ABSTRACT
Exercise reduces the risk of inflammatory disease by modulating a variety of tissue and cell types, including those within the gastrointestinal tract. Recent data indicates that exercise can also alter the gut microbiota, but little is known as to whether these changes affect host function. Here, we use a germ-free (GF) animal model to test whether exercise-induced modifications in the gut microbiota can directly affect host responses to microbiota colonization and chemically-induced colitis. Donor mice (n = 19) received access to a running wheel (n = 10) or remained without access (n = 9) for a period of six weeks. After euthanasia, cecal contents were pooled by activity treatment and transplanted into two separate cohorts of GF mice. Two experiments were then conducted. First, mice were euthanized five weeks after the microbiota transplant and tissues were collected for analysis. A second cohort of GF mice were colonized by donor microbiotas for four weeks before dextran-sodium-sulfate was administered to induce acute colitis, after which mice were euthanized for tissue analysis. We observed that microbial transplants from donor (exercised or control) mice led to differences in microbiota β-diversity, metabolite profiles, colon inflammation, and body mass in recipient mice five weeks after colonization. We also demonstrate that colonization of mice with a gut microbiota from exercise-trained mice led to an attenuated response to chemical colitis, evidenced by reduced colon shortening, attenuated mucus depletion and augmented expression of cytokines involved in tissue regeneration. Exercise-induced modifications in the gut microbiota can mediate host-microbial interactions with potentially beneficial outcomes for the host.
KEYWORDS: exercise, microbiome, gut, microbiota, colitis, germ-free, transplant, colonization inflammation, voluntary wheel running
Introduction
Participation in regular, moderate exercise is beneficial for numerous tissues including muscle, adipose and brain, and can reduce the incidence of metabolic and inflammatory disease in humans and in animal models.1,2 Recent studies have also revealed that exercise training can influence the gut and its associated gut microbiome in rodents.3-6 Importantly, changes in the gut microbiome induced by exercise training have been associated with parallel changes in host physiology, including alterations in metabolism, immunity, and behavior.7,8 Other studies have indicated that exercise-induced changes in the gut microbiome may also be involved in the modulation of disease states. For instance, exercise can affect the gut microbiome of mice during exposure to a high fat-diet,8,9 experimental diabetes,10 and toxin-induced dysbiosis.5 However, these findings are correlational and fail to address the direct role of the gut microbiota in mediating host physiology. Therefore, a more definitive approach is warranted to determine whether exercise-induced changes in the gut microbiota can impart meaningful changes in host physiology.
Experiments using germ-free (GF) mice colonized with known bacterial communities (i.e. gnotobiotic mice) provide an opportunity to study environmental regulation of host-gut microbiota interactions. Indeed, a growing number of studies have indicated direct relationships between the gut microbiota (e.g. through diet and antibiotic use) and host function.11,12 Such studies have revealed that environmentally-induced changes in gut microbiota and gut microbiota-derived metabolites (e.g. short chain fatty acids) can directly and robustly influence the structure of the mucus layer and the development of the immune system after gut microbiota colonization in GF animals.13,14 Exercise training can also modify levels of gut microbiota-derived short chain fatty acids within the intestines of conventionally-raised animals.6 Short-chain fatty acids (SCFAs) are derived through gut microbial fermentation of complex polysaccharides or endogenous mucins and have numerous downstream effects on the host.15 SCFAs have been shown to contribute to increased energy harvest,16 enhance the production of satiety peptides,17 and reduce inflammation18 within the gut. Despite this, little is known as to whether exercise training-induced increases in SCFAs may modulate host processes.
