Abstract
Several evidences indicate that aging negatively affects the effectiveness of influenza vaccination. Although it is well established that immunosenescence has an important role in vaccination response, the molecular pathways underlying this process are largely unknown. Given the importance of epigenetic remodeling in aging, here we analyzed the relationship between responsiveness to influenza vaccination and DNA methylation profiles in healthy subjects of different ages. Peripheral blood mononuclear cells were collected from 44 subjects (age range: 19 – 90 years old) immediately before influenza vaccination. Subjects were subsequently classified as responders or non-responders according to hemagglutination inhibition assay 4–6 weeks after the vaccination. Baseline whole genome DNA methylation in peripheral blood mononuclear cells was analyzed using the Illumina® Infinium 450k microarray. Differential methylation analysis between the two groups (responders and nonresponders) was performed through an analysis of variance, correcting for age, sex and batch. We identified 83 CpG sites having a nominal p-value < 0.001 and absolute difference in DNA methylation of at least 0.05 between the two groups. For some CpG sites, we observed age-dependent decrease or increase in methylation, which in some cases was specific for the responders and non-responders groups. Finally, we divided the cohort in two subgroups including younger (age < 50) and older (age ≥ 50) subjects and compared DNA methylation between responders and non-responders, correcting for sex and batch in each subgroup. We identified 142 differentially methylated CpG sites in the young subgroup and 305 in the old subgroup, suggesting a larger epigenetic remodeling at older ages. Interestingly, some of the differentially methylated probes mapped in genes involved in immunosenescence (CD40) and in innate immunity responses (CXCL16, ULK1, BCL11B, BTC). In conclusion, the analysis of epigenetic landscape can shed light on the biological basis of vaccine responsiveness during aging, possibly providing new appropriate biomarkers of this process.
Keywords: DNA methylation, Influenza vaccination, Aging, Immunosenescence
1. Introduction
Influenza is an important public health challenge in our countries, with yearly epidemics responsible of significant mortality, morbidity and loss of productivity (Paules and Subbarao, 2017). Specific populations such as very young children, individuals aged 65 years and older, or subjects with pre-existent conditions (immunocompromised states, cardiovascular or cerebrovascular diseases, diabetes, chronic respiratory failure, pregnancy) are particularly vulnerable to this infection and at greater risk for complications.
Vaccination is the most effective method to prevent influenza infection. Annual vaccination with an injectable trivalent inactivated vaccine is recommended, especially for individuals aged 65 years or older. However, the protection delivered by these vaccines is incomplete. Rates of protective immune response to vaccination are frequently low in vaccinated subjects, with worsened responses in older adults (Jefferson et al., 2010; Osterholm et al., 2012).
Part of the poor vaccine efficacy in the elderly is due to immunosenescence (Haralambieva et al., 2015; Kennedy et al., 2016; Targonski et al., 2007) but molecular pathways associated with impaired vaccine responses remain incompletely understood. Identification of the mechanisms associated with the development of a protective immunity is of central importance in vaccinology, in order to improve our capacity to predict response to vaccination or develop potential interventions to improve the immune responses.
To date, several studies have been conducted to identify genome-wide changes in transcriptional profiles that correlate with clinical response to influenza vaccination (Bucasas et al., 2011; Nakaya et al., 2011, 2015; Obermoser et al., 2013; Thakar et al., 2015; Tsang et al., 2014; Zhu et al., 2010), by assessing genome-wide gene expression with microarrays before and/or after vaccination of subjects. These molecular signatures, associated with better antibody responses, were frequently enriched in immune pathways, especially with type I interferon signaling, antigen presentation pathways or B-cell proliferation. Thakar et al. identified a dysregulation in this gene signature in older adults, specially in frail subjects who were nonresponders to vaccination (Thakar et al., 2015). Other large-scale profiling studies have tried to identify further relevant biomarkers that could predict vaccine response: in this attempt, Furman et al. identified nine immunological baseline predictors of protective immunity, with two of these variables involved in apoptosis (Furman et al., 2013). Finally, models integrating and combining transcriptomic data with additional data types to predict response to vaccination have been developed recently (Tsang et al., 2014; Zimmermann et al., 2017).
While transcriptomic data have been deeply studied in this field, few reports have been published regarding epigenetic aspects. DNA methylation has an important role in several biological processes, especially in aging (Sen et al., 2016), and is therefore an interesting candidate to be investigated. Furthermore, DNA methylation measures tend to be more stable than transcriptomic data within the short period (days-weeks) and are more reproducible from a technical point of view. Lu et al. discovered two relevant epigenetic variations in poor-responders to the vaccine directed against Hepatitis B virus (Lu et al., 2014). Concerning influenza vaccine, one recent study has identified numerous CpG sites showing associations with gene expression and other ones associated with the induction of the humoral immune response (Zimmermann et al., 2016).
To complete these findings and evaluate the effect of age on vaccination response, here we investigated baseline (that is, immediately before vaccination) genome-wide DNA methylation in peripheral blood mononuclear cells (PBMC) from 44 healthy donors, ranging from 19 to 90 years, who received influenza vaccination and were classified as responders and non-responders according to hemagglutination inhibition assays (HIA) after 28 days.
2. Materials and Methods
2.1 Participants
Participants (age range: 19 – 90 years) were recruited at the University of Miami Miller School of Medicine. Experiments were conducted using peripheral blood. Enrolled participants received the influenza vaccine in the pandemic season 2009 and in the season 2010–2011. Participants enrolled in the pandemic 2009 season received the subunit vaccine containing the A/California/7/2009 (H1N1) strain, whereas those enrolled in the 2010–2011 season received the Trivalent Inactivated influenza vaccine containing the following viral strains: A/California/7/2009 (H1N1), A/Perth/16/2009 (H3N2), B/Brisbane/60/2008. Whole blood samples were collected immediately before vaccination. PBMC were collected using Vacutainer CPT tubes (BD 362761). Cells were washed and cryopreserved. Appropriate signed informed consent was obtained from each subject prior to enrollment. The study was approved with IRB protocol #20070481. Each participant was asked questions regarding demographics, health behaviors, presence of symptoms associated with inflammatory conditions or respiratory infections at the time of enrollment. No one reported subclinical inflammatory conditions and/or had respiratory tract infections at the time of enrollment, nor was on any anti-inflammatory treatment or on medications known to alter the immune response. Participants were excluded if they had diseases known to alter the immune response.
