Abstract
Periodic food shortages are a major challenge faced by organisms in natural habitats. Cave-dwelling animals must withstand long periods of nutrient deprivation, as—in the absence of photosynthesis—caves depend on external energy sources such as seasonal floods1. Here we show that cave-adapted populations of the Mexican tetra, Astyanax mexicanus, have dysregulated blood glucose homeostasis and are insulin-resistant compared to river-adapted populations. We found that multiple cave populations carry a mutation in the insulin receptor that leads to decreased insulin binding in vitro and contributes to hyperglycaemia. Hybrid fish from surface–cave crosses carrying this mutation weigh more than non-carriers, and zebrafish genetically engineered to carry the mutation have increased body weight and insulin resistance. Higher body weight may be advantageous in caves as a strategy to cope with an infrequent food supply. In humans, the identical mutation in the insulin receptor leads to a severe form of insulin resistance and reduced lifespan. However, cavefish have a similar lifespan to surface fish and do not accumulate the advanced glycation end-products in the blood that are typically associated with the progression of diabetes-associated pathologies. Our findings suggest that diminished insulin signalling is beneficial in a nutrient-limited environment and that cavefish may have acquired compensatory mechanisms that enable them to circumvent the typical negative effects associated with failure to regulate blood glucose levels.
The fish species A. mexicanus consists of interfertile river-dwelling and cave-dwelling populations (Fig. 1a), here referred to as ‘surface fish’ and ‘cavefish’, respectively, that experience markedly different nutrient availablility2. Cavefish are resistant to starvation; when food-deprived, cavefish lose a smaller fraction of their body weight compared to surface fish3. Several factors have been identified that contribute to starvation resistance, including reduced metabolic circadian rhythm4, decreased metabolic rate5 and elevated body fat3. The genetic changes underlying these adaptations remain largely unknown.
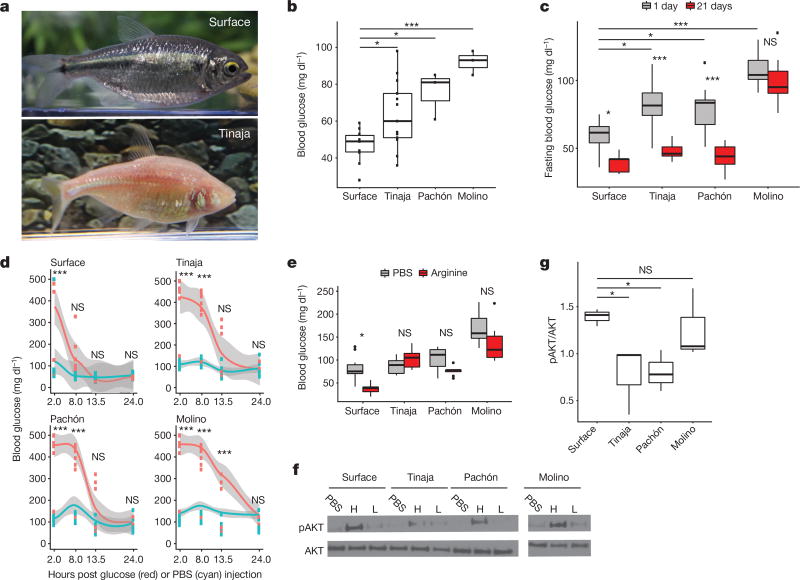
Figure 1. Altered glucose homeostasis in cave-adapted A. mexicanus populations.
a, Surface fish and Tinaja cavefish of A. mexicanus. b, Blood glucose (1 h postprandial) in surface fish compared to cavefish (n = 10, 13, 3 and 3, respectively, for surface fish, Tinaja, Pachón and Molino cavefish). c, Fasting blood glucose at day 1 versus day 21 (n = 20 per population and condition). d, Glucose tolerance test. Blood glucose after intraperitoneal injection of glucose (red) or PBS (blue). Data points represent values for individual fish and grey shade indicates 95% confidence interval for polynomial regression. e, Blood glucose 5 h after intraperitoneal injection of arginine (n = 10 per population and condition). f, Western blot: cell lysates probed with pAKT (ser473) and AKT antibodies. Lysates produced from skeletal muscle treated ex vivo with PBS, a high (H, 9.5–11.5 µg ml−1) or a low (L, 0.95–1.15 µg ml−1) level of insulin. g, Quantification of bands by densitometry of highest concentration treatment (n = 3 per population). For box plots, median, 25th, 50th and 75th percentiles are represented by horizontal bars, and vertical bars represent 1.5× interquartile ranges. Significance calculated using one-way ANOVA with Tukey’s honest significant difference (HSD) post hoc test. NS, P > 0.05; * P < 0.05; * * P < 0.005; * * * P < 0.0005. For gel source data, see Supplementary Fig. 1.
In this study, we focused on three cavefish populations, which are named for the caves they inhabit (Tinaja, Pachón and Molino), that have evolved independently from two different stocks of surface fish that invaded caves millions of years ago6. Tinaja and Pachón cavefish originated from a more ancient surface population compared to Molino cavefish7.
A critical aspect of metabolic homeostasis is blood glucose regulation8. We compared blood glucose levels of Tinaja, Pachón and Molino cavefish with those of laboratory-raised surface fish, and found that the cave populations had significantly higher postprandial blood glucose levels (means of 64, 76 and 92 versus 47 mg dl−1, respectively; Fig. 1b). We investigated the dynamics of glucose homeostasis during short- and long-term fasting (Fig. 1c). Cavefish had significantly higher blood glucose levels after 24 hours of fasting (mean of 80, 80 and 107 mg dl−1 for Tinaja, Pachón and Molino populations, respectively, versus 59 mg dl−1 for surface fish, P < 0.05, one-way ANOVA). After 21 days, we observed a marked decrease in blood glucose levels in Tinaja and Pachón cavefish; at 1 and 21 days, respectively, Tinaja cavefish showed mean blood glucose levels of 80 versus 48 mg dl−1 (P < 0.0001) and Pachón cavefish showed mean levels of 80 versus 44 mg dl−1 (P < 0.0001). Surface fish experienced a minor decrease from 59 to 40 mg dl−1 (at 1 and 21 days, respectively, P = 0.02), and there was no significant change in Molino cavefish (107 versus 100 mg dl−1 at 1 and 21 days, respectively, P = 0.89). Molino cavefish maintain elevated blood glucose levels, highlighting what appears to be a fundamental difference between their metabolic adaptation and the adaptations of the other cavefish populations that we investigated. However, our results suggest that dysregulated glucose homeostasis is a common feature of cavefish populations.
To further test this hypothesis, we compared acute control of glucose homeostasis using a glucose tolerance test (Fig. 1d). We injected glucose into the intraperitoneal cavity of surface fish and cavefish, transiently raising blood glucose levels to over 400 mg dl−1 in most fish (Fig. 1d). Eight hours after injection, blood glucose levels of the surface fish were the same as those of PBS-injected controls (mean 126 versus 120 mg dl−1, respectively; Fig. 1d) whereas blood glucose levels remained highly elevated in all cavefish populations (mean 374, 432 and 411 mg dl−1 in Tinaja, Pachón and Molino populations, respectively, P < 0.0005, one-way ANOVA compared to PBS-injected controls; Fig. 1d). In Tinaja and Pachón cavefish, blood glucose levels remained elevated at 13.5 h after injection, but were not significantly different from those of PBS-injected controls (Tinaja cavefish, mean 157 versus 79 mg dl−1 in glucose-injected and PBS-injected fish, respectively, P = 0.47; in Pachón cavefish, mean 159 versus 105 mg dl−1 in glucose-injected and PBS-injected fish, respectively, P = 0.65). In Molino cavefish, blood glucose remained significantly elevated at 13.5 h (315 mg dl−1 versus 144 mg dl−1 in PBS-injected controls, P = 0.000001) and returned to the same levels as PBS-injected controls after 24 h (132 versus 133 mg dl−1 in glucose-injected versus PBS-injected controls, respectively). Our results suggest that cavefish have impaired glucose clearance.
