NFκBlow-NPC.
Upon exogenous NF-κB activation in NFκBlow-NPC we saw a significant increase in energy production by OXPHOS. Metabolic reprogramming, PPP, and pyruvate production have all been shown to be influenced by canonical NF-κB (p65/p50) signaling in a variety of diverse contexts (García-Nogales et al., 1999, Mauro et al., 2011). The increased PPP activity and decreased lactate release observed in the NFκBhigh-NPC is consistent with the typical glycolysis-to-PPP switch previously described in differentiated neurons (Herrero-Mendez et al., 2009). Although there was no change observed in glycolysis, the decreased lactate release reflects increased pyruvate oxidation by mitochondria in NFκBhigh-NPC. Energy production via OXPHOS results in the production of ROS, specifically SO, and NO that is potentially toxic to cells at elevated concentrations but crucially also act as important second messengers in signal transduction (Le Belle et al., 2011). The induction of OXPHOS by NF-κB stimulation in NFκBlow-NPC implied a consequential increase in ROS and NO levels. As anticipated, we did observe either a direct or indirect oxidative stress response to NF-κB activity. ROS and NO have been shown to induce cell signaling pathways in normal neural development without inducing cell death (Tsatmali et al., 2006). When NO is present alongside SO, peroxynitrite acts as a pro-oxidant agent able to activate protective antioxidant pathways, including the PPP (García-Nogales et al., 2003), which may explain the absence of apoptosis in our cultures. Interestingly, both increased ROS and NO have been shown to induce p21-mediated cellular senescence through independent mechanisms (Kibbe et al., 2000, Owusu-Ansah et al., 2008). Furthermore, two recent studies have shown in ESCs and NSCs that, when expressed, p21 binds the SOX2 promoter to prevent expression and induce differentiation (Marqués-Torrejón et al., 2013, Yamamizu et al., 2014). These studies corroborate our data showing decreased SOX2 and increased p21 expression in NFκBhigh-NPC, and the upregulation of p21 observed in stimulation of NFκBlow-NPC. Collectively, these data support a hypothesis that a cellular maturation program is combining both metabolic and cell-cycle changes.
NFκBhigh-NPC demonstrated higher levels of autophagic and lysosomal proteins LC3-II and LAMP1 compared with NFκBlow-NPC but showed reduced autophagic flux. There is some ambiguity in the literature with regard to the role of autophagic proteins in neural differentiation. An increase in the levels of LC3-II is indicative of either elevated levels of autophagosome synthesis or a reduction in the turnover in autophagosomes. Treatment with Baf confirms that in our NFκBhigh-NPC, LC3-II increase is due to a decline in autophagosome turnover.
Our data fit with the autophagic profile observed in in vivo and in vitro neurogenesis models. In vivo analysis of autophagy proteins in LC3-GFP transgenic mice indicates that autophagy is differentially regulated in organs. In the brain, neurons are resistant to autophagic flux irrespective of nutrient availability (Mizushima et al., 2004). Moreover, Zeng and Zhou (2008) showed that upon treatment with retinoic acid, the levels of LC3-II increase mouse neuroblastoma cells. We noticed a similar increase in LC3-II/I ratio in NFκBhigh-NPC, without the requirement for exogenous maturation stimulants. Lemasters (2005) analyzed the potential association between autophagic flux and OXPHOS and noted that an increase in oxidative phosphorylation resulted in simultaneous elevation in the levels of LC3-II protein. We have shown that NF-κB links OXPHOS and autophagy during neural specification; we demonstrated that NF-κB induces an upregulation of key autophagy protein p62, which has recently been linked in a positive feedback loop with NF-κB in a cancer model (Yang et al., 2017).
Although the increase in mTOR activity observed in NFκBhigh-NPC was consistent with the reduction in autophagic flux, we also observed an upregulation in TFEB activity. However, a recent study by Li et al. describes an mTORC1-independent mechanism of TFEB-mediated lysosomal biogenesis via PKC (Li et al., 2016) and it is plausible that NF-κB/PKC is stimulating lysosomal activity in an mTORC1-independent manner.
