ABSTRACT
The biological changes that occur during pregnancy in the female mammal include shifts in hormonal regulation in preparation for parturition and lactation, and changes in energy metabolism. In women, studies have also shown that during pregnancy there is a reduction in bacterial species richness in the gut. In the current experiment rats were used to model the interaction of diet, reproductive status, and intestinal bacterial microbiota during pregnancy and lactation. In Experiment 1 rats were exposed to either standard chow or high-fat chow (60%) and were divided into two groups: unmated (NULL) or mated (RE). In Experiment 2, both NULL and RE rats were exposed to high-fat chow for a 30-day period. High-throughput sequencing of the 16S rRNA gene revealed that pregnancy impacted the gut microbiota in a similar manner to humans. The impact of reproductive status on microbiota composition, however, was stronger in rats fed a high-fat (HF) diet. Diet-induced changes replicated some of the changes observed in humans, such as increasing the Firmicutes/Bacteroidetes ratio. However, in contrast to humans, pregnancy in rats did not increase β-diversity between microbiota from different animals. These results indicate that during pregnancy in rats, the gut microbiota is altered in a similar manner to that which occurs in women, and that these changes are further exaggerated by exposure to a HF diet. Thus, the rat may allow modelling the effects of consumption of HF food during pregnancy and enable future studies to determine the risks of HF diets during pregnancy and its consequences on the offspring.
KEYWORDS: high-fat diet; lactation, microbiota; pregnancy; 16S amplicon sequencing
Introduction
Both maternal under- and overnutrition (obesity) during pregnancy affects not only the mother herself1 but her offspring's health after birth2,3 and into adulthood.4–7 Studies have shown a myriad of effects in adults including alterations in metabolic function (e.g., type 2 diabetes)8,9 cardiovascular function (e.g., stroke),10 immune function (e.g., infection, autoimmunity),11 and emotional behavior (e.g., anxiety-like behaviors, mood disorders).12–17 A diet rich in fat, specifically during pregnancy, can cause anxiety-like behavior in juvenile female offspring in nonhuman primates18 and lead to learning impairment in rats.19
High-fat diets and obesity may not only have long-lasting physiological effects on the individual and offspring, but impact the intestinal gut ecology in both humans20–22 and animals.23–27 Taxonomic changes,28 reduced α-diversity23 and an increased ability to extract energy from the diet29 have been reported. Changes in the intestinal microbiota similar to those that occur in obesity and in individuals consuming HF diets also occur during normal pregnancy. In pregnancy, body fat increases, insulin sensitivity is reduced and immune changes similar to metabolic inflammation following obesity are observed.30 These metabolic changes however are beneficial to pregnant females in preparing for lactation.31 The intestinal microbiota also changes during normal pregnancy; an increase in β-diversity among individuals and reduced taxonomic richness has been reported,32 but it is unknown whether such changes are a result of pregnancy, changes in dietary habits or both. High body mass index (BMI) during pregnancy has been shown to exacerbate changes in the microbial ecology of the gut with increased abundance of potentially pathogenic species.33,34
The objective of the current series of experiments was to compare the microbiota of virgin, nulliparous (NULL) female rats with reproductively experienced (pregnant and lactating, RE) rats in order to determine if the gut microbiota changes in rats during pregnancy as occurs in women.32 With these experiments, we wished to assess to what extent pregnant and lactating rats can model the gut ecology in women.
Results
Experiment 1: Comparison of NULL rats on a short-term high-fat diet to RE rats fed the high-fat diet throughout pregnancy and lactation.
This experiment was designed to investigate the impact of diet on the microbiota of rats. Specifically, we were interested in the effect of lactation and pregnancy, and to assess to what extent changes in the gut environment observed in women can be modeled in rat dams.
Body weight
As expected, body weight increased over time in the NULL group (Fig. 1A). There were significant main effects of both diet (standard chow vs HF, F1,17 = 5.826, p = .027) and time (day 0 vs day 10, F1,17 = 34.622, p < .001) with a significant interaction between the two factors (F1,17 = 4.838, p = .042). Body weight significantly increased in each diet group compared to day 0 of Diet (standard chow, p = .016; HF, p < .001) with the HF diet gaining significantly more than the standard chow group (p = .024).
Figure 1.
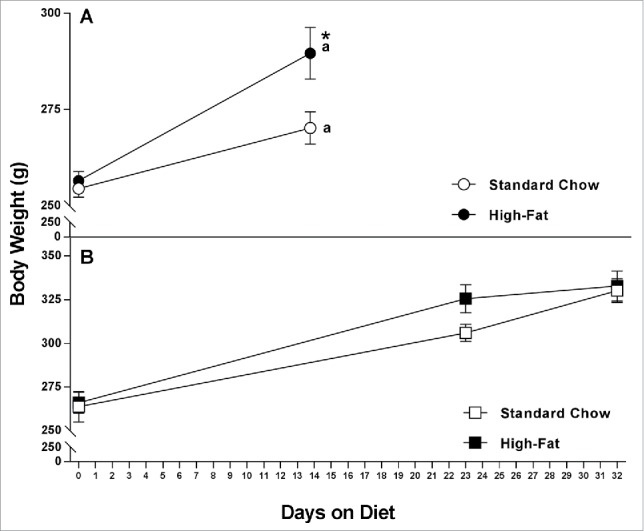
(A) Mean body weight (±SEM, g) of nulliparous (NULL) rats fed either a standard chow or HF diet for 13–14 days. *Significantly different from the standard chow group. aSignificantly different from the day 0 of the diet (p < .05); (B) Mean body weight (±SEM, g) of reproductive experience (RE) rats fed either a standard chow or HF diet on gestation day 1 (day 0 of the diet s), lactation day 1 (day 23 of the diet) and day of euthanasia (day 32 of the diet). There was a significant main effect of time (see text).
Again, as expected, body weight increased over time in the RE animals (Fig. 1B). There was a significant main effect of time (F2,20 = 141.672, p < .001); no main effect of diet (F1,10 = 1892, p = .351) and no significant interaction (F2,20 = 1.659, p = .215) between time and diet. Rats on either the standard chow or HF diet weighed significantly more on lactation day 1 (Day 23 of Diet) and at euthanasia (Day 32 of Diet) compared to day 1 of gestation (Day 0 of Diet; both p's = < .001).