The influence of the gut microbiota on host function is also critically important to study in the context of inflammatory bowel diseases, such as ulcerative colitis (UC).19 UC is an inflammatory condition in which open lesions and ulcers develop throughout the epithelial and mucosal layers of the colon.20 The etiology of UC is not completely understood, but it appears to arise from an aberrant immune response to the gut microbiota as a result of a disrupted mucus and epithelial barrier,21-23 Though current treatments, such as anti-inflammatory aminosalicylates, corticosteroids, and targeted monoclonal antibiotic therapy (e.g. anti-TNF), are sometimes successful in preventing symptomology, their use is often associated with drug resistance, inconsistent long-term efficacy, and undesirable side-effects.24,25 Therefore, it is vital to investigate adjunctive therapeutic approaches for preventing and attenuating UC. Epidemiological studies in humans have shown that exercise training may reduce the symptoms of disease and improve the quality of life for patients with UC.26,27 There is also a strong body of epidemiological evidence indicating that exercise also minimizes the risk for developing colorectal cancer,28 a disease that can originate from chronic UC. Research from our laboratory and others have shown that voluntary exercise training can attenuate the inflammatory insult and disease symptomology in mouse models of colitis,29,30 further providing evidence that moderate exercise can beneficially affect the gastrointestinal tract and disease outcomes. Unfortunately, a mechanistic understanding as to how exercise may modulate intestinal diseases and syndromes is not well established.
In this report, we investigated whether exercise-induced changes in the gut microbiota of conventionally-raised donor mice could modulate colon physiology of recipient, sterilely-raised gnotobiotic mice after five weeks of a gut microbiota colonization. In a second experiment, we explored whether a transplanted gut microbiota from donor-exercised or donor-control mice could attenuate the clinical symptomology and inflammatory insult of dextran-sodium sulfate (DSS)-induced colitis in recipient mice, which we and others have previously shown possible in conventionally raised animals.29,30 We hypothesized that exercise-induced modifications in the gut microbiota of donor mice would lead to reduced colon inflammation in recipient mice after five weeks of colonization. Additionally, we hypothesized that a microbiota transplant from donor exercise-trained mice to recipient, gnotobiotic mice would attenuate the clinical symptomology and the inflammatory response to DSS-induced colitis when compared to mice that received a microbiota from control animals.
Results
Figure 1 depicts the research design of two independent experiments.
Figure 1.
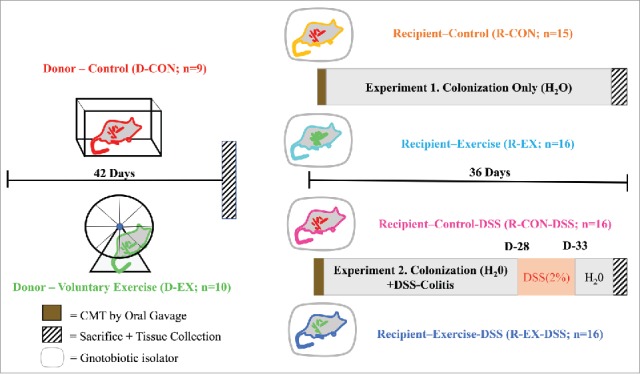
Experimental Design. Graphical representation of the designs of Experiment 1 (colonization experiment) and Experiment 2 (acute colitis experiment). CMT = cecal microbial transplant, DSS = dextran sodium sulfate.
Experiment 1: Host response to colonization
Exercise training-induced modifications of the gut microbiota in conventionally-raised donor mice persist in recipient, gnotobiotic mice after five weeks of a gut microbiota colonization
We first confirmed that exercise could significantly modify the gut microbiota, as previously reported by our laboratory and others.3-5 Principle coordinates analysis (PCoA) revealed that exercise training (i.e. voluntary wheel running) for 6 weeks led to differences in the gut microbiota community structure in donor mice as determined by Unweighted Unifrac (Fig. 2A) and Weighted Unifrac (Fig. S1A). These exercise-induced differences were also evident at the genus level of taxonomic analysis (Table S1). Notably, the gut microbiota of D-EX mice contained higher relative abundances of genera: Anaerostipes spp, Akkermansia spp, Family Lachnospiraceae Genus unclassified, Ruminococcus spp, and Parabacteroides spp, and a lower abundance of Prevotella spp. compared to the D-CON mice (FDR p < 0.05).
Figure 2.