2.2 Assessment of response to vaccination
Immunogenicity of influenza vaccine in subjects was assessed by hemagglutination inhibition assays (HIA). For this purpose, blood samples were collected immediately before vaccination (baseline) and 4–6 weeks after to evaluate the in vivo response and identify responders and non-responders. Responders had at least a 4-fold increase in the reciprocal of the titers in response to the whole vaccine, which was the A/California/7/2009 (H1N1) strain in the 2009 season and the Trivalent Inactivated Influenza vaccine (containing A/California/7/2009 (H1N1), A/Perth/16/2009 (H3N2), B/Brisbane/60/2008) in the 2010–2011 season. Briefly, sera were pretreated with receptor destroying enzyme (RDE, Denke Seiken Co Ltd) for 20 hours at 37°C; in order to inactivate this enzyme, sera were then heated at 56°C for 60 minutes. Two-fold serial dilutions were done; 25 µL of diluted sera were incubated with an equal volume of 4 HA units of the 2009 vaccine or of the 2010–2011 vaccine, for 1 hour at room temperature and then 50 µL of a 1.25 % suspension of chicken red blood cells were added. After 2 hours of incubation at room temperature titers were determined.
2.3 Genome-wide DNA methylation analysis
Genome-wide DNA methylation analysis was performed on PBMC collected immediately before vaccination (baseline) and cryopreserved. DNA methylation patterns are generally stable, highly reproducible and only slightly affected by freezing (Bulla et al., 2016). Total genomic DNA was extracted from PBMC using the AllPrep DNA/RNA Mini kit (Qiagen), according to manufacturer’s instructions. DNA concentrations were determined using NanoDrop spectrophotometer. DNA was bisulfite-converted using the EZ DNA Methylation Kit (Zymo Research Corporation®) and analyzed on the Infinium HumanMethylation450 BeadChip (Illumina®) following manufacturer's instructions (Bibikova et al., 2011). Arrays were scanned by HiScan (Illumina®) and signal intensities were extracted from .idat files using the minfi Bioconductor package (Aryee et al., 2014). Data were normalized using the preprocessQuantile function of the package minfi. Probes on the X and Y chromosomes were removed, as well as probes associated to a SNP. Identification of CpG sites with differential methylation between responders and non-responders to influenza vaccination was performed through an analysis of variance (ANOVA) model, correcting for sex and batch. CpG sites differentially methylated between responders and non-responders were defined as having a nominal p-value inferior to 0.01 and an absolute difference between values of responders and non-responders of at least 0.05. Figures were generated using R.
2.4 Epigenetic age estimation
DNAm age, also referred to as epigenetic age, was calculated as described by Horvath (Horvath 2013), using the online age calculator freely available at the website: https://dnamage.genetics.ucla.edu.
3. Results
3.1 DNA methylation differences in responders and non-responders to influenza vaccination
Forty-four subjects were included in the present study: 23 were responders to influenza vaccine, while 21 were considered as non-responders according to HIA assay. Characteristics of the subjects are summarized in Table 1.
Table 1.
Demographic characteristics of the subjects involved in the study.
| N of subjects | Sex | N of subjects aged ≥ 50 years |
Age in the Young group (mean and sd) |
Age in the Old group (mean and sd) |
|
|---|---|---|---|---|---|
| Responders | 23 | 10M / 13F | 11 | 28.4 (8.0) | 64.1 (12.5) |
| Non-responders | 21 | 10M / 11F | 12 | 33.4 (6.6) | 63.2 (7.4) |
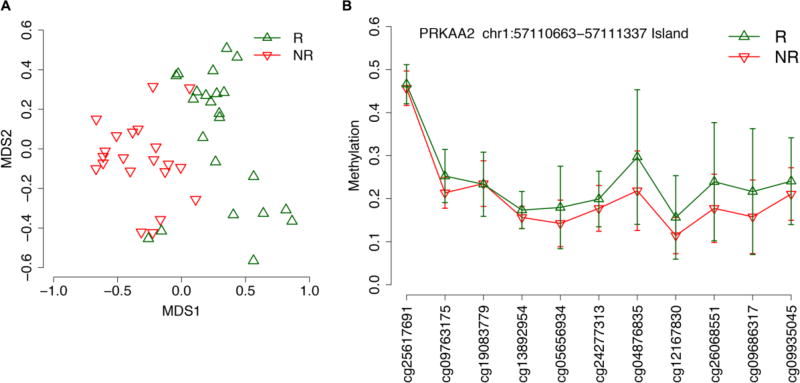
We used the Illumina® Infinium 450k microarray to generate genome wide DNA methylation data from PBMC collected pre-vaccination (baseline). As a first step in our analysis, we compared DNA methylation between responders and non-responders to influenza vaccination by ANOVA, correcting for age, sex and batch. No CpG sites withstood Benjamini-Hochberg false discovery rate correction for multiple testing (significant threshold: q-value <0.05). We then considered less stringent criteria (uncorrected p-value inferior to 0.01 and absolute difference between mean methylation values of responders and non-responders of at least 0.05). In this way, we identified 83 differentially methylated CpG sites between the two groups of subjects, mapping in 52 annotated genes (Tables 2 and 3; Supplementary File 1), that were able to distinguish responders and non-responders in a multidimensional scaling (MDS) plot (Figure 1A). When looking at the genomic localization of these probes, we found 3 genomic regions in which multiple adjacent CpG probes showed differential methylation between responders and non-responders: 3 probes mapped within the chr1:57110663–57111337 island associated to PRKAA2 gene (Figure 1B), 2 mapped within the chr6:160209941–160212015 island associated to TCP1/MRPL18 genes and 2 mapped in TRAPPC9 gene, one in the chr8:141348840–141349195 island and one in the shore of the chr8:141467218–141467927 island. The presence of multiple differentially methylated probes within the same region indicates a general epigenetic remodeling of the genomic tract, which is likely to be biologically relevant (Bacalini et al., 2015; Wessely and Emes, 2012).
Table 2.
Number of differentially methylated CpG sites between responders and non-responders to influenza vaccination, identified according to criteria described in Methods.
| Number of differentially methylated CpG sites between responders and non-responders |
|
|---|---|
| All subjects | 83 |
| Young subjects (Age < 50 years) | 142 |
| Old subjects (Age ≥ 50 years) | 305 |
Table 3.
Top 10 of the 83 differentially methylated CpG sites between responders (R) and non-responders (NR) to influenza vaccination.
| Illumina ID |
Chromosome | UCSC_RefGene_Name | UCSC_CpG_Islands_Name | Relation_to_UCSC_CpG_Island | Uncorrected p-value |
Difference between mean methylation values of R and NR |
|---|---|---|---|---|---|---|
| cg13235976 | 3 | chr3:193858770-193859695 | S_Shelf | 3.04E-06 | −0.0659 | |
| cg01281776 | 4 | chr4:20253276-20256868 | Island | 0.00016 | −0.0562 | |
| cg27559562 | 16 | CLUAP1 | chr16:3550773-3551274 | N_Shore | 0.00025 | −0.0535 |
| cg19492632 | 19 | SLC7A9 | chr19:33350719-33350932 | N_Shelf | 0.00028 | 0.0637 |
| cg19497709 | 3 | COPG | chr3:128997382-128997600 | N_Shelf | 0.00057 | −0.0544 |
| cg15219393 | 5 | PPARGC1B | chr5:149109570-149111750 | Island | 0.00062 | 0.0582 |
| cg05146089 | 5 | chr5:926586-927401 | Island | 0.00068 | −0.0507 | |
| cg22385827 | 2 | C2orf67 | chr2:211035478-211036637 | Island | 0.00072 | 0.0501 |
| cg24845595 | 1 | NTRK1; INSRR | chr1:156814881-156815792 | N_Shore | 0.00073 | −0.0513 |
| cg17177528 | 7 | chr7:123672063-123673691 | N_Shelf | 0.00074 | −0.0509 |
Figure 1.