Fecal microbiota
β diversity between microbiota was visualized separately for each diet using PCoA (Fig. 2). There was no apparent clustering of microbiota samples following the standard chow diet (Fig. 2A); however, microbiota samples from the HF group clearly clustered according to reproductive experience (Fig. 2B). Although ANOSIM35 returned a significant overall result when all standard chow samples are tested (p = 0.04), none of three the pairwise comparisons between reproductive groups were significant (p> 0.08). In contrast, for rats on the HF diet for at least 4 days (NULL – day 7 and 13/14 of Diet; RE – days 8, 14, 21, 26 and 30 of Diet), microbiota significantly clustered by reproductive group (p<0.001) and all pairwise comparisons were significant according to ANOSIM (Table 1).
Figure 2.
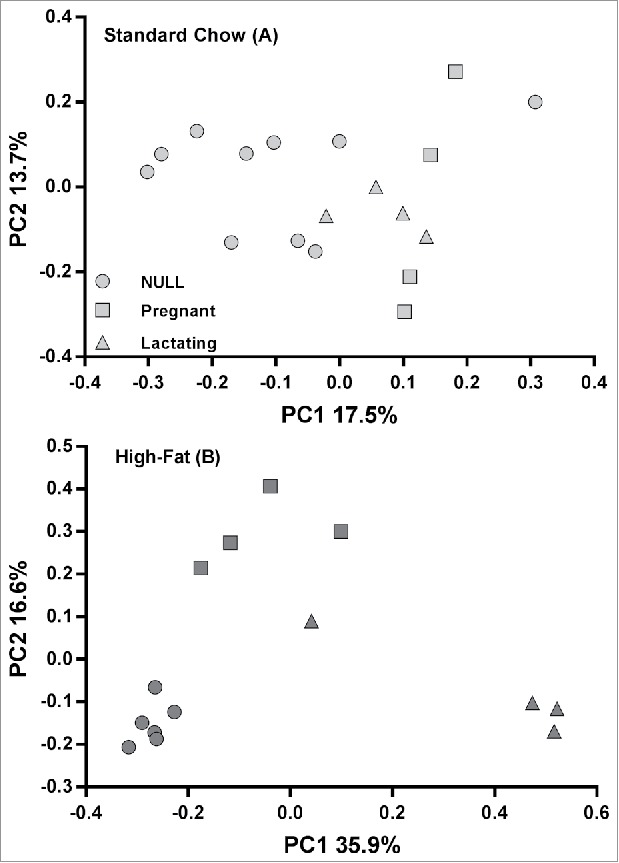
Principal Coordinate Analysis of fecal microbiota from NULL, pregnant and lactating rats 1 on a standard chow (panel A) or HF diet(panel B). Composition of gut microbiota segregates according to reproductive status in rats fed a diet high in fat but not in rats on a standard chow diet. HF diet: n = 14 total; low-fat diet: n = 18 total.
Table 1.
ANOSIM analysis of the effect of reproductive status on bacterial gut microbiota in rats fed a standard chow and high-fat diet in experiment 1.
| Diet |
||
|---|---|---|
| Comparison | Standard Chow (n = 18) | High-fat (n = 14) |
| NULL vs Pregnant vs Lactating | 0.22a (0.04b) | 0.66a (<0.001b) |
| NULL vs Pregnant | 0.25 (0.08) | 0.56 (0.003) |
| NULL vs Lactating | 0.20 (0.16) | 0.80 (0.004) |
| Pregnant vs Lactating | 0.17 (0.18) | 0.70 (0.024) |
ANOSIM R
p value
The relative impact of diet and reproductive experience on the composition of the fecal microbiota was examined using Variation Partitioning Analysis.36 The analysis based on 32 samples comprising all diet and reproductive groups (see Fig. 2A, 2B) showed that microbiota composition was significantly impacted by diet which was associated with 13% of explained variation. Reproductive experience was also significant, explaining 11% of total explained variation. Both effects are statistically significant at p = 0.001 (data not shown). When the analogous variation partitioning analysis was applied to sequence data from the animals on a HF diet for 4 days or longer (n = 14; Fig. 2A), reproduction was associated with more than half (60.1%) of explained variation, whereas the number of days on HF diet was associated only with 7.8% of explained variation (Table 2). Since the latter analysis included samples from rats which had been fed HF diet for ≥4 days, the small diet effect indicates that the bacterial microbiota changed rapidly in response to the new diet and by day 4 had essentially stabilized. This observation is likely explained by a diminishing response of the microbiota to the high-fat diet, as the bacterial community adapts to the diet and reaches a new equilibrium.
Experiment 2: Comparison of NULL and RE on a 30-day high-fat diet.
Table 2.
Variation partitioning of the relative effect of diet and reproductive experience on the composition of bacterial microbiota on rats fed either standard chow or a high-fat diet (experiment 1).
| Fraction | Variation | % Explained | p |
|---|---|---|---|
| RE | 0.5004 | 60.1 | 0.002 |
| Days on diet | 0.065 | 7.8 | 0.002 |
| Shared | 0.267 | 32.1 | 0.004 |
| Total Explained | 0.832 | 100 | |
| Total Variation | 2.1 |
In light of Experiment 1 results showing a significant effect of reproductive status on the intestinal microbiota of dams fed HF diet, a second experiment was designed focusing exclusively on cycling, pregnant and lactating rats maintained on HF diet. As in the first experiment, the sequence data were analyzed to assess the impact of diet and reproductive status on the intestinal microbiota. Instead of controls fed LF diet, the effect of diet was quantified by monitoring changes in the microbiome in function of the time the animals were fed a HF diet.
Body weight
As in experiment 1, body weight increased over time (Fig. 3). There were significant main effects of both reproductive group (NULL vs RE, F1,32 = 20062.45, p < .001) and time (F8,256 = 370.53, p < .001) with a significant interaction between the two factors (F8,256 = 85.62, p < .001). In the NULL group, body weight significantly increased at each time point until day 22 on the diet (all p's < .05). Body weight on diet days 22 and 26 reached a temporary plateau with no further significant weight gain until day 30 (p < .05). During pregnancy, the rats gained weight compared to day 1 of pregnancy at each time point until the periparturitional period (day 20 of the diet, all p's < .05). After parturition, body weight was not different from that recorded on day 10 of diet (all p's > .05). Comparing across reproductive groups, body weight was significantly higher in the reproductive group at every time point (all p's < .05).
Figure 3.