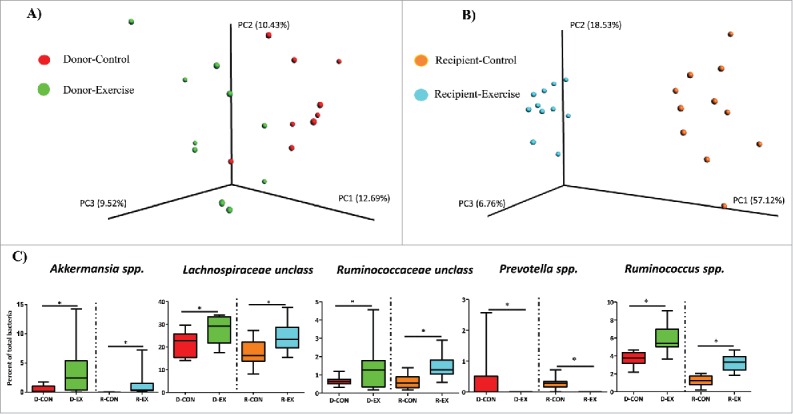
Comparisons of gut microbiome β-diversity in donor and recipient mice. (Experiment 1). (A) Exercise training leads to differences in β-diversity of the fecal microbiome (Unweighted UniFrac) of donor mice after 6 weeks of exercise training; red circles = Donor-Control (D-CON), green circles = Donor-Exercise (D-EX); PERMANOVA p<0.05. (B) Community structure of the fecal microbiome (Unweighted UniFrac) of recipient, gnotobiotic mice 5 weeks after colonization by donor cecal microbiota; orange circles = Recipient-Control (R-CON), teal circles = Recipient-Exercise (R-EX); PERMANOVA p<0.01. (C) Abundance (% of total bacteria) of bacterial genera that were differentially represented by exercise group in donor mice and that remained differentially abundant (*) in recipient mice after transplant. *FDR p < 0.05.
We then tested if the gut microbiota changes induced by exercise training could persist in recipient gnotobiotic mice after five weeks of gut microbial colonization. In accordance with our hypothesis, PCoA of both the unweighted (Fig. 2B) and weighted Unifrac (Fig. S1B) distance metrics revealed that gut microbiota communities clustered by exercise-transfer status in the recipient mice 35 days after colonization. The clustering by exercise status was even more evident in recipient than in donor mice, highlighting an overall effect of colonization on the gut microbiota communities despite relatively high transfer efficiency (as measured by presence of taxa) from donor to recipient mice at the genus and OTU level of analysis (Table S2). Despite this colonization effect, many of the genera differences observed between activity groups in the donor mice were maintained in the recipient mice (Fig. 2C). For instance, Akkermansia spp., Lachnospiraceae unclassified, and Ruminococcus spp. remained significantly more abundant while Prevotella spp. remained significantly less represented in recipient-exercise (R-EX) versus recipient-control (R-CON) mice. Some of the exercise-induced genera differences apparent in donor mice did not persist in recipient mice (i.e. Anaerostipes spp. and Parabacteroides spp, Table S1), while numerous other treatment genera differences (not evident in donor mice) manifested in recipient mice (see Table S1).
Gut microbiota transfer from exercise-trained mice leads to attenuated inflammation and modified histological features in colons of recipient mice
To further probe the host response to colonization, we analyzed the expression of inflammatory and anti-inflammatory/regenerative cytokines in the distal colon of recipient mice. We show that colonization with an ‘exercised gut microbiota’ led to significantly lower colonic gene expression of innate inflammatory mediators, interleukin-1β (IL-1β), indoleamine 2,3 dioxygenase (IDO1), and interleukin-23 (IL-23) in R-EX versus R-CON mice (Fig. 3A; p < 0.05). No statistically significant differences in expression of IL-6, tumor necrosis factor (TNF)-α, or IL-17a were observed (Fig. 3A; p > 0.05). R-EX also exhibited significantly lower transforming growth factor (TGF)-β gene expression (Fig. 3B; p < 0.05). No statistically significant differences in IL-10, FoxP3 and IL-22 gene expression were observed (Fig. 3B; p > 0.05).
Figure 3.