Comparison of DNA methylation profiles between responders (R) and nonresponders (NR) to influenza vaccination. (A) Multidimensional scaling plots of DNA methylation data of 83 differentially methylated CpG sites between R (green) and NR (red). (B) Line plot of mean methylation values +/− standard deviation in R (green) and NR (red) for each CpG site mapping in the CpG island chr1:57110663–57111337 (PRKAA2 gene).
3.2 Age-associated epigenetic determinants of vaccination response
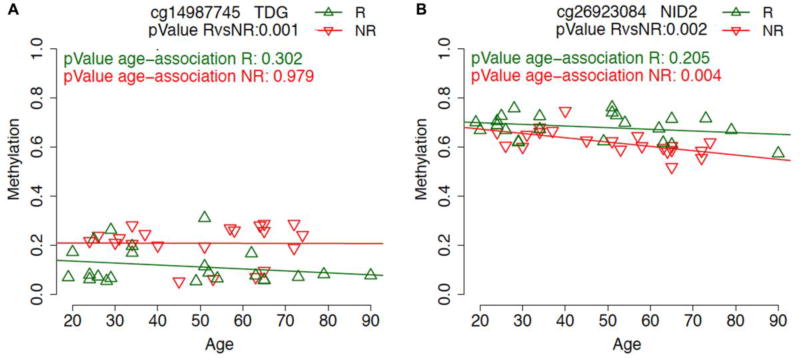
In order to assess the effect of age on vaccination response, for each of the CpG probes identified as differentially methylated in the previous analysis we calculated the association with age, considering separately responders and non-responders to influenza vaccine (Supplementary Files 1 and 2). For some CpG probes, such as cg14987745 in TDG gene, methylation differences between responders and non-responders were not affected by age (Figure 2A). On the contrary, others CpG sites showed age-association specifically in one of the two groups. For example, the probe cg26923084 in the gene NID2 showed an age-dependent decrease in methylation only in non-responders, with larger DNA methylation differences after the age of 50 years old (Figure 2B).
Figure 2.
Scatter plots of methylation values (y-axis) according to age of the subjects (x-axis) for the CpG probes in TDG (A) and NID2 (B) genes. Values indicate the pValue of the association between methylation and age in responders (R) (green) and non-responders (NR) (red) subjects.
To better identify the epigenetic determinants of vaccine responsiveness that are dependent on the age of the subjects, we divided the cohort in two groups including younger (age < 50) and older (age ≥ 50) subjects. Within each subgroup, we compared DNA methylation between responders and non-responders, correcting for sex and batch. Also in this case, no CpG site survived the multiple test correction. Using the same selection criteria described above, in the Young group we identified 142 differentially methylated CpG sites, mapping in 98 annotated genes (Tables 2 and 4; Supplementary Files 3 and 4). Four loci had multiple differentially methylated probes: C5orf33 (2 CpG probes in the chr5:36241003–36242664 island), HCG4P6 (3 CpG probes in the chr6:29894140–29895117 island and its shore), TRAF3 (2 CpG probes in the body of the gene) and RPTOR (4 CpG probes in the body of the gene, in the chr17:78863569b78863813 island and in the adjacent shore). In the Old subgroup, 305 CpG probes turned out to be differentially methylated, mapping in 206 genes (Tables 2 and 5; Supplementary Files 3 and 5). Eight loci contained multiple differentially methylated probes between old responders and non-responders: KIF5C (2 CpG probes, one in the chr2:149632682–149633882 island and one in the shore of chr2:149645536–149645834 island), COL7A1 (2 CpG probes in the chr3:48631882–48632901 island), MIR886 (2 CpG probes in the chr3:48631882–48632901 island), TRAPPC9 (2 CpG probes, one in the chr8:141348840–141349195 island and one in the shore of chr8:141467218–141467927 island), NKX2-3 (2 CpG probes in the chr10:101294443–101297263 island), MCF2L (2 CpG probes, one in the chr13:113687421–113687828 island and one in the shore of the chr13:113714784–113715160 island), MYOD (2 CpG probes, one in the shore of chr17:30822917–30823210 island and one in the chr17:30845903–30846702 island) and RGL3 (2 CpG probes, one in the chr19:11531278–11531590 island and one in its shore). No CpG probes were identified as differentially methylated both in the Young and in the Old group; however, when considering the genes in which the CpG probes map, COL7A1 and PAPLN turned out to be present in both the comparisons. For both the genes, the CpG probes identified as differentially methylated in the two comparisons mapped in near positions along the chromosome (within 1000) bp) and showed consistent age-dependent trends (for both the genes, and age-related hypomethylation specifically in the non-responders group), suggesting a general epigenetic remodeling of the two genomic regions.
Table 4.
Top 10 of the 142 differentially methylated CpG sites between responders (R) and non-responders (NR) to influenza vaccination (young subjects with age < 50 years).
| Illumina ID |
Chromosome | UCSC_RefGene_Name | UCSC_CpG_Islands_Name | Relation_to_UCSC_CpG_Island | Uncorrected p-value |
Difference between mean methylation values of R and NR |
|---|---|---|---|---|---|---|
| cg21690945 | 4 | chr4:170695989-170696274 | S_Shelf | 0.00011 | −0.0520 | |
| cg10420952 | 7 | TWIST1 | chr7:19156050-19158042 | Island | 0.00012 | −0.0591 |
| cg25622597 | 3 | RHOA; TCTA | chr3:49448861-49449965 | Island | 0.00024 | 0.0626 |
| cg10033725 | 7 | HEATR2 | chr7:821689-822634 | S_Shelf | 0.00029 | −0.0662 |
| cg24233211 | 6 | 0.00031 | −0.1159 | |||
| cg07681156 | 19 | ZNF581 | chr19:56154791-56155117 | Island | 0.00038 | −0.0551 |
| cg14562060 | 6 | TCP11 | chr6:35108801-35109499 | Island | 0.00040 | −.0553 |
| cg05696877 | 1 | IFI44L | 0.00046 | 0.0573 | ||
| cg23696891 | 6 | TNFAIP3 | chr6:138187825-138189141 | Island | 0.00052 | −0.0631 |
| cg17014647 | 6 | ATXN1 | 0.00067 | 0.0589 |
Table 5.