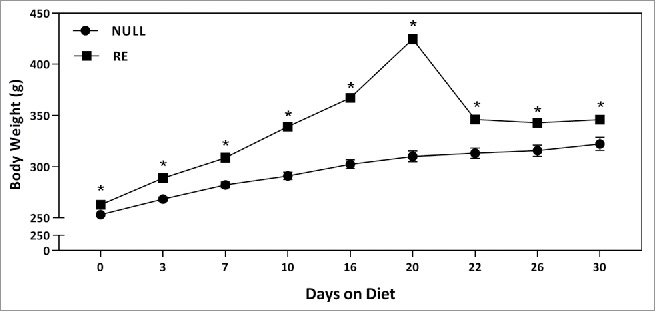
Mean body weight (±SEM, g) of either NULL or RE rats fed a HF diet for 30 days. *Significantly different between NULL and RE groups (p's < .05).
Fecal microbiota
The effect of a HF diet on gut microbiota composition in both NULL and RE animals is presented in Figure 4. As observed in Experiment 1, PCoA based on pairwise weighted Unifrac distances between 57 microbiota samples originating from animals in both reproductive groups which had consumed a HF diet for 10 days or longer showed a clear separation into two reproductive clusters, NULL and RE. In contrast to studies in human cohorts which showed increasing β diversity in the last trimester of pregnancy,32 pairwise Unifrac distances among RE animals (741 comparisons) and among NULL animals (990 comparisons) did not differ (Mann-Whitney test, p = 0.905).
Figure 4.
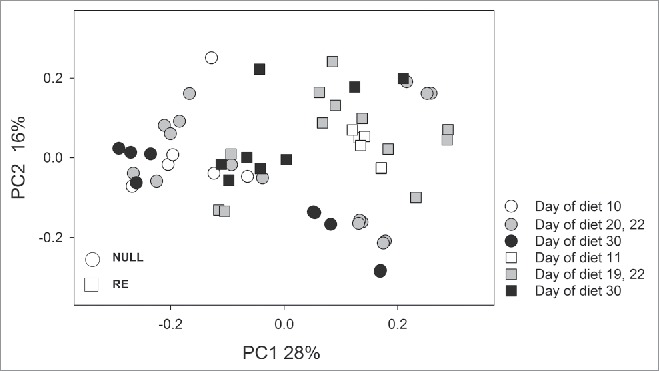
Principal Coordinate Analysis of fecal microbiota from NULL, pregnant and lactating rats 10 days or longer on a HF diet in experiment 2 (n = 57).
As in Experiment 1, the impact of reproductive experience on the gut microbiota was tested using ANOSIM (Table 3). Similar to experiment 1, the 3-way comparison (NULL vs. pregnant vs. lactating) indicated a significant effect of reproductive experience on the gut microbiota (p = 0.03). Unlike in experiment 1, of the three pairwise comparisons, NULL vs. pregnant microbiota were not significantly different (p = 0.35), whereas microbiota of lactating animals were significantly different from the NULL and pregnant groups (p = 0.01, p<0.01, respectively).
Table 3.
ANOSIM of experiment 2 microbiota of NULL, pregnant, and lactating rats on a high-fat diet in for 10 days or longer.
| Groups | Comparison | R | p |
|---|---|---|---|
| 3 Groups (n = 57) | NULL vs Pregnant vs Lactating- | 0.10 | 0.03 |
| NULL vs Pregnant | 0.02 | 0.35 | |
| NULL vs Lactating | 0.14 | 0.01 | |
| Pregnant vs Lactating | 0.02 | <0.001 | |
| 2 Groups (n = 32) | Pregnant vs Lactating- | 0.36 | <0.001 |
We examined the relative abundance ratio of Firmicutes to Bacteroidetes in response to the HF diet. We observed an increase with the pre-diet ratio ranging from 0.2 – 4.0 (n = 14, mean = 0.9) and peaking on day of diet 22 with a range of 3.8 – 59.2 (n = 16, mean = 22.0) (Fig. 5). The same trend in the Firmicutes:Bacteroidetes ratio was observed in both reproductive groups.
Figure 5.
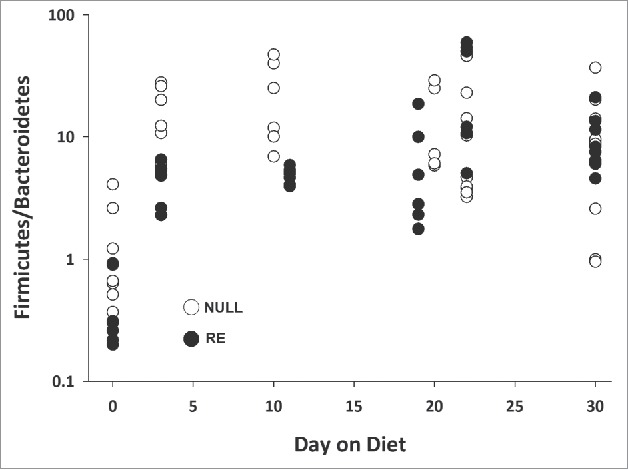
Firmicutes-to-Bacteroidetes abundance ratio increases with time on HF diet. Empty circles, nulliparous (NULL) rats; full circles, reproductive experience (RE) rats.
As described above for Experiment 1, the impact of diet and reproductive experience on the composition of the fecal microbiota was examined using Variation Partition Analysis. Since all animals were fed a HF diet for the duration of the experiment, the variable Diet was expressed as number of days the animals consumed the diet. In this designation, Day 0 indicates the day the animals were switched from standard chow to HF diet. Table 4 shows the results of a Variation Partitioning Analysis and the associated significance tests. The analysis was applied to all microbiota samples (n = 84), to 57 microbiota samples originating from animals which had consumed HF diet for 10 days or longer and to 44 samples from animals fed a HF diet 19 days or longer. This analysis demonstrates that Days on Diet explains more variation (89.2%, 78.1% and 63.8% for n = 84, n = 57 and n = 44, respectively) than reproductive experience (12.3%, 33.7% and 55.8%, respectively), as visible in column “% of explained”. However, consistent with results from Experiment 1, with increasing time on the HF diet, the impact of reproductive experience on gut microbiota increased from 12.3% to 55.8%. Also, as observed in Experiment 1, this result likely indicates a relatively rapid adjustment of the microbiota to the HF diet. After 19 days on diet, this process is however not complete, as time on diet, from 19 to 30 days, is still a significant variable (p = 0.001).
Table 4.
