Abstract
Human hematopoiesis involves cellular differentiation of multipotent cells into progressively more lineage-restricted states. While the chromatin accessibility landscape of this process has been explored in defined populations, single-cell regulatory variation has been hidden by ensemble averaging. We collected single-cell chromatin accessibility profiles across 10 populations of immunophenotypically-defined human hematopoietic cell types and constructed a chromatin accessibility landscape of human hematopoiesis to characterize differentiation trajectories. We find variation consistent with lineage-bias toward different developmental branches in multipotent cell-types. We observe heterogeneity within common myeloid progenitors (CMPs) and granulocyte-macrophage progenitors (GMPs), and develop a strategy to partition GMPs along their differentiation trajectory. Furthermore, we integrated scRNA-seq data to associate transcription factors to chromatin accessibility changes, and regulatory elements to target genes through correlations of expression and regulatory element accessibility. Overall, this work provides a framework for integrative exploration of complex regulatory dynamics in a primary human tissue at single-cell resolution.
Introduction
In 1957, Conrad Waddington developed an influential analogy for developmental cell biology by conceptualizing cellular differentiation as a ball rolling down a bifurcating three-dimensional surface (Goldberg et al., 2007; Waddington, 1960). This developmental landscape defines a descriptive path a cell might follow, choosing different developmental fates as it reaches saddle points that separate different, increasingly restricted, cellular states. The shape of this landscape is largely defined by transcription factors (“guy-wires”), which recruit chromatin effectors to reconfigure chromatin (Calo and Wysocka, 2013; Long et al., 2016) and promote new cellular phenotypes (Graf and Enver, 2009; Takahashi and Yamanaka, 2006). These concepts – the first a descriptive notion of development (Figure S1A), and the second a mechanistic description of the molecular actors that drive state changes (Figure S1B) – have provided a conceptual framework for understanding cell fate choices. Recent technological advances in single-cell epigenomic assays (Kelsey et al., 2017) now provide the opportunity to ascribe epigenomic features to this landscape by quantifying overall epigenomic similarity of individual cells during a normal differentiation process, as well as the activity of master regulators that influence cell fate decisions.
Hematopoietic differentiation serves as an ideal model for exploring the nature of multipotent cell fate decisions (Laurenti and Göttgens, 2018; Orkin and Zon, 2008). The hematopoietic system is maintained by the activity of a small number of self-renewing, long-lived hematopoietic stem cells (HSCs) capable of giving rise to the majority of blood cell lineages (Becker et al., 1963; Laurenti and Göttgens, 2018; Orkin and Zon, 2008) whereby multipotent cells transit multiple decision points while becoming increasingly lineage-restricted (Figure 1A). The human hematopoietic system is an extensively characterized adult stem cell hierarchy with diverse cell types capable of phenotypic isolation with multi-parameter fluorescence activated cell sorting (FACS) (Corces et al., 2016; Laurenti and Göttgens, 2018). This capacity for phenotypic isolation has enabled measurement of the epigenomic and transcriptional dynamics associated with sorted human progenitors across differentiation providing a foundation for the dissection of regulatory variation in normal multi-lineage cellular differentiation (Chen et al., 2014; Corces et al., 2016; Farlik et al., 2016; Novershtern et al., 2011). Furthermore, recent work measuring single-cell transcriptomes has revealed significant transcriptional heterogeneity in isolated progenitors (Notta et al., 2015) and across differentiation (Laurenti and Göttgens, 2018; Velten et al., 2017). These observations set the stage for single-cell epigenomic measurements that may define cis- and trans- regulatory mechanisms underlying transcriptional and cell fate commitment heterogeneity in hematopoiesis.
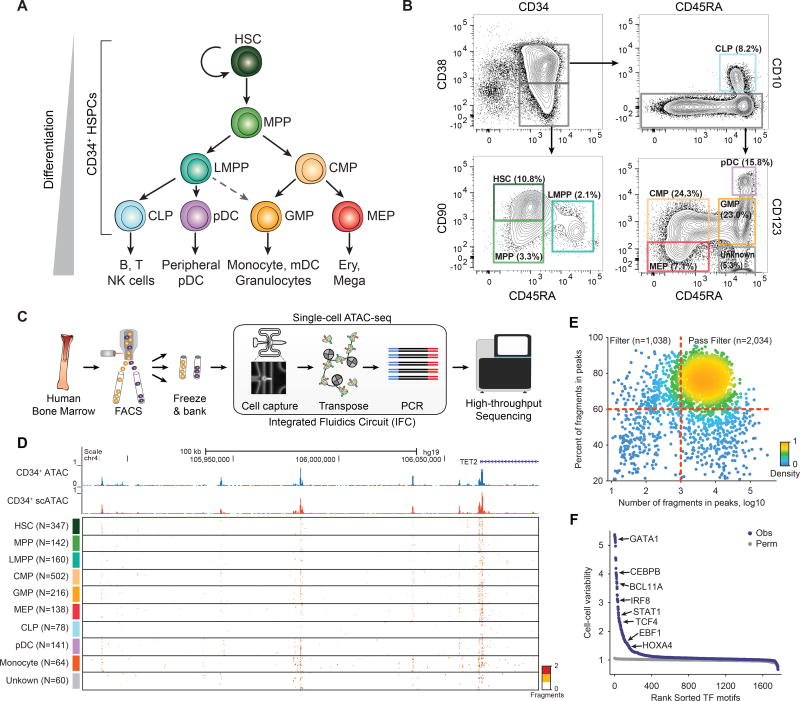
Figure 1. Single-cell ATAC-seq profiles chromatin accessibility within single hematopoietic progenitors.
(A) A schematic of human hematopoietic differentiation. (B) Sorting strategy for CD34+ cells. (C) Single-cell ATAC-seq workflow used in this study. (D) Single-cell epigenomic profiles along the TET2 locus. (E) Percent fragments in peaks by number of fragments in peaks, red lines show cutoffs used for determining which cells pass filter; points are colored by density. (F) TF motif variability analysis in all single-cell epigenomic profiles collected for this study.
To define a single-cell chromatin accessibility landscape of this developmental hierarchy, we applied a single-cell assay for transposase-accessible chromatin by sequencing (scATAC-seq) (Buenrostro et al., 2015b) to 10 sortable populations in human bone marrow or blood comprising multipotent and lineage restricted progenitors. We find that the regulatory landscape of human hematopoiesis is continuous, with cell surface markers reflecting “basins” within this landscape. This single-cell analysis also uncovered substantial heterogeneity within immunophenotypically-defined cellular populations, including variability within multipotent progenitors strongly correlated along the dimensions of hematopoietic differentiation – an observation consistent with lineage priming at the level of chromatin accessibility. We observe especially strong variability within populations of immunophenotypically-defined common myeloid progenitor (CMP) and granulocyte-macrophage progenitor (GMP) cell types. Using ATAC-seq and RNA-seq, we confirm that GMPs are substantially heterogeneous on both epigenomic and transcriptomic levels, and demonstrate a strategy to enrich for sub-populations within GMPs at different developmental stages of a myeloid differentiation trajectory. Last, we generate scRNA-seq data and integrate these data with scATAC-seq to associate expression changes of transcription factors (TFs) to changes in chromatin accessibility at cis-regulatory elements. Using these integrated data, we also link changes at cis-regulatory elements to changes in the expression of nearby genes. These methods for assaying and analyzing single-cell epigenomics data provides the opportunity for de novo discovery of cell types and states, define regulatory variability within immunophenotypically-pure populations, and capture the cis- and trans-regulatory dynamics across a cell-resolved regulatory landscape of differentiation.
Results
Single-cell chromatin accessibility of distinct hematopoietic cell types
We used FACS to isolate 8 distinct cellular populations from CD34+ human bone marrow, which included cell types spanning the myeloid, erythroid, and lymphoid lineages (Figures 1A and 1B). In addition, we also profiled a CD34+CD38−CD45RA+CD123− subset which has not been well characterized (Manz et al., 2002). Cells analyzed after sorting and cells cryopreserved after sorting provided comparable data quality and yield (Figures S1C–E) and therefore we performed all further scATAC-seq measurements on cryopreserved cells (Figure 1C). Together, this sorting strategy captures ~97% of all CD34+ cells (Figure S1F) and using post-sort analysis, we found that sorted cell types were on average 97% pure by cell surface marker immunophenotype (Corces et al., 2016). Using this approach, we profiled the chromatin accessibility landscapes (CALs) across a total of 30 independent single-cell experiments representing 6 human donors, with each progenitor population assayed from two or more donors (Figure S1G). We did not profile CD34- bone marrow stem cells, as they are rare and less well described (Matsuoka et al., 2015).
Aggregated single-cell chromatin accessibility profiles closely resemble bulk CD34+ ATAC-seq profiles (Figure 1D and Figures S1H–I). Including previously published scATAC-seq data from LMPPs and monocytes (Corces et al., 2016), this data set comprised 3,072 single-cell CALs across 32 integrated fluidic circuits (IFCs). Single-cell profiles were of consistent high-quality with 2,034 cells passing stringent quality filtering, yielding a median of 8,268 fragments per cell with 76% of those fragments mapping to peaks, resulting in a median of 6,442 fragments in peaks per cell (Figure 1E; see methods).
TF activity inference using ChromVAR
We applied ChromVAR to calculate TF motif associated CAL changes and identify potential regulators of epigenomic variability (Buenrostro et al., 2015b; Schep et al., 2017). This approach quantifies accessibility variation across single-cells by aggregating accessible regions containing a specific TF motif, then compares the observed accessibility of all peaks containing a TF motif to a background set of peaks normalizing for known technical confounders. ChromVAR identifies high-variance TF motifs across CALs representing known master regulators of hematopoiesis such as GATA1, BATF and CEBPB (Figure 1F). Notably, TFs of the same family often share a similar motif and thus are difficult to disambiguate, therefore TF motifs highlighted throughout the text are representative TF motifs which may encompass the individual activities of multiple expressed TFs.
Hierarchical clustering of single-cell profiles using TF z-scores generally classifies single-cells by their immunophenotypically-defined cell type identity (Figure 2A). Interestingly, despite the overall high quality of the HSC profiles (Figure S1J–L), HSCs exhibit low TF z-scores for lineage specifying TF motifs. Furthermore, the HOX TF motif was most enriched in HSCs, previously shown to regulate stem cell activity (Lawrence et al., 1997; Magnusson et al., 2007), however, this TF motif also exhibited relatively low-level activity compared to lineage defining TFs in more-differentiated cells. Low level expression of lineage specifying TFs in other multipotent cell systems has been described (Grün et al., 2016) and is hypothesized to generally promote multipotency (Graf and Enver, 2009), which may also explain the low level TF z-scores in this analysis of HSCs.
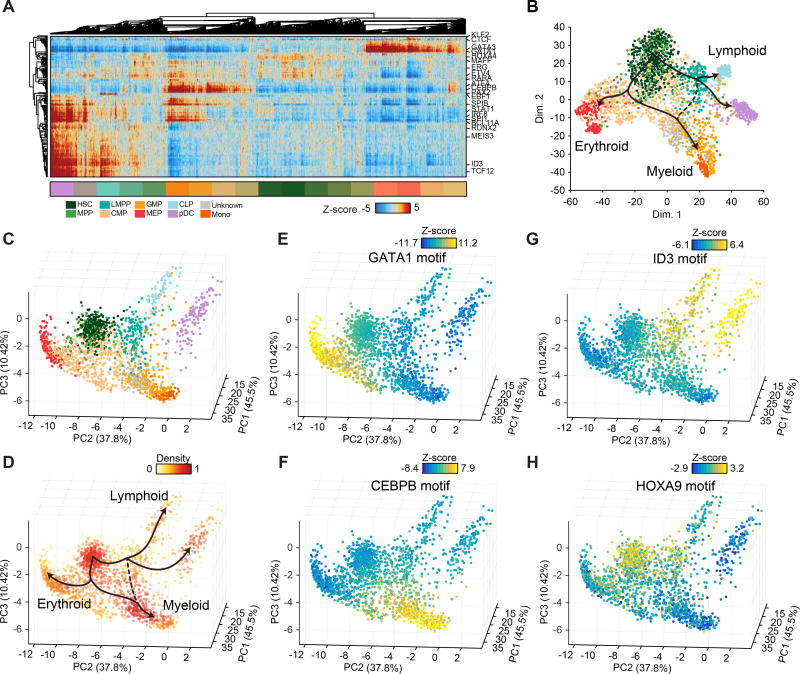
Figure 2. Lineage projection of human hematopoietic progenitors.
(A) (top) Hierarchical clustering of single-cell epigenomic profiles (columns) and TF motif accessibility z-scores (rows), (bottom) single cell profiles colored by their sorted immunophenotype identity. (B) t-SNE of TF z-scores shown in (A), cells are colored by their sorted immunophenotype identify. (C,D) Single-cell epigenomic landscape defined by PCA projection (see methods) colored by (C) cell type identity using immunophenotype and (D) density (see methods) overlaid with nominal trajectories expected from the literature, as shown in Figure 1A. PC projection colored by (E) GATA, (F) CEBPB, (G) ID3 and (H) HOXA9 TF motif accessibility z-scores.