Top 10 of the 305 differentially methylated CpG sites between responders (R) and non-responders (NR) to influenza vaccination (old subjects with age ≥ 50 years).
| Illumina ID |
Chromosome | UCSC_RefGene_Name | UCSC_CpG_Islands_Name | Relation_to_UCSC_CpG_Island | Uncorrected p-value |
Difference between mean methylation values of R and NR |
|---|---|---|---|---|---|---|
| cg14273502 | 12 | chr12:113013099-113013529 | Island | 6.99E-05 | −0.0575 | |
| 113013529 | 113013529 | 113013529 | 113013529 | 113013529 | 113013529 | 113013529 |
| cg11644052 | 2 | NFU1 | chr2:69664291-69664816 | S_Shore | 0.00012 | −0.0567 |
| cg26923084 | 14 | NID2 | chr14:52534581-52536722 | S_Shore | 0.00013 | −0.0899 |
| cg23301925 | 14 | 0.00021 | −0.0501 | |||
| cg22256027 | 4 | MSX1 | chr4:4864456-4864834 | N_Shore | 0.00024 | 0.0586 |
| cg13617280 | 12 | SLC15A4; MGC16384 | 0.00029 | 0.0825 | ||
| cg24927800 | 2 | DES | chr2:220283200-220283750 | Island | 0.00031 | −0.0710 |
| cg16956133 | 2 | chr2:863930-865091 | Island | 0.00035 | −0.0861 | |
| cg21126344 | 17 | SOX9 | chr17:70116274-70119998 | Island | 0.00036 | −-0.0816 |
| cg10857729 | 8 | chr8:976015-976416 | N_Shelf | 0.00043 | 0.0622 |
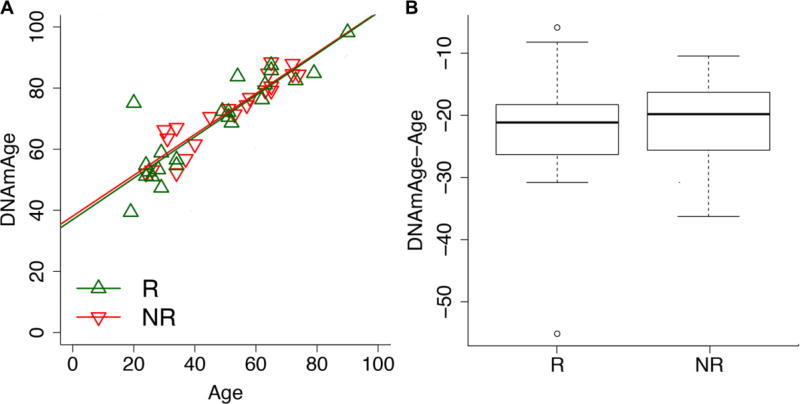
Finally, we investigated if the responsiveness to vaccination was associated to the epigenetic age of the subjects. In the last few years, DNA methylation-based biomarkers have gained particular relevance in the field of aging research and have been proposed as proxy of the biological age of an individual. The most popular epigenetic age estimator is Horvath’s epigenetic clock (Horvath, 2013) which has been shown to correlate with mortality (Chen et al., 2016; Christiansen et al., 2016; Marioni et al., 2015a; Perna et al., 2016), physical and cognitive fitness (Marioni et al., 2015b) and to detect a biological age-acceleration in frail subjects (Breitling et al., 2016). Frailty is of particular importance from our perspective, since it has been observed that this status impacts susceptibility to influenza and responsiveness to influenza vaccine (Yao et al., 2011). Here we used Horvath’s epigenetic clock to estimate the DNA methylation age (DNAmAge) in our cohort. Then for each individual, we calculated the delta between chronological age and DNAmAge, in order to detect deceleration (the subject is epigenetically younger than his/her chronological age) or acceleration effects (the subject is epigenetically older than his/her chronological age). No significant differences in delta values were observed between responders and non-responders, nor when we divided the cohort in the young and old subgroups (Figure 3).
Figure 3.
Estimation of Horvath’s epigenetic clock in responders (R) and non-responders (NR) to influenza vaccination. (A) Scatter plot of DNAmAge (y axis) versus chronological age (x axis) in R and NR to influenza vaccination. (B) Boxplot of differences between DNAmAge and chronological age in R and NR.
4. Discussion
Understanding mechanisms associated with impaired vaccine responses is an important goal and this is particularly true in the context of aging. Age-associated alterations in immune responses, named immunosenescence, are in part responsible of the poor response after vaccination in elderly (Goronzy and Weyand, 2013). Role of DNA methylation in this phenomenon has been investigated in subjects aged 50 – 74 years old vaccinated against influenza. The authors identified multiples CpG sites associated with age and with strong correlations with immunosenescence markers (Kennedy et al., 2016), highlighting the possible implication of epigenetic regulation as an active mechanism that shapes the immune response during aging.
In this study, we analyzed baseline whole genome DNA methylation profiles of 44 subjects ranging 19–90 years and receiving influenza vaccination. To the best of our knowledge, this is the first study to investigate methylation data in vaccinated subjects with such a wide range of ages, making it possible to identify age-related determinants of influenza vaccination response. Accordingly, we identified 83 differentially methylated probes when correcting for the age of the subjects, 142 in the young subgroup and 305 in the old subgroup. This suggests that DNA methylation differences between responders and non-responders are larger at older ages compared to younger ones. Non-responders did not show epigenetic age acceleration effects compared to responders, according to Horvath’s epigenetic clock.
To our knowledge, none of the CpG sites we identified was previously found differentially methylated in studies investigating DNA methylation profiles in human vaccinology, and no significant enrichment in KEGG pathways was observed (data not shown). However, it is interesting that we identified differentially methylated probes mapping in genes that are involved in immunosenescence (CD40) (Metcalf et al., 2015; Toapanta and Ross, 2009) and in innate immunity responses during viral infection: CXCL16 (Piqueras et al., 2006), ULK1 (Prantner et al., 2017), BCL11B (Yu et al., 2015), BTC (Al-Yahya et al., 2015).
Senescence of the adaptive compartment of immunity, which is involved in the classic immunological memory (Crotty and Ahmed, 2004), has been widely studied and most of the cellular markers studied in the report published by Kennedy et al. are related to B or T lymphocytes (Kennedy et al., 2016). However, during last years, evidence has emerged that also innate immune cells have a sort of memory, called “trained immunity”. This phenomenon could be a contributor to the protective effects of vaccination, independently of T and B cells, mostly observed in bacteriological vaccines, such as BCG (Garly et al., 2003; Kleinnijenhuis et al., 2012; Tribouley et al., 1978; van ’t Wout et al., 1992). Whereas immunological memory associated to adaptive immunity relies upon genetic recombination and mutations for the development of antigen specific receptors, mechanistic studies have brought evidence that trained immunity does not involve these types of permanent genetic changes but is largely based on epigenetic remodeling in innate immune cells (such as myeloid cells (monocytes, macrophages), natural killer cells or innate lymphoid cells) (Netea et al., 2016). The epigenetic shift, mostly based on histone modifications but also on microRNAs and DNA methylation changes, follows modifications in cellular metabolism driven by the antigenic stimulation (Arts et al., 2016) and can shape the cellular response and remains over time. Identification of differentially methylated CpG sites between responders and non-responders after vaccination is an important step to gain knowledge on these processes, for the identification of genes that could be potential targets for functional analysis and for the development of new strategies of vaccination.