Variation partitioning of the relative effect of diet and reproductive experience on the composition of bacterial microbiota in rats on a high-fat diet (experiment 2).
| Days on Diet | Fraction | Variation | % Explained | p |
|---|---|---|---|---|
| ≥10 | RE | 0.159 | 33.7 | 0.001 |
| Days on diet | 0.369 | 78.1 | 0.001 | |
| Shared | −0.06 | −11.8 | 0.001 | |
| Total Explained Variation | 0.473 | 100 | ||
| Total Variation | 2.521 | |||
| ≥19 | RE | 0.169 | 55.8 | 0.001 |
| Days on diet | 0.192 | 63.8 | 0.001 | |
| Shared | −0.59 | −19.6 | 0.001 | |
| Total Explained Variation | 0.302 | 100 | ||
| Total Variation | 3.713 |
We identified OTUs which were present at different relative abundance in NULL and RE animals in Experiment 2. Supplementary Table 1 depicts the taxa which differed significantly between experimental groups. Among the 67 significantly different taxa, 50 belonged to the phylum Firmicutes, 10 to the phylum Bacteroidetes, one each were classified as Verrucomicrobia, Proteobacteria and Actinobacteria, and four were unclassified at the phylum level using a 70% probability cutoff (Table S1, Fig. S1). Eighteen OTUs were overrepresented in the NULL animals and 49 in the RE animals. No differences in the proportion of OTU phylum-level classification was found; Firmicute OTUs represented 81% of classified OTUs in the NULL group, whereas the corresponding fraction in the RE group was 84% (p > .05). The OTU representing the species Akkermanisa muciniphila was significantly more abundant in the RE group. This OTU had the lowest type I error probability (p = 5.2 × 10−9) among all OTUs flagged as significantly different by LEfSe.37
As lower microbiota diversity has been associated with obesity in rodents and humans32, we analyzed whether in our rat model weight gain correlated with α diversity. Weight gain was calculated by subtracting the mean of the last two weight measurements taken on day 26 and 30 of Diet from the mean of the first two days measured on day 0 and 3. Microbiota diversity was estimated on day 30 of diet. These calculations were performed for the NULL and the RE animals. Figure 6 shows a plot of diversity as a function of weight gain for the NULL animals and indicates that lower α diversity is indeed associated with higher weight gain (Inverse Simpson diversity, R2 = 0.82, p = 0.001).
Figure 6.
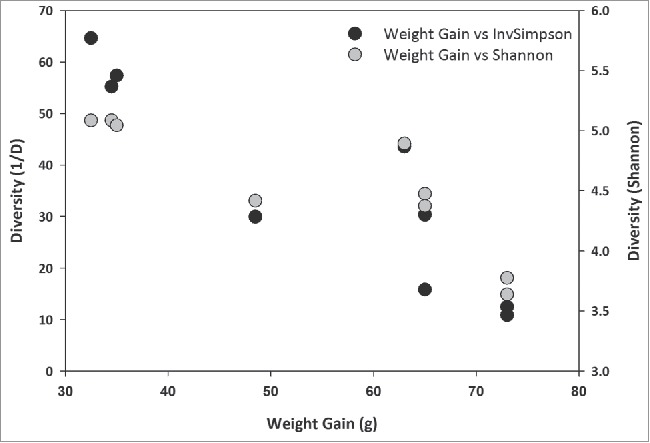
Weight gain of rats on a HF diet correlates with lower α diversity. α diversity was estimated using the Shannon index and the reverse Simpson (1/D) index and plotted against weight gain (n = 9).
Methods
Animals
Nulliparous (virgin) Sprague-Dawley female rats (225–250 g; CRL:CD (SD)BR) were purchased from Charles River Laboratories, Inc. (Kingston, NY, USA). The females were triply-housed in polypropylene cages (45 × 25 × 20 cm) that contained approximately 1.5-liter medium grade wooden flakes and nesting material (Enviro-dri, Shepherd Specialty Papers, Milford, NJ, USA). Food (Envigo, Teklad Rodent Diet, Madison, WI, USA) and water were available ad libitum in light (on 0500–1900 h) – and temperature (21–25°C) – and humidity (30-70%) controlled rooms. All animals were maintained in accordance with the guidelines of the Lab Animal Medicine Service at Cummings School of Veterinary Medicine at Tufts University, which follows the procedures for animal care issued by the Committee on the Care and Use of Laboratory Animal Resources, National Research Council. This research was approved by the Cummings Institutional Animal Care and Use Committee (IACUC).
Experimental design (see Fig. 7)
Figure 7.
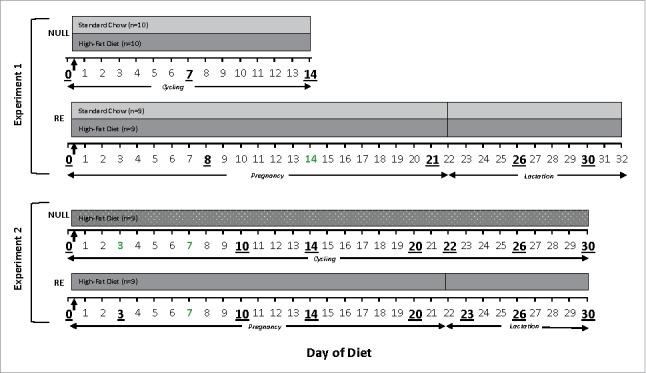
Research design for experiments 1 and 2. The horizontal axis shows the time scale, where Day 0 is the day the animals in the HF groups were switched to HF diet. The same time scale is used throughout the text. Days underlined and larger are the approximate times that fecal samples were obtained. Vertical arrows indicate when the HF diet group was placed on the diet. Null: nulliparous, cycling females; RE: reproductively experienced females.
Experiment 1: Comparison of NULL rats on a short-term high-fat diet to RE rats fed the high-fat diet throughout pregnancy and lactation.
NULL group: One week after arrival, the NULL animals were randomly divided into two groups of 10 animals each: a standard chow (Teklad 2018) group and HF chow (Teklad TD.06414 Adjusted Calories Diet) group (Fig. 7). Sixty percent of the total calories of the high-fat chow came from fat (37% saturated, 47% monosaturated, and 16% polyunsaturated). Fecal samples were collected throughout the experiment (see below). The animals were euthanized on days 13–14 of the diet.
RE group: One week after arrival the nulliparous females were placed with experienced males. On the day that sperm was present in the vaginal lavage (Day 0 of Diet) half of these subjects were fed HF diet (n = 9), while the other half received standard chow (n = 9) (Fig. 7). Two days before parturition the females were singly housed. The day of parturition was considered day 0 of lactation (Day 22 of Diet). On day 1 of lactation the body weight of the dams was recorded and the litter was culled to four females and four males. Fecal samples were collected during the 22 days of gestation (see Fig. 7 and below). The animals were euthanized on Day 32 of Diet.
Experiment 2: Comparison of NULL and RE rats on a 30-day high-fat diet.