Glucose homeostasis requires the balanced release of insulin and glucagon that instructs tissues to absorb glucose from the blood or produce glucose from stored glycogen9. We found that at ten days post fertilization the first pancreatic islet is visible and does not differ in the number of cells producing glucagon (54 versus 50 in surface fish and cavefish, respectively, P = 0.678) or insulin (54 versus 52 in surface fish and cavefish, respectively, P = 0.275), which suggests that the pancreas develops similarly in surface fish and cavefish (n = 5 per population; Extended Data Fig. 1). In 1–2-year-old adults, we did not detect a difference in circulating glucagon levels after a 24 h fast (Extended Data Fig. 2). Circulating insulin levels tended to be higher in Tinaja cavefish, but the results were not significant (n = 24 per population; Extended Data Fig. 3). Nonetheless, we found evidence of diminished insulin response in cavefish. We injected the fish with arginine, which stimulates the simultaneous release of glucagon and insulin10, and observed that while surface fish experienced a significant decrease in blood glucose level (mean 80 versus 38 mg dl−1, PBS-injected versus arginine-injected fish, P = 0.006), cavefish blood glucose levels did not change (Fig. 1e). In addition, we found that injection of recombinant human insulin caused a significant drop in blood glucose after 60 min in surface fish, but not in Tinaja cavefish (Extended Data Fig. 4). Our combined observations that glucagon and insulin levels do not differ between surface and cavefish, and that cavefish do not decrease blood glucose levels in response to arginine or insulin, suggest that cavefish may be insulin resistant.
Insulin-stimulated glucose uptake proceeds through phosphorylation of AKT at serine 473 (pAKT)11. We compared the ratio of pAKT to AKT in freshly dissected skeletal muscle treated with recombinant insulin (Fig. 1f, g). Consistent with the apparent dysregulation of glucose homeostasis, we observed lower pAKT levels in Tinaja cavefish than in surface fish (mean 0.775 versus 1.39 pAKT:AKT ratio, respectively, P = 0.017; Fig. 1f, g), which suggests that Tinaja cavefish are indeed insulin resistant relative to surface fish. Pachón cavefish muscle also had lower pAKT levels in response to insulin (mean 0.806 pAKT:AKT ratio, P = 0.027; Fig. 1f, g); however, despite having elevated blood glucose (Fig. 1b), Molino cavefish pAKT levels were equivalent to surface fish (mean 1.26 pAKT:AKT ratio, P = 0.99; Fig. 1f, g). Our results suggest that Tinaja and Pachón fish have evolved altered blood glucose regulation and insulin resistance in parallel, whereas Molino fish may have evolved their altered glucose metabolism through a different mechanism.
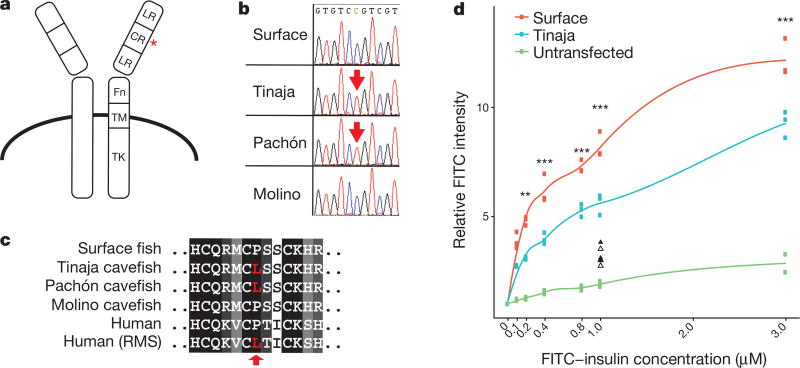
To investigate the genetic mechanism underlying insulin resistance in cavefish, we examined the sequences of all known genes in the insulin pathway using the available genome sequence12 (Supplementary Information 1). Notably, we found a coding difference in the insulin receptor gene (insra) between surface fish and cavefish that affects a conserved proline in the extracellular cysteine-rich domain (P211L; Fig. 2a–c). The presence of the mutation correlates with insulin resistance, as Tinaja and Pachón populations carry the mutation whereas Molino cavefish have the wild-type allele (Fig. 2b, c). Notably, the same genetic alteration is implicated in at least two known cases of Rabson–Mendenhall syndrome13,14, a form of severe insulin resistance in humans (Fig. 2c). The biochemical effect of the mutation has not previously been explored, but the position in the cysteine-rich domain suggests a role in insulin binding15. To test this, we generated transgenic HEK293T (Flp-In-293) cell lines that stably express the full-length surface fish or Tinaja cavefish insra and incubated the cells with different concentrations of fluorescein isothiocyanate (FITC)-labelled human insulin. We measured fluorescence as a readout for binding efficiency using an image-based cytometry approach (Imagestream X Mark II) and found that cells that expressed the cavefish receptor displayed significantly lower binding at all but the lowest concentrations of insulin (Fig. 2d). Our results suggest that the P211L mutation of insra affects insulin signalling by altering insulin binding efficiency.
Figure 2. Coding mutation in the cavefish insulin receptor leads to decreased insulin binding.
a, Schematic of the insulin receptor (adapted from ref. 24). Red asterisk depicts position of the P211L mutation. LR, leucine-rich repeats; CR, cysteine-rich domain; Fn, fibronectin type III domain; TM, transmembrane domain; TK, tyrosine-kinase domain. b, Sequence chromatogram of the mutation in Astyanax. c, Amino acid alignment of the insulin receptor P211L mutation with patients with Rabson–Mendelhall syndrome (‘Human (RMS)’). d, Relative FITC intensity of cells stably transfected with Flag-tagged surface-fish or Tinaja-cavefish insulin receptor and incubated with FITC-labelled insulin. Each point represents mean FITC intensity of >2,500 live cells normalized to the mean intensity of untreated cells. Lines represent results from local polynomial regression fitting. Triangles (surface fish, filled; Tinaja cavefish, unfilled) represent data from competitive binding assay in which cells were incubated with 10 µM unlabelled insulin. Significance calculated using one-way ANOVA (between surface fish and Tinaja cavefish) with Tukey’s HSD post hoc test, * P < 0.05; * * P < 0.005; * * * P < 0.0005.
We next tested for the presence and frequency of the P211L mutation in wild-caught fish. We genotyped 71 surface fish from different localities, and 51 cavefish from 6 different caves (Supplementary Information 2; Fig. 3a). Consistent with our observations above, the mutation was absent in Molino fish (n = 8) but present in all other tested cave populations (Tinaja, Yerbaniz, Pachón, Japonés and Arroyo, combined n = 36). Notably, the cave populations carrying this mutation are all derived from the same ancestral stock of surface fish16. Although the mutation was present in heterozygote conditions in some of the caves, we did not find any cavefish homozygous for the surface allele (Fig. 3a). Our findings suggest that there is active selection for the mutation in the caves, and a partially dominant effect of the cave allele. We did not observe the cave allele in any of the surface fish, which suggests that the mutation either appeared de novo in the cave populations, represents a rare variant not detected by our sampling frequency or is absent in the current surface population but was present in the ancestral surface fish stocks16 (Supplementary Information 3).
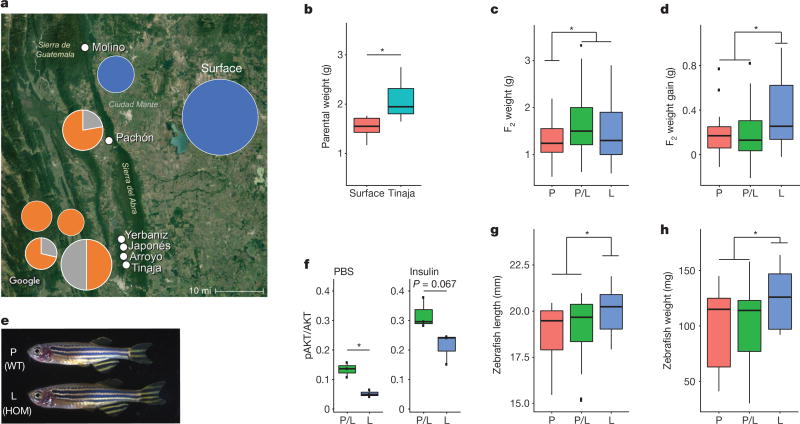
Figure 3. The P211L mutation of insra is overrepresented in cave environments and is associated with higher body weight in surface–cave hybrids.