In this study we analyzed total PBMC samples, like in most published system vaccinology studies which used whole blood or PBMC. One important advantage of this approach is that blood and PBMC can be easily obtained from individuals, but an important limit is the presence multiple diverse cell subsets. Indeed, the heterogeneity in cell populations in the analysis can possibly hide differences in DNA methylation patterns in individual cell types. Hoek et al. previously investigated the transcriptomic and proteomic profiles from PBMC and from sorted immune cells after influenza vaccination (T cells, B cells, natural killer cells, myeloid dendritic cells, monocytes and neutrophils). They described significantly different RNA and protein expression profiles between the groups (Hoek et al., 2015).
Another possible limitation in our work is the lack of data regarding pre-existing immune status of the subjects that could affect vaccine response. It is known that older subjects are frequently non-responders to vaccine and that a 4-fold increase in HIA titers is more frequently found in young subjects as compared to old ones: in a meta-analysis, Goodwin et al. generated an adjusted odds-ratio of responses in elderly vs young adults of 0.24 to 0.59 in terms of seroconversion and seroprotection against 3 different influenza antigens (H1N1, H3N2 and B) (Goodwin et al., 2006). Weakened serological response to vaccination in elderly subjects are related to the effect of immunosenescence, but previous exposures to influenza antigens (via vaccinations or infections) have also an influence on the response to the new vaccination. In general, subjects with repeated previous influenza vaccinations and influenza specific antibodies at baseline have a reduced humoral response and so lower post-vaccination titers (Bucasas et al., 2011; Gulati et al., 2005; Huang et al., 2017; Ng et al., 2013; Sasaki et al., 2008; Thakar et al., 2015; Tsang et al., 2014). However, it has also recently been demonstrated in a study conducted on 136 young and 122 elderly individuals that young individuals respond better than elderly individuals to the first vaccination, but after subsequent vaccinations the difference in response between young and elderly individuals declines rapidly, suggesting that different prior vaccination history and/or infection histories must also be taken in consideration when influenza vaccination is examined (Mosterín Höpping et al., 2016).
In this study, we evaluated the association of baseline DNA methylation patterns and response to the vaccination at different ages. Future studies should systematically evaluate the epigenetic response to influenza vaccination in subjects of different ages, analyzing DNA methylation and gene expression at different time points post-vaccination and correlating changes in epigenetic patterns with the antibody response. Although it has been demonstrated that vaccination does not profoundly alter DNA methylation profiles in PBMC (Zimmermann et al., 2016), further studies are necessary in this field, considering the impact of the age of the individuals and analyzing different cell types separately. Furthermore, DNA methylation signatures should be explored and validated in other independent datasets considering other influenza strains.
5. Conclusion
In conclusion, in this paper we identified possible age-related DNA methylation contributors to vaccine responsiveness. Further studies on larger independent cohorts are needed to define epigenetic biomarkers that can predict the vaccine efficacy. These models could be used to develop novel strategies to achieve optimal protective immune responses, specially in the elderly.
Supplementary Material
Supplementary File 1: List of the differentially methylated CpG probes between responders (R) and non-responders (NR).
Supplementary File 2: Scatter plots of methylation values (y-axis) according to age of the subjects (x-axis) for the 83 CpG probes identified as differentially methylated between responders (R) (green) and non-responders (NR) (red) subjects. Values indicate the p-values of the association between methylation and age in responders (R) (green) and non-responders (NR) (red) subjects.
Supplementary File 3: Lists of the differentially methylated CpG probes between responders (R) and non-responders (NR) in young and old subjects.
Supplementary File 4: Scatter plots of methylation values (y-axis) according to age of the subjects (x-axis) for the 142 CpG probes identified as differentially methylated between in responders (R) (green) and non-responders (NR) (red) subjects in the young subgroup. Values indicate the p-values of the association between methylation and age in responders (R) (green) and non-responders (NR) (red) subjects.
Supplementary File 5: Scatter plots of methylation values (y-axis) according to age of the subjects (x-axis) for the 305 CpG probes identified as differentially methylated between in responders (R) (green) and non-responders (NR) (red) subjects in the old subgroup. Values indicate the p-values of the association between methylation and age in responders (R) (green) and non-responders (NR) (red) subjects.
Highlights.
Response to flu vaccination is associated to baseline DNA methylation differences
Some differentially methylated sites show age-associated trends
Genes involved in immunosenescence and innate response show differential methylation
Acknowledgments
Funding: This study was supported by NIH AG-32576 (BBB), AI096446, AG042826 and AG032576 (BBB and DF); by the European Union’s Seventh Framework Programme to CF (grant number 602757, HUMAN); by the European Union’s H2020 Project to CF and PG (grant number 634821, PROPAG-AGING); by JPco-fuND to CF (ADAGE); by the CARIPLO foundation (“Humoral innate immunity in the regulation of tissue repair and metabolism in aging” project) to CF.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of interest: none
References
- Al-Yahya S, Mahmoud L, Al-Zoghaibi F, Al-Tuhami A, Amer H, Almajhdi FN, Polyak SJ, Khabar KSA. Human Cytokinome Analysis for Interferon Response. J. Virol. 2015;89:7108–7119. doi: 10.1128/JVI.03729-14. https://doi.org/10.1128/JVI.03729-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arts RJW, Joosten LAB, Netea MG. Immunometabolic circuits in trained immunity. Semin. Immunol. 2016;28:425–430. doi: 10.1016/j.smim.2016.09.002. https://doi.org/10.1016/j.smim.2016.09.002. [DOI] [PubMed] [Google Scholar]