In summary, we have identified a key role for NF-κB signaling during neural specification of hESCs in vitro. It will be important to further substantiate our data in vivo by tracking of NF-κB activity during early neurogenesis in order to distinguish how our NPC populations map onto the developing brain. Moreover, it would be fascinating to selectively modulate NF-κB activity in CNS organoid culture to model the consequences of prolonged NF-κB upregulation on CNS development. It would be of great value to assess the effects on neurogenesis in specific CNS lineages, but this was beyond the limitations of our current model. A more complete understanding of the role played by NF-κB in NPC commitment and terminal neural differentiation, as well as the mechanistic contribution of canonical NF-κB signaling to metabolic reprogramming, could result in the establishment of better in vitro models of human neural development. Finally, targeting NFκB-mediated neurogenesis could represent a mechanism of preventing depletion of the NSC pool associated with chronic inflammation during normal aging or dementia.
Experimental Procedures
Maintenance of Cell Lines, MEFs, and Shef3 hESCs
The Shef3 and Shef6 hESC line was obtained from the UK Stem Cell Bank under the project SCSC10-48. HEK293T cells were a kind gift from Dr. Steve Howe (Institute of Child Health, University College London) and mouse embryonic fibroblasts (MEFs) were from Cambridge Bioscience (CBA-310). MEFs were cultured in complete medium (DMEM supplemented with 10% [v/v] fetal bovine serum, 1× [v/v] penicillin/streptomycin, and 1× [v/v] non-essential amino acids [NEAA]). MEFs were mitotically inactivated with mitomycin C (0.1 μg/mL) (all from Sigma). MEFs were seeded at a density of 5 × 104 cells/cm2 on 0.1% (w/v) gelatin-coated tissue culture dishes. hESCs were maintained on MEFs in hESC medium (DMEM/F12 supplemented with NEAA, 20% knockout serum replacement [Life Technologies], 0.1 mM β-mercaptoethanol [Sigma], and 10 ng/mL FGF2 [Peprotech]). Colonies were mechanically passaged every 5–7 days. hESCs were conditioned to feeder-free culture by TrypLE Express (Gibco) enzyme dissociation and plating at high density (1:1) onto human purified fibronectin (20 μg/mL, Millipore)-coated flasks in mTESR (STEMCELL Technologies). Contaminating inactivated MEFs were diluted out within three passages.
Neural Differentiation of hESCs
Feeder-free hESCs (Shef3/Shef6) were replated in neural induction medium (NIM; DMEM/F12, 1× NEAA, 1× N2 supplement [all from Life Technologies], 4 mg/mL polyvinyl alcohol [Sigma], 2 μg/mL heparin sodium salt [Sigma], and 20 ng/mL FGF2) in uncoated, V-shaped 96-well plates at a density of 1 × 104 cells/well. Within 24 hr, neuroepithelial clusters formed. Medium was replaced daily for 5 days; on day 6 aggregates were replated in NIM onto 20 μg/mL laminin (Milipore)-coated dishes to allow neural rosette formation (2–3 days). Neural rosette clusters were mechanically isolated and replated onto laminin-coated dishes in neural expansion medium (NIM plus 1× B27 supplement [Life Technologies]). Early- and late-passage NPCs were cultured in the same conditions. All cells were cultured in atmospheric oxygen. NPCs were passaged 1:3 using TrypLE. To direct terminal differentiation of neural cells, we replated NPCs onto a thin layer of Matrigel at a density of 1.3 × 104 cells/cm2. Medium was replaced 48 hr later with neural maturation medium (DMEM/F12 [Life Technologies], 1× B27, L-glutamine, and 1% horse serum [both from Sigma]).
Transcriptomic Microarray
RNA was isolated from three biological repeats at the ESC, NEC, NRPC, and NPC stages of differentiation. The whole-genome gene expression array (Human HT-12 v4 Expression BeadChip Kit [Illumina]) was processed with quality control assessment by Dr. Jayne Dennis (School of Biological and Chemical Sciences, Queen Mary University of London). The GPLOTS package in R studio generated heatmaps of normalized data where log2(x) was plotted; mean was set to 0 and SD to 1. Public microarray data obtained from Affymetrix Mouse Genome 430 2.0 chips (accession number GEO: GPL1261) were downloaded from the NCBI GEO ftp site. Custom Unix/Linux and R version 2.0 scripts were developed, filtered, and analyzed by packages built by Bioconductor (R) to access the data. The scripts were designed to isolate microarray data obtained from experiments with keywords such “neural stem,” “neural progenitor,” or “neuron.” Raw data (cel files) were corrected for background and normalized using the Bioconductor “GCRMA” package. The R studio “cor” function was used to generate correlation matrices using the Pearson method. Subsequent data from the meta-analyses and normalized data from the Illumina microarray were assessed using Geneontology.org, the PANTHER classification system KEGG 2015, and EnrichR.