Both NULL (n = 18) and RE (n = 16) groups were generated as described in experiment 1. One week after arrival (Day 0 of Diet) both groups were placed on the HF diet as shown in Figure 7. Body weights were recorded and fecal samples collected throughout the diet. Both groups were euthanized on Day 30 of Diet.
Fecal sample collection
As indicated in Figure 7 with underlined numbers, fecal samples were collected from NULL rats before initiating the diet (day 0) and on days 7 and 13–14 while on either the standard chow or HF diet. Since females were triply housed, 6 fecal samples from different cages were used for sequencing for the NULL group microbiota. During the 22 days of gestation, 4 fecal samples were collected from different cages and analyzed. Fecal samples were also collected on days 3 and 6 of lactation (Day of diet 26 and 30). Samples were collected immediately after defecation using sterile forceps, then placed in sterile, nuclease-free, microcentrifuge tubes and stored at −20°C until processed for DNA extraction.
Extraction of fecal DNA and preparation of 16S amplicon libraries
DNA was extracted from 200 µl of fecal homogenate. Three cycles of thawing and freezing was applied and fecal DNA extracted with the HighPure PCR template preparation kit (Roche Diagnostics, Indianapolis, Indiana, USA). Eluted DNA was stored at – 20°C.
PCR primers 27F and 338R38 were used to amplify the approximately 320 nucleotide (nt) V1V2 variable region of the 16S ribosomal RNA (rRNA) gene as previously described.39 Briefly, the V1V2 region was pre-amplified with the above mentioned primers using 20 PCR cycles. A 1 μL volume of the primary reaction was added to a second PCR during which Illumina TrueSeq (Illlumina, San Diego, CA, USA) adaptors, a unique 6-nt barcode and a sequence complementary to the barcode sequencing primer were incorporated into each amplicon using an additional 20 temperature cycles. The enzymatic reagent ExoSAP-IT (Affymetrix, Santa Clara, CA, USA) was used to remove excess primers and nucleotides from final PCR products. Multiple barcoded V1V2 amplicons were pooled in a combined amplicon library and sequenced with an Illumina MiSeq sequencer operated by the Tufts University Genomics core facility (tucf.org).
Analysis of sequence data
Unless stated otherwise, programs in the open-source software mothur40 were used for curating and analyzing 16S sequence data. Random subsamples of 104 sequences per sample were used. V1V2 sequences were trimmed to 200 nucleotides to eliminate 3′ end sequence with a mean Phred quality score <30. Trimmed sequences were aligned using Clustal Omega.41 Aligned sequences were screened to remove sequences which did not align or were unusually short or long. Putative chimeric sequences were detected with UChime42 and removed. The fraction of chimeric sequences detected by UChime was below 2%. FastTree43 was used to build phylogenetic trees. The phylogenetic distance between samples was quantified using the weighted Unifrac distance (D),44 a measure of β diversity. Matrices of pairwise Unifrac distances were imported into GenAlex45 and distances between samples visualized using Principal Coordinate Analysis (PCoA).
Sequences curated as described in the previous paragraph were grouped into Operational Taxonomic Units (OTUs) using the average neighbor distance and a distance cutoff of 3%. OTUs were classified using the Greengenes reference taxonomy (greengenes. secondgenome.com) using 70% as minimum probability value for taxonomic assignment. OTUs present at a significantly different abundance were identified using LEfSe37 as implemented in mothur.
To assess the magnitude of technical variation in the sequence data, three randomly chosen fecal DNA samples were amplified twice and the duplicated amplicons tagged with different barcodes. The average weighted Unifrac distance between duplicates37 was 0.083 (range 0.024 – 0.140). In comparison, the average distance between non-replicated samples was 0.460 (range 0.036 – 0.750). These data indicate that technical variation was significantly smaller than experimental variation (Mann-Whitney Rank Sum test, p = 0.003).
Additional statistical analysis
Body weight across time was analyzed for each reproductive group (NULL and RE) separately because of the different days on diet with a two-factor mixed design analysis of variance (ANOVA, between factor, diet [standard chow vs HF], within factor, time). Multiple comparisons were performed using the Fisher's LSD Test. Differences were considered significant if p < 0.05.
Discussion
Effect of HF diet on the intestinal microbiota of pregnant and lactating rats
As observed in overweight women during pregnancy,32–34 the present results demonstrate that the composition of the gut microbiota changes in pregnant and lactating rats fed a HF diet. Representation of pairwise β diversity values by PCoA shows a clear clustering of microbiota depending upon reproductive status (NULL, pregnant and lactating). This effect was particularly pronounced in animals on the HF diet, which prompted us to further analyze in a second experiment the effect of reproductive status on the microbiota in a HF environment. Sequence data indicate that the longer the rats were on the HF diet, the larger was the relative effect of reproductive experience. In fact, in Experiment 1, the diet effect became smaller than the effect of reproductive status when samples from animals which had consumed HF diet for at least four days were analyzed. When all time points were included, the effect was reversed, i.e., the diet effect was larger than the effect of reproductive status. The same trend of decreasing diet effect was observed in Experiment 2; the diet effect decreased from 89.2% to 63% as samples from animals maintained on HF diet for a short time were excluded. The diminishing diet effect we observed is consistent with a rapid adaptation of the intestinal bacterial ecosystem to diet.
Diet, reproductive status and weight
While NULL rats on a HF diet gained more weight than standard chow fed rats, in the RE group there were, surprisingly, no significant differences in weight gain during pregnancy and lactation between the diet groups. Maternal obesity and/or maternal HF diet studies are usually focused on the consequences to the offspring.46–48 These studies vary greatly in terms of the mothers' diet before pregnancy, the duration and the amount of fat in the diet. Similar to our study, Tamashiro et al49 exposed rats to a 60% HF diet starting on day 2 of gestation, measured body weight on days 14–21, and found no significant weight gain. Leturgue et al50 started rats on a 55% fat diet on day 0 of gestation and also found no significant differences in weight gain. Animal studies that model maternal obesity, rather than administering HF diets during gestation, usually feed the HF diet many weeks before mating. Pereira et al51 started female rats on a 45% fat diet 4 weeks before mating and found that they gained significantly more weight during pregnancy than control females, while Mayer et al46 had female mice on a 42% fat diet for over 3 months before mating, and found that the animals lost a significant amount of weight compared to standard fed females. Gohir et al52 found no differences in weight gain after eight weeks of a 45% fat diet or during pregnancy. These results indicate that rodents fed a HF diet during pregnancy do not necessarily gain significant amounts of weight.