- Aryee MJ, Jaffe AE, Corrada-Bravo H, Ladd-Acosta C, Feinberg AP, Hansen KD, Irizarry RA. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinforma. Oxf. Engl. 2014;30:1363–1369. doi: 10.1093/bioinformatics/btu049. https://doi.org/10.1093/bioinformatics/btu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacalini MG, Boattini A, Gentilini D, Giampieri E, Pirazzini C, Giuliani C, Fontanesi E, Remondini D, Capri M, Del Rio A, Luiselli D, Vitale G, Mari D, Castellani G, Di Blasio AM, Salvioli S, Franceschi C, Garagnani P. A meta-analysis on age-associated changes in blood DNA methylation: results from an original analysis pipeline for Infinium 450k data. Aging. 2015;7:97–109. doi: 10.18632/aging.100718. https://doi.org/10.18632/aging.100718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibikova M, Barnes B, Tsan C, Ho V, Klotzle B, Le JM, Delano D, Zhang L, Schroth GP, Gunderson KL, Fan J-B, Shen R. High density DNA methylation array with single CpG site resolution. Genomics. 2011;98:288–295. doi: 10.1016/j.ygeno.2011.07.007. https://doi.org/10.1016/j.ygeno.2011.07.007. [DOI] [PubMed] [Google Scholar]
- Breitling LP, Saum K-U, Perna L, Schöttker B, Holleczek B, Brenner H. Frailty is associated with the epigenetic clock but not with telomere length in a German cohort. Clin. Epigenetics. 2016;8:21. doi: 10.1186/s13148-016-0186-5. https://doi.org/10.1186/s13148-016-0186-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucasas KL, Franco LM, Shaw CA, Bray MS, Wells JM, Niño D, Arden N, Quarles JM, Couch RB, Belmont JW. Early patterns of gene expression correlate with the humoral immune response to influenza vaccination in humans. J. Infect. Dis. 2011;203:921–929. doi: 10.1093/infdis/jiq156. https://doi.org/10.1093/infdis/jiq156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulla A, De Witt B, Ammerlaan W, Betsou F, Lescuyer P. Blood DNA Yield but Not Integrity or Methylation Is Impacted After Long-Term Storage. Biopreservation Biobanking. 2016;14:29–38. doi: 10.1089/bio.2015.0045. https://doi.org/10.1089/bio.2015.0045. [DOI] [PubMed] [Google Scholar]
- Chen BH, Marioni RE, Colicino E, Peters MJ, Ward-Caviness CK, Tsai P-C, Roetker NS, Just AC, Demerath EW, Guan W, Bressler J, Fornage M, Studenski S, Vandiver AR, Moore AZ, Tanaka T, Kiel DP, Liang L, Vokonas P, Schwartz J, Lunetta KL, Murabito JM, Bandinelli S, Hernandez DG, Melzer D, Nalls M, Pilling LC, Price TR, Singleton AB, Gieger C, Holle R, Kretschmer A, Kronenberg F, Kunze S, Linseisen J, Meisinger C, Rathmann W, Waldenberger M, Visscher PM, Shah S, Wray NR, McRae AF, Franco OH, Hofman A, Uitterlinden AG, Absher D, Assimes T, Levine ME, Lu AT, Tsao PS, Hou L, Manson JE, Carty CL, LaCroix AZ, Reiner AP, Spector TD, Feinberg AP, Levy D, Baccarelli A, van Meurs J, Bell JT, Peters A, Deary IJ, Pankow JS, Ferrucci L, Horvath S. DNA methylation-based measures of biological age: meta-analysis predicting time to death. Aging. 2016;8:1844–1865. doi: 10.18632/aging.101020. https://doi.org/10.18632/aging.101020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiansen L, Lenart A, Tan Q, Vaupel JW, Aviv A, McGue M, Christensen K. DNA methylation age is associated with mortality in a longitudinal Danish twin study. Aging Cell. 2016;15:149–154. doi: 10.1111/acel.12421. https://doi.org/10.1111/acel.12421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotty S, Ahmed R. Immunological memory in humans. Semin. Immunol. 2004;16:197–203. doi: 10.1016/j.smim.2004.02.008. https://doi.org/10.1016/j.smim.2004.02.008. [DOI] [PubMed] [Google Scholar]
- Furman D, Jojic V, Kidd B, Shen-Orr S, Price J, Jarrell J, Tse T, Huang H, Lund P, Maecker HT, Utz PJ, Dekker CL, Koller D, Davis MM. Apoptosis and other immune biomarkers predict influenza vaccine responsiveness. Mol. Syst. Biol. 2013;9:659. doi: 10.1038/msb.2013.15. https://doi.org/10.1038/msb.2013.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garly M-L, Martins CL, Balé C, Baldé MA, Hedegaard KL, Gustafson P, Lisse IM, Whittle HC, Aaby P. BCG scar and positive tuberculin reaction associated with reduced child mortality in West Africa. A non-specific beneficial effect of BCG? Vaccine. 2003;21:2782–2790. doi: 10.1016/s0264-410x(03)00181-6. [DOI] [PubMed] [Google Scholar]
- Goodwin K, Viboud C, Simonsen L. Antibody response to influenza vaccination in the elderly: a quantitative review. Vaccine. 2006;24:1159–1169. doi: 10.1016/j.vaccine.2005.08.105. https://doi.org/10.1016/j.vaccine.2005.08.105. [DOI] [PubMed] [Google Scholar]
- Goronzy JJ, Weyand CM. Understanding immunosenescence to improve responses to vaccines. Nat. Immunol. 2013;14:428–436. doi: 10.1038/ni.2588. https://doi.org/10.1038/ni.2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulati U, Kumari K, Wu W, Keitel WA, Air GM. Amount and avidity of serum antibodies against native glycoproteins and denatured virus after repeated influenza whole-virus vaccination. Vaccine. 2005;23:1414–1425. doi: 10.1016/j.vaccine.2004.08.053. https://doi.org/10.1016/j.vaccine.2004.08.053. [DOI] [PubMed] [Google Scholar]
- Haralambieva IH, Painter SD, Kennedy RB, Ovsyannikova IG, Lambert ND, Goergen KM, Oberg AL, Poland GA. The impact of immunosenescence on humoral immune response variation after influenza A/H1N1 vaccination in older subjects. PloS One. 2015;10:e0122282. doi: 10.1371/journal.pone.0122282. https://doi.org/10.1371/journal.pone.0122282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoek KL, Samir P, Howard LM, Niu X, Prasad N, Galassie A, Liu Q, Allos TM, Floyd KA, Guo Y, Shyr Y, Levy SE, Joyce S, Edwards KM, Link AJ. A cell-based systems biology assessment of human blood to monitor immune responses after influenza vaccination. PloS One. 2015;10:e0118528. doi: 10.1371/journal.pone.0118528. https://doi.org/10.1371/journal.pone.0118528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013;14:R115. doi: 10.1186/gb-2013-14-10-r115. https://doi.org/10.1186/gb-2013-14-10-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K-YA, Chang S-C, Huang Y-C, Chiu C-H, Lin T-Y. Antibody Responses to Trivalent Inactivated Influenza Vaccine in Health Care Personnel Previously Vaccinated and Vaccinated for The First Time. Sci. Rep. 2017;7 doi: 10.1038/srep40027. https://doi.org/10.1038/srep40027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson T, Di Pietrantonj C, Al-Ansary LA, Ferroni E, Thorning S, Thomas RE. Vaccines for preventing influenza in the elderly. Cochrane Database Syst. Rev. 2010:CD004876. doi: 10.1002/14651858.CD004876.pub3. https://doi.org/10.1002/14651858.CD004876.pub3. [DOI] [PubMed]