Metabolic Analysis Using Cell Titer Glo Assay and Seahorse XFp Platform
The Cell Titer Glo Luminescent Assay (Promega) measuring ATP production was used as described previously (Hawkins et al., 2016). Oxygen consumption rate (OCR) in NFκBlow and NFκBhigh-NPCs was measured using the Seahorse XFp Bioanalyzer (Agilent) Cell Mito Stress Test Kit. Spare respiratory capacity was calculated by normalizing to baseline OCR.
Lactate Production, Glycolytic Flux, and PPP Analysis
PPP flux was measured in 8-cm2 flasks of adherent cells at 60%–70% confluence containing a central microcentrifuge tube with either 0.8 mL of benzethonium hydroxide (Sigma) for 14CO2 equilibration or 1 mL of H2O for 3H2O equilibration. Incubations were carried out in Krebs Ringer Phosphate buffer (145 mM NaCl, 5.7 mM Na2HPO4, 4.86 mM KCl, 0.54 mM CaCl2, 1.22 mM 33 MgSO4 [pH 7.4]) containing 5 mM D-glucose (KPRG buffer) at 37°C. To ensure adequate oxygen supply for oxidative metabolism throughout the incubation period, we filled flasks with oxygen before sealing them.
To measure the carbon flux from glucose through the PPP, we incubated cells in KPRG buffer supplemented with 0.5 μCi D-[1-14C]glucose or [6-14C]glucose for 90 min as previously described (Herrero-Mendez et al., 2009, Rodriguez-Rodriguez et al., 2013). Cellular respiration was then terminated by the addition of 0.2 mL of 20% perchloric acid (Merck Millipore) for 40 min before the benzethonium hydroxide (containing 14CO2) was removed and the radioactivity measured by liquid scintillation counting. PPP flux was calculated as the difference between 14CO2 production from [1-14C]glucose (which decarboxylates through the 6-phosphogluconate dehydrogenase-catalyzed reaction) and that of [6-14C]glucose (which decarboxylates through the TCA cycle) (Herrero-Mendez et al., 2009, Larrabee, 1990). Glycolytic flux was measured by assaying the rate of 3H2O production from [3-3H]glucose using a similar method but instead incubating cells with 5 μCi D-[3-3H]glucose in KPRG buffer per flask for 120 min as previously described (Herrero-Mendez et al., 2009, Rodriguez-Rodriguez et al., 2013). Respiration was then terminated with perchloric acid and by incubating the cells for 96 hr with a microcentrifuge tube containing H2O suspended above the cells to allow 3H2O equilibration. The 3H2O was then removed and measured by liquid scintillation counting. Under these experimental conditions, 75% of the produced CO2 or 28% of the produced 3H2O was recovered, as previously established (Rodriguez-Rodriguez et al., 2013).
ROS and RNS Detection
IKK2/TNFα-treated or control cells were plated in a 96-well format at a density of 1 × 104 cells/well before quantifying ROS/RNS (reactive nitrogen species) by colorimetric assay (Enzo Life Sciences) according to the manufacturer's instructions. Non-overlapping fluorescent detection probes discriminate between global levels of ROS (excitation [Ex] 490 nm, emission [Em] 525 nm) and superoxide (Ex 550, Em 620 nm). Cellular NO was quantified by fluorescence absorbance (Ex 495 nm, Em 515 nm) as the non-fluorescent precursor diaminofluorescein diacetate (DAF-2-DA) is converted to the fluorescent triazole derivative, DAF-2T in the presence of NO and O2.