Using the vector of TF z-scores as features, we visualize hematopoietic differentiation within these data using t-SNE, which clearly displays the expected branching into four distinct differentiated final states representing erythroid, myeloid, lymphoid and pDC differentiation (Figure 2B and Figure S2A–D). In past work we also used ChIP-seq data as annotations to explore chromatin accessibility differences in single-cell profiles (Buenrostro et al., 2015b). Here, we found that K562, a cell line described as an erythroid progenitor model, ChIP-seq data discriminated between cells at different stages of erythroid differentiation, however, failed to capture the variance associated with myeloid and lymphoid trajectories (Figure S2E). Therefore, due to the relatively paucity of TF ChIP-seq in primary bone-marrow derived CD34+ cells or in early myeloid and lymphoid cell models, we chose to use TF motifs in downstream analyses.
Mapping single profiles on hematopoietic principal components
The TF z-scores can be used to cluster single-cell profiles, however, this unsupervised analysis may not distinguish chromatin accessibility changes associated with differentiation from changes associated with other biological phenomenon such as the cell cycle or niche-dependent cell-cell signaling (Crane et al., 2017). Furthermore, the use of t-SNE and TF z-scores for clustering makes relative cell-cell distances difficult to interpret. We reasoned a reference guided approach that utilizes accessibility co-variance of regulatory elements in bulk hematopoietic samples, combining previously published (Corces et al., 2016) and additional reference profiles (see methods), would provide a natural and intuitive subspace for dimensionality reduction of single-cell data. To achieve this we implemented a computational strategy, similar to recent methods for single-cell RNA-seq analysis (Li et al., 2017), that first identifies principal components (PCs) of variation in bulk ATAC-seq samples (Figure S2F) (Corces et al., 2016), then scores each single-cell by the contribution of each PC. Cells are subsequently clustered using the Pearson correlation coefficients between these normalized PCs scores and all other cells (Figure S2G). For low-dimensional visual representation, we performed PCA on this correlation matrix (Figures 2C and 2D) and display the first 3 principal components, which represent 93.7% of the total subspace variance (Figure S2H–M).
We validated this computational approach by down sampling bulk profiles to 104 fragments and find that the PCA-projection approach closely follows sample clustering using the bulk data set (Figure S2N). We next down-sampled ensemble single-cell profiles to match the sequence depths observed in single-cells to quantify the expected mean error to be 1.95%, 1.70% and 3.1% per cell of the total signal for PCs 1–3 respectively (Figures S2O,P). To test our sensitivity for identifying intermediate cell states, we created synthetic mixtures from ensemble profiles, down-sampled to 104 fragments and found the synthetic mixtures to closely follow the expected paths (Figures S2Q,R). Last, we found this approach to be robust to expected experimental confounders in single-cell data (Figure S2S–V). Overall, this visual representation of the data provides a reference-guided landscape of differentiation with similarities to Waddington’s developmental landscape (Figure S1A), furthermore layering TF z-scores onto this representation provides insight into the “guy wires” that may underlie epigenomic changes during differentiation (Figure S1B).
The continuous landscape of human hematopoiesis
Using this computational approach, we find the CAL of human hematopoiesis radiates away from a common basin of early hematopoietic progenitors (Figure 2C). HSC/MPP (left) and LMPP (right) localize at the center of the projection, followed by CMP and GMP cells that comprise a large and diverse basin. Differentiation into CLP (lymphoid), MEP (erythroid) and monocytes (myeloid) appear as distinct differentiation trajectories that swoop away from the central HSC basin (Figure 2D). Furthermore, motifs associated with master lineage regulators ID3, CEBPB and GATA1 (Orkin and Zon, 2008) show continuous gradients of activity across lymphoid, myeloid and erythroid development, while the HSC and LMPP compartment show higher accessibility associated with the HOX motif (Figures 2E–H). We also observe examples of FACS misclassification of cell types, particularly between CMP:MPP, GMP:LMPP (separated by CD38) and GMP:CLP (separated by CD10), likely due to the continuous nature of the cell surface markers (Figure S3A–D).
CMP, GMP and MEP profiles appear markedly heterogeneous in this projected space. We quantified the statistical significance of this observed heterogeneity by comparing the observed variability to down-sampled aggregate profiles and found CMPs to be the most heterogeneous cell type (p < 10−111), with all cell types displaying statistically significant heterogeneity (Figure S3E). To further assess the statistical significance of this heterogeneity, we permuted peaks matched in mean accessibility and GC content (see methods) and find that variability across all cell types, with the exception of monocytes (p = 0.35), remained statistically significant (Figures S3F,G). Finally, single-cell TF z-scores (Schep et al., 2017), which are calculated without using bulk ATAC-seq data as a reference, also exhibit significant variability for all cell types (Figure S3H). Thus, rather than identifying a series of discrete cellular states (Figure 1A), these results suggest that the CAL in early hematopoietic differentiation (HSC, MPP, LMPP and CMP) comprise a fairly broad basin of allowable states, while paths of later differentiation become more canalized into distinct and continuous differentiation trajectories (see DataS1, single-cell CALs can be further explored using our web resource: http://schemer.buenrostrolab.com/).
De novo identification of uncharacterized chromatin states
Given the observed limitations of our sort markers, we sought to define hematopoietic cell types de novo by applying k-medoids clustering on the first five principal components from this PC projection approach. We defined 14 unique clusters (Figures 3A and 3B; see methods) that largely overlap with previously-defined cell surface marker-based definitions of human hematopoietic subsets (Figure 3C) and changes in the accessibility of TF motifs associated with hematopoiesis (Figure 3D). This analysis identified key hematopoietic regulators de novo, including motifs associated with well-described master regulators GATA1 (erythroid), CEBPD (myeloid) and EBF1 (lymphoid) lineage-specifying factors (Orkin and Zon, 2008). Notably, we also find a specific HSC cluster of TFs that include HOX, ERG and MAFF motifs.
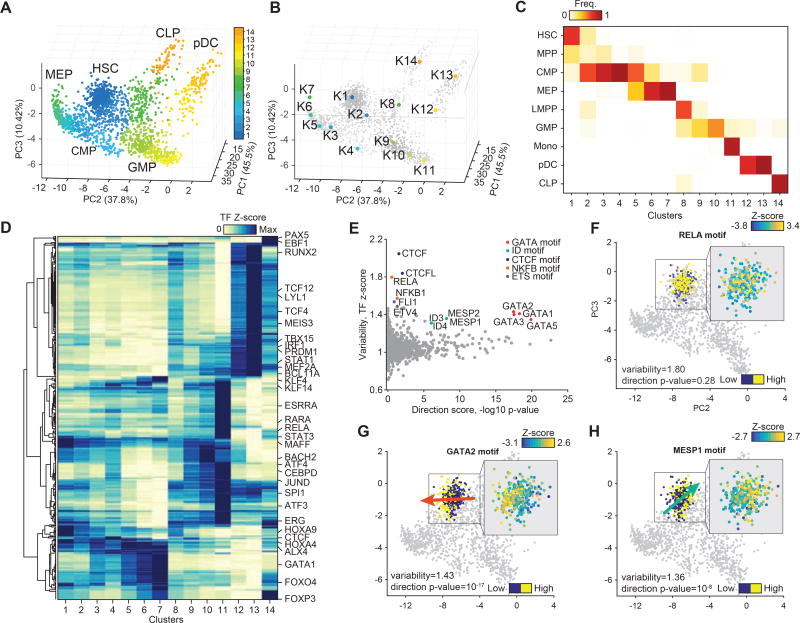
Figure 3. Molecular characterization of data-defined clusters.
(A) Single-cell epigenomic landscape defined by PCA projection, colored by data-driven cluster number. (B) Medoids of data-driven centroids depicted on the PCA sub-space. (C) Confusion matrix of data-driven clusters representing the percent frequency of immunophenotypically defined cell types. (D) TF motif accessibility z-scores averaged across data defined clusters and hierarchically clustered. Scores are normalized by the max value of each TF motif. (E) TF motif variability and direction – log10 p-value for each TF motif for the HSC EIPP cluster, TFs sharing a similar motif are highlighted. (F-H) TF motif accessibility z-scores of HSC profiles for (F) RELA, (G) GATA2 and (H) MESP1 motifs, arrows denote the direction of the signal bias and are colored by the target cell type.
Using this clustering approach, we find that CMPs separate into 4 clusters denoted here as clusters 2–5, which includes a cluster with mixed contribution from CMP and MEP cells (cluster 5). We observe that the 4 CMP clusters within our data show significant variability across motifs associated with GATA1, BCL11A and SPI1 (PU.1), TFs implicated in myeloid/erythroid specification (Figure S3I). We identify 1,801 differentially accessible regions across these CMP clusters, including two previously validated erythroid enhancers (Fulco et al., 2016) regulating GATA1 expression (Figure S3I,J). Given these differences, we assigned CMP clusters as CMP-K3 (early erythroid), CMP-K5 (late erythroid), CMP-K4 (unknown) and CMP-K2 (myeloid primed). These strong chromatin accessibility differences further validate recent work describing functional and transcriptional heterogeneity within mouse (Paul et al., 2015; Perié et al., 2015) and human (Notta et al., 2015) CMPs, and strongly suggest that CMPs can be partitioned into myeloid and erythroid committed progenitors. In addition, we also find that MEP (K5-7), GMP (K9,10) and pDCs (K12,13) predominantly separate as two or more distinct clusters each, likely representing early- and late-stage progenitor differentiation.
Chromatin accessibility variability within data driven clusters
We next sought to measure chromatin accessibility differences within stringently-defined HSC and LMPP progenitors, populations previously described as primed toward different lineage fates (Busch et al., 2015; Karamitros et al., 2018; Laurenti and Göttgens, 2018; Naik et al., 2013; Pei et al., 2017). To quantify this variability, we created stringent cluster definitions that required cells to be both CAL cluster-pure and immunophenotypically (FACS identity) pure populations, which we call epigenomically and immunophenotypically pure (EIPP) clusters. We then computed TF z-scores (Schep et al., 2017) for cells within each EIPP group and found substantial heterogeneity within these subsets (Figure S3K). To explore the relationship of TF-associated variability within HSCs to directions of differentiation we categorized individual EIPP HSCs by their TF z-scores (high or low), computed the distance between high/low centroids in the PC space, and calculated statistical significance by comparing high/low distances to permuted HSC EIPP profiles (Figure 3E and Figure S3L–N). Using this analysis approach, we find CTCF, NFKB (represented by the RELA motif) and ETS motifs to be significantly variable in HSCs, however uncorrelated with any specific direction of differentiation (Figure 3F). Interestingly, NFkB signaling (inflammatory signaling) has been implicated in mouse HSC stem-cell maintenance (Zhao et al., 2012) and HSC emergence mediated by neutrophil secretion of TNFa (Espín-Palazón et al., 2014; Sawamiphak et al., 2014). In contrast, we find the GATA (p=10−17) and MESP/ID (p=10−8) motif (represented by GATA2 and MESP1 motifs) TF z-scores to be significantly correlated to erythroid and lymphoid trajectories respectively (Figures 3G–H and Figures S3N). The direction of this TF bias is consistent with previous studies that lineage trace HSCs which suggest that HSCs exhibit oligo-lineage bias toward erythroid-myeloid or lymphoid cell fates (Pei et al., 2017).
We next turned to LMPPs, a subset previously shown to demonstrate lineage priming toward dendritic (pDC), myeloid (GMP:Monocytes) and lymphoid (CLP:B-cell) fates in mice (Busch et al., 2015; Naik et al., 2013) and in human (Karamitros et al., 2018). Interestingly, we found motifs associated with the TFs TCF4, STAT1 and CEBPE to be significantly correlated with directionality toward CLP, pDC and GMP differentiation, respectively (Figures S3O–R), notably the TCF4 motif is similar to the TCF3, ID3 and MESP1 motifs. TCF4 and CEBPE motif accessibility were anti-correlated with each other, and each defined a unique direction toward CLP (lymphoid) or GMP (myeloid) differentiation respectively, suggesting antagonism between myeloid/lymphoid differentiation programs. Separately, the STAT1 motif accessibility appeared to be directed towards CLP (lymphoid) and pDC (dendritic) cell fates. Overall, this reference-guided computational approach provides a statistical framework for assigning TF motif associated variability to lineage-associated CAL variation providing a resource for identifying molecular factors that may be involved in lineage priming across multipotent cell populations.
Heterogeneous cell types can be further divided along developmental trajectories
We next sought to order cells along continuous differentiation trajectories across branches of hematopoietic development. To achieve this, we first determined the shortest path between cluster centroids and assigned cells to the closest point along that path, a similar approach has been described for analyzing scRNA-seq data (Shin et al., 2015). This approach aligned cells to well-defined lineage pathways (Orkin and Zon, 2008) producing an ordering of single cells along continuous erythroid (K1,3,5,6,7), lymphoid (K1,2,8,14), pDC (K1,2,8,12,13) and myeloid (K1,2,9,10,11) differentiation trajectories (Figure 4A–D and Figure S4A–D). These trajectories allow for interpretation of CAL heterogeneity within progenitors and provide methods to further parse cellular sub states across differentiation.
