Background
Barcoded read sequencing allows short-reads to carry long-range information by virtue of read “barcodes”, and has several advantages (including significantly reduced cost and lower error rates) over long-read sequencing. Here we introduce a two-tiered statistical binning approach, EMerAld—or EMA for short—to barcoded read sequence alignment, an essential component of any barcoded sequencing pipeline, and as a result improve downstream genotyping and phasing. Our method enables the probabilistic placement of reads between different read clouds [1], and also in a single cloud that spans homologous elements. The two tiers consist of: (i) a novel latent variable model to probabilistically assign reads to possible source fragments; and (ii) newly exploiting expected read coverage (read density) to resolve the difficult case of multiple repetitive alignments of reads within a single read cloud. These ambiguous alignments account for a large fraction of the rare variants that currently cannot be resolved and are of great interest to biologists [2].
Methods
Current linked-read alignment methods first perform a standard all-mapping, then partition the resulting alignments into groups of nearby reads with a common barcode called “read clouds”. Reads are then assigned to one of their possible clouds by optimizing a global score function that takes into account edit distance, mate pairs, read clouds, etc. Our two main conceptual advances are as follows. Intuitively, rather than assigning each read to just one of its possible alignments at any given time, we make use of probabilistic assignments of reads to clouds and employ a latent variable model to determine final alignment probabilities; thereby, we select the most likely cloud (and thus alignment) for each read. During the cloud alignment process, we also utilize a disjoint-set data structure over read clouds to normalize alignment probabilities in a physically sensible way. Once reads are assigned to clouds, we propose a different statistical binning optimization approach to better handle the ubiquitous repetitive regions of the genome. Whereas currently-used methods simply pick the lowest edit distance alignment of a read in a given cloud, we instead optimize a combination of edit distance and “read density”, which takes into account the read density distribution over fragments. This two-tiered process can be interpreted as statistical binning first in assigning reads to clouds and then within clouds. The EMA pipeline is shown in Fig. 1.
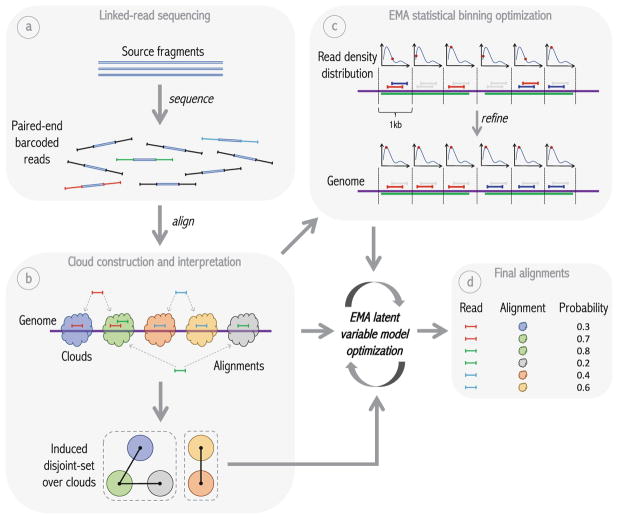
Fig. 1.
Overview of EMA pipeline. (a) Idealized model of linked-read sequencing, wherein some number of unknown source fragments in a single droplet are sheared, barcoded and sequenced to produce linked-reads. (b) EMA’s “read clouds” are constructed by grouping near-by-mapping reads sharing the same barcode; these clouds represent possible source fragments. EMA then partitions the clouds into a disjoint-set induced by the alignments, where two clouds are connected if there is a read aligning to both; connected components in this disjoint-set (enclosed by dashed boxes) correspond to alternate possibilities for the same unknown source fragment. EMA’s latent variable model optimization is subsequently applied to each of these connected components individually. (c) EMA applies a novel statistical binning optimization algorithm to clouds containing multiple alignments of the same read to pick out the most likely alignment, by optimizing a combination of alignment edit distances and read densities within the cloud. In the figure, the green regions of the genome are homologous, thereby resulting in multi-mappings within a single cloud. (d) While the statistical binning optimization operates within a single cloud, EMA’s latent variable model optimization determines the best alignment of a given read between different clouds, and produces not only the final alignment for each read, but also interpretable alignment probabilities.
Results
EMA is much faster and less memory intensive compared to other tools. EMA’s overhead over the initial run of an all-mapper is virtually negligible, and EMA is at least 1.5× faster than Lariat (the current 10x alignment tool [1]), which translates into days faster for the user. In addition, we show that genotypes called from EMA’s alignments contain over 30% fewer false positives than those called from Lariat’s, with a fewer number of false negatives, on 10x WGS datasets of NA12878 and NA24385, as compared to NIST GIAB gold standard variant calls. We also demonstrate that EMA’s alignments improve phasing performance over Lariat’s in both NA12878 and NA24385, producing fewer switch/mismatch errors and larger phased blocks on average.
Moreover, we demonstrate that EMA is able to effectively resolve alignments in regions containing nearby homologous elements—a particularly challenging problem in read mapping—through the introduction of our novel statistical binning optimization framework, which enables us to find variants in the pharmacogenomically important CYP2D region that go undetected when using Lariat or BWA. This enhanced capability addresses one of the major weaknesses of linked-read sequencing as compared to long-read sequencing, where only a relatively small subset of the original source fragment is observed—and more specifically, that the order of reads within the fragment is not known—making it difficult to produce accurate alignments if the fragment spans homologous elements.
Discussion
Our advance is a general framework applicable to many barcoded sequencing problems. It is likely to be of interest to any developers, and even users, of barcoded or linked-read sequencing technologies that come along. We highlight that 10x sequencing is just an instance of general “barcoded read sequencing”, and other technologies that make use of the same paradigm already exist and are likely to emerge in the future, given its numerous advantages over long-read sequencing. Several technologies already employ barcoded sequencing in addition to 10x Genomics’, such as Illumina’s TruSeq SLR platform (formerly Moleculo), and Complete Genomics’ Long Fragment technology. Our framework should apply to these (and similar) technologies as well. Due to their substantial improvements over existing methods for aligning and interpreting linked-read data, the algorithms employed by EMA are likely to be a fundamental component of read cloud-based methods in the future.
Acknowledgments
We thank Chris Whelan, Chad Nusbaum, Eric Banks, as well as the rest of the SV Group from the Broad Institute for providing us with data samples and many valuable suggestions. Also, we thank Jian Peng and Lillian Zhang for their helpful suggestions.
Funding A.S., I.N. and B.B. are partially funded by NIH grant GM108348.
References
- 1.Bishara A, et al. Read clouds uncover variation in complex regions of the human genome. Genome Res. 2015;25(10):1570–1580. doi: 10.1101/gr.191189.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sekar A, et al. Schizophrenia risk from complex variation of complement component 4. Nature. 2016;530:177. doi: 10.1038/nature16549. [DOI] [PMC free article] [PubMed] [Google Scholar]