In Vitro Lentiviral Transcription Factor-Activated Reporter Assay
Production of high-titer NF-κB TFAR lentivirus was performed in HEK293T cells as previously described (Buckley et al., 2015). hESCs and NRPCs were transduced at an MOI of 25 with the NF-κB TFAR and an MOI of 2.5 of the constitutively active LNT-SFFV-Vluc construct (expressing vargula luciferase) (Figure 3A). These constructs are described in Hawkins et al. (2016). Conditioned supernatant was collected after 6 hr. Twenty microliters of sample was combined with 20 μL of luciferase assay buffer (25 mM Tris phosphate [pH 7.8], 1 mM DTT, 1 mM EDTA, 1% Triton X-100, 8 mM 6MgCl2, 30% glycerol, 2% BSA) in a white-bottomed plate with technical repeats in triplicate. Samples were injected with either0.5 μg/mL coelenterazine (GoldBio) or 5 nM cypridina luciferin (Nanolight), and photon emissions were measured at 460 nM.
pCMV-IKK2 Plasmid Transfection
pLNT NFκB-NanoLuc/EGFP/pLNT-SFFV-Vluc transduced NPCs were transfected using 0.1 mM PEI (PEI/DNA ratio of 1:1) with pCMV-IKK2 (Addgene) and a control, pCMV-FLAG. To detect NF-κB activity, we analyzed the medium by luminometry at the following time points: 24 hr, 48 hr, and 72 hr.
Immunocytochemistry and Live MitoTracker Imaging
For fluorescence immunocytochemistry and confocal microscopy, cells were fixed in 4% paraformaldehyde (PFA) or 100% ice-cold MeOH (for confocal). Samples were blocked in 2% BSA in PBS + 0.05% (v/v) Tween 20 (all from Sigma). Antibodies were diluted in blocking buffer (SOX2 1:100 [#AF2018], β-III-tubulin 1:200 [#MAB1195], Nestin 1:200 [#MAB1295] [all R&D Systems], LAMP1 1:200 [Abcam #Ab24170], p21 1:400 [Cell Signaling Technologies #2947]) and incubated overnight at 4°C. Secondary antibodies (1:500, all Alexa Fluor) were applied for 1 hr at room temperature. Immunostained cells were mounted with or without DAPI and analyzed using an Olympus IX81 inverted fluorescence microscope or the Leica TCS SP5 X for confocal. Mitochondria were imaged using MitoTracker probes (25 nM) (Life Technologies), following the manufacturer's instructions.
qRT-PCR
Total RNA was isolated from cells using the RNeasy kit (Qiagen) according to the manufacturer's instructions. Random hexamer primers and M-MLV reverse transcriptase (both Promega) were used to reverse transcribe RNA, as per the manufacturer's instructions. Triplicate samples were analyzed by the Bio-Rad CFX Connect Real-Time PCR detection system using Fast SYBR Green master mix (Thermo Fisher Scientific) with the appropriate primer sets (0.4 μM, Table S1). Transcript levels were normalized to PABPC4. This target was chosen because in our microarray it showed no normalized alteration in ESC, NEC, NRPC, NPC, and neuronal cultures (our unpublished data).
Western Blotting
Total protein was quantified using the Bio-Rad Protein Assay. Lysates were resolved on SDS-PAGE and electrotransferred onto Hybond-enhanced chemiluminescence (ECL) nitrocellulose membrane (Amersham Biosciences). The membrane was blocked in 5% BSA in PBS + 0.05% Tween 20 and incubated overnight in primary antibody p21 (1:1,000, Cell Signaling #2947), β-actin (1:4,000, Sigma #A2228), pP70S6K (1:1,000, Cell Signaling #108D2) anti-P70S6K (1:1,000, Stressgen #KAP-CC035), LC3 (1:1,000 Novus Biologicals #NB100-2220), p62 (1:1,000, Abcam #Ab56416) at 4°C. Blots were incubated with the appropriate horseradish peroxidase-conjugated secondary antibody (Dako, 1:2,000) in blocking buffer for 1 hr at room temperature. The membrane was developed using ECL (Pierce/Amersham) according to the manufacturer's instructions. Densitometric quantification was performed on unsaturated images in ImageJ (NIH).