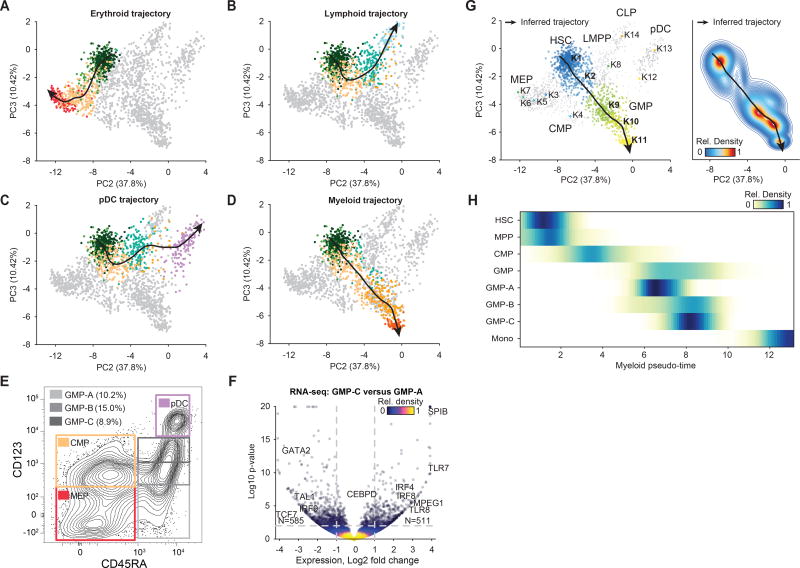
Figure 4. Identifying continuous differentiation trajectories.
(A–D) PC2 by PC3 projection of single-cells highlighting cells progressing through the inferred (A) erythroid, (B) lymphoid, (C) pDC and (D) myeloid developmental trajectory (black line), cells used for inference are colored by sorted identity, all other cells are shown in grey. (E) Sorting schema for different GMP progenitors defined by CD123 expression, marked by CD123 low (GMP-A, light-grey), CD123 medium (GMP-B, grey) and CD123 high (GMP-C, dark-grey). (F) Bulk RNA-seq log2-fold-change and -(log p-value) for expressed genes comparing GMP-C and GMP-A. (G) Single-cells used for the myeloid trajectory colored by (left) their cluster identity (cluster colors as in Figure 3) or (right) their density along the trajectory. (H) Density of myeloid progression scores for immunophenotypically defined cell types, including the GMP subsets.
We examined variability within two clusters (K9 and K10) of GMPs, which show significant differences in accessibility among myeloid-defining factors SPI1 (PU.1) and CEBP associated motifs across the myeloid developmental trajectory (Figure S4E). To further partition this population, we sought to identify cell surface markers that may differentially enrich for K9 and K10 GMPs. We hypothesized that CD123 expression may correlate with early and late GMP differentiation for two reasons: (i) the UNK population, which is CD123−/lo, is enriched in the GMP K9 cluster and (ii) CD123, also known as IL3RA, is a high affinity receptor for the myeloid promoting cytokine IL3. We therefore performed scATAC-seq, bulk ATAC-seq (Buenrostro et al., 2013; 2015a) and bulk RNA-seq on cells from three distinct bins of CD123 expression (Figure 4E). Bulk ATAC-seq and RNA-seq data revealed substantial chromatin accessibility and transcriptomic differences across the GMP-A and GMP-C populations (Figure 4F and Figure S4F–H). The list of differentially expressed genes included important developmental regulators, including down regulation of HSPC TFs GATA2 and TAL1 and up regulation of myeloid genes SPIB, IRF8, TLR7 and MPEG1 in the GMP-C cell population (Figure 4F). In addition, projection of the scATAC-seq data from the three cell fractions revealed that this strategy provides strong separation of early (GMP-A) and late (GMP-C) stages of myeloid differentiation (Figures 4G,H and Figure S4I–K). Altogether, we validate the heterogeneity within GMPs and more generally demonstrate a data driven approach for defining cell populations from single-cell epigenomic data.
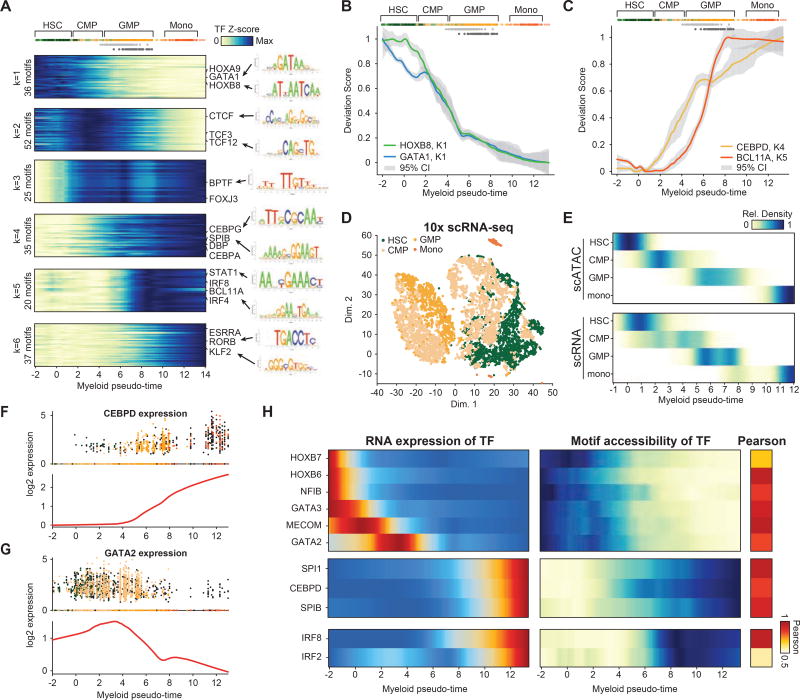
Motif accessibility dynamics along myeloid cell differentiation
The myeloid trajectory described above transits two heterogeneous cell populations (CMP and GMP), as such, regulatory analysis of myeloid differentiation has been previously obscured in bulk studies due to the limitations of the immunophenotypic markers of these populations. We therefore sought to characterize TF dynamics across myeloid development by mapping TF z-scores to cells along the continuous myeloid differentiation trajectory (Figure S4L–N). Using this approach, we find 6 clusters (see methods) of TF z-score profiles during myeloid development (Figure 5A and Figure S5A–C). Accessibility at TF motifs associated with regulators HOXB8 and GATA1 (cluster 1) is high in HSCs and decreases through differentiation to CMPs. Interestingly, loss of GATA motif accessibility (represented by the GATA1 motif) begins within the HSC compartment, while HOX motif accessibility (represented by the HOXB8 motif) is lost at the transition of HSC to CMP differentiation, suggesting that loss of GATA motif accessibility may be an early event in lineage commitment within HSCs (Figure 5B). We also observe two distinct modes of activation for myeloid-associated TF motifs; cluster 4 TFs (CEBPD and SPIB associated motifs) display early and gradual gain in activity beginning within CMPs, while cluster 5 TFs (STAT1, IRF8 and BCL11A associated motifs) increase sharply in activity across the GMP-A to GMP-C transition, implicating the CEBP family of TFs (represented by the CEBPD motif) as an initiating factor for myeloid-erythroid specification (Figure 5C). In addition to the activity patterns associated with canonical myeloid-defining factors, we also identify a pulse of activity within CMPs from cluster 2 TFs (TCF3/12 associated TF motifs up regulated in CLP/pDC), which may reflect transient activation of a lymphoid program within pre-committed myeloid-biased CMPs (Figure S5C).
Figure 5. Transcription factor dynamics across myeloid differentiation.
(A) K-medoids clustering of TF motif accessibility (left) and PWM logos (right) for dynamic TF motif profiles across myeloid development. (B,C) Smoothed profiles of TF motif accessibility z-scores in myeloid progression for (B) HSC active TFs GATA1 (blue) and HOXB8 (green), and (C) monocyte active regulators CEBPD (yellow) and BCL11A (red), error bars (grey) denote 95% confidence intervals. (D) t-SNE of scRNA-seq data showing HSC, CMP, GMP and Monocyte cells. (E) Density of myeloid pseudo-time scores for (top) scATAC-seq and (bottom) computationally matched scRNA-seq profiles (see methods). (F,G) Log2 mean expression profiles for TFs (F) CEBPD and (G) GATA2 across myeloid pseudo-time, (top) individual cells are colored by their sorted identity, CD34+ cells are shown in black and (bottom) smoothed profiles are shown in red. (H) (left) Expression and (right) TF motif accessibility dynamics across myeloid pseudo-time for correlated (R > 0.5) gene-motif pairs.
Matching transcriptomes with chromatin accessibility
We next aimed to develop a means to pair single-cell epigenomic and transcriptomic measurements, with the goal of linking chromatin accessibility changes associated with DNA sequence motifs to expressed TFs, as well as linking accessibility changes at putative enhancers to expression changes at target genes. We first performed single-cell RNA-seq (10X genomics platform) across HSC, CMP and GMPs, collecting a total of 7,818 cells passing filter (2,268, 4,454 and 1,096 respectively; Figure 5D). In addition, we included publically available scRNA-seq data from CD34+ and CD14+ monocyte cells (Zheng et al., 2017), altogether analyzing transcriptional dynamics of 14,432 cells across myeloid differentiation. Using these data, we developed a reference-guided approach to pair scATAC-seq and scRNA-seq profiles (see DataS2). To do this, we first fit a linear model to match the measured bulk ATAC-seq PCs, which measure global variation in chromatin accessibility, to changes in gene expression as measured by bulk RNA-seq across sorted populations (Figures S5D–F). We then used this map between ATAC-seq PCs and gene expression to assign “inferred transcriptomes” to each cell in the scATAC-seq data set (Figure S5G). Finally, to pair each scRNA-seq profile to a scATAC-seq cell, we assigned scRNA-seq profiles to the most correlated scATAC-seq “inferred transcriptome” (Figure S5H). Using this approach, we found that the sorted identity of scRNA-seq profiles were enriched for the corresponding matched sorted identity for scATAC-seq profiles (Figure S5I). Furthermore, by pairing single-cell RNA-seq to scATAC-seq cells we found scRNA-seq profiles of FACS sorted CMPs associated with the four scATAC-seq defined CMP clusters discussed above (Figure S5J). Further validating the recent reports of heterogeneity in mouse (Paul et al., 2015; Perié et al., 2015) and human (Notta et al., 2015) CMPs, we found expression heterogeneity of known hematopoietic regulators in CMPs, which included HOXA5, GATA1 and CEBPB (Figure S5K,L).
Our approach provides a computational method to fit gene expression changes across bulk ATAC-seq and RNA-seq “anchor points” generated from well-defined sorted populations, providing a reference for analysis of single-cell gene expression and chromatin changes spanning these anchor points to resolve continuous regulatory changes in cell differentiation. This reference-guided strategy resulted in a total of 9,312 scRNA-seq cells positioned across myeloid pseudo-time with high concordance in the enrichment of immunophenotypically-defined cells across the trajectories (Figure 5E). Using this unified lineage order we mapped expression dynamics across myeloid cell differentiation and found expected patterns across known regulators of myelopoesis (Figures 5F,G). To further validate this pairing approach, we compared the ATAC:RNA paired lineage order with ordering scRNA-seq cells using diffusion pseudotime (DPT) (Haghverdi et al., 2016). In this comparison, we find that the two approaches for cell ordering are overall highly correlated (R=0.86; Figure S5M). However, we find that unsupervised ordering of cells using DPT was more correlated to the number of genes detected than the ATAC:RNA pairing approach described above (R=0.68 versus R=0.14), suggesting computational ordering of scRNA-seq data with DPT may be more sensitive to dropout (Figure S5N,O). This may be expected as DPT does not explicitly model cell-cell differences in dropout (zero counts for genes) and further suggests that computational tools for joint analysis of scATAC-seq and scRNA-seq may be more robust to technical confounders. Most importantly, the gene expression trajectories (Figure S5P) are largely similar between the two approaches, validating the ATAC:RNA pairing approach.
Linking TF expression with associated accessibility variation in binding motif
In effort to disambiguate TFs that bind the same or similar motif and thus assign expression of TFs to downstream changes in chromatin accessibility at TF motifs, we correlated the expression of TFs with the TF motif z-scores across myeloid pseudo-time. We then filtered for motif accessibility-TF expression correlations of R>0.5, this approach yielded 11 TFs that defined different stages of myeloid development (Figure 5H), including loss in the expression of HOX factors (HOXB7 and HOXB8) (Argiropoulos and Humphries, 2007) and activation of well-known master regulators of myeloid cell development including SPI1 (PU.1) and IRF8 (Satpathy et al., 2012). Resolution of the developmental order of these activated TFs across myeloid differentiation has been previously obscured in bulk studies, in part, due to the cellular heterogeneity within CMPs and GMPs. Interestingly, we also observed a strong correlation between GATA3 expression and GATA motif accessibility, deletion of GATA3 has been shown to promote self-renewal in HSCs (Frelin et al., 2013), together leading to the hypothesis that GATA3 may be associated with HSC lineage priming. Here, single-cell chromatin accessibility, paired with single-cell transcriptomics, resolves the temporal dynamics of master regulator expression and associated chromatin changes in myeloid cell development, providing a resource for further functional studies and for the analysis of regulatory changes associated with differentiation.