a, Map of the region, overlain with genotyping results of wild-caught samples. Pie charts indicate percentage of fish homozygous for surface allele (blue), cave allele (orange) or heterozygous (grey). Size of pie chart roughly indicates the number of fish genotyped (Molino, n = 8; Surface, n = 71; Pachón, n = 9; Yerbaniz, n = 8; Japonés, n = 5; Arroyo, n = 7; Tinaja, n = 14; position of pie charts corresponds to location and vertical order of population name on the map). The P211L allele is absent in all wild-caught surface fish and Molino cavefish (descended from a more-recent surface-fish lineage). The P211L mutation is present in all sampled cavefish populations descended from the more-ancient surface-fish lineage. Tinaja, Yerbaniz, Japonés and Arroyo are geographically close and believed to represent a single invasion event; Pachón represents an independent invasion25. Map source: Imagery ©2017 Landsat/Copernicus, Map data ©2017 Google, INEGI. b, Weight of Tinaja males (n = 6) and surface males (n = 5) on a nutrient-limited diet. c, Weight of 18-month-old F2 male Tinaja–surface hybrids genotyped for the P211L mutation. P-homozygous (P) surface fish, n = 22; L-homozygous (L) cavefish, n = 27; heterozygotes (P/L), n = 53. d, Change in weight of F2 Tinaja–surface hybrid males on fixed diet. n = 21 (P), 39 (P/L) and 20 (L). e, Images of wild-type (WT) and homozygous P211L mutant (HOM) zebrafish siblings. f, Ratio of pAKT:AKT in adult zebrafish skeletal muscle treated ex vivo with PBS or insulin (n = 3 per genotype and condition). g, h, Length and weight of wild-type zebrafish (n = 13 (P)) and heterozygous (n = 22 (P/L)) and homozygous (n = 11 (L)) P211L mutant zebrafish. In box plots the median, 25th, 50th, and 75th percentiles are represented by horizontal bars and vertical bars represent 1.5× interquartile ranges. Significance calculated using two-tailed students t-test, * P < 0.05.
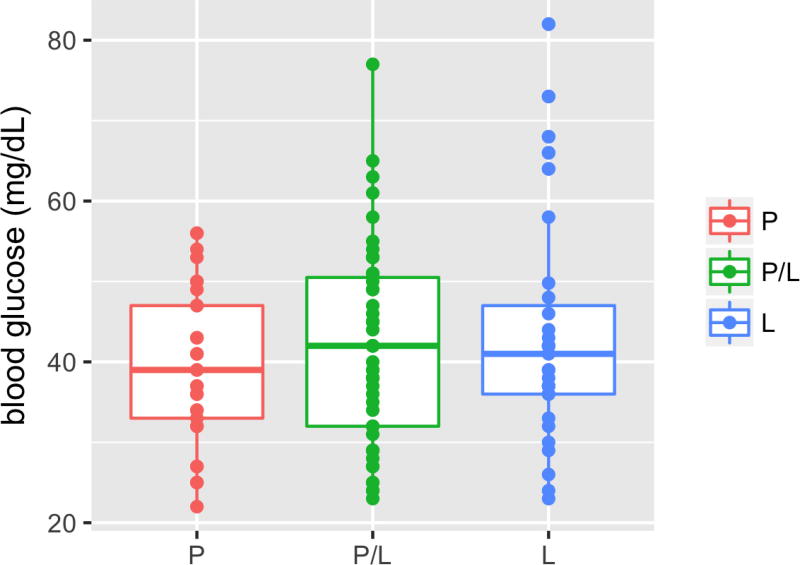
To investigate whether the P211L mutation contributes to altered glucose regulation, we measured fasting blood glucose levels in 192 F2 fish from a surface fish–Tinaja cavefish cross fed ad libitum their entire lives (Extended Data Fig. 5). We found that elevated blood glucose—defined as levels that exceed the surface fish mean (59 mg dl−1) or maximum (75 mg dl−1)—is a non-Mendelian trait in our cross: only 4.7%, rather than the expected 25%, of fish inherited elevated blood glucose, which indicates that the trait is not monogenic. Although F2 fish with the P211L mutation did not differ significantly in blood glucose levels from those without the mutation, we found that only F2 fish carrying the P211L mutation have elevated blood glucose levels exceeding the surface fish mean (n = 9) or maximum (n = 3), which suggests a necessary but not sufficient role for the mutation in altered blood glucose regulation.
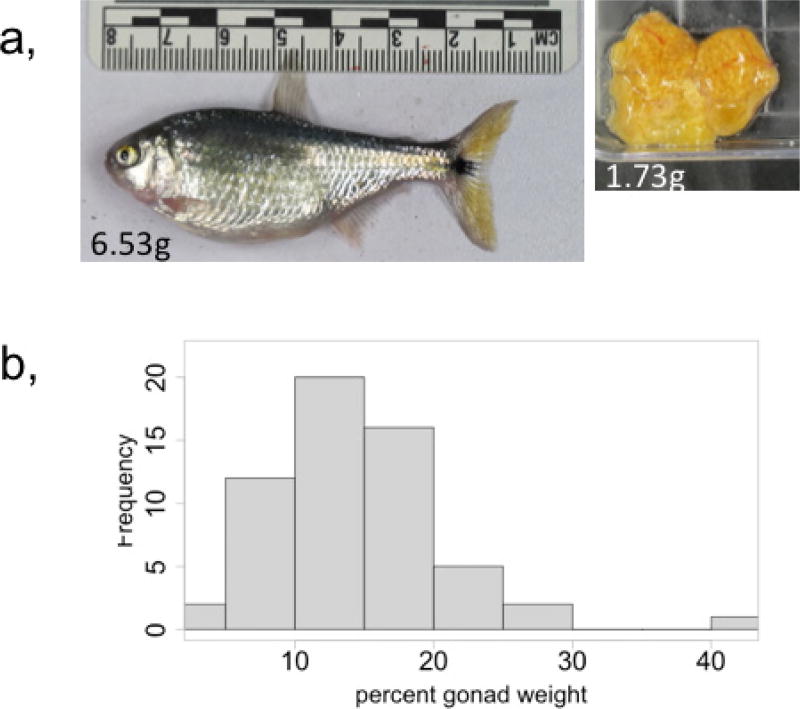
We found that cavefish weigh more than surface fish on a nutrient- limited diet (mean 2.08 versus 1.52 g, respectively, P = 0.02; Fig. 3b). To investigate whether the insra mutation influences weight, we genotyped and weighed 124 surface–Tinaja male F2 fish at approximately 1.5 years of age that were fed ad libitum. We focused on males, as egg mass varies between individual females and can account for as much as 41% of female body weight (Extended Data Fig. 6), representing a confounding variable if included in our analysis. We found that males carrying one or two copies of the cave P211L insra allele weigh on average 27% more than hybrids that carry only the surface allele (mean 1.63 versus 1.28 g, respectively, P = 0.006; Fig. 3c). Cavefish have an increased appetite that is associated with a mutation in the melanocortin 4 receptor3. Although this mutation segregates independently from the insra mutation in the F2 hybrid population, it is possible that another locus in cis to the P211L mutation influences appetite. To eliminate the effect of appetite on our analysis of the insra allele, we housed F2 fish of approximately the same starting weight (< 2 g) individually and ensured they ate a diet of 6 mg of food per day for 4 months. We found that homozygous P211L fish gained significantly more weight (mean 0.37 g, n = 20) than fish that did not carry the cave allele (0.19 g, n = 60, P = 0.02) consistent with the idea that variation at the insra locus influences weight gain independent of appetite regulation.
To determine whether the insulin resistance and weight gain associated with the P211L mutation in cavefish are indeed due to alteration of the insra gene, we used CRISPR gene editing to introduce the mutation into zebrafish (Danio rerio) via homology directed repair17 (Extended Data Fig. 7). We found that zebrafish homozygous for the P211L mutation have a lower ratio of pAKT to AKT in their skeletal muscle compared to heterozygous siblings, in both untreated (0.05 versus 0.13 in P211L-homozygous and heterozygous fish, respectively, P = 0.016) and insulin-stimulated conditions (0.13 versus 0.32 in P211L-homozygous and heterozygous fish, respectively, P = 0.067, n = 3 per genotype and condition) (Fig. 3f). In addition, we found that zebrafish homozygous for the cave allele are longer (mean 20.0 mm, versus 18.4 mm in heterozygous fish, P = 0.0046) and weigh more (mean 124.6 mg, versus 99.7 mg in heterozygous fish, P = 0.022) than their siblings when raised under the same conditions (Fig. 3e, g, h). Our findings show that the P211L mutation contributes to both the increased weight and insulin resistance observed in Tinaja and Pachón cavefish. Increased body weight in fish homozygous for the insulin-receptor mutation is unexpected, considering that in mammals, full loss-of-function mutations in the insulin receptor are associated with retarded growth and lower levels of body fat18. Our results suggest that diminished insulin signalling has an opposite effect in fish, but the mechanisms leading to the difference remain unclear.