Flow Cytometry
Cell cycle was assayed in control or IKK-2/TNFα-treated NPCs using propidium iodide (PI) (Sigma). Cells were dissociated in TrypLE, resuspended in PBS, and centrifuged at 900 rpm for 8 min. Supernatant was discarded and 1 mL of fixative (70% ethanol) was added drop-wise while vortexing. Cells were incubated in fixative for 30 min at 4°C. After centrifugation cells were washed with PBS before the supernatant was discarded and 100 μL of a 100 μg/mL stock solution of Rnase A was used to resuspend the cells. Four hundred microliters of a 50 μg/mL stock of PI was added and the reaction was incubated in darkness for 10 min prior to analysis. PI staining was analyzed using a Becton Dickinson (BD Bioscience) FACScalibur flow cytometer and CellQuest Pro analysis software (BD Bioscience). For detection of neural markers in LNT NFκB-NanoLuc/EGFP transduced cells, cultures were trypsinized, washed in PBS, and resuspended in 4% PFA for 20 min at room temperature. Samples were permeabilized in 0.3% Triton X-100 and blocked in 2% BSA (Sigma-Aldrich) + 0.05% (v/v) Tween 20. Antibodies were diluted in blocking buffer (SOX10 1:100 [#AF2864], β-III-tubulin 1:200 [#MAB 1195], Nestin 1:200 [#MAB1259] [all R&D Systems]), and incubated for 1 hr at room temperature. Untransduced NPCs and immunoglobulin G from mouse and goat serum were used as negative controls (Sigma #I5381, #I5006). Secondary antibodies (1:500, all Alexa Fluor #647) were applied for 1 hr at room temperature and samples were washed prior to FACS analysis.
Senescence-Associated β-Galactosidase Assay
NPCs were fixed for 5 min with 0.5% (v/v) glutaraldehyde (pH 7.2), washed in PBS supplemented with 1 mM MgCl2, and incubated in β-galactosidase solution (120 μM potassium ferricyanide/potassium ferrocyanide [pH 7.0] and 1 mg/mL X-gal) at 37°C overnight. After incubation, cells were washed in an appropriate volume of PBS and analyzed by bright-field microscopy.
Statistical Analysis
Statistical analysis of in vitro vector analysis data, qPCR, and metabolic assays was performed using an unpaired Student's two-tailed t test. Two-way ANOVA was employed for Figure S4B. All data are expressed as mean values ± SEM, with each sample being measured at least in triplicate.
Author Contributions
The project was conceived by T.R.M., L.M.F., and S.N.W., and experiments were carried out by L.M.F., K.E.H., J.M.K.M.D., and L.B. Metabolic assays were designed and carried out by E.F. and J.P.B., and autophagy experiments were designed and carried out by C.S. and D.L.M. A.N. scripted and performed meta-analyses. The manuscript was written by L.M.F., J.B.P., D.L.M., and T.R.M.
Acknowledgments
We would like to acknowledge Dr. Jayne Dennis for bioinformatics analysis of microarray data. We would also like to thank Aldevron for de novo synthesis and plasmid manufacture. L.M.F. and K.E.H. were partly funded by St. George's, University of London. T.R.M., J.M.K.M.D., and S.N.W. were funded by ERC grant “Somabio” (260862) and the NC3Rs (NC/L001780/1). J.P.B. is funded by MINECO (SAF2016-78114-R) CIBERFES (CB16/10/00282), SP3-People-MC-ITN program (608381). L.M.F., T.R.M., C.S., D.L.M., and J.P.B. are all funded by Horizon 2020 grant BATCure (666918). S.N.W. received funding from UK Medical Research Council grants (G1000709 and MR/N026101/1, MR/R015325/1, MR/P026494/1, and MR/N019075/1) and from SPARKS (17UCL01).
Published: April 19, 2018
Footnotes
Supplemental Information includes six figures and two tables and can be found with this article online at https://doi.org/10.1016/j.stemcr.2018.03.015.