Regulatory element and gene activation across myelopoiesis
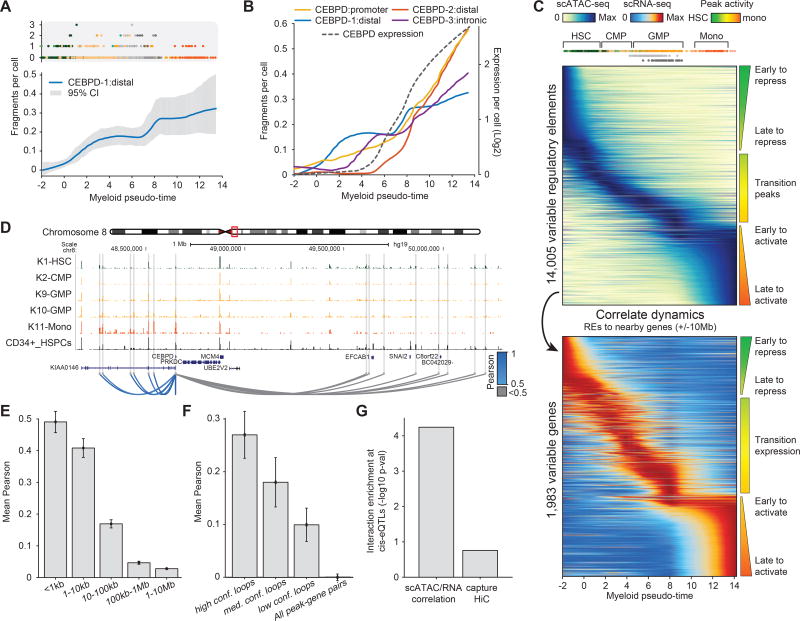
We next sought to characterize locus-specific cis-regulatory dynamics during myeloid differentiation. We first filtered for regulatory elements with high fragment counts and with significant variability across the ordered cells identifying 14,005 cis-regulatory elements for analysis (see methods). These regulatory elements exhibited highly heterogeneous patterns of accessibility changes (Figure 6A–C and Figures S6A–C) – suggesting that a limited number of TF motif accessibility patterns (k=6) could induce a surprising level of variation of chromatin accessibility at individual regulatory elements. For example, within the regulatory elements surrounding the myeloid regulator CEBPD (numbered for simplicity, see Figure 6B), the distal element CEBPD-1 was “fast-to-activate” and showed step-wise gains of activity while the distal element CEBPD-2 was “slow-to-activate” and showed a more discrete pulse of activity (Figure 6B).
Figure 6. Regulatory element dynamics links distal elements to genes.
(A) Fragments per cell for a CEBPD distal element ordered by myeloid pseudo-time, (top) cells are colored by their sorted identity and (bottom) values are smoothed (blue), error bars (grey) denotes 95% confidence intervals. (B) Cis-regulatory and expression dynamics across four regulatory elements near the myeloid regulator CEBPD. (C) Accessibility (top) and expression (bottom) dynamics across myeloid pseudo-time, rows are sorted by their peak intensity in the myeloid trajectory. (D) Regulatory profiles surrounding the CEBPD gene, dynamic enhancers are highlighted in grey with significant (blue) and non-significant (grey) correlated peak-gene pairs shown as loops. (E,F) Mean Pearson correlation coefficients binned by (E) genomic distance to the gene and (F) loop confidence, error bars represent 1 standard deviation on the estimate of the mean. (G) P-value of enriched peak-gene correlation or promoter capture HiC at cis-eQTLs overlapping dynamic enhancers.
To visualize the complete repertoire of dynamic regulatory profiles we ordered elements based on their accessibility changes over this trajectory (Figure 6C and Figure S6). This analysis reveals multiple broad classes of regulatory element behaviors, ranging from fast- to slow-to-repress HSC regulatory elements and fast- to slow-to-activate monocyte regulatory elements (Figure 6C). We also observe a collection of “transition” cis-regulatory elements that exhibit peak accessibility at intermediate stages of myeloid development, as well as “reactivation” elements that are initially lost and subsequently reactivated in later stages of myeloid differentiation (Figures S6D,E). Thus, from a small number of discrete clusters of TF motif accessibility, highly diverse cis-regulatory profiles likely arise from the combinatorial control of trans-factor binding to their target regulatory elements (Figure S6F).
We reasoned that correlation between dynamically activated patterns of distal regulatory elements with nearby expressed genes may be used to connect enhancers to target genes (Figure 6C). Indeed, we found dynamic regulatory elements surrounding CEBPD were highly correlated with CEBPD expression (Figure 6D). More generally, we calculated the correlation of regulatory elements to dynamic genes within 10 Mb of annotated transcription start sites (“peak-gene pairs”) and found that proximal regulatory elements (<100 kb) were significantly more correlated to the expression of nearby genes than distant elements (>100 kb) (Figure 6E). Further validating this approach, we also found that the correlation of regulatory elements to target genes improved as a function of loop confidence within promoter capture HiC (PCHiC) data (Figure 6F), here defined by PCHiC loops from both CD34+ (Mifsud et al., 2015) and monocyte (Javierre et al., 2016) cells. Importantly, we find loop interactions at this resolution do not necessarily define correlated peak-gene pairs. In our analysis, only 45% of dynamic enhancers within high confidence loops are called as correlated to the expression of PCHiC defined target genes (Figure S6G). This observation may be due to the fact that PCHiC loops link relatively large genomic regions, often encompassing multiple regulatory elements, which may independently regulate downstream genes.
We next sought to test whether previously defined cis-linked expression quantitative trait loci (cis-eQTLs) overlapped enhancer-gene interactions identified using these integrated single-cell data. We reasoned that correlated peak-gene pairs could be used to functionally connect relevant genetic variation at regulatory elements to consequences in gene expression important in normal monocyte function. We therefore collected previously-published cis-eQTLs, derived from interferon-γ and lipopolysaccharide stimulation of monocytes (Fairfax et al., 2014), and filtered for SNPs within developmentally dynamic regulatory elements linked to dynamic genes (n=370 peak-gene pairs). To directly compare enrichment of either correlated peak-gene pairs or PCHiC loops at cis-eQTL defined peak-gene interactions, we determined significant enrichment of each data set by normalizing to a background set of peak-genes matched for distance (Figure S6H). We found that cis-eQTLs were strongly enriched for scATAC/scRNA-seq correlated peak-gene pairs (p=4.9×10−5) and observed only a modest enrichment PCHiC loop interactions (p=0.19) (Figure 6G and Figure S6I). Thus, statistical linkage between single-cell chromatin accessibility and gene expression can serve as a means to functionally link enhancers to target gene promoters.
Discussion
We used single-cell chromatin accessibility and transcriptomic analysis to identify regulatory heterogeneity and continuous differentiation trajectories in early human hematopoiesis by developing a broadly applicable computational framework for analysis of these single-cell data. This framework includes a means for visualizing single-cell chromatin accessibility, and computationally pairing these data with single-cell RNA-seq, by using bulk data as a reference. With this approach, we find that immunophenotypically-defined cell populations often flow from one state to another and further we dissociate TF motif activity variability within these populations as correlated or uncorrelated to axis of differentiation. In this effort, we find the activity of TF motifs, such as the GATA motif in HSCs, may represent indicators of lineage priming pulling cells toward different developmentally committed states. While this reference-guided approach enabled us to pair scATAC-seq and scRNA-seq data along a common lineage trajectory, this approach may be generalized to pair cells along more-diverse cell fate transitions. Notably, methods for computationally pairing multi ‘–omic’ profiles have advantages over experimentally coupled approaches, for example, a computational approach may provide i) more flexible experimental workflows, ii) allow pairing data across experimental methods that may not be easily combined, and iii) the reanalysis of the large repertoire of scRNA-seq data already or soon-to-be collected (Regev et al., 2017). As such, the data generated here and associated computational methods may be broadly adapted to further develop computational tools to pair different single-cell data types.
Furthermore, single-cell CALs can be aggregated to define unique cis-regulatory elements active at different stages of differentiation. The intersection of genetic variants with these regulatory elements may provide new insights into cell types or stages of differentiation relevant to disease (Corces et al., 2016; Guo et al., 2016). Current experimental methods that aim to associate non-coding genetic variation to changes in gene expression generally measure either physical interactions using chromatin conformation capture approaches (Javierre et al., 2016) or direct genetic perturbation (Fulco et al., 2016). Here, we show that correlation of naturally occurring regulatory heterogeneity across single-cells can be used to pair regulatory elements to target genes. This single-cell inference approach for linking regulatory elements to genes may be particularly useful for inferring enhancer-gene interactions in rare cells or across cells states where FACS markers are not well defined. We expect future studies will combine an integrated single-cell inference approach with physical interaction or genetic perturbation maps for improved linking of enhancers to target genes, providing a single-cell resolved interaction landscape of noncoding genetic variation.
Overall this work has defined one representation of the epigenomic states underlying hematopoiesis, reminiscent of Waddington’s landscape of differentiation. However, given the static snapshot of the CAL profiles we have quantified it remains uncertain to what degree density of this landscape might allow inference of cell state transition kinetics and potential. Joint measures with the emerging repertoire of CRISPR-based tools for lineage tracing (Woodworth et al., 2017) will be essential for quantifying the epigenomic contribution of lineage priming on cell fate decisions over time. It also remains to be seen to what extent lineage priming is reflected in transcriptional diversity within HSCs, and whether the lineage-associated CAL variability we observe within HSCs is tightly coupled with transcriptional changes (Yu et al., 2016). We expect future work to couple single-cell epigenomic, transcriptomic, proteomic and lineage measures may reveal important insights into the molecular details and temporal order of initiating regulatory factors governing multipotent cell fate transitions. Altogether, we expect further improvements in experimentally or computationally integrating single-cell data to unravel a dynamic regulatory landscape underlying developmental or disease cell fate decisions.
STAR Methods
Contact for reagent and resource sharing
Experimental model and subject details
-
-
Cell collection and isolation
Method details
-
-
Single-cell ATAC-seq
-
-
Single-cell RNA-seq
-
-
Bulk ATAC-seq and RNA-seq
Quantification and statistical analysis
-
-
Data pre-processing and TF scores
-
-
PCA projection
-
-
Clustering, K-medoids and computing density
-
-
Lineage bias analysis
-
-
Significantly differential CMP peaks
-
-
Ordering cells for pseudo-time and smoothing
-
-
Filtering and regulatory element analysis
-
-
Bulk and single cell RNA-Seq analysis
-
-
Matching scRNA-seq to scATAC-seq
-
-
Integration of promoter capture Hi-C data
-
-
Monocyte cis-eQTL data
Data and software availability
Key resources table
Additional resources
Contact for reagent and resource sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, William Greenleaf (wjg@stanford.edu).
Experimental model and subject details
Cell collection and isolation
Human bone marrow was sorted as previously described (Corces et al., 2016). In addition to the cell types previously described, we also isolated plasmacytoid dendritic cells (pDCs), an unknown population (UNK) and megakaryocytes. To isolate pDCs from human bone marrow using FACS we gated for live, lineage negative, CD34+ CD38+ CD10- CD45RA+ CD123+ cells. To isolate UNK cells from human bone marrow by FACS, we gated for live, lineage negative, CD34+ CD38+ CD10- CD45RA+ CD123-. Megakaryocytes were isolated using in vitro differentiation of bone marrow derived CD34+ cells to megakaryocytes. To do this, CD34+ cells were cultured in StemSpan SFEM with Megakaryocyte Expansion Supplement (Stem Cell Technologies) for 14 days, yielding approximately 100-fold expansion in cell number. After cell isolation of all populations using FACS, 15,000 single-cells were resuspended in 100 µl of BAMBANKER™ serum-free cell freezing medium (Wako Chemicals, 302-14681) and cryopreserved in liquid nitrogen. Samples were collected commercially from allcells. Further information about the donors profiled and FACS protocols can be found in SupplementaryTable1.
Method details
Single-cell ATAC-seq and single-cell RNA-seq
Single-cells not cryopreserved after FACS (fresh) were assayed as previously described (Buenrostro et al., 2015b). To assay cells cryopreserved after FACS (frozen), cells were allowed to recover for 10 min at 37°C in IMDM with 10% FBS. After recovery, cells were washed twice in cold 1× PBS and once in with the C1 DNA Seq Cell Wash Buffer (Fluidigm). Cells were then resuspended in 6 µl of C1 DNA Seq Cell Wash Buffer, and were combined with 4 µl of C1 Cell Suspension Reagent, 7 µl of this cell mix was loaded onto the Fluidigm IFC. Single-cells were then assayed using scATAC-seq as previously described (Buenrostro et al., 2015b).