Evaluation of the rat model
The primary motivation of this study was to assess whether pregnant and lactating rats can model the interaction between diet, reproductive status and gut microbiota observed in women. Such comparisons are difficult to make because human studies are limited by the heterogeneity of human cohorts and a paucity of studies comparing pregnant and non-pregnant individuals. For obvious reasons, dietary interventions in human cohorts are problematic. Our results, however, are comparable to changes found in women during pregnancy. Collado et al,33 using flow cytometry of in situ labeled bacteria, found a general increase in bacterial abundance during pregnancy. Overweight pregnant women had significantly higher abundance of bacteria belonging to the genus Clostridium. Santacruz et al34 found that at 24 weeks of gestation Bifodobacterium and Bacteroides were more abundant in normal-weight woman as compared to overweight women, while overweight women had higher levels of Enterobacteriaceae, Staphylococcus, and E. coli. Koren et al32 found significant changes in gut microbiota, both in composition and abundance, in women in their third trimester compared with the first trimester. These authors also found that β diversity among first trimester samples was similar to diversity among controls from the Human Microbiome Project. The increase in β diversity later in pregnancy may be related to changing dietary habits, a variable that cannot be eliminated from human cohort studies and is difficult to emulate in animals. In the present study, we compared rats during pregnancy and lactation (RE) with virgin, cycling rats (NULL), which can be considered comparable to nonpregnant rats, and similarly found a significant effect of reproductive status on the microbiota.
Similar to our study, Gohir et al52 assessed the impact of diet and pregnancy on the gut microbiota, but using mice. The conclusion that diet and reproductive status impact the composition of the microbiota is similar to the results we obtained with rats. As we observed, the effect of diet was more important than the effect of reproductive status. Gohir et al also found that upon conception the relative abundance of mucin-degrading bacteria increased, which is in agreement with the significant increase in relative abundance of A. muciniphila in RE animals we found in our study (Supplemental Table S1). In contrast to Gohir's observation, we did not find an increase in Bifidobacteria and did not detect any sequences belonging to the class Actinobacteria.
The fact that diet influenced the gut microbiota to a greater extent than reproductive status correlates well with the concept that the changes in the microbiota that normally occur during pregnancy mimic the changes that occur following a high fat diet in both humans53 and animals.26 The five most abundant phyla in the normal gut are Firmicutes, Bacteroidetes, Actinobacteria, Proteobacteria, and Verrucomicrobia54,55 with Firmicutes and Bacteroidetes dominating in both mice53 and humans.55 The ratio between these phyla, especially Firmicutes to Bacteroidetes, can change due to environmental influence and health.56,57 An obesogenic diet can change this ratio with Firmicutes increasing relative to Bacteroidetes.20,53 The present results confirm those findings and support the use of rats to model the gut microbiota in response to HF diets.
Conclusions
Rats fed a HF diet during pregnancy demonstrate similar changes in their gut microbiota as maternally obese humans. Comparison of data from rodent studies with results from human cohorts are needed to further determine the validity of the rat model, and to assess if changes in the intestinal ecosystem affect the offspring as described in mice,11 humans,58 and nonhuman primates.18,59 In light of the effect of intestinal dysbiosis on health, the rat model could be valuable to explore the effect diet on the dam and her offspring. For instance, a recent study in mice, Seki et al., 2017, demonstrated a link between maternal diet and gene regulation in the offspring.48 The refinement of animal models will facilitate research into mechanisms linking diet, microbiota, and metabolic changes in mother and offspring.
Supplementary Material
Disclosure of potential conflicts of interest
The authors declare no conflict of interest.
Acknowledgments
Our thanks to Albert Tai and the staff at Tufts Genomics Core. In addition, we thank Dr. Robert Bridges for his help and support.
Funding
This study was supported by a pilot grant from Tufts University Cummings School of Veterinary Medicine and a Collaborates grant also from Tufts University. G.W. The authors acknowledge financial support for the NIAID (grant R21 AI125891).
References
- [1].Tsoi E, Shaikh H, Robinson S, Teoh TG. Obesity in pregnancy: a major healthcare issue. Postgrad Med J. 2010;86:617–23. doi: 10.1136/pgmj.2010.098186. [DOI] [PubMed] [Google Scholar]
- [2].Dessi A, Atzori L, Noto A, Visser GH, Gazzolo D, Zanardo V, Barberini L, Puddu M, Ottonello G, Atzei A, et al.. Metabolomics in newborns with intrauterine growth retardation (IUGR): urine reveals markers of metabolic syndrome. J Matern Fetal Neonatal Med. 2011;24(Suppl 2):35–9. [DOI] [PubMed] [Google Scholar]
- [3].Lakshmy R. Metabolic syndrome: role of maternal undernutrition and fetal programming. Rev Endocr Metab Disord. 2013;14:229–40. doi: 10.1007/s11154-013-9266-4. [DOI] [PubMed] [Google Scholar]
- [4].Barker DJ. The origins of the developmental origins theory. J Intern Med. 2007;261:412–7. doi: 10.1111/j.1365-2796.2007.01809.x. [DOI] [PubMed] [Google Scholar]
- [5].Galliano D, Bellver J. Female obesity: short- and long-term consequences on the offspring. Gynecol Endocrinol. 2013;29:626–31. doi: 10.3109/09513590.2013.777420. [DOI] [PubMed] [Google Scholar]
- [6].Gluckman PD, Hanson MA, Cooper C, Thornburg KL. Effect of in utero and early-life conditions on adult health and disease. N Engl J Med. 2008;359:61–73. doi: 10.1056/NEJMra0708473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Williams L, Seki Y, Vuguin PM, Charron MJ. Animal models of in utero exposure to a high fat diet: a review. Biochimica et biophysica acta. 2014;1842:507–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Barker DJ. The fetal origins of type 2 diabetes mellitus. Annals of Internal Medicine. 1999;130:322–4. doi: 10.7326/0003-4819-130-4-199902160-00019. [DOI] [PubMed] [Google Scholar]