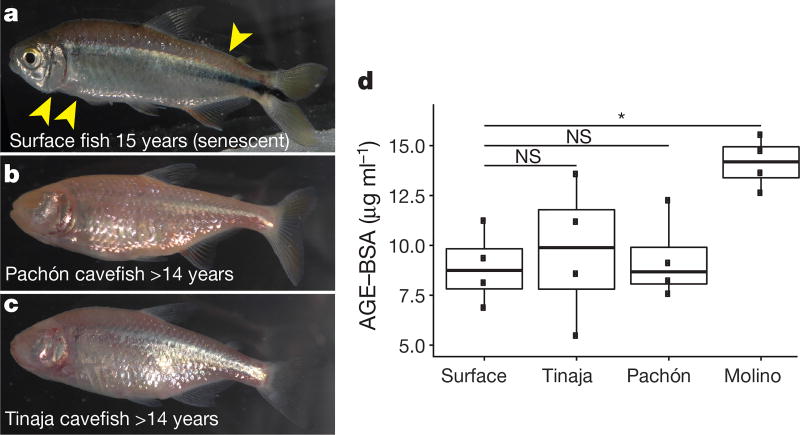
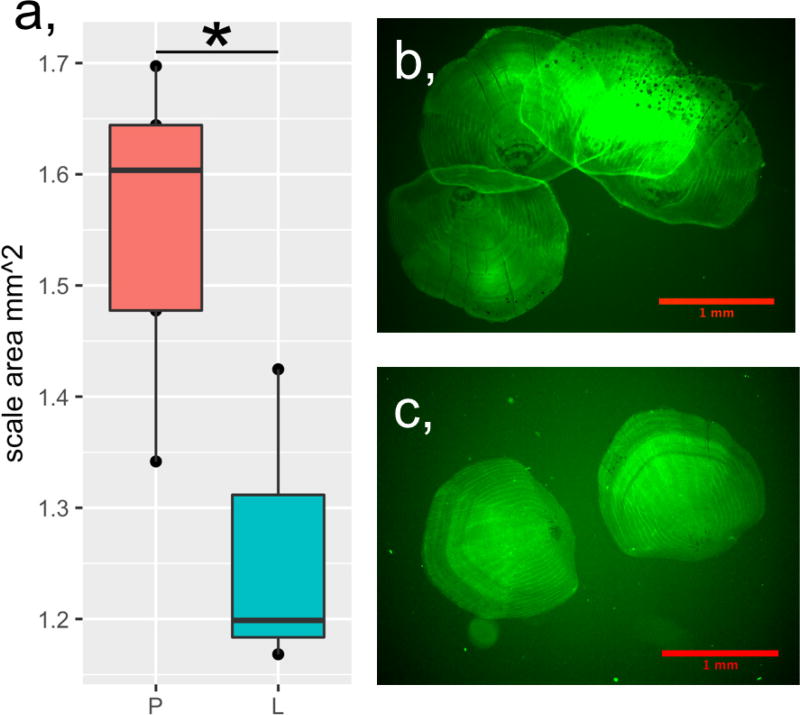
Cavefish are insulin resistant and hyperglycaemic; in humans these phenotypes precede and define type 2 diabetes, respectively. In addition, cavefish have a fatty liver3, which is also frequently associated with type 2 diabetes. Transgenic zebrafish carrying the insra mutation show a reduction in scale size (Extended Data Fig. 8), as previously described in other hyperglycaemic zebrafish models19 and a similar phenotype has recently been described in cavefish20. In principle, these data could suggest an evolutionary tradeoff in which physiological health is sacrificed to reap the benefits of starvation resistance. However, whereas surface fish begin to exhibit signs of ageing such as sunken skin, tattered fins and a hunched back21 (Fig. 4a) by age 15, Tinaja and Pachón cavefish can live in excess of 14 years without these indications of senescence (Fig. 4b, c) and with no difference in fertility decline relative to surface fish. Cavefish may have evolved compensatory mechanisms enabling them to remain healthy despite potentially deleterious metabolic changes.
Figure 4. Despite elevated blood glucose levels and insulin resistance, Tinaja and Pachón cavefish do not show signs of senescence and do not accumulate advanced glycation end-products in the blood.
a–c, Surface (a), Pachón (b) and Tinaja (c) fish kept in the laboratory for the indicated duration and fed ad libitum. Cavefish were wild-caught; ages represent minimum age. Surface fish (a) shows signs of ageing, such as loose skin and bent tails (yellow arrows), that are absent in cavefish at comparable ages (b, c). d, Quantification of advanced glycation end-products in serum (AGE–BSA) from approximately two-year-old fish after a three-day fast (n = 4 for each population). * P < 0.05, one-way ANOVA with Tukey’s HSD post hoc test. For box plots, median, 25th, 50th and 75th percentiles are represented by horizontal bars and vertical bars represent 1.5× interquartile ranges
A major cause of morbidity in diabetic patients is tissue damage caused by excessive non-enzymatic glycation of proteins in the blood, generating advanced glycation end-products (AGEs)22. AGEs are closely associated with diabetes-induced vascular damage, cardiovascular disease and ageing23. We compared the level of AGEs in the serum of two-year-old fish that had been fed ad libitum their entire lives (Fig. 4b). We did not detect a difference in the levels of AGEs between Tinaja and Pachón cavefish relative to surface fish (mean of 9.7, 9.3 and 8.9 µg ml−1, respectively, P = 0.99, 0.95), despite the elevated blood glucose levels of the cavefish (Fig. 1b). These two populations may have mechanisms for reducing protein glycation, rendering them impervious to the damaging effects of elevated blood glucose. Notably, the Molino cavefish do have elevated levels of AGEs (mean 14.1 µg ml−1, versus 8.9 µg ml−1 in surface fish, P = 0.03). It remains to be determined whether the health and longevity of the Molino population is influenced by accumulation of AGEs, but our results suggest they may have evolved altered blood glucose homeostasis through a different mechanism than did Tinaja and Pachón cavefish.
Our findings establish cavefish as a model with which to investigate resistance to pathologies of diabetes-like dysregulation of glucose homeostasis. Moreover, our results highlight the extreme physiological measures that can evolve in critical metabolic pathways to accommodate exceptional environmental challenges.
Online Content Methods, along with any additional Extended Data display items and Source Data, are available in the online version of the paper; references unique to these sections appear only in the online paper.
Methods
No statistical methods were used to predetermine sample size. The experiments were not randomized. Investigators were blinded to fish genotypes during experiments when possible.
Fish husbandry and diet
Fish husbandry was performed as previously described26. Unless stated otherwise, fish were fed ad libitum with a combination of New Life Spectrum TheraA + small fish formula and Artemia and housed at densities of less than or equal to two adult fish per litre of water. F2 hybrids were housed individually in 1.5 l tanks and fed three pellets (~6 mg) of New Life Spectrum TheraA + small fish formula once per day for over 4 months. For the starvation experiment, fish were moved to individual containers and water was changed daily.
Blood glucose, glucose tolerance and arginine tolerance
Blood was collected from the caudal tail vein using a U-100 insulin needle and glucose was measured using FreeStyle Lite blood glucose meter and test strips.