Single-cell RNA-seq
Single-cell RNA-seq data was collected using the recommended protocol for the 3’ scRNA-seq 10X genomics platform using v2 chemistry. DNA shearing used the Covaris S220, libraries were sequenced on a NextSeq with 26×8×98 read lengths.
Bulk ATAC-seq and RNA-seq
ATAC-seq and RNA-seq libraries were generated as previously described (Buenrostro et al., 2015b; Corces et al., 2016) with slight modifications for frozen cells. One vial of 15,000 frozen cells in 100 µl of BAMBANKER™ freezing medium was quickly thawed at 37°C. 70 µl for bulk ATAC-seq and 30 µl for bulk RNA-seq were then added to 500 µl of warm IMDM with 10% FBS. For bulk ATAC-seq, the cells were split into 2 tubes of 5,000 cells used as technical replicates. Cells were washed twice in 1× PBS, all supernatant was carefully removed without disturbing the cell pellet, and cells were resuspended in 40 µl of transposition mix (20 µl of 2× TD buffer, 2 µl of TDE1, 0.2 µl of 2% digitonin, 13.33 µl of 1× PBS, and 4.47 µl of nuclease-free water) (Illumina, FC-121-1030; Promega, G9441), here the transposition reactions were scaled down to compensate for cell loss during washes. Transposition reactions were incubated at 37 °C for 30 min in an Eppendorf ThermoMixer with agitation at 300 rpm. The transposed DNA fragments were purified and amplified as described (Corces et al., 2016).
For bulk RNA-seq, cells were split into 2 technical replicates. RNA was isolated using the Qiagen RNeasy Plus Micro kit, and RNA was eluted in 10 µl of RNase-free water. 5 µl of total RNA was used as input for NuGen Ovation V2 cDNA synthesis kit. The yield of purified SPIA-amplified cDNA was measured using Qubit dsDNA HS Assay kit. 50 ng of SPIA cDNA was fragmented using Nextera DNA library preparation kit (Illumina, FC-121-1030). Fragmented SPIA cDNA was then purified using Qiagen MinElute Reaction Cleanup Kit, and purified DNA was eluted in 10 µl of elution buffer (10mM Tris-HCl, pH 8). Purified SPIA cDNA fragments was amplified and purified as previously described for ATAC-seq (Corces et al., 2016). ATAC-seq and RNA-seq libraries were quantified using qPCR, amplified libraries were sequenced using paired-end, dual-index sequencing on a NextSeq 500 instrument with 76 bp read lengths.
Quantification and statistical analysis
Data pre-processing and TF scores
Single-cell and bulk ATAC-seq alignment, quality filtering and peak calling was performed as previously described (Corces et al., 2016), with one exception. For single-cell profiles, any fragment that occurred in two cells from the same experiment (a single 96-cell IFC) was removed from further analysis. Using the previously described approach (Corces et al., 2016), we defined a peak list using all bulk hematopoietic data analyzed here, resulting in 491,437 500bp nonoverlapping peaks which we use for the remainder of this study. To count the number of fragments per peak in a sample or cell, for computing TF z-scores per cell and for determining background peak sets matched in GC and peak intensity, we used the default settings in the chromVAR package (Schep et al., 2017). Single-cells were filtered for quality requiring at least 60% of fragments in peaks and requiring greater than 1,000 fragments passing quality filters, quality filters are previously described (Corces et al., 2016) which includes removal of mitochondrial reads and low alignment quality (Q30). Bulk counts were normalized as previously described (Corces et al., 2016) using quantile normalization and the CQN package.
PCA projection
To calculate PCs on the bulk data sets, later used for projecting single-cell profiles, we first removed peaks in annotated promoters or aligning to chrX, peaks associated with chrY and unmapped contigs were filtered out in the preprocessing steps described above. 455,057 peaks remained after filtering and were used for the PCA projection analysis. To normalize the bulk count matrix by library size, we identified 19,287 low variance promoters, using a MATLAB implementation of a previously described approach (Brennecke et al., 2013), across all bulk samples and normalized each sample by the mean fragment counts within the low variance promoters. We subsequently took the mean counts of all normalized bulk sample replicates (HSC, MPP, LMPP, CMP, GMP, MEP, Mono, CD4, CD8, NK, NKT, B, CLP, Ery, UNK, pDC and Megakaryocyte) and performed PCA-SVD, resulting in 15 principal components.
To score single-cells by the activity for each component, we first centered the counts for each cell by dividing each peak by the mean fragment counts in peaks for a given cell. We then used the weighted coefficients for each peak and PC (determined using PCA-SVD of the bulk data above) to take the product of the weighted PC coefficients by the centered count values for each cell, taking the sum of this value resulted in a matrix of cells by PCs. Last, we calculate a cell-cell similarity matrix using Pearson correlation and perform PCA on the similarity matrix of correlation values. To assess this computational approach, we repeated this procedure for the bulk data, down sampled bulk data, mean single-cell profiles and synthetic mixtures. Notably, we found a patient-specific batch effect in the HSC single-cell profiles in the PCA projected subspace, this batch signal was strongly correlated with Jun/Fos TF z-scores. We therefore normalized each HSC batch to the mean value of all HSC profiles, and in addition, we blacklisted all motifs correlated with Jun/Fos accessibility. Correlated Jun/Fos motifs were determined by calculating motif similarity (Pearson) (Schep et al., 2017) and removing all motifs with an R>0.8. We use these corrected PC values and filtered TF motifs for all subsequent visualizations of these data.
To determine significant cell-cell variability in the PC projected sub-space, we either down sampled or permuted peaks by their GC content and mean accessibility. To permute peaks that match GC% and mean accessibility, we take the sum accessibility of all cells of a given immunophenotypically defined cell type (e.g. HSCs) and use the ChromVAR function “get_background_peaks” with the default settings. Notably, ChromVAR samples background peaks with replacement and may select the observed peak as a background peak, and therefore provides a conservative estimation of excess variability.
Clustering, K-medoids and computing density
All hierarchical clustering performed used Pearson correlation as the distance function. All k-medoids clustering shown in this work is performed using 10 replicates with the distance function Pearson correlation. For k-mediods clustering throughout this work, the gap statistic is used to determine the appropriate number of clusters, here the optimal cluster number is determined wherein the minimum K satisfies the follow criteria: the gap value for a given K is greater than the gap value of K+1 minus the standard error of the clustering solution for K+1. The first 5 PCs were chosen for clustering cells by their projected PC values. All metrics of cell data density shown in this work are weighted by the expected in vivo frequency of each cell type, as measured from flow cytometry data. 2D data density is calculated using KDE weighted by the in vivo frequency.
Lineage bias analysis
To compute TF variability within epigenome and immunophenotype pure (EIPP) cells, we first determined the most represented cell type for each of the 14 k-medoids determined clusters. We defined EIPP clusters as cells that were immunophenotypically-marked by this most-represented type, and also were within one of the k-medoids clusters. We then collected cluster-pure and immunophenotype-pure profiles for HSCs (k1 cluster) and LMPPs (k8 cluster), and proceeded to compute variability and TF z-scores using ChromVAR (Schep et al., 2017). To determine the magnitude and direction of the TF lineage bias, we first partitioned TF z-scores as greater than zero (high) or less than zero (low). We then computed the mean of the first 5 PCs from the PCA projection for the cells assigned to the high or low TF z-score, distance of the high and low centroids was calculated using Euclidean distance. We determined significance using “direction z-scores”, whereby we repeated the analysis described above for PCs calculated using 50 background peak permutations matching GC and mean accessibility, peaks were determined using ChromVAR (Schep et al., 2017). Direction z-scores were computed comparing the observed to the 50 permuted distances.
Significantly differential CMP peaks
To determine differential CMP peaks, aggregate CMP profiles for each k-medoids cluster were collected. Pairwise binomial tests were performed for each aggregate profile (4 CMP clusters) for each peak. Peaks with a p-value of <10−5 in one or more comparisons was used for further analysis (n = 1,801 peaks). For clustering and visualization, counts were normalized using column z-scores and clustered using k-medoids, the gap statistic (as described above) was used to determine a K of 6.
Ordering cells for pseudo-time and smoothing
To determine continuous differentiation trajectories, we developed a supervised approach, similar to a recently published method (Shin et al., 2015). To do this, we first fit a line through each cluster centroid for the relevant clusters using linear interpolation across the first 5 PCs from the PC projection described above. These relevant clusters were determined using prior literature describing functional cell differentiation trajectories. To assign each cell to a given trajectory, we determined the closest point of each cell within the implicated clusters to the interpolated fit line that connected each cluster centroid across the 5 projected PCs (the closest point was determined using Euclidean distance). The pseudo-time values represented in the main figures represent Euclidean distance along the interpolated line across the 5 PCs from the projected PC space, all values are relative to the mean of HSCs (defined as the start point for all trajectories) and thus represent the value 0 in all pseudo-time trajectories. To order cells by myeloid development, only cells within clusters K1, K2, K9, K10, K11 were considered. For the erythroid, lymphoid and pDC trajectories, only the following clusters were considered: erythroid (K1,3,5,6,7), lymphoid (K1,2,8,14) and pDC (K1,2,8,12,13). To determine the TF motif accessibility dynamics across this inferred trajectory, we smoothed the TF z-scores along myeloid progression with a lowess function with a span of 200, implemented within the MATLAB function smooth. Continuous profiles were normalized by their min/max value for plotting and downstream clustering. The accessibility of individual peaks were smoothed and normalized as was done with TFs, however, with the notable difference of smoothing using a span of 500. To determine error in the smoothed TF z-score profiles and cis-regulatory elements we computed 95% confidence intervals by resampling cells (n=100 permutations) with replacement. We next repeated the smoothing for each permutation and used the MATLAB function paramci to determine the 95% confidence interval per TF or regulatory element.
Filtering and regulatory element analysis
To determine TFs that were significantly variable within the smoothed myeloid trajectories, we compared the standard deviation of the observed smoothed scores to a set of similarly smoothed, permuted, TF scores generated by randomly permuting the myeloid cell order. Selecting TFs with a standard deviation greater than 0.5 provided 205 motifs with an FDR < 1%. We used two criteria to determine highly variable regulatory elements, to do this we first filtered regulatory elements with greater than a mean accessibility of 0.01 fragments per cell and subsequently filtered for regulatory elements with a coefficient of variation of greater than 0.5 (n = 3,403), resulting in a high-quality list. This list was used for all analysis of regulatory elements except for quantifying enrichment at cis-eQTLs. For cis-eQTL analysis we reduced the coefficient of variation filter to 0 to yield a larger list of peaks for analysis (n=14,005).
To determine motif enrichment across dynamic accessible peaks, we first collected log-PWM scores per peak using ChromVAR (Schep et al., 2017) for motifs that were selected in the TF analysis described above. Peak-to-peak distances were computed using Euclidean distance of the PC scores determined by PCA of the min/max normalized accessibility dynamics across the myeloid trajectory. For each peak, the mean log-PWM score was computed for the nearest 300 peaks.
Bulk and single cell RNA-Seq analysis
Raw RNA-Seq reads were aligned to hg19 and quantified per cell barcode using CellRanger v1.2.0 (https://support.10xgenomics.com/single-cell-gene-expression/software/downloads/latest). While the full quantification pipeline is automated and reproducible through CellRanger, we briefly describe the workflow here. First, raw sequencing data was demultiplexed using Illumina bcl2fastq v2.17.1.14. Raw sequencing reads per cell barcode were aligned using STAR version 2.5.1b (Dobin et al., 2013) before duplicates were removed based on unique molecular indicies (UMIs). Per cell, per transcript counts were aggregated, and cells with fewer than 1,000 UMIs/gene counts were excluded. To augment our sorted scRNA-Seq data, we downloaded processed monocytes and CD34+ scRNA-Seq data (as previously described (Zheng et al., 2017)) from the 10X website.
To match the exact feature set quantified in the scRNA-Seq pipeline, all bulk RNA-Seq (including samples originally described in our previous study (Corces et al., 2016) and new samples described in this study) were aligned and quantified using STAR version 2.5.1b (Dobin et al., 2013) and the same gene transfer format file provided in the CellRanger v1.2.0 distribution. Read counts for biological replicates were summed over each transcript to provide a single transcript count per sorted cell type.