- [9].Armitage JA, Poston L, Taylor PD. Developmental origins of obesity and the metabolic syndrome: the role of maternal obesity. Front Horm Res. 2008;36:73–84. [DOI] [PubMed] [Google Scholar]
- [10].Barker DJ, Osmond C. Infant mortality, childhood nutrition, and ischaemic heart disease in England and Wales. Lancet. 1986;1:1077–81. [DOI] [PubMed] [Google Scholar]
- [11].Myles IA, Fontecilla NM, Janelsins BM, Vithayathil PJ, Segre JA, Datta SK. Parental dietary fat intake alters offspring microbiome and immunity. J Immunol. 2013;191:3200–9. doi: 10.4049/jimmunol.1301057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Lahti J, Raikkonen K, Heinonen K, Pesonen AK, Kajantie E, Forsen T, Osmond C, Barker DJ, Eriksson JG. Body size at birth and socio-economic status in childhood: implications for Cloninger's psychobiological model of temperament at age 60. Psychiatry Res. 2008;160:167–74. doi: 10.1016/j.psychres.2007.07.016. [DOI] [PubMed] [Google Scholar]
- [13].Lahti J, Raikkonen K, Kajantie E, Heinonen K, Pesonen AK, Jarvenpaa AL, Strandberg T. Small body size at birth and behavioural symptoms of ADHD in children aged five to six years. J Child Psychol Psychiatry. 2006;47:1167–74. doi: 10.1111/j.1469-7610.2006.01661.x. [DOI] [PubMed] [Google Scholar]
- [14].Lahti J, Raikkonen K, Pesonen AK, Heinonen K, Kajantie E, Forsen T, Osmond C, Barker DJ, Eriksson JG. Prenatal growth, postnatal growth and trait anxiety in late adulthood – the Helsinki Birth Cohort Study. Acta Psychiatr Scand. 2010;121:227–35. doi: 10.1111/j.1600-0447.2009.01432.x. [DOI] [PubMed] [Google Scholar]
- [15].Lahti J, Raikkonen K, Sovio U, Miettunen J, Hartikainen AL, Pouta A, Taanila A, Joukamaa M, Järvelin MR, Veijola J. Early-life origins of schizotypal traits in adulthood. Br J Psychiatry. 2009;195:132–7. doi: 10.1192/bjp.bp.108.054387. [DOI] [PubMed] [Google Scholar]
- [16].Peleg-Raibstein D, Luca E, Wolfrum C. Maternal high-fat diet in mice programs emotional behavior in adulthood. Behav Brain Res. 2012;233:398–404. doi: 10.1016/j.bbr.2012.05.027. [DOI] [PubMed] [Google Scholar]
- [17].Thompson C, Syddall H, Rodin I, Osmond C, Barker DJ. Birth weight and the risk of depressive disorder in late life. The British journal of psychiatry; the journal of mental science. 2001;179:450–5. doi: 10.1192/bjp.179.5.450. PMID:11689404 [DOI] [PubMed] [Google Scholar]
- [18].Sullivan EL, Grayson B, Takahashi D, Robertson N, Maier A, Bethea CL, Smith MS, Coleman K, Grove KL. Chronic consumption of a high-fat diet during pregnancy causes perturbations in the serotonergic system and increased anxiety-like behavior in nonhuman primate offspring. J Neurosci: the official journal of the Society for Neuroscience. 2010;30:3826–30. doi: 10.1523/JNEUROSCI.5560-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Rodriguez JS, Rodriguez-Gonzalez GL, Reyes-Castro LA, Ibanez C, Ramirez A, Chavira R, Larrea F, Nathanielsz PW, Zambrano E. Maternal obesity in the rat programs male offspring exploratory, learning and motivation behavior: prevention by dietary intervention pre-gestation or in gestation. International journal of developmental neuroscience: the official journal of the International Society for Developmental Neuroscience. 2012;30:75–81. doi: 10.1016/j.ijdevneu.2011.12.012. [DOI] [PubMed] [Google Scholar]
- [20].Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology – Human gut microbes associated with obesity. Nature. 2006;444:1022–3. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- [21].Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–31. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- [22].Rothe M, Blaut M. Evolution of the gut microbiota and the influence of diet. BenefMicrobes. 2013;4:31–7. [DOI] [PubMed] [Google Scholar]
- [23].Turnbaugh PJ, Backhed F, Fulton L, Gordon JI. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell HostMicrobe. 2008;3:213–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Li W, Dowd SE, Scurlock B, Acosta-Martinez V, Lyte M. Memory and learning behavior in mice is temporally associated with diet-induced alterations in gut bacteria. Physiology & Behavior. 2009;96:557–67. doi: 10.1016/j.physbeh.2008.12.004. [DOI] [PubMed] [Google Scholar]
- [25].Sefcikova Z, Kmet V, Bujnakova D, Racek L, Mozes S. Development of gut microflora in obese and lean rats. Folia Microbiol (Praha). 2010;55:373–5. doi: 10.1007/s12223-010-0061-2. [DOI] [PubMed] [Google Scholar]
- [26].Mujico JR, Baccan GC, Gheorghe A, Diaz LE, Marcos A. Changes in gut microbiota due to supplemented fatty acids in diet-induced obese mice. Br J Nutr. 2013;110:711–20. doi: 10.1017/S0007114512005612. [DOI] [PubMed] [Google Scholar]
- [27].Pyndt Jorgensen B, Hansen JT, Krych L, Larsen C, Klein AB, Nielsen DS, Josefsen K, Hansen AK, Sørensen DB. A possible link between food and mood: dietary impact on gut microbiota and behavior in BALB/c mice. PLoS One. 2014;9:e103398. doi: 10.1371/journal.pone.0103398. PMID:25133574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Million M, Lagier JC, Yahav D, Paul M. Gut bacterial microbiota and obesity. Clin Microbiol Infect. 2013;19:305–13. doi: 10.1111/1469-0691.12172. [DOI] [PubMed] [Google Scholar]
- [29].Turnbaugh PJ, Gordon JI. The core gut microbiome, energy balance and obesity. J Physiol. 2009;587:4153–8. doi: 10.1113/jphysiol.2009.174136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Hellmuth C, Lindsay KL, Uhl O, Buss C, Wadhwa PD, Koletzko B, Entringer S. Association of maternal prepregnancy BMI with metabolomic profile across gestation. International Journal of Obesity. 2017;41:159–69. doi: 10.1038/ijo.2016.153. [DOI] [PubMed] [Google Scholar]
- [31].Nelson SM, Matthews P, Poston L. Maternal metabolism and obesity: modifiable determinants of pregnancy outcome. Hum Reprod Update. 2010;16:255–75. doi: 10.1093/humupd/dmp050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Koren O, Goodrich JK, Cullender TC, Spor A, Laitinen K, Backhed HK, Werner JJ, Angenent LT, Knight R, Bäckhed F, et al.. Host remodeling of the gut microbiome and metabolic changes during pregnancy. Cell. 2012;150:470–80. doi: 10.1016/j.cell.2012.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Collado MC, Isolauri E, Laitinen K, Salminen S. Distinct composition of gut microbiota during pregnancy in overweight and normal-weight women. American Journal of Clinical Nutrition. 2008;88:894–9. [DOI] [PubMed] [Google Scholar]