- Kennedy RB, Ovsyannikova IG, Haralambieva IH, Oberg AL, Zimmermann MT, Grill DE, Poland GA. Immunosenescence-Related Transcriptomic and Immunologic Changes in Older Individuals Following Influenza Vaccination. Front. Immunol. 2016;7:450. doi: 10.3389/fimmu.2016.00450. https://doi.org/10.3389/fimmu.2016.00450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinnijenhuis J, Quintin J, Preijers F, Joosten LAB, Ifrim DC, Saeed S, Jacobs C, van Loenhout J, de Jong D, Stunnenberg HG, Xavier RJ, van der Meer JWM, van Crevel R, Netea MG. Bacille Calmette-Guerin induces NOD2-dependent nonspecific protection from reinfection via epigenetic reprogramming of monocytes. Proc. Natl. Acad. Sci. U. S. A. 2012;109:17537–17542. doi: 10.1073/pnas.1202870109. https://doi.org/10.1073/pnas.1202870109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Cheng Y, Yan W, Nardini C. Exploring the molecular causes of hepatitis B virus vaccination response: an approach with epigenomic and transcriptomic data. BMC Med. Genomics. 2014;7:12. doi: 10.1186/1755-8794-7-12. https://doi.org/10.1186/1755-8794-7-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marioni RE, Shah S, McRae AF, Chen BH, Colicino E, Harris SE, Gibson J, Henders AK, Redmond P, Cox SR, Pattie A, Corley J, Murphy L, Martin NG, Montgomery GW, Feinberg AP, Fallin MD, Multhaup ML, Jaffe AE, Joehanes R, Schwartz J, Just AC, Lunetta KL, Murabito JM, Starr JM, Horvath S, Baccarelli AA, Levy D, Visscher PM, Wray NR, Deary IJ. DNA methylation age of blood predicts all-cause mortality in later life. Genome Biol. 2015a;16:25. doi: 10.1186/s13059-015-0584-6. https://doi.org/10.1186/s13059-015-0584-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marioni RE, Shah S, McRae AF, Ritchie SJ, Muniz-Terrera G, Harris SE, Gibson J, Redmond P, Cox SR, Pattie A, Corley J, Taylor A, Murphy L, Starr JM, Horvath S, Visscher PM, Wray NR, Deary IJ. The epigenetic clock is correlated with physical and cognitive fitness in the Lothian Birth Cohort 1936. Int. J. Epidemiol. 2015b;44:1388–1396. doi: 10.1093/ije/dyu277. https://doi.org/10.1093/ije/dyu277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metcalf TU, Cubas RA, Ghneim K, Cartwright MJ, Grevenynghe JV, Richner JM, Olagnier DP, Wilkinson PA, Cameron MJ, Park BS, Hiscott JB, Diamond MS, Wertheimer AM, Nikolich-Zugich J, Haddad EK. Global analyses revealed age-related alterations in innate immune responses after stimulation of pathogen recognition receptors. Aging Cell. 2015;14:421–432. doi: 10.1111/acel.12320. https://doi.org/10.1111/acel.12320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosterín Höpping A, McElhaney J, Fonville JM, Powers DC, Beyer WEP, Smith DJ. The confounded effects of age and exposure history in response to influenza vaccination. Vaccine. 2016;34:540–546. doi: 10.1016/j.vaccine.2015.11.058. https://doi.org/10.1016/j.vaccine.2015.11.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaya HI, Hagan T, Duraisingham SS, Lee EK, Kwissa M, Rouphael N, Frasca D, Gersten M, Mehta AK, Gaujoux R, Li G-M, Gupta S, Ahmed R, Mulligan MJ, Shen-Orr S, Blomberg BB, Subramaniam S, Pulendran B. Systems Analysis of Immunity to Influenza Vaccination across Multiple Years and in Diverse Populations Reveals Shared Molecular Signatures. Immunity. 2015;43:1186–1198. doi: 10.1016/j.immuni.2015.11.012. https://doi.org/10.1016/j.immuni.2015.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaya HI, Wrammert J, Lee EK, Racioppi L, Marie-Kunze S, Haining WN, Means AR, Kasturi SP, Khan N, Li G-M, McCausland M, Kanchan V, Kokko KE, Li S, Elbein R, Mehta AK, Aderem A, Subbarao K, Ahmed R, Pulendran B. Systems biology of vaccination for seasonal influenza in humans. Nat. Immunol. 2011;12:786–795. doi: 10.1038/ni.2067. https://doi.org/10.1038/ni.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netea MG, Joosten LAB, Latz E, Mills KHG, Natoli G, Stunnenberg HG, O’Neill LAJ, Xavier RJ. Trained immunity: A program of innate immune memory in health and disease. Science. 2016;352:aaf1098. doi: 10.1126/science.aaf1098. https://doi.org/10.1126/science.aaf1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng S, Ip DKM, Fang VJ, Chan K-H, Chiu SS, Leung GM, Peiris JSM, Cowling BJ. The effect of age and recent influenza vaccination history on the immunogenicity and efficacy of 2009–10 seasonal trivalent inactivated influenza vaccination in children. PloS One. 2013;8:e59077. doi: 10.1371/journal.pone.0059077. https://doi.org/10.1371/journal.pone.0059077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obermoser G, Presnell S, Domico K, Xu H, Wang Y, Anguiano E, Thompson-Snipes L, Ranganathan R, Zeitner B, Bjork A, Anderson D, Speake C, Ruchaud E, Skinner J, Alsina L, Sharma M, Dutartre H, Cepika A, Israelsson E, Nguyen P, Nguyen Q-A, Harrod AC, Zurawski SM, Pascual V, Ueno H, Nepom GT, Quinn C, Blankenship D, Palucka K, Banchereau J, Chaussabel D. Systems scale interactive exploration reveals quantitative and qualitative differences in response to influenza and pneumococcal vaccines. Immunity. 2013;38:831–844. doi: 10.1016/j.immuni.2012.12.008. https://doi.org/10.1016/j.immuni.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterholm MT, Kelley NS, Sommer A, Belongia EA. Efficacy and effectiveness of influenza vaccines: a systematic review and meta-analysis. Lancet Infect. Dis. 2012;12:36–44. doi: 10.1016/S1473-3099(11)70295-X. https://doi.org/10.1016/S1473-3099(11)70295-X. [DOI] [PubMed] [Google Scholar]
- Paules C, Subbarao K. Influenza. Lancet Lond. Engl. 2017 doi: 10.1016/S0140-6736(17)30129-0. https://doi.org/10.1016/S0140-6736(17)30129-0. [DOI] [PubMed]