Matching scRNA-seq to scATAC-seq
scRNA-seq profiles were matched to scATAC-seq profiles by first linking bulk ATAC-seq PCs to bulk RNA-seq expression profiles. To do this, we first normalized the projected PC scores per cell by the sum-of-squares, which effectively normalizes to differences in the number of reads per cell, and determined the mean projected PC score (in scATAC-seq space) for each immunophenotypically defined cell type with matched bulk RNA-seq. We then trained a linear model, using step-wise regression (MATLAB implementation), to fit the log2 expression of all bulk gene expression profiles across sorted populations as the linear combination of ATAC-seq PC’s. Using the fit coefficients from the step-wise regression, we inferred the expression of all genes for each scATAC-seq profile to produce a reference-assisted “inferred transcriptome”. To match scRNA-seq data to scATAC-seq cells, we first selected for genes that were both variable (defined by a standard deviation across cells greater than 3) and well described by the regression model of bulk PCs (R > 0.9) resulting in a total of 853 genes. To improve matching by denoising profiles, we performed PCA on the expression level inferred for these variable genes, then scored profiles for both scATAC-seq “inferred transcriptomes” and scRNA-seq transcriptomes by the PC loadings from this PCA. Next, we calculated the correlation (Pearson), using the top 20 PCs, between each scATAC-seq “inferred transcriptome” and each cell from the scRNA-seq data set. Single-cell RNA-seq profiles were then matched to ATAC-seq data by selecting the single-cell ATAC-seq cell with the maximum correlation coefficient. ScRNA-seq cells with low correlation values (R<0.9) were discarded from further analysis.
Integration of promoter capture Hi-C data
Processed promoter-capture Hi-C loops for CD34+ (Mifsud et al., 2015) and monocytes (Javierre et al., 2016) were downloaded from the supplemental resources associated with each of the original publications. While the authors reported only significant loops for the CD34+ data, we filtered loops from the full dataset, thresholding the interaction score to > 5 as recommended by the authors. In instances where the promoter bait spanned two or more defined gene promoters (as reported in the original data file), loops were considered for each promoter gene separately. In total, 275,848 significant loops for CD34+ and 443,980 significant loops for monocytes were considered in our downstream analyses, 36,962 loop were overlapping (same target promoter; same distal regulatory annotation).
Monocyte cis-eQTL data
A list of statistically significant cis-eQTL associations were downloaded from the supplemental materials from Fairfax et al. (Fairfax et al., 2014). Only cis-eQTLs within the longer list of dynamic regulatory elements were considered (n = 14,005), not other filtering was performed. The filter for variable genes was reduced to generate a longer gene list (std. > 2, n = 1,983). Using this expanded list 370 cis-eQTLs with associated dynamic peak-gene pairs were available for analysis.
Data and software availability
The GEO accession number for the sequencing data reported in this paper is: GSE96772. Processed scATAC-seq data, which include PC values and TF scores per cell can be found in DataS1. The software developed for analyzing these data, which includes projecting scATAC-seq profiles onto bulk hematopoietic PCs and pairing single-cell ATAC-seq with single-cell RNA-seq, as well as processed data from this manuscript can be found in DataS2.
Additional resources
The data can be visualized in the UCSC genome browser using the track hubs representing bulk data (https://s3.amazonaws.com/JasonBuenrostro/scATAC_heme_label/hub.txt) and clusters of single-cell ATAC-seq data (https://s3.amazonaws.com/JasonBuenrostro/scATAC_heme/hub.txt). This manuscript is accompanied by a web resource for visualizing the single-cell ATAC-seq data, which can be found at: http://schemer.buenrostrolab.com/.
Supplementary Material
DataS1: TF z-scores and PCA values for each cell, related to Figure 2. PC projected scores and TF scores for single-cell ATAC-seq data.
DataS2: Code for projecting data onto bulk PCs and pairing with scRNA-seq, related to Figure 2. MATLAB code for reproducing the PCA projection of single-cell ATAC-seq data onto the hematopoietic PCs and pairing single-cell RNA-seq with single-cell ATAC-seq data.
Groups of similar motifs.
(A,B) Waddington landscape representing (A) the sinuous epigenetic landscape wherein a cell (ball) can roll down different cell fates, and (B) “guy-wires” that shape the epigenetic landscape. (C,D) Comparison of scATAC-seq from ‘fresh’ (blue) and cells frozen after FACS sorting (red), only cells passing quality filtering are shown. Profiles from HSCs showing the (C) fragment yield per cell and (D) fraction of fragments in peaks. (E) Comparison of the log2 accessibility between donor-matched fresh and frozen aggregate accessibility profiles, R=0.88. (F) Fraction of immunophenotypically defined cell types from CD34+ cells for each bone marrow donor, ungated cells are marked in grey. (G) The measured (blue) and average in vivo (yellow) frequency of cells in the data set. (H) Fragment size (bp) distribution and (I) enrichment at transcription start sites (TSSs) for bulk (blue) and aggregate single-cell (red) profiles. (J) Number of cells passing filter for each cell type assayed. (K–L) Mean (K) fragment counts and (L) fraction of reads in peaks of cells passing filter for each cell type assayed.
(A–D) t-SNE embedding of cells colored by the TF z-score activity of (A) GATA1 and (B) CEBPD motifs, or the quality metrics (C) log10 fragment counts and (D) fraction of reads in peaks. (E) t-SNE embedding of single-cell profiles using ENCODE ChIP-seq of K562s as a feature vector, instead of the TF motifs shown above, cells are colored by their cell identity. (F) PC1 and PC2 from PCA of fragments in peaks for bulk samples (grey) and their centroids (red). (G) (left) Hierarchical clustering of the correlation of single-cell epigenomic profiles scored by bulk PCs, (right) profiles colored by their sorted immunophenotype identity. (H) Percent variance explained for each PC from (top) PCA derived from bulk data using fragments in peaks and (bottom) PCA of the PCA projected subspace (see methods). (I–M) PC projection of single-cell ATAC-seq data showing cells scored by PC components (I) PC1 and PC2, (J) PC2 and PC3, (K) PC3 and PC4, (L) PC4 and PC5, and (M) PC5 and PC6. (N) Bulk sample-sample correlation matrix determined by correlation of (left) fragments in peaks, (middle) PCA projection values and (right) PCA projection values after down sampling data to 10,000 fragments per sample. Far left represents the sorted immunophenotype of each bulk sample. (O,P) PCA projection of mean single-cell profiles of immunophenotypically defined cell types down-sampled to (O) 10,000 or (P) matched fragment counts to the observed single-cell data set. (Q,R) Synthetic mixtures of two immunophenotypically defined single-cell profiles down sampled to 10,000 fragments with varying mixtures of (Q) CMP/GMP and (R) LMPP/CLP cell types, unmixed cell types from panel (O) are shown for reference. (S–V) PCA projection of single-cells colored by (S) log10 fragment counts, (T) fraction of reads in peaks, (U) fresh HSC versus frozen profiles, and (V) donor.
(A–B) PCA projection of highlighted cell types for (A) MPP and GMP, and (B) CMP and LMPP. (C,D) Flow cytometry back gating of (C) CMPs and (D) GMPs to show that a subset of cells exhibit CD90 and CD45RA cell surface marker expression without significant CD38 signal. These potentially mis-gated CMPs localize to the MPP gate while mis-gated GMPs localize to the LMPP gate. (E) Fold variance of the PCA projection over the variability expected due to count noise, determined by down-sampling counts from the mean of each immunphenotypically defined cell type to matched sequencing depths of the observed single-cell profiles. Error bars represent 1 standard deviation estimated using bootstrap sampling (1,000 iterations) of cells. (F) Peaks are permuted by their GC content and the mean fragment count for each aggregate immunophenotypically defined cell type, and permuted single-cell profiles are projected onto the PC subspace, un-permuted cells shown in grey for reference. (G) Fold variance over expected for each cell type, quantified as described in panel (E) using the permuted scores shown in (F). (H) TF motif z-score variability sorted by the rank score for each cell type. (I) (left) Differential motifs and (right) regulatory elements across CMP clusters (K2-5), motifs are normalized by max-min values and regulatory elements are normalized as z-scores and clustered using k-medoids. (J) Accessibility at GATA1 locus across the CMP clusters highlighting (grey) two validated (Fulco et al., 2016) enhancers of GATA1. (K) Cell-cell TF motif variability within each EIPP cluster (see methods). (L) Peaks were permuted by their GC content and mean peak fragment count for each aggregate single-cell profile, single cell profiles were then projected onto the PC subspace. (M) (left) Schematic for determining direction p-value using permuted PCA scores (n=50) described in panel (F and L), (right) TF motif variability and direction −log10 p-value for each TF motif for the HSC EIPP cluster. (N,P) Hierarchical clustering of single-cell (N) HSC and (P) LMPP EIPP profiles (columns) for TF motifs appearing as highly variable and directional (rows). (O–R) PC2 and PC3 projection of single LMPP profiles colored by high (yellow) or low (blue) TF motif accessibility z-scores for (O) TCF4, (Q) STAT1 and (R) CEBPE motifs, arrows denote the direction of the signal bias.
(A–D) Histogram denoting number of cells within each point in the (A) myeloid, (B) erythroid, (C) lymphoid and (D) pDC by pseudo-temporal developmental ordering. (E) TF motifs sorted by their rank difference in the mean TF z-score between GMP K10 and K9 clusters, important regulators are highlighted. (F,G) Scatter plots colored by density denoting log2 fragments in individual peaks across samples for (F) UNK versus GMP-A and (G) GMP-A versus GMP-C profiles. (H) Genome browser track highlighting two differentially accessible regions surrounding the GATA2 gene. (I) PCA projection of (light grey) GMP-A, (grey) GMP-B and (dark grey) GMP-C single-cell profiles. (J) Frequency of single-cell profiles from differing GMP sorted immunophenotypes within previously defined data-driven epigenomic clusters. (K) Single-cell profiles colored by their immunophenotypic cell type identity rank sorted by myeloid developmental progression. (L-N) Single-cell myeloid progression and TF z-scores for (L) BCL11A, (M) CEBPD and (N) GATA1 motifs, smoothed motif accessibility trajectories are shown in red.
(A) The standard deviation of smoothed TF accessibility scores, as shown above, for observed scores (blue) and cells randomly permuted (red), dotted line represents an FDR less than 1%. (B) Gap values for 1 to 20 k-medoids clusters, with optimal cluster number 6 highlighted. (C) Centroids for each k-medoid TF cluster for (left) early and (right) late acting TFs, cells ordered by myeloid development (top) are shown for reference. (D) Computational workflow for pairing scATAC-seq with scRNA-seq profiles using bulk data as reference. (E) Stepwise regression fit coefficients linking the activity of scATAC-seq PCs to the expression of CEBPD. (F) Fit and observed expression values (Log2) for bulk transcriptomes of sorted populations (R=0.996). (G) PCA projection of scATAC-seq cell profiles colored by inferred expression of CEBPD. (H) Hierarchical clustering of scATAC-seq by scRNA-seq profiles, values represent Pearson correlation coefficient (see methods). (I,J) Fraction of cell assignments for (I) scRNA-seq profiles to immunophenotypically defined cell types in scATAC-seq data and (J) CMP scATAC-seq of different donors or scRNA-seq profiles to defined clusters as shown in Figure 3,B. (K) scRNA-seq counts of CMP cells matching to cluster 2 or cluster 3 scATAC-seq cells. (L) Hierarchical clustering of genes for scRNA-seq CMPs matched to the four scATAC-seq defined CMP clusters showing (top) significantly variable genes or (bottom) known marker genes. (M) Correlation of transcriptome profiles ordered by their developmental trajectory, comparing scATAC and scRNA pairing (as described in the main text) with diffusion pseudo-time (DPT) of cells (HSCs in green, CMP in yellow and GMP in orange). (N,O) Pseudotime order and number of genes detected for HSCs using (N) ATAC:RNA pairing and (O) DPT, cells are colored by the number of genes detected. (P) Log2 mean expression profiles for TFs (left) HOXA9 and (right) CEBPD across myeloid pseudo-time determined using (top) ATAC:RNA pairing or (bottom) DPT, cells are colored by their sorted identity and smoothed profiles are shown in red.
(A,B) Example cis-regulatory dynamics across myeloid development for (A) HSC active peaks and (B) a reactivated peak near IL7. (C) Hierarchical clustering of k-medoids centroids across all peaks, values (left) denote number of peaks per cluster and labels (right) denote categorization into different global patterns. (D,E) t-SNE plots of dynamic enhancer profiles highlighting (D) reactivation or transition elements, points are colored by the max accessibility of the element in myeloid pseudo-time, and (E) colored by accessibility in monocytes. (F) (left) Schematic for determining motif enrichment across similar peak profiles, (right) raw log PWM score or averaged (see methods) local PWM score for a representative subset of dynamic TF motifs across myeloid development for the t-SNE embedding shown in Figure 5. (G) Fraction of correlated peaks (R>0.5) as a function of loop confidence. Error bars represent 1 standard deviation on the estimate of the mean. (H) Distribution of distances of cis-eQTLs (purple) or background peak-gene pairs (grey) to the predicted target gene. (I) Regulatory profiles surrounding the ITGAX gene (also known as CD11c), dynamic enhancers are highlighted in grey with significant (blue) and non-significant (grey) correlated peak-gene pairs shown as loops, cis-eQTL rs1978487 is highlighted.
Donor information and quality control metrics for cell isolation and single-cell ATAC-seq data.
Highlights.