Glucose (2.5 mg/gram fish), arginine (6.6 µM/gram fish), or PBS was injected into the intraperitoneal cavity using a U-100 insulin needle.
pAKT quantification
We quantified pAKT level in fillets of skeletal muscle taken directly after fish decapitation. For A. mexicanus, skeletal muscle was cut into three equal strips per fish. Strips were incubated in PBS, 0.1× or 1× concentration of recombinant human insulin for 25 min (Sigma product I9278; 1× concentration = 9.5–11.5 µg/ml insulin). The tissues were rinsed in PBS and then homogenized and lysed in RIPA buffer (Sigma) with protease and phosphatase inhibitor (Pierce Protease and Phosphatase Inhibitor Mini Tablets, EDTA Free) for 30 min. Protein concentration was measured via BCA (Pierce). Lysate protein concentrations were then equalized and run on 4–12% Bis-Tris protein gel and transferred on a nitrocellulose membrane. Blots were probed for AKT (Cell Signaling). Following stripping, blots were probed for pAKT (ser473) (Cell Signaling). ImageJ was used for densitometry measurements. For D. rerio two fillets of skeletal muscle were removed from both sides of fish directly after decapitation. Fillets were rinsed in PBS and then incubated in PBS or 10 µg/ml human recombinant insulin (Sigma, I0908) for approximately 40 min. The skin was then removed from the skeletal muscle and the muscle was finely minced using a scalpel. We quantified the ratio of pAKT:AKT using the Akt(pS473) + total Akt ELISA Kit (Abcam ab126433) according to the manufacturers’ protocol. We used 200 µl lysis buffer per sample and loaded 85 µl of lysate per well.
insra P211L genotyping in A. mexicanus
Genomic DNA from tail fin clips was diluted fivefold and used as target DNA to amplify the insra locus using the following oligonucleotide primers: insra_f: GCACCCTTACACCCTTACATGA; insra_r: TACCGCTCAGCACTAATTTGGA; product size: 700 bp. PCR reactions were carried out in 12.5-µl volume containing 1× LA PCR Buffer II (Clontech), 2.5 mM MgCl2, 0.4 mM dNTP mix, 0.4 µM of each forward and reverse primer and 0.05 units of TaKaRa LA Taq DNA Polymerase (Clontech). The PCR cycling conditions were as follows: initial denaturation at 94 °C for 2 min, followed by 35 cycles of 94 °C for 30 s, annealing temperature 52 °C for 30 s and 72 °C for 1 min. A final 5-min elongation step was performed at 72 °C. The PCR products were diluted tenfold and sequenced directly on a 3730XL DNA Analyzer (Applied Biosystems) using the sequencing primer: GGTGGAGTTGATGGTGGTATAG.
Selection scans at the insra locus
We examined the insra locus with data that are a part of an ongoing genome-wide selection and demography companion study (A. Herman et al., manuscript in preparation). Methods are explained in greater detail in this demographic companion study; in brief, we used Illumina Hiseq 2000 to sequence 100-bp reads from 6–10 individuals from each population of Tinaja cave, Molino cave, Pachón cave, Rascon surface and Río Choy surface populations, (total n = 43) and two individuals from the sister taxa Astyanax aeneus. Individuals were sequenced with v3 chemistry. Reads were cleaned with Trimmomatic v.0.3027 and cut-adapt v.1.2.128 and aligned to the reference Pachón cavefish genome using bwa-mem algorithm in bwa-0.7.129 resulting in an aligned coverage depth of ~7–12×. Variants were called using the Genome Analysis Toolkit v.3.3.0 (GATK)30 and Picard v.1.83 (http://broadinstitute.github.io/picard/). Outlier scan metrics (π, DXY, FST and Tajima’s D) were conducted using VCFtools v.0.1.1331 and custom scripts. HSCAN (https://messerlab.org/resources/) and hapFLK32 were also used to examine insra for outliers. Metrics were dense-ranked across the genome and the ranking position of insra was used to determine whether it was exceptionally divergent between cave and surface populations relative to the rest of the genome.
Glucagon and insulin quantification
The number of cells producing insulin or glucagon was determined using the following protocol: 10–11 days post fertilization fish were euthanized with an overdose of tricaine and fixed in 4% paraformaldehyde overnight at 4 °C. Fish were washed in PBST, transferred to water for 1 min, acetone for 10 min at −20 °C, water for 1 min, and then washed in PBST. Blocking in 5% donkey serum, 1% DMSO and 0.2% BSA was done for 1 h at room temperature. Fish were incubated with primary antibodies (1:200 sheep anti-glucagon (Abcam), 1:200 guinea pig anti-insulin (DAKO)) and then secondary antibodies (1:400 donkey anti-sheep 488, 1:400 goat anti-guinea pig 647) overnight at room temperature in glass vials, then washed with PBST, stained with DAPI and imaged. Images were collected at 63× using a 1.0 µm z-stack on a Zeiss 780 confocal microscope. Nuclei surrounded by insulin or only glucagon were counted manually using Fiji cell counter.
To quantify circulating glucagon level, we collected serum from the caudal tail vein of fish that were approximately 2-years-old and had fasted for 24 h (n = 12 for each population). The serum was used for a glucagon radioimmunoassay according to the manufacturer’s protocol (MGL-32K; Millipore). To quantify circulating insulin levels, we collected plasma from the caudal tail vein of 2-year-old and 1-year-old fish and blotted the serum onto a nitrocellulose membrane using a Bio-dot sf device (Bio-Rad). Ponceau protein staining was used to verify equal protein loading, after which insulin was probed using anti-insulin (DAKO). Quantification of insulin levels was done using Fiji.
Insulin-binding experiment
The full-length A. mexicanus insra protein-coding sequence was amplified from surface-fish and Tinaja-cavefish cDNA and cloned into a modified pcDNA3.1/Hygro vector providing an N-terminal Flag epitope tag33. To generate stable cell lines, the Flag-tagged insra cassettes were cloned into pcDNA5/FRT vector (Invitrogen, catalogue number V601020) allowing for Flp recombinase-mediated integration into the Flp-In-293 cell line according to the manufacturer’s procedures. The FLP-In-293 cell line was originally purchased from Invitrogen/Thermo Fisher Scientific (catalogue number R75007), then cell-banked internally at the Stowers Institute. Cell line authentication was performed by Promega/ATCC, using short tandem repeat (STR) profiling. Cell lines were tested for mycoplasma contamination using the Universal Mycoplasma Detection Kit, ATCC 30-1012. One positive clone from each surface-fish and Tinaja-cavefish cell line was selected and used for the insulin-binding assays using a protocol derived from a previously published protocol34. In brief, 100-mm plates were seeded at 30% confluency and cultured in DMEM, 10% FBS + 1× Glx medium for 48 h. The plates were then changed to insulin-free FreeStyle 293 Expression Medium (catalogue number 12338018) and incubated for an additional 24 h. Plates at ~70–80% confluency were pre-chilled for 30 min at 4 °C and the medium was replaced with 5 ml of cold FreeStyle 293 Expression Medium containing 42 mM HEPES pH7.5 and human FITC-labelled insulin (Sigma, catalogue number I3661) at final concentrations of 0, 0.1, 0.2, 0.4, 0.8, 1 and 3 µM. For binding competition, 10 µM of unlabelled human insulin (Sigma, catalogue number I9278) and 1 µM of human FITC-labelled insulin were added. After one hour of incubation in the dark at 4 °C, the medium was removed and the plates were washed with 5 ml of cold 1× PBS. Cells were dissociated in 2 ml of 0.5 mM EDTA in PBS at 37 °C for 7 min, transferred into Eppendorf tubes, pelleted for 5 min at 200g at 4 °C and resuspended in 1 ml of cold 1× PBS. To stain dead cells, 1 µl of Fixable Viability Dye (FVD eFluor450, Invitrogen, catalogue number 65-0863) was added to each 1 ml of cell suspension and incubated on ice in the dark for 20 min. Subsequently, the cells were washed once with cold 1× PBS and fixed in 1 ml of 4% formaldehyde. After two more washes with cold 1× PBS, the cells were resuspended in 150 µl of cold 1× PBS, filtered through a 70-µm cell strainer (Filcons 070-67-S) and transferred into a round-bottom 96-well plate. Binding data were acquired on an ImagestreamX MarkII (EMD Millipore) at 40×. FITC was excited with 150 mW 488 nm. Fixable live/dead was excited with 12 mW 405 nm. Single colour controls were used for colour compensation. Bright field was collected on different channels. Analysis was performed in IDEAS v.6.2 and fluorescence intensity was reported as integrated intensity within an adaptive erode mask for bright field.