- Perna L, Zhang Y, Mons U, Holleczek B, Saum K-U, Brenner H. Epigenetic age acceleration predicts cancer, cardiovascular, and all-cause mortality in a German case cohort. Clin. Epigenetics. 2016;8:64. doi: 10.1186/s13148-016-0228-z. https://doi.org/10.1186/s13148-016-0228-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piqueras B, Connolly J, Freitas H, Palucka AK, Banchereau J. Upon viral exposure, myeloid and plasmacytoid dendritic cells produce 3 waves of distinct chemokines to recruit immune effectors. Blood. 2006;107:2613–2618. doi: 10.1182/blood-2005-07-2965. https://doi.org/10.1182/blood-2005-07-2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prantner D, Perkins DJ, Vogel SN. AMP-activated Kinase (AMPK) Promotes Innate Immunity and Antiviral Defense through Modulation of Stimulator of Interferon Genes (STING) Signaling. J. Biol. Chem. 2017;292:292–304. doi: 10.1074/jbc.M116.763268. https://doi.org/10.1074/jbc.M116.763268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki S, He X-S, Holmes TH, Dekker CL, Kemble GW, Arvin AM, Greenberg HB. Influence of prior influenza vaccination on antibody and B-cell responses. PloS One. 2008;3:e2975. doi: 10.1371/journal.pone.0002975. https://doi.org/10.1371/journal.pone.0002975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen P, Shah PP, Nativio R, Berger SL. Epigenetic Mechanisms of Longevity and Aging. Cell. 2016;166:822–839. doi: 10.1016/j.cell.2016.07.050. https://doi.org/10.1016/j.cell.2016.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Targonski PV, Jacobson RM, Poland GA. Immunosenescence: role and measurement in influenza vaccine response among the elderly. Vaccine. 2007;25:3066–3069. doi: 10.1016/j.vaccine.2007.01.025. https://doi.org/10.1016/j.vaccine.2007.01.025. [DOI] [PubMed] [Google Scholar]
- Thakar J, Mohanty S, West AP, Joshi SR, Ueda I, Wilson J, Meng H, Blevins TP, Tsang S, Trentalange M, Siconolfi B, Park K, Gill TM, Belshe RB, Kaech SM, Shadel GS, Kleinstein SH, Shaw AC. Aging-dependent alterations in gene expression and a mitochondrial signature of responsiveness to human influenza vaccination. Aging. 2015;7:38–52. doi: 10.18632/aging.100720. https://doi.org/10.18632/aging.100720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toapanta FR, Ross TM. Impaired immune responses in the lungs of aged mice following influenza infection. Respir. Res. 2009;10:112. doi: 10.1186/1465-9921-10-112. https://doi.org/10.1186/1465-9921-10-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tribouley J, Tribouley-Duret J, Appriou M. [Effect of Bacillus Callmette Guerin (BCG) on the receptivity of nude mice to Schistosoma mansoni] CRSeances Soc. Biol. Fil. 1978;172:902–904. [PubMed] [Google Scholar]
- Tsang JS, Schwartzberg PL, Kotliarov Y, Biancotto A, Xie Z, Germain RN, Wang E, Olnes MJ, Narayanan M, Golding H, Moir S, Dickler HB, Perl S, Cheung F, Baylor HIPC Center, CHI Consortium Global analyses of human immune variation reveal baseline predictors of postvaccination responses. Cell. 2014;157:499–513. doi: 10.1016/j.cell.2014.03.031. https://doi.org/10.1016/j.cell.2014.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van ’t Wout JW, Poell R, van Furth R. The role of BCG/PPD-activated macrophages in resistance against systemic candidiasis in mice. Scand. J. Immunol. 1992;36:713–719. doi: 10.1111/j.1365-3083.1992.tb03132.x. [DOI] [PubMed] [Google Scholar]
- Wessely F, Emes RD. Identification of DNA methylation biomarkers from Infinium arrays. Front. Genet. 2012;3:161. doi: 10.3389/fgene.2012.00161. https://doi.org/10.3389/fgene.2012.00161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao X, Hamilton RG, Weng N, Xue Q-L, Bream JH, Li H, Tian J, Yeh S-H, Resnick B, Xu X, Walston J, Fried LP, Leng SX. Frailty is associated with impairment of vaccine-induced antibody response and increase in post-vaccination influenza infection in community-dwelling older adults. Vaccine. 2011;29:5015–5021. doi: 10.1016/j.vaccine.2011.04.077. https://doi.org/10.1016/j.vaccine.2011.04.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Wang C, Clare S, Wang J, Lee S-C, Brandt C, Burke S, Lu L, He D, Jenkins NA, Copeland NG, Dougan G, Liu P. The transcription factor Bcl11b is specifically expressed in group 2 innate lymphoid cells and is essential for their development. J. Exp. Med. 2015;212:865–874. doi: 10.1084/jem.20142318. https://doi.org/10.1084/jem.20142318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W, Higgs BW, Morehouse C, Streicher K, Ambrose CS, Woo J, Kemble GW, Jallal B, Yao Y. A whole genome transcriptional analysis of the early immune response induced by live attenuated and inactivated influenza vaccines in young children. Vaccine. 2010;28:2865–2876. doi: 10.1016/j.vaccine.2010.01.060. https://doi.org/10.1016/j.vaccine.2010.01.060. [DOI] [PubMed] [Google Scholar]
- Zimmermann MT, Kennedy RB, Grill DE, Oberg AL, Goergen KM, Ovsyannikova IG, Haralambieva IH, Poland GA. Integration of Immune Cell Populations, mRNA-Seq, and CpG Methylation to Better Predict Humoral Immunity to Influenza Vaccination: Dependence of mRNA-Seq/CpG Methylation on Immune Cell Populations. Front. Immunol. 2017;8:445. doi: 10.3389/fimmu.2017.00445. https://doi.org/10.3389/fimmu.2017.00445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann MT, Oberg AL, Grill DE, Ovsyannikova IG, Haralambieva IH, Kennedy RB, Poland GA. System-Wide Associations between DNA-Methylation, Gene Expression, and Humoral Immune Response to Influenza Vaccination. PloS One. 2016;11:e0152034. doi: 10.1371/journal.pone.0152034. https://doi.org/10.1371/journal.pone.0152034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary File 1: List of the differentially methylated CpG probes between responders (R) and non-responders (NR).
Supplementary File 2: Scatter plots of methylation values (y-axis) according to age of the subjects (x-axis) for the 83 CpG probes identified as differentially methylated between responders (R) (green) and non-responders (NR) (red) subjects. Values indicate the p-values of the association between methylation and age in responders (R) (green) and non-responders (NR) (red) subjects.
Supplementary File 3: Lists of the differentially methylated CpG probes between responders (R) and non-responders (NR) in young and old subjects.
Supplementary File 4: Scatter plots of methylation values (y-axis) according to age of the subjects (x-axis) for the 142 CpG probes identified as differentially methylated between in responders (R) (green) and non-responders (NR) (red) subjects in the young subgroup. Values indicate the p-values of the association between methylation and age in responders (R) (green) and non-responders (NR) (red) subjects.
Supplementary File 5: Scatter plots of methylation values (y-axis) according to age of the subjects (x-axis) for the 305 CpG probes identified as differentially methylated between in responders (R) (green) and non-responders (NR) (red) subjects in the old subgroup. Values indicate the p-values of the association between methylation and age in responders (R) (green) and non-responders (NR) (red) subjects.