Single-cell chromatin accessibility reveals a heterogeneous hematopoietic landscape
TF motif-associated chromatin variability in HSCs follow erythroid/lymphoid paths
Characterization of two GMP subsets with chromatin and transcriptome differences
Integrative analysis enables regulatory insights into cis and trans acting factors
Acknowledgments
This work was supported by National Institutes of Health (NIH) P50HG007735 (to H.Y.C. and W.J.G.), Stinehart-Reed Foundation (R.M. and H.Y.C.), U19AI057266 (to W.J.G.) and the Rita Allen Foundation (to W.J.G.) and the Baxter Foundation Faculty Scholar Grant and the Human Frontiers Science Program (to W.J.G). W.J.G is a Chan Zuckerberg Biohub investigator. J.D.B. acknowledges support from the Harvard Society of Fellows and Broad Institute Fellowship. R.M. is a Leukemia and Lymphoma Society Scholar. M.R.C is a Fellow of The Leukemia & Lymphoma Society. We thank members of Greenleaf, Chang, Majeti and Buenrostro labs for valuable discussions. We acknowledge the C. Bustamante lab for help with sequencing.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
J.D.B., M.R.C., H.Y.C, and W.J.G. conceived the project. M.R.C. and R.M. performed cell sorting. J.D.B. performed ATAC-seq and scATAC-seq data analysis, and oversaw scATAC-seq library generation and protocol optimization performed by B.W. B.W. generated the scATAC, bulk ATAC-seq, bulk RNA-seq and scRNA-seq data, C.A.L. performed the RNA-seq, PCHi-C data analysis. A.N.S developed the TF motif analysis tools and C.A.L. developed the associated web resource. J.D.B. and W.J.G. wrote the manuscript with input from all authors.
Declaration of Interests
Stanford University has filed a provisional patent on ATAC-seq, J.D.B., H.Y.C., and W.J.G. are named as inventors. H.Y.C. and W.J.G. are founders of Epinomics.
ITI
Integrative analysis of single cell transcriptomics and chromatin accessibility provides insights into regulatory features and dynamics in human hematopoiesis.
References
- Argiropoulos B, Humphries RK. Hox genes in hematopoiesis and leukemogenesis. Oncogene. 2007;26:6766–6776. doi: 10.1038/sj.onc.1210760. [DOI] [PubMed] [Google Scholar]
- Becker AJ, McCulloch EA, Till JE. Cytological demonstration of the clonal nature of spleen colonies derived from transplanted mouse marrow cells. Nature. 1963;197:452–454. doi: 10.1038/197452a0. [DOI] [PubMed] [Google Scholar]
- Brennecke P, Anders S, Kim JK, Kołodziejczyk AA, Zhang X, Proserpio V, Baying B, Benes V, Teichmann SA, Marioni JC, et al. Accounting for technical noise in single-cell. RNA-seq experiments. 2013;10:1093–1095. doi: 10.1038/nmeth.2645. [DOI] [PubMed] [Google Scholar]
- Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Wu B, Chang HY, Greenleaf WJ. ATAC-seq: A Method for Assaying Chromatin Accessibility Genome-Wide. Curr Protoc Mol Biol. 2015a;109 doi: 10.1002/0471142727.mb2129s109. 21.29.1–.29.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Wu B, Litzenburger UM, Ruff D, Gonzales ML, Snyder MP, Chang HY, Greenleaf WJ. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature. 2015b;523:486–490. doi: 10.1038/nature14590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch K, Klapproth K, Barile M, Flossdorf M, Holland-Letz T, Schlenner SM, Reth M, Höfer T, Rodewald H-R. Fundamental properties of unperturbed haematopoiesis from stem cells in vivo. Nature. 2015;518:542–546. doi: 10.1038/nature14242. [DOI] [PubMed] [Google Scholar]
- Calo E, Wysocka J. Modification of Enhancer Chromatin: What, How, and Why? Molecular Cell. 2013;49:825–837. doi: 10.1016/j.molcel.2013.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Kostadima M, Martens JHA, Canu G, Garcia SP, Turro E, Downes K, Macaulay IC, Bielczyk-Maczynska E, Coe S, et al. Transcriptional diversity during lineage commitment of human blood progenitors. 2014;345:1251033–1251033. doi: 10.1126/science.1251033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corces MR, Buenrostro JD, Wu B, Greenside PG, Chan SM, Koenig JL, Snyder MP, Pritchard JK, Kundaje A, Greenleaf WJ, et al. Lineage-specific and single-cell chromatin accessibility charts human hematopoiesis and leukemia evolution. Nature Genetics. 2016;48:1193–1203. doi: 10.1038/ng.3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crane GM, Jeffery E, Morrison SJ. Adult haematopoietic stem cell niches. Nature Reviews Immunology 2017 17: 9. 2017;17:573–590. doi: 10.1038/nri.2017.53. [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espín-Palazón R, Stachura DL, Campbell CA, García-Moreno D, Del Cid N, Kim AD, Candel S, Meseguer J, Mulero V, Traver D. Proinflammatory Signaling Regulates Hematopoietic Stem Cell Emergence. Cell. 2014;159:1070–1085. doi: 10.1016/j.cell.2014.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairfax BP, Humburg P, Makino S, Naranbhai V, Wong D, Lau E, Jostins L, Plant K, Andrews R, McGee C, et al. Innate Immune Activity Conditions the Effect of Regulatory Variants upon Monocyte Gene Expression. Science. 2014;343:1246949–1246949. doi: 10.1126/science.1246949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farlik M, Halbritter F, Müller F, Choudry FA, Ebert P, Klughammer J, Farrow S, Santoro A, Ciaurro V, Mathur A, et al. DNA Methylation Dynamics of Human Hematopoietic Stem Cell Differentiation. Cell Stem Cell. 2016;19:808–822. doi: 10.1016/j.stem.2016.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frelin C, Herrington R, Janmohamed S, Barbara M, Tran G, Paige CJ, Benveniste P, Zúñiga-Pflücker J-C, Souabni A, Busslinger M, et al. GATA-3 regulates the self-renewal of long-term hematopoietic stem cells. Nature Immunology. 2013;14:1037–1044. doi: 10.1038/ni.2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulco CP, Munschauer M, Anyoha R, Munson G, Grossman SR, Perez EM, Kane M, Cleary B, Lander ES, Engreitz JM. Systematic mapping of functional enhancer-promoter connections with CRISPR interference. Science. 2016;354:aag2445–aag2773. doi: 10.1126/science.aag2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg AD, Allis CD, Bernstein E. Epigenetics: A Landscape Takes Shape. Cell. 2007;128:635–638. doi: 10.1016/j.cell.2007.02.006. [DOI] [PubMed] [Google Scholar]
- Graf T, Enver T. Forcing cells to change lineages. Nature. 2009;462:587–594. doi: 10.1038/nature08533. [DOI] [PubMed] [Google Scholar]
- Grün D, Muraro MJ, Boisset J-C, Wiebrands K, Lyubimova A, Dharmadhikari G, van den Born M, van Es J, Jansen E, Clevers H, et al. De Novo Prediction of Stem Cell Identity using Single-Cell Transcriptome Data. Cell Stem Cell. 2016;19:266–277. doi: 10.1016/j.stem.2016.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo MH, Nandakumar SK, Ulirsch JC, Zekavat SM, Buenrostro JD, Natarajan P, Salem RM, Chiarle R, Mitt M, Kals M, et al. Comprehensive population-based genome sequencing provides insight into hematopoietic regulatory mechanisms. Proceedings of the National Academy of Sciences of the United States of America. 2016 doi: 10.1073/pnas.1619052114. 201619052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haghverdi L, Büttner M, Wolf FA, Buettner F, Theis FJ. Diffusion pseudotime robustly reconstructs lineage branching. Nat. Methods. 2016;13:845–848. doi: 10.1038/nmeth.3971. [DOI] [PubMed] [Google Scholar]
- Javierre BM, Burren OS, Wilder SP, Kreuzhuber R, Hill SM, Sewitz S, Cairns J, Wingett SW, Várnai C, Thiecke MJ, et al. Lineage-Specific Genome Architecture Links Enhancers and Non-coding Disease Variants to Target Gene Promoters. Cell. 2016;167:1369–1384. e19. doi: 10.1016/j.cell.2016.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karamitros D, Stoilova B, Aboukhalil Z, Hamey F, Reinisch A, Samitsch M, Quek L, Otto G, Repapi E, Doondeea J, et al. Single-cell analysis reveals the continuum of human lympho-myeloid progenitor cells. Nature Immunology. 2018;19:85–97. doi: 10.1038/s41590-017-0001-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelsey G, Stegle O, Reik W. Single-cell epigenomics: Recording the past and predicting the future. Science. 2017;358:69–75. doi: 10.1126/science.aan6826. [DOI] [PubMed] [Google Scholar]
- Laurenti E, Göttgens B. From haematopoietic stem cells to complex differentiation landscapes. Nature. 2018;553:418. doi: 10.1038/nature25022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence HJ, Helgason CD, Sauvageau G, Fong S, Izon DJ, Humphries RK, Largman C. Mice bearing a targeted interruption of the homeobox gene HOXA9 have defects in myeloid, erythroid, and lymphoid hematopoiesis. Blood. 1997;89:1922–1930. [PubMed] [Google Scholar]
- Li H, Courtois ET, Sengupta D, Tan Y, Chen KH, Goh JJL, Kong SL, Chua C, Hon LK, Tan WS, et al. Reference component analysis of single-cell transcriptomes elucidates cellular heterogeneity in human colorectal tumors. Nature Genetics. 2017;49 doi: 10.1038/ng.3818. ng.3818–718. [DOI] [PubMed] [Google Scholar]
- Long HK, Prescott SL, Wysocka J. Ever-Changing Landscapes: Transcriptional Enhancers in Development and Evolution. Cell. 2016;167:1170–1187. doi: 10.1016/j.cell.2016.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnusson M, Brun ACM, Lawrence HJ, Karlsson S. Hoxa9/hoxb3/hoxb4 compound null mice display severe hematopoietic defects. Experimental Hematology. 2007;35:1421.e1–1421.e9. doi: 10.1016/j.exphem.2007.05.011. [DOI] [PubMed] [Google Scholar]
- Manz MG, Miyamoto T, Akashi K, Weissman IL. Prospective isolation of human clonogenic common myeloid progenitors. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:11872–11877. doi: 10.1073/pnas.172384399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka Y, Sumide K, Kawamura H, Nakatsuka R, Fujioka T, Sasaki Y, Sonoda Y. Human cord blood-derived primitive CD34-negative hematopoietic stem cells (HSCs) are myeloid-biased long-term repopulating HSCs. Blood Cancer J. 2015;5:e290. doi: 10.1038/bcj.2015.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mifsud B, Tavares-Cadete F, Young AN, Sugar R, Schoenfelder S, Ferreira L, Wingett SW, Andrews S, Grey W, Ewels PA, et al. Mapping long-range promoter contacts in human cells with high-resolution capture Hi-C. Nature Genetics. 2015;47:598–606. doi: 10.1038/ng.3286. [DOI] [PubMed] [Google Scholar]
- Naik SH, Perié L, Swart E, Gerlach C, van Rooij N, de Boer RJ, Schumacher TN. Diverse and heritable lineage imprinting of early haematopoietic progenitors. Nature. 2013;496:229–232. doi: 10.1038/nature12013. [DOI] [PubMed] [Google Scholar]
- Notta F, Gan OI, Wilson G, Kaufmann KB, Mcleod J, Laurenti E, Dunant CF, John D, Stein LD, Dror Y, et al. Distinct routes of lineage development reshape the human blood hierarchy across ontogeny. Science. 2015:1–16. doi: 10.1126/science.aab2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novershtern N, Subramanian A, Lawton LN, Mak RH, Haining WN, McConkey ME, Habib N, Yosef N, Chang CY, Shay T, et al. Densely interconnected transcriptional circuits control cell states in human hematopoiesis. Cell. 2011;144:296–309. doi: 10.1016/j.cell.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orkin SH, Zon LI. Hematopoiesis: An Evolving Paradigm for Stem Cell Biology. Cell. 2008;132:631–644. doi: 10.1016/j.cell.2008.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul F, Arkin Y, Giladi A, Jaitin DA, Kenigsberg E, Keren-Shaul H, Winter D, Lara-Astiaso D, Gury M, Weiner A, et al. Transcriptional Heterogeneity and Lineage Commitment in Myeloid Progenitors. Cell. 2015;163:1663–1677. doi: 10.1016/j.cell.2015.11.013. [DOI] [PubMed] [Google Scholar]
- Pei W, Feyerabend TB, Rössler J, Wang X, Postrach D, Busch K, Rode I, Klapproth K, Dietlein N, Quedenau C, et al. <i>Polylox</i>barcoding reveals haematopoietic stem cell fates realized<i>in vivo</i>. Nature. 2017;548:456–460. doi: 10.1038/nature23653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perié L, Duffy KR, Kok L, de Boer RJ, Schumacher TN. The Branching Point in Erythro-Myeloid Differentiation. Cell. 2015;163:1655–1662. doi: 10.1016/j.cell.2015.11.059. [DOI] [PubMed] [Google Scholar]
- Regev A, Teichmann SA, Lander ES, Amit I, Benoist C, Birney E, Bodenmiller B, Campbell PJ, Carninci P, Clatworthy M, et al. Science Forum: The Human Cell Atlas. eLife Sciences. 2017;6:e27041. doi: 10.7554/eLife.27041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satpathy AT, Wu X, Albring JC, Murphy KM. Re(de)fining the dendritic cell lineage. Nature Immunology. 2012;13:1145–1154. doi: 10.1038/ni.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawamiphak S, Kontarakis Z, Stainier DYR. Interferon Gamma Signaling Positively Regulates Hematopoietic Stem Cell Emergence. Developmental Cell. 2014;31:640–653. doi: 10.1016/j.devcel.2014.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schep AN, Wu B, Buenrostro JD, Greenleaf WJ. chromVAR: Inferring transcription factor variation from single-cell epigenomic data. bioRxiv. 2017:110346. doi: 10.1038/nmeth.4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J, Berg DA, Zhu Y, Shin JY, Song J, Bonaguidi MA, Enikolopov G, Nauen DW, Christian KM, Ming G-L, et al. Single-Cell RNA-Seq with Waterfall Reveals Molecular Cascades underlying Adult Neurogenesis. Cell Stem Cell. 2015;17:360–372. doi: 10.1016/j.stem.2015.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Velten L, Haas SF, Raffel S, Blaszkiewicz S, Islam S, Hennig BP, Hirche C, Lutz C, Buss EC, Nowak D, et al. Human haematopoietic stem cell lineage commitment is a continuous process. Nature Cell Biology. 2017;19:271–281. doi: 10.1038/ncb3493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddington C. The Strategy of the Genes: a discussion of some aspects of theoretical biology. The American Biology Teacher. 1960;22:47–47. [Google Scholar]
- Woodworth MB, Girskis KM, Walsh CA. Building a lineage from single cells: genetic techniques for cell lineage tracking. Nature Reviews Genetics. 2017;18:230–244. doi: 10.1038/nrg.2016.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu VWC, Yusuf RZ, Oki T, Wu J, Saez B, Wang X, Cook C, Baryawno N, Ziller MJ, Lee E, et al. Epigenetic Memory Underlies Cell-Autonomous Heterogeneous Behavior of Hematopoietic Stem Cells. Cell. 2016;167:1310–1322. e1317. doi: 10.1016/j.cell.2016.10.045. [DOI] [PubMed] [Google Scholar]
- Zhao C, Xiu Y, Ashton J, Xing L, Morita Y, Jordan CT, Boyce BF. Noncanonical NF-κB signaling regulates hematopoietic stem cell self-renewal and microenvironment interactions. - PubMed - NCBI. Stem Cells. 2012;30:709–718. doi: 10.1002/stem.1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng GXY, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R, Ziraldo SB, Wheeler TD, McDermott GP, Zhu J, et al. Massively parallel digital transcriptional profiling of single cells. Nature Communications 2017 8:Null. 2017;8 doi: 10.1038/ncomms14049. ncomms14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
DataS1: TF z-scores and PCA values for each cell, related to Figure 2. PC projected scores and TF scores for single-cell ATAC-seq data.