Genome editing in zebrafish using the CRISPR–Cas9 technology
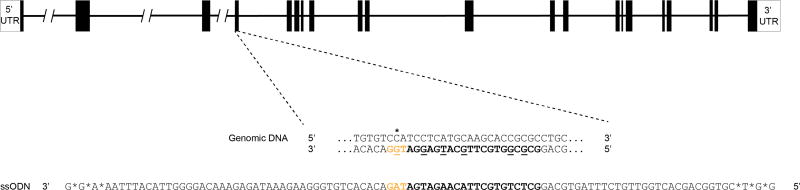
We introduced the cavefish P211L coding alteration into D. rerio by causing a precise C632T base pair exchange on the third exon of the insra gene (Extended Data Fig. 7). We designed the guide RNA target site using the web tool of the MIT Zhang laboratory (http://crispr.mit.edu). We then validated the target region and checked for naturally occurring SNPs by PCR and sequencing of genomic DNA using the following oligonucleotide forward, reverse and sequencing primers, respectively: (1) TGAGGTGTGTCGAGTGTTCT; (2) TGTGCGTGTTCGTTGAGTGT, product size of 598 bp; and (3) CAGCCCTTGAAGGTGTAGAA. The PCR reaction components and procedures followed that described in ‘insra P211L genotyping in A. mexicanus’.
We used Cas9 protein from PNABio and 2-part Alt-R guide RNAs from IDT. Single-stranded oligodeoxynucleotides (ssODNs) were ordered as ultramers from IDT for generating the SNP mutations. The ssODNs included 100 bp of homology arms as well as silent SNP mutations in the guide RNA target site to prevent re-annealing of the guide RNA following homologous recombination. We also protected both ends of the ssODNs with three phosphorothioate bonds to inhibit exonuclease degradation in the cell. To form the guide RNA complex, we first annealed the specific CRISPR RNA (crRNA) with trans-activating crRNA (1 µM), followed by hybridization with the Cas9 protein (6.6 µM) to form a ribonucleoprotein complex. This mixture was incubated for 10 min. Next, we combined the ribonucleoprotein complex with the ssODNs (50 ng/µl) to form a total volume of 15 µl of injection mix, which was then injected into the cell at the 1-cell stage in at least 200 and up to 1,200 zebrafish embryos.
insra P211L genotyping in zebrafish
We designed a genotypic screening assay to confirm the expected location of the P211L SNP mutation at various points through development. First, after injections we divided the embryos into two groups; group 1 to screen at the embryo stage and group 2 to raise if the mutation was detected in group 1. Out of the 93 embryos screened, 34 fish exhibited a mixture of insertion and deletion mutations (35.5%), and at least 1 fish showed the exact SNP mutation (1.1%). Once these fish matured, we then identified adult mosaic individuals. Out of the 64 screened F0 adult fish, 17 fish carried the intended P211L SNP mutation with the corresponding silent SNP changes (26.6%), but only 5 fish showed germline transmission (7.81%) when outcrossed to wild-type zebrafish. Next, we genotyped the F1 embryos to calculate the germline transmission rate of each F0 mosaic founder. The germline transmission rate ranged from 8.3% to 45.8% in the 5 F0 fish that showed positive germline transmission. We then raised the progenies of these mosaic founders. Lastly, we genotyped these F1 progenies upon maturity and maintained the line by outcrossing heterozygous mutants to wild-type fish in each subsequent generation. To test whether the P211L mutation contributes to zebrafish weight, we paired heterozygous fish with successful germline transmission and weighed their progeny shortly before maturity (55 days post fertilization) to avoid gonadal effects on weight.
Quantification of advanced glycation end-products
We used Oxiselect Advanced Glycation End Product (AGE) Competitive ELISA Kit according to the manufacturer’s protocol to measure AGE level in serum from two-year-old fish that were fasted for three days.
Statistics and figure preparation
Graphical data and statistics were produced using R35 and ggplot236 package. We used Shapiro–Wilk to test for normality. We tested significance in normally distributed data using a t-test and non-parametric data using Mann–Whitney U-test. For non-parametric data with multiple ties we used a t-test. For comparison of more than two groups we used a one-way ANOVA with Tukey’s HSD post hoc test.
Animal experiment statement
Research and animal care were approved by the Institutional Animal Care and Use Committees of the laboratories involved.
Data availability
Original data underlying this manuscript can be accessed from the Stowers Original Data Repository at http://www.stowers.org/research/publications/libpb-1205 and/or are available from the corresponding authors on reasonable request.
Extended Data
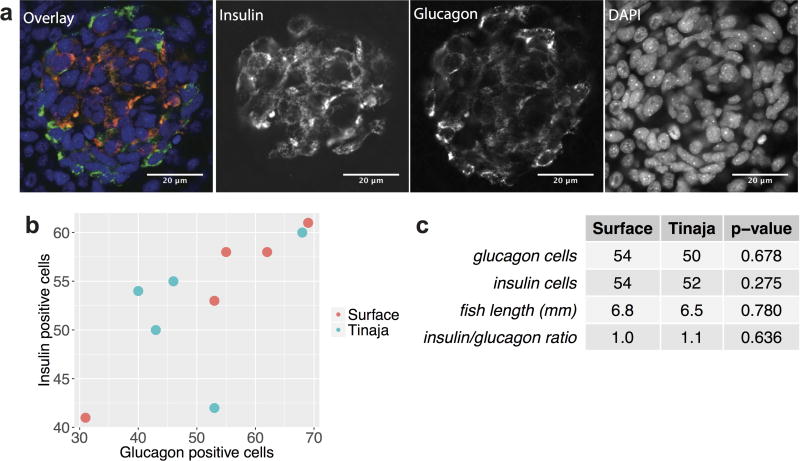
Extended Data Figure 1. Numbers of insulin- and glucagon-positive cells in the developing pancreas are unchanged in Tinaja cavefish relative to surface fish.
a, Whole-mount immunohistochemical detection of insulin- and glucagon-positive cells in Tinaja larvae at 10 days post fertilization. b, Number of glucagon- and insulin-positive cells in surface and Tinaja larvae at 10–11 days post fertilization (n = 5 fish per population,). c, Average number of glucagon- and insulin-positive cells, fish length, ratio of insulin to glucagon positive cells and P value comparing the surface and Tinaja values (determined using Student’s t-test)
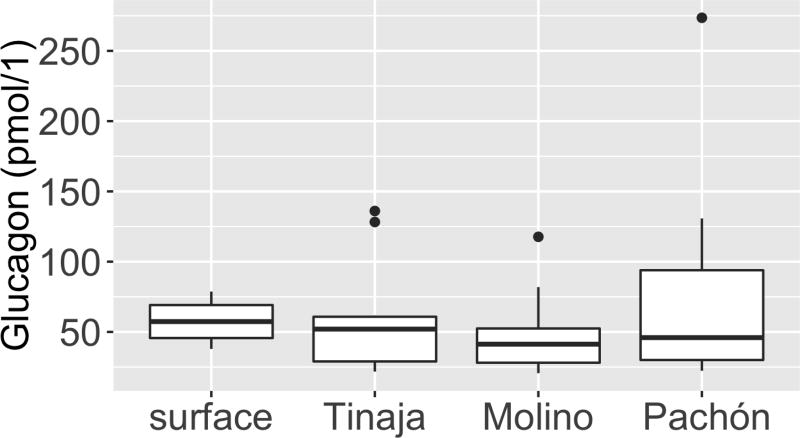
Extended Data Figure 2. Serum glucagon levels are comparable between the different populations.