Supplemental Information
s
References
- Buckley S.M., Delhove J.M., Perocheau D.P., Karda R., Rahim A.A., Howe S.J., Ward N.J., Birrell M.A., Belvisi M.G., Arbuthnot P. In vivo bioimaging with tissue-specific transcription factor activated luciferase reporters. Sci. Rep. 2015;5:11842. doi: 10.1038/srep11842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burridge P.W., Thompson S., Millrod M.A., Weinberg S., Yuan X., Peters A., Mahairaki V., Koliatsos V.E., Tung L., Zambidis E.T. A universal system for highly efficient cardiac differentiation of human induced pluripotent stem cells that eliminates interline variability. PLoS One. 2011;6:e18293. doi: 10.1371/journal.pone.0018293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cañón E., Cosgaya J.M., Scsucova S., Aranda A. Rapid effects of retinoic acid on CREB and ERK phosphorylation in neuronal cells. Mol. Biol. Cell. 2004;15:5583–5592. doi: 10.1091/mbc.E04-05-0439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denis-Donini S., Caprini A., Frassoni C., Grilli M. Members of the NF-kappaB family expressed in zones of active neurogenesis in the postnatal and adult mouse brain. Brain Res. Dev. Brain Res. 2005;154:81–89. doi: 10.1016/j.devbrainres.2004.10.010. [DOI] [PubMed] [Google Scholar]
- Denis-Donini S., Dellarole A., Crociara P., Francese M.T., Bortolotto V., Quadrato G., Canonico P.L., Orsetti M., Ghi P., Memo M. Impaired adult neurogenesis associated with short-term memory defects in NF-kappaB p50-deficient mice. J. Neurosci. 2008;28:3911–3919. doi: 10.1523/JNEUROSCI.0148-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dottori M., Pera M.F. Neural differentiation of human embryonic stem cells. Methods Mol. Biol. 2008;438:19–30. doi: 10.1007/978-1-59745-133-8_3. [DOI] [PubMed] [Google Scholar]
- Fimia G.M., Stoykova A., Romagnoli A., Giunta L., Di Bartolomeo S., Nardacci R., Corazzari M., Fuoco C., Ucar A., Schwartz P., Gruss P. Ambra1 regulates autophagy and development of the nervous system. Nature. 2007;447:1121–1125. doi: 10.1038/nature05925. [DOI] [PubMed] [Google Scholar]
- García-Nogales P., Almeida A., Fernández E., Medina J.M., Bolaños J.P. Induction of glucose-6-phosphate dehydrogenase by lipopolysaccharide contributes to preventing nitric oxide-mediated glutathione depletion in cultured rat astrocytes. J. Neurochem. 1999;72:1750–1758. doi: 10.1046/j.1471-4159.1999.721750.x. [DOI] [PubMed] [Google Scholar]
- García-Nogales P., Almeida A., Bolaños J.P. Peroxynitrite protects neurons against nitric oxide-mediated apoptosis. A key role for glucose-6-phosphate dehydrogenase activity in neuroprotection. J. Biol. Chem. 2003;278:864–874. doi: 10.1074/jbc.M206835200. [DOI] [PubMed] [Google Scholar]
- Hatano E., Bennett B.L., Manning A.M., Qian T., Lemasters J.J., Brenner D.A. NF-kappaB stimulates inducible nitric oxide synthase to protect mouse hepatocytes from TNF-alpha- and Fas-mediated apoptosis. Gastroenterology. 2001;120:1251–1262. doi: 10.1053/gast.2001.23239. [DOI] [PubMed] [Google Scholar]
- Hansen T.O., Rehfeld J.F., Nielsen F.C. Cyclic AMP-induced neuronal differentiation via activation of p38 mitogen-activated protein kinase. J. Neurochem. 2000;75:1870–1877. doi: 10.1046/j.1471-4159.2000.0751870.x. [DOI] [PubMed] [Google Scholar]
- Hawkins K.E., Joy S., Delhove J.M., Kotiadis V.N., Fernandez E., Fitzpatrick L.M., Whiteford J.R., King P.J., Bolanos J.P., Duchen M.R. NRF2 orchestrates the metabolic shift during induced pluripotent stem cell reprogramming. Cell Rep. 2016;14:1883–1891. doi: 10.1016/j.celrep.2016.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero-Mendez A., Almeida A., Fernández E., Maestre C., Moncada S., Bolaños J.P. The bioenergetic and antioxidant status of neurons is controlled by continuous degradation of a key glycolytic enzyme by APC/C–Cdh1. Nat. Cell Biol. 2009;11:747. doi: 10.1038/ncb1881. [DOI] [PubMed] [Google Scholar]