DataS2: Code for projecting data onto bulk PCs and pairing with scRNA-seq, related to Figure 2. MATLAB code for reproducing the PCA projection of single-cell ATAC-seq data onto the hematopoietic PCs and pairing single-cell RNA-seq with single-cell ATAC-seq data.
Groups of similar motifs.
(A,B) Waddington landscape representing (A) the sinuous epigenetic landscape wherein a cell (ball) can roll down different cell fates, and (B) “guy-wires” that shape the epigenetic landscape. (C,D) Comparison of scATAC-seq from ‘fresh’ (blue) and cells frozen after FACS sorting (red), only cells passing quality filtering are shown. Profiles from HSCs showing the (C) fragment yield per cell and (D) fraction of fragments in peaks. (E) Comparison of the log2 accessibility between donor-matched fresh and frozen aggregate accessibility profiles, R=0.88. (F) Fraction of immunophenotypically defined cell types from CD34+ cells for each bone marrow donor, ungated cells are marked in grey. (G) The measured (blue) and average in vivo (yellow) frequency of cells in the data set. (H) Fragment size (bp) distribution and (I) enrichment at transcription start sites (TSSs) for bulk (blue) and aggregate single-cell (red) profiles. (J) Number of cells passing filter for each cell type assayed. (K–L) Mean (K) fragment counts and (L) fraction of reads in peaks of cells passing filter for each cell type assayed.
(A–D) t-SNE embedding of cells colored by the TF z-score activity of (A) GATA1 and (B) CEBPD motifs, or the quality metrics (C) log10 fragment counts and (D) fraction of reads in peaks. (E) t-SNE embedding of single-cell profiles using ENCODE ChIP-seq of K562s as a feature vector, instead of the TF motifs shown above, cells are colored by their cell identity. (F) PC1 and PC2 from PCA of fragments in peaks for bulk samples (grey) and their centroids (red). (G) (left) Hierarchical clustering of the correlation of single-cell epigenomic profiles scored by bulk PCs, (right) profiles colored by their sorted immunophenotype identity. (H) Percent variance explained for each PC from (top) PCA derived from bulk data using fragments in peaks and (bottom) PCA of the PCA projected subspace (see methods). (I–M) PC projection of single-cell ATAC-seq data showing cells scored by PC components (I) PC1 and PC2, (J) PC2 and PC3, (K) PC3 and PC4, (L) PC4 and PC5, and (M) PC5 and PC6. (N) Bulk sample-sample correlation matrix determined by correlation of (left) fragments in peaks, (middle) PCA projection values and (right) PCA projection values after down sampling data to 10,000 fragments per sample. Far left represents the sorted immunophenotype of each bulk sample. (O,P) PCA projection of mean single-cell profiles of immunophenotypically defined cell types down-sampled to (O) 10,000 or (P) matched fragment counts to the observed single-cell data set. (Q,R) Synthetic mixtures of two immunophenotypically defined single-cell profiles down sampled to 10,000 fragments with varying mixtures of (Q) CMP/GMP and (R) LMPP/CLP cell types, unmixed cell types from panel (O) are shown for reference. (S–V) PCA projection of single-cells colored by (S) log10 fragment counts, (T) fraction of reads in peaks, (U) fresh HSC versus frozen profiles, and (V) donor.
(A–B) PCA projection of highlighted cell types for (A) MPP and GMP, and (B) CMP and LMPP. (C,D) Flow cytometry back gating of (C) CMPs and (D) GMPs to show that a subset of cells exhibit CD90 and CD45RA cell surface marker expression without significant CD38 signal. These potentially mis-gated CMPs localize to the MPP gate while mis-gated GMPs localize to the LMPP gate. (E) Fold variance of the PCA projection over the variability expected due to count noise, determined by down-sampling counts from the mean of each immunphenotypically defined cell type to matched sequencing depths of the observed single-cell profiles. Error bars represent 1 standard deviation estimated using bootstrap sampling (1,000 iterations) of cells. (F) Peaks are permuted by their GC content and the mean fragment count for each aggregate immunophenotypically defined cell type, and permuted single-cell profiles are projected onto the PC subspace, un-permuted cells shown in grey for reference. (G) Fold variance over expected for each cell type, quantified as described in panel (E) using the permuted scores shown in (F). (H) TF motif z-score variability sorted by the rank score for each cell type. (I) (left) Differential motifs and (right) regulatory elements across CMP clusters (K2-5), motifs are normalized by max-min values and regulatory elements are normalized as z-scores and clustered using k-medoids. (J) Accessibility at GATA1 locus across the CMP clusters highlighting (grey) two validated (Fulco et al., 2016) enhancers of GATA1. (K) Cell-cell TF motif variability within each EIPP cluster (see methods). (L) Peaks were permuted by their GC content and mean peak fragment count for each aggregate single-cell profile, single cell profiles were then projected onto the PC subspace. (M) (left) Schematic for determining direction p-value using permuted PCA scores (n=50) described in panel (F and L), (right) TF motif variability and direction −log10 p-value for each TF motif for the HSC EIPP cluster. (N,P) Hierarchical clustering of single-cell (N) HSC and (P) LMPP EIPP profiles (columns) for TF motifs appearing as highly variable and directional (rows). (O–R) PC2 and PC3 projection of single LMPP profiles colored by high (yellow) or low (blue) TF motif accessibility z-scores for (O) TCF4, (Q) STAT1 and (R) CEBPE motifs, arrows denote the direction of the signal bias.
(A–D) Histogram denoting number of cells within each point in the (A) myeloid, (B) erythroid, (C) lymphoid and (D) pDC by pseudo-temporal developmental ordering. (E) TF motifs sorted by their rank difference in the mean TF z-score between GMP K10 and K9 clusters, important regulators are highlighted. (F,G) Scatter plots colored by density denoting log2 fragments in individual peaks across samples for (F) UNK versus GMP-A and (G) GMP-A versus GMP-C profiles. (H) Genome browser track highlighting two differentially accessible regions surrounding the GATA2 gene. (I) PCA projection of (light grey) GMP-A, (grey) GMP-B and (dark grey) GMP-C single-cell profiles. (J) Frequency of single-cell profiles from differing GMP sorted immunophenotypes within previously defined data-driven epigenomic clusters. (K) Single-cell profiles colored by their immunophenotypic cell type identity rank sorted by myeloid developmental progression. (L-N) Single-cell myeloid progression and TF z-scores for (L) BCL11A, (M) CEBPD and (N) GATA1 motifs, smoothed motif accessibility trajectories are shown in red.
(A) The standard deviation of smoothed TF accessibility scores, as shown above, for observed scores (blue) and cells randomly permuted (red), dotted line represents an FDR less than 1%. (B) Gap values for 1 to 20 k-medoids clusters, with optimal cluster number 6 highlighted. (C) Centroids for each k-medoid TF cluster for (left) early and (right) late acting TFs, cells ordered by myeloid development (top) are shown for reference. (D) Computational workflow for pairing scATAC-seq with scRNA-seq profiles using bulk data as reference. (E) Stepwise regression fit coefficients linking the activity of scATAC-seq PCs to the expression of CEBPD. (F) Fit and observed expression values (Log2) for bulk transcriptomes of sorted populations (R=0.996). (G) PCA projection of scATAC-seq cell profiles colored by inferred expression of CEBPD. (H) Hierarchical clustering of scATAC-seq by scRNA-seq profiles, values represent Pearson correlation coefficient (see methods). (I,J) Fraction of cell assignments for (I) scRNA-seq profiles to immunophenotypically defined cell types in scATAC-seq data and (J) CMP scATAC-seq of different donors or scRNA-seq profiles to defined clusters as shown in Figure 3,B. (K) scRNA-seq counts of CMP cells matching to cluster 2 or cluster 3 scATAC-seq cells. (L) Hierarchical clustering of genes for scRNA-seq CMPs matched to the four scATAC-seq defined CMP clusters showing (top) significantly variable genes or (bottom) known marker genes. (M) Correlation of transcriptome profiles ordered by their developmental trajectory, comparing scATAC and scRNA pairing (as described in the main text) with diffusion pseudo-time (DPT) of cells (HSCs in green, CMP in yellow and GMP in orange). (N,O) Pseudotime order and number of genes detected for HSCs using (N) ATAC:RNA pairing and (O) DPT, cells are colored by the number of genes detected. (P) Log2 mean expression profiles for TFs (left) HOXA9 and (right) CEBPD across myeloid pseudo-time determined using (top) ATAC:RNA pairing or (bottom) DPT, cells are colored by their sorted identity and smoothed profiles are shown in red.
(A,B) Example cis-regulatory dynamics across myeloid development for (A) HSC active peaks and (B) a reactivated peak near IL7. (C) Hierarchical clustering of k-medoids centroids across all peaks, values (left) denote number of peaks per cluster and labels (right) denote categorization into different global patterns. (D,E) t-SNE plots of dynamic enhancer profiles highlighting (D) reactivation or transition elements, points are colored by the max accessibility of the element in myeloid pseudo-time, and (E) colored by accessibility in monocytes. (F) (left) Schematic for determining motif enrichment across similar peak profiles, (right) raw log PWM score or averaged (see methods) local PWM score for a representative subset of dynamic TF motifs across myeloid development for the t-SNE embedding shown in Figure 5. (G) Fraction of correlated peaks (R>0.5) as a function of loop confidence. Error bars represent 1 standard deviation on the estimate of the mean. (H) Distribution of distances of cis-eQTLs (purple) or background peak-gene pairs (grey) to the predicted target gene. (I) Regulatory profiles surrounding the ITGAX gene (also known as CD11c), dynamic enhancers are highlighted in grey with significant (blue) and non-significant (grey) correlated peak-gene pairs shown as loops, cis-eQTL rs1978487 is highlighted.
Donor information and quality control metrics for cell isolation and single-cell ATAC-seq data.