Box plot comparing serum glucagon levels between surface, Tinaja, Molino and Pachón fish after 24-h fast. n = 12 fish per population, average of 57.87, 59.76, 79.66 and 48.89 respectively. P = 0.52, one-way ANOVA. Box plots show 25th, 50th and 75th percentiles (horizontal bars), and 1.5× interquartile ranges (error bars), dots represent outliers
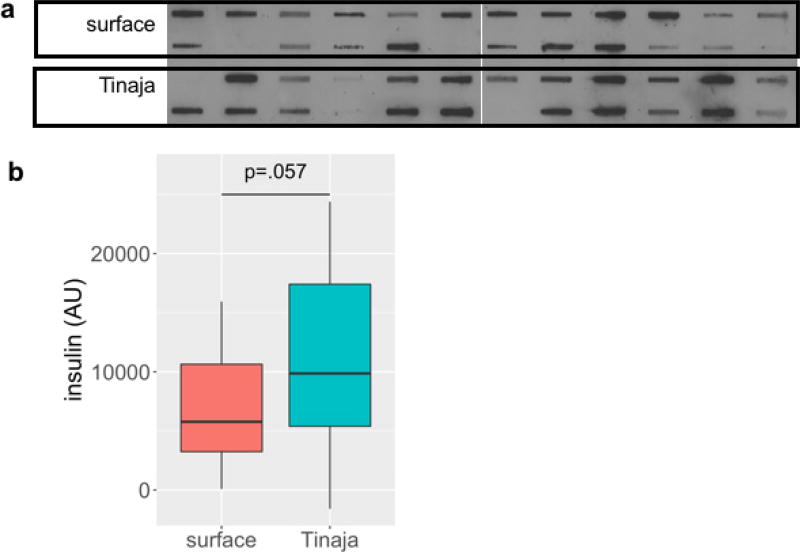
Extended Data Figure 3. Serum insulin levels are comparable between surface and Tinaja fish.
a, Serum blotted onto nitrocellulose membrane using Bio-Dot SF microfiltration apparatus (Bio-Rad, catalogue number 1706542) probed with anti-insulin antibody (DAKO). Each blot represents an individual fish between 1- and 2-years-old (n = 24 fish per population). b, Quantification of insulin level measured by densitometry of blots.
AU, artificial units; median, 25th, 50th and 75th percentiles (horizontal bars) and error bars at 1.5× interquartile ranges. Tinaja cavefish insulin levels (mean = 10,770) tended to be higher than those of surface fish (mean = 7,194) but the results are not significant (P = 0.057, Student’s two-sample t-test)
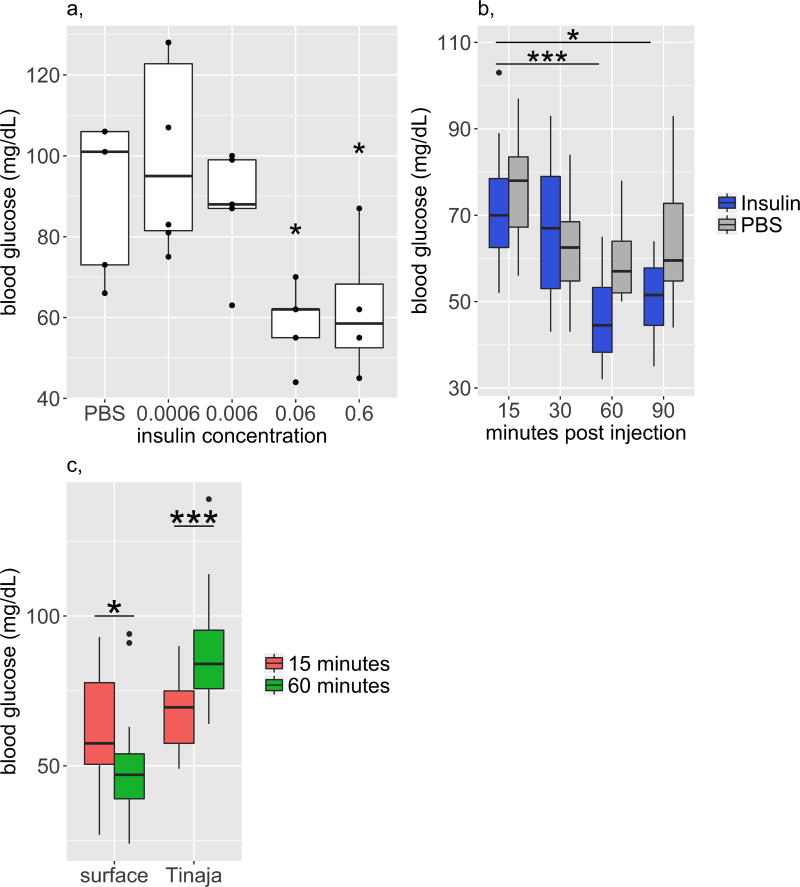
Extended Data Figure 4. Insulin decreases blood glucose level in surface fish.
a, We injected different concentrations of human recombinant insulin (Sigma, product I9278, stock 9.5–11.5 mg ml−1) into the intraperitoneal cavity of surface fish to determine the effective dosage for subsequent experiments. Blood glucose levels are significantly lower after injection of approximately 0.6 or 0.06 g insulin per mg of fish weight compared to 0.0006 g (30 min after injection, n = 4 fish per dosage, dots represent individual fish, significance calculated using one-way ANOVA with Tukey’s HSD post hoc test, * P < 0.05). We used 0.06 g insulin per mg of fish in subsequent experiments. b, Blood glucose levels of surface fish over time, after injection of PBS or insulin. Blood glucose levels are significantly lower at 60 and 90 min compared to 15 min after insulin injection (n = 10 fish per time point and condition, significance calculated using one-way ANOVA with Tukey’s HSD post hoc test, * P < 0.05). Therefore, we focused on the 60-min time point for comparisons with Tinaja cavefish. c, Blood glucose levels at 15 and 60 min after insulin injection in surface fish and Tinaja cavefish. Surface fish display a significant decrease in blood glucose levels, whereas cavefish display a significant increase in blood glucose levels (significance calculated using two-tailed Student’s t-test, * P < 0.05, * * * P < 0.0005). Tinaja cavefish blood glucose levels may increase owing to the stress of being injected; stress hormones, such as catecholamines, ACTH and epinephrine, cause transient increases in blood glucose in humans and mice, an effect that cannot be mitigated in the absence of insulin signalling. Although both cavefish and surface fish probably undergo a stress response upon injection, this is overcome in the surface fish, which have wild-type insulin activity, but not in the Tinaja cavefish, which have reduced insulin signalling
Extended Data Figure 5. An elevated fasting blood glucose level correlates with the presence of the P211L allele in F2 hybrids.
Blood glucose levels of 192 F2 surface–Tinaja hybrids of the indicated genotype 24 h after feeding. All of the fish with elevated blood glucose (greater than 60 mg dl−1) carry the P211L allele. Median, 25th, 50th and 75th percentiles (horizontal bars), and error bars at 1.5× interquartile ranges
Extended Data Figure 6. Egg mass is a confounding variable in female fish.
a, Image of an F2 surface–Tinaja hybrid female and removed gonad, of indicated weights. b, Histogram displaying per cent gonad weight (gonad weight/total weight, multiplied by 100) of 62 F2 surface–Tinaja hybrid females fed 6 mg per day for 4 months (minimum = 3.57, 1st quartile = 10.64, median = 13.96, mean = 13.96, 3rd quartile = 17.57 and maximum = 41.86)
Extended Data Figure 7. Genome editing strategy.
CRISPR–Cas9 mediated genome editing strategy in exon 3 of the insulin receptor a (insra) zebrafish gene. The guide RNA target sequence is emphasized in bold in both the reverse strand of the wild-type genomic DNA and in the ssODN. The intended SNP exchanges are underlined, and the specific C632T to alter the P211 to L is denoted with a star. Both ends of the ssODN are protected by three phosphorothioate bonds, denoted with asterisks. The protospacer adjacent motif is shown in orange
Extended Data Figure 8. Scale growth is impaired in the insra zebrafish mutant.
a, Quantification of scale size in zebrafish of the indicated genotype; each point represents the mean scale size of an individual fish based on the measurement of 10–14 scales removed from the left side of the body from the posterior edge of the dorsal fin to the posterior edge of the ventral fin by gentle scraping with a scalpel. Wild type (P), n = 4, P211L mutant (L), n = 3. b, c, Representative images of scales stained with 0.005% calcein with contrast and brightness adjusted to show scale edges. Significance calculated using two-way t-test of mean values, * P < 0.05.
Supplementary Material
Acknowledgments
We thank Y. Chinchore and C. Sengel for technical advice; X. Gao for bioinformatics support; Z. Zakibe for photographs of the fish; the Aquatics facility at Stowers for fish maintenance and support; the cell culture core at Stowers for cell line maintenance and advice; the molecular biology core at Stowers for design, execution and validation of the CRISPR constructs; the proteomics core; M. Levy for advice and computational modelling of the insulin receptor; A. Herman for help with the genome scan; the Microscopy Resources on the North Quad (MicRoN) core at Harvard Medical School; M. Miller for illustration; and S. Williams, F. Damen, S. Xiong, E. Kingsley and K. Fox for feedback on the manuscript text. This work was supported by a grant from the NIH to C.J.T. (HD089934) and institutional funding to N.R. M.R.R. was supported by a National Research Service Award (DK108495) and R.P. was supported by a grant from the Deutsche Forschungsgemeinschaft (PE 2807/1-1).