- [34].Santacruz A, Collado MC, Garcia-Valdes L, Segura MT, Martin-Lagos JA, Anjos T, Martí-Romero M, Lopez RM, Florido J, Campoy C, et al.. Gut microbiota composition is associated with body weight, weight gain and biochemical parameters in pregnant women. British Journal of Nutrition. 2010;104:83–92. doi: 10.1017/S0007114510000176. [DOI] [PubMed] [Google Scholar]
- [35].Clarke KR. Nonparametric multivariate mnalyses of changes in community structure. Aust J Ecol. 1993;18:117–43. doi: 10.1111/j.1442-9993.1993.tb00438.x. [DOI] [Google Scholar]
- [36].Borcard D, Legendre P, Drapeau P. Partialling out the Spatial Component of Ecological Variation. Ecology. 1992;73:1045–55. doi: 10.2307/1940179. [DOI] [Google Scholar]
- [37].Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C, Lopez R, McWilliam H, Remmert M, Söding J, et al.. Metagenomic biomarker discovery and explanation. Genome biology. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. PMID:21702898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Hamady M, Walker JJ, Harris JK, Gold NJ, Knight R. Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nature Methods. 2008;5:235–7. doi: 10.1038/nmeth.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Ras R, Huynh K, Desoky E, Badawy A, Widmer G. Perturbation of the intestinal microbiota of mice infected with Cryptosporidium parvum. Int J Parasitol. 2015;45:567–73. doi: 10.1016/j.ijpara.2015.03.005. [DOI] [PubMed] [Google Scholar]
- [40].Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, et al.. Introducing mothur: Open-Source, Platform-Independent, Community-Supported Software for Describing and Comparing Microbial Communities. Applied and Environmental Microbiology. 2009;75:7537–41. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li WZ, Lopez R, McWilliam H, Remmert M, Söding J, et al.. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539-44. PMID:21988835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Price MN, Dehal PS, Arkin AP. FastTree 2-Approximately Maximum-Likelihood Trees for Large Alignments. Plos One. 2010;5. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Lozupone C, Hamady M, Knight R. UniFrac – An online tool for comparing microbial community diversity in a phylogenetic context. Bmc Bioinformatics. 2006;7:371-84. doi: 10.1186/1471-2105-7-371. PMID:16893466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Peakall R, Smouse PE. GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics. 2012;28:2537–9. doi: 10.1093/bioinformatics/bts460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Mayor RS, Finch KE, Zehr J, Morselli E, Neinast MD, Frank AP, Hahner LD, Wang J, Rakheja D, Palmer BF, et al.. Maternal high-fat diet is associated with impaired fetal lung development. Am J Physiol Lung Cell Mol Physiol. 2015;309:L360−8. doi: 10.1152/ajplung.00105.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Cunha Fda S, Dalle Molle R, Portella AK, Benetti Cda S, Noschang C, Goldani MZ, Silveira PP. Both food restriction and high-fat diet during gestation induce low birth weight and altered physical activity in adult rat offspring: the “Similarities in the Inequalities” model. PLoS One. 2015;10:e0118586. doi: 10.1371/journal.pone.0118586. PMID:25738800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Seki Y, Suzuki M, Guo X, Glenn AS, Vuguin PM, Fiallo A, Du Q, Ko YA, Yu Y, Susztak K, et al.. In Utero Exposure to a High-Fat Diet Programs Hepatic Hypermethylation and Gene Dysregulation and Development of Metabolic Syndrome in Male Mice. Endocrinology. 2017;158:2860–72. doi: 10.1210/en.2017-00334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Tamashiro KL, Terrillion CE, Hyun J, Koenig JI, Moran TH. Prenatal stress or high-fat diet increases susceptibility to diet-induced obesity in rat offspring. Diabetes. 2009;58:1116–25. doi: 10.2337/db08-1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Leturque A, Burnol AF, de Saintaurin MA, Penicaud L, Girard J. Effect of feeding a high-fat diet during pregnancy on glucose metabolism in the rat. Metabolism. 1987;36:66–70. doi: 10.1016/0026-0495(87)90065-5. [DOI] [PubMed] [Google Scholar]
- [51].Pereira TJ, Fonseca MA, Campbell KE, Moyce BL, Cole LK, Hatch GM, Doucette CA, Klein J, Aliani M, Dolinsky VW, et al.. Maternal obesity characterized by gestational diabetes increases the susceptibility of rat offspring to hepatic steatosis via a disrupted liver metabolome. J Physiol. 2015;593:3181–97. doi: 10.1113/JP270429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Gohir W, Whelan FJ, Surette MG, Moore C, Schertzer JD, Sloboda DM. Pregnancy-related changes in the maternal gut microbiota are dependent upon the mother's periconceptional diet. Gut Microbes. 2015;6:310–20. doi: 10.1080/19490976.2015.1086056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Ley RE, Backhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc Natl Acad Sci U S A. 2005;102:11070–5. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Tap J, Mondot S, Levenez F, Pelletier E, Caron C, Furet JP, Ugarte E, Muñoz-Tamayo R, Paslier DL, Nalin R, et al.. Towards the human intestinal microbiota phylogenetic core. Environ Microbiol. 2009;11:2574–84. doi: 10.1111/j.1462-2920.2009.01982.x. [DOI] [PubMed] [Google Scholar]
- [55].Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–8. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Flint HJ, Duncan SH, Scott KP, Louis P. Links between diet, gut microbiota composition and gut metabolism. Proc Nutr Soc. 2015;74:13–22. doi: 10.1017/S0029665114001463. [DOI] [PubMed] [Google Scholar]
- [57].Flint HJ, Scott KP, Louis P, Duncan SH. The role of the gut microbiota in nutrition and health. Nat Rev Gastroenterol Hepatol. 2012;9:577–89. doi: 10.1038/nrgastro.2012.156. [DOI] [PubMed] [Google Scholar]
- [58].Catalano PM, Shankar K. Obesity and pregnancy: mechanisms of short term and long term adverse consequences for mother and child. BMJ. 2017;356:j1. doi: 10.1136/bmj.j1. PMID:28179267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Ma J, Prince AL, Bader D, Hu M, Ganu R, Baquero K, Alan Harris R, Frias AE, Grove KL, Aagaard KM. High-fat maternal diet during pregnancy persistently alters the offspring microbiome in a primate model. NatCommun. 2014;5:3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