- Iliopoulos D., Hirsch H.A., Struhl K. An epigenetic switch involving NF-kappaB, Lin28, Let-7 MicroRNA, and IL6 links inflammation to cell transformation. Cell. 2009;139:693–706. doi: 10.1016/j.cell.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawauchi K., Araki K., Tobiume K., Tanaka N. p53 regulates glucose metabolism through an IKK-NF-kappaB pathway and inhibits cell transformation. Nat. Cell Biol. 2008;10:611–618. doi: 10.1038/ncb1724. [DOI] [PubMed] [Google Scholar]
- Kibbe M.R., Li J., Nie S., Watkins S.C., Lizonova A., Kovesdi I., Simmons R.L., Billiar T.R., Tzeng E. Inducible nitric oxide synthase (iNOS) expression upregulates p21 and inhibits vascular smooth muscle cell proliferation through p42/44 mitogen-activated protein kinase activation and independent of p53 and cyclic guanosine monophosphate. J. Vasc. Surg. 2000;31:1214–1228. doi: 10.1067/mva.2000.105006. [DOI] [PubMed] [Google Scholar]
- Larrabee M.G. Evaluation of the pentose phosphate pathway from 14CO2 data. Fallibility of a classic equation when applied to non-homogeneous tissues. Biochem. J. 1990;272:127–132. doi: 10.1042/bj2720127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Belle J.E., Orozco N.M., Paucar A.A., Saxe J.P., Mottahedeh J., Pyle A.D., Wu H., Kornblum H.I. Proliferative neural stem cells have high endogenous ROS levels that regulate self-renewal and neurogenesis in a PI3K/Akt-dependant manner. Cell Stem Cell. 2011;8:59–71. doi: 10.1016/j.stem.2010.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemasters J.J. Selective mitochondrial autophagy, or mitophagy, as a targeted defense against oxidative stress, mitochondrial dysfunction, and aging. Rejuvenation Res. 2005;8:3–5. doi: 10.1089/rej.2005.8.3. [DOI] [PubMed] [Google Scholar]
- Li J., Tang Y., Cai D. IKKbeta/NF-kappaB disrupts adult hypothalamic neural stem cells to mediate a neurodegenerative mechanism of dietary obesity and pre-diabetes. Nat. Cell Biol. 2012;14:999–1012. doi: 10.1038/ncb2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Xu M., Ding X., Yan C., Song Z., Chen L., Huang X., Wang X., Jian Y., Tang G. Protein kinase C controls lysosome biogenesis independently of mTORC1. Nat. Cell Biol. 2016;18:1065. doi: 10.1038/ncb3407. [DOI] [PubMed] [Google Scholar]
- López-Carballo G., Moreno L., Masiá S., Pérez P., Barettino D. Activation of the phosphatidylinositol 3-kinase/Akt signaling pathway by retinoic acid is required for neural differentiation of SH-SY5Y human neuroblastoma cells. J. Biol. Chem. 2002;277:25297–25304. doi: 10.1074/jbc.M201869200. [DOI] [PubMed] [Google Scholar]
- Marqués-Torrejón M.Á., Porlan E., Banito A., Gómez-Ibarlucea E., Lopez-Contreras A.J., Fernández-Capetillo O., Vidal A., Gil J., Torres J., Fariñas I. Cyclin-dependent kinase inhibitor p21 controls adult neural stem cell expansion by regulating SOX2 gene expression. Cell Stem Cell. 2013;12:88–100. doi: 10.1016/j.stem.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauro C., Leow S.C., Anso E., Rocha S., Thotakura A.K., Tornatore L., Moretti M., De Smaele E., Beg A.A., Tergaonkar V. NF-kappaB controls energy homeostasis and metabolic adaptation by upregulating mitochondrial respiration. Nat. Cell Biol. 2011;13:1272–1279. doi: 10.1038/ncb2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N., Yamamoto A., Matsui M., Yoshimori T., Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol. Biol. Cell. 2004;15:1101–1111. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan A.M., Collins L.M., Wyatt S.L., Gutierrez H., O'Keeffe G.W. The neurite growth inhibitory effects of soluble TNFalpha on developing sympathetic neurons are dependent on developmental age. Differentiation. 2014;88:124–130. doi: 10.1016/j.diff.2014.12.006. [DOI] [PubMed] [Google Scholar]