Footnotes
Supplementary Information is available in the online version of the paper.
Author Contributions M.R.R., A.C.A., C.J.T. and N.R. conceived the project and designed research with additional contributions from K.G., R.P. and A.C.B. M.R.R., A.C.A., K.G., R.P., J.Y.S., B.M., M.P., A.C.B., J.A.T., S.M., R.B. and N.R. performed the research. M.R.R., A.C.A., C.J.T. and N.R. wrote the paper.
The authors declare no competing interests.
References
- 1.Culver DC, Pipan T. The Biology of Caves and Other Subterranean Habitats. Oxford Univ. Press; 2009. [Google Scholar]
- 2.Horst Wilkens US. Evolution in the Dark, Darwin’s Loss Without Selection. Springer; 2017. [Google Scholar]
- 3.Aspiras AC, Rohner N, Martineau B, Borowsky RL, Tabin CJ. Melanocortin 4 receptor mutations contribute to the adaptation of cavefish to nutrient-poor conditions. Proc. Natl Acad. Sci. USA. 2015;112:9668–9673. doi: 10.1073/pnas.1510802112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moran D, Softley R, Warrant EJ. Eyeless Mexican cavefish save energy by eliminating the circadian rhythm in metabolism. PLoS ONE. 2014;9:e107877. doi: 10.1371/journal.pone.0107877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hüppop K. Oxygen consumption of Astyanax fasciatus (Characidae, Pisces): a comparison of epigean and hypogean populations. Environ. Biol. Fishes. 1986;17:299–308. [Google Scholar]
- 6.Gross JB. The complex origin of Astyanax cavefish. BMC Evol. Biol. 2012;12:105. doi: 10.1186/1471-2148-12-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bradic M, Teotónio H, Borowsky RL. The population genomics of repeated evolution in the blind cavefish Astyanax mexicanus. Mol. Biol. Evol. 2013;30:2383–2400. doi: 10.1093/molbev/mst136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saltiel AR, Kahn CR. Insulin signalling and the regulation of glucose and lipid metabolism. Nature. 2001;414:799–806. doi: 10.1038/414799a. [DOI] [PubMed] [Google Scholar]
- 9.Rines AK, Sharabi K, Tavares CD, Puigserver P. Targeting hepatic glucose metabolism in the treatment of type 2 diabetes. Nat. Rev. Drug Discov. 2016;15:786–804. doi: 10.1038/nrd.2016.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Navarro I, et al. Insights into insulin and glucagon responses in fish. Fish Physiol. Biochem. 2002;27:205–216. [Google Scholar]
- 11.Lizcano JM, Alessi DR. The insulin signalling pathway. Curr. Biol. 2002;12:R236–R238. doi: 10.1016/s0960-9822(02)00777-7. [DOI] [PubMed] [Google Scholar]
- 12.McGaugh SE, et al. The cavefish genome reveals candidate genes for eye loss. Nat. Commun. 2014;5:5307. doi: 10.1038/ncomms6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Atray A, et al. Rabson Mendenhall Syndrome; a case report. J. Diabetol. 2013;2:2. [Google Scholar]
- 14.Carrera P, et al. Substitution of Leu for Pro-193 in the insulin receptor in a patient with a genetic form of severe insulin resistance. Hum. Mol. Genet. 1993;2:1437–1441. doi: 10.1093/hmg/2.9.1437. [DOI] [PubMed] [Google Scholar]
- 15.Taylor SI, et al. Mutations in insulin-receptor gene in insulin-resistant patients. Diabetes Care. 1990;13:257–279. doi: 10.2337/diacare.13.3.257. [DOI] [PubMed] [Google Scholar]
- 16.Bradic M, Beerli P, García-de León FJ, Esquivel-Bobadilla S, Borowsky RL. Gene flow and population structure in the Mexican blind cavefish complex (Astyanax mexicanus) BMC Evol. Biol. 2012;12:9. doi: 10.1186/1471-2148-12-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Albadri S, Del Bene F, Revenu C. Genome editing using CRISPR/Cas9-based knock-in approaches in zebrafish. Methods. 2017;121–122:77–85. doi: 10.1016/j.ymeth.2017.03.005. [DOI] [PubMed] [Google Scholar]
- 18.Savage DB, Semple RK. Recent insights into fatty liver, metabolic dyslipidaemia and their links to insulin resistance. Curr. Opin. Lipidol. 2010;21:329–336. doi: 10.1097/MOL.0b013e32833b7782. [DOI] [PubMed] [Google Scholar]
- 19.Suzuki N, Kitamura KI, Hattori A. Fish scale is a suitable model for analyzing determinants of skeletal fragility in type 2 diabetes. Endocrine. 2016;54:575–577. doi: 10.1007/s12020-016-1153-9. [DOI] [PubMed] [Google Scholar]
- 20.Simon V, et al. Comparing growth in surface and cave morphs of the species Astyanax mexicanus: insights from scales. Evodevo. 2017;8:23. doi: 10.1186/s13227-017-0086-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hayes AJ, et al. Spinal deformity in aged zebrafish is accompanied by degenerative changes to their vertebrae that resemble osteoarthritis. PLoS ONE. 2013;8:e75787. doi: 10.1371/journal.pone.0075787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yan SF, Ramasamy R, Schmidt AM. Mechanisms of disease: advanced glycation end-products and their receptor in inflammation and diabetes complications. Nat. Clin. Pract. Endocrinol. Metab. 2008;4:285–293. doi: 10.1038/ncpendmet0786. [DOI] [PubMed] [Google Scholar]
- 23.Prasad A, Bekker P, Tsimikas S. Advanced glycation end products and diabetic cardiovascular disease. Cardiol. Rev. 2012;20:177–183. doi: 10.1097/CRD.0b013e318244e57c. [DOI] [PubMed] [Google Scholar]
- 24.De Meyts P, Whittaker J. Structural biology of insulin and IGF1 receptors: implications for drug design. Nat. Rev. Drug Discov. 2002;1:769–783. doi: 10.1038/nrd917. [DOI] [PubMed] [Google Scholar]
- 25.Borowsky R. Restoring sight in blind cavefish. Curr. Biol. 2008;18:R23–R24. doi: 10.1016/j.cub.2007.11.023. [DOI] [PubMed] [Google Scholar]
- 26.Elipot Y, Legendre L, Père S, Sohm F, Rétaux S. Astyanax transgenesis and husbandry: how cavefish enters the laboratory. Zebrafish. 2014;11:291–299. doi: 10.1089/zeb.2014.1005. [DOI] [PubMed] [Google Scholar]
- 27.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17 http://dx.doi.org/10.14806/ej.17.1.200. [Google Scholar]
- 29.Li H, Durbin R. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Van der Auwera GA, et al. From FastQ data to high-confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr. Protoc. Bioinform. 2013;43:11.10.1–11.10.33. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Danecek P, et al. The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fariello MI, Boitard S, Naya H, SanCristobal M, Servin B. Detecting signatures of selection through haplotype differentiation among hierarchically structured populations. Genetics. 2013;193:929–941. doi: 10.1534/genetics.112.147231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tomomori-Sato C, et al. A mammalian mediator subunit that shares properties with Saccharomyces cerevisiae mediator subunit Cse2. J. Biol. Chem. 2004;279:5846–5851. doi: 10.1074/jbc.M312523200. [DOI] [PubMed] [Google Scholar]
- 34.Murphy RF, Powers S, Verderame M, Cantor CR, Pollack R. Flow cytofluorometric analysis of insulin binding and internalization by Swiss 3T3 cells. Cytometry. 1982;2:402–406. doi: 10.1002/cyto.990020608. [DOI] [PubMed] [Google Scholar]
- 35.R Development Core Team. A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; 2016. [Google Scholar]
- 36.Wickham H. ggplot2: Elegant Graphics for Data Analysis. Springer; 2009. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Original data underlying this manuscript can be accessed from the Stowers Original Data Repository at http://www.stowers.org/research/publications/libpb-1205 and/or are available from the corresponding authors on reasonable request.