- Owusu-Ansah E., Yavari A., Mandal S., Banerjee U. Distinct mitochondrial retrograde signals control the G1-S cell cycle checkpoint. Nat. Genet. 2008;40:356–361. doi: 10.1038/ng.2007.50. [DOI] [PubMed] [Google Scholar]
- Qiao J., Paul P., Lee S., Qiao L., Josifi E., Tiao J.R., Chung D.H. PI3K/AKT and ERK regulate retinoic acid-induced neuroblastoma cellular differentiation. Biochem. Biophys. Res. Commun. 2012;424:421–426. doi: 10.1016/j.bbrc.2012.06.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Rodriguez P., Almeida A., Bolaños J.P. Brain energy metabolism in glutamate-receptor activation and excitotoxicity: role for APC/C-Cdh1 in the balance glycolysis/pentose phosphate pathway. Neurochem. Int. 2013;62:750–756. doi: 10.1016/j.neuint.2013.02.005. [DOI] [PubMed] [Google Scholar]
- Rolls A., Shechter R., London A., Ziv Y., Ronen A., Levy R., Schwartz M. Toll-like receptors modulate adult hippocampal neurogenesis. Nat. Cell Biol. 2007;9:1081–1088. doi: 10.1038/ncb1629. [DOI] [PubMed] [Google Scholar]
- Tojima T., Kobayashi S., Ito E. Dual role of cyclic AMP-dependent protein kinase in neuritogenesis and synaptogenesis during neuronal differentiation. J. Neurosci. Res. 2003;74:829–837. doi: 10.1002/jnr.10754. [DOI] [PubMed] [Google Scholar]
- Tsatmali M., Walcott E.C., Makarenkova H., Crossin K.L. Reactive oxygen species modulate the differentiation of neurons in clonal cortical cultures. Mol. Cell. Neurosci. 2006;33:345–357. doi: 10.1016/j.mcn.2006.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worlitzer M.M., Schwamborn J.C. The Notch co-repressor protein NKAP is highly expressed in adult mouse subventricular zone neural progenitor cells. Neuroscience. 2014;266:138–149. doi: 10.1016/j.neuroscience.2014.02.019. [DOI] [PubMed] [Google Scholar]
- Yamamizu K., Schlessinger D., Ko M.S. SOX9 accelerates ESC differentiation to three germ layer lineages by repressing SOX2 expression through P21 (WAF1/CIP1) Development. 2014;141:4254–4266. doi: 10.1242/dev.115436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanishi E., Yoon K., Alberi L., Gaiano N., Mizutani K. NF-kappaB signaling regulates the generation of intermediate progenitors in the developing neocortex. Genes Cells. 2015;20:706–719. doi: 10.1111/gtc.12267. [DOI] [PubMed] [Google Scholar]
- Yang S., Qiang L., Sample A., Shah P., He Y.Y. NF-kB signaling activation induced by chloroquine requires autophagosome, p62 protein, and C-Jun N-terminal kinase (JNK) signaling and promotes tumor cell resistance. J. Biol. Chem. 2017;292:3379–3388. doi: 10.1074/jbc.M116.756536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng M., Zhou J.N. Roles of autophagy and mTOR signaling in neuronal differentiation of mouse neuroblastoma cells. Cell. Signal. 2008;20:659–665. doi: 10.1016/j.cellsig.2007.11.015. [DOI] [PubMed] [Google Scholar]
- Zhang X.Q., Zhang S.C. Differentiation of neural precursors and dopaminergic neurons from human embryonic stem cells. Methods Mol. Biol. 2010;584:355–366. doi: 10.1007/978-1-60761-369-5_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G., Li J., Purkayastha S., Tang Y., Zhang H., Yin Y., Li B., Liu G., Cai D. Hypothalamic programming of systemic ageing involving IKK-beta, NF-kappaB and GnRH. Nature. 2013;497:211–216. doi: 10.1038/nature12143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Liu L., Wang R., Tuo H., Guo Y., Yi L., Wang J., Wang D. MiR-199a-5p promotes migration and tube formation of human cytomegalovirus-infected endothelial cells through downregulation of SIRT1 and eNOS. Arch. Virol. 2013;158:2443–2452. doi: 10.1007/s00705-013-1744-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
s