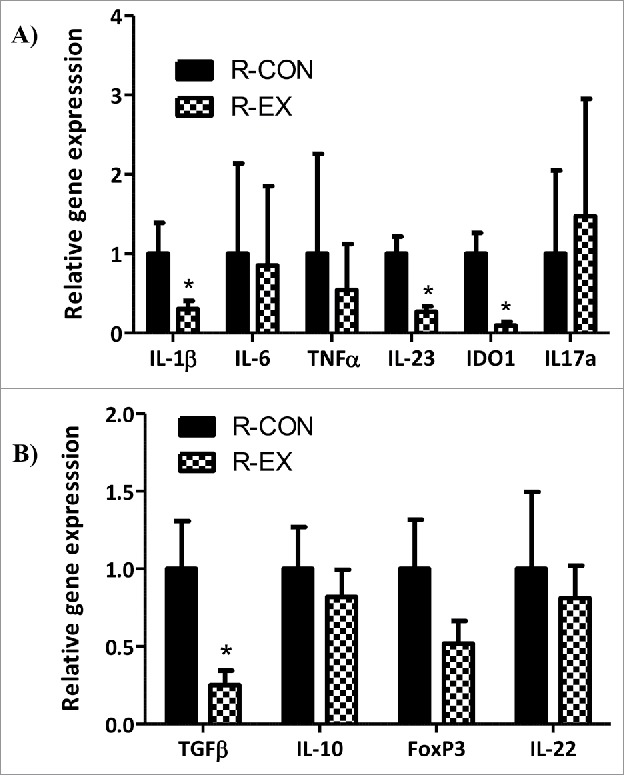
Distal colon cytokine gene expression in recipient mice after colonization (Experiment 1). (A) Pro-inflammatory and, (B) Anti-inflammatory cytokines after 5 weeks of colonization (n = 15-16/group). *denotes significant difference, ns = not significantly different at p < 0.05.
In agreement with the reduction in inflammatory transcripts, there was an attenuated inflammatory cell infiltration in R-EX mice compared to R-CON mice as indicated by histology (Fig. 4Ai-ii and 4Bi). In addition, we observed reduced goblet cell mucus depletion in the colons of R-EX versus R-CON (Fig. 4Ai-ii and 4Bii) and an attenuated overall histology score (Fig. 4Ai-ii and 4Biii). No statistically significant differences were observed between groups for destruction of architecture or crypt abscesses (data not shown). No differences were observed in colon length, an indicator of inflammation, between recipient groups (R-CON 74 ± 1.4 mm vs. R-EX 73.9 ± 0.8 mm, respectively; p = 0.91).
Figure 4.
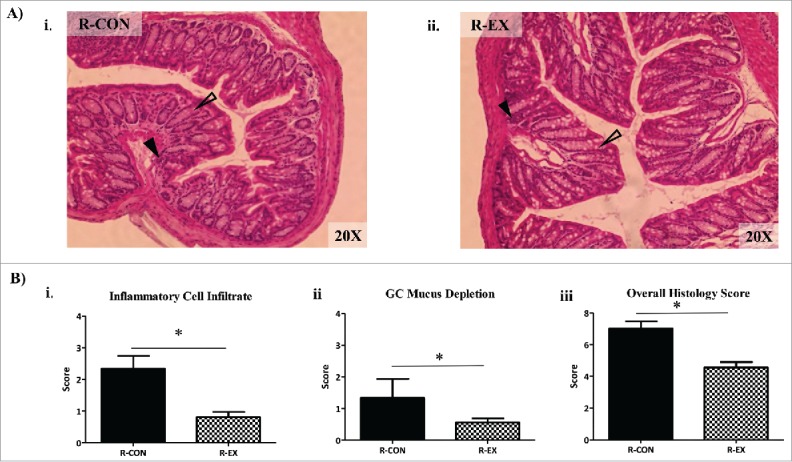
Distal colon histology of recipient mice after colonization (Experiment 1). (A) Representative hemotoxylin and eosin (H&E) stains (20x) of distal colons from i) R-CON and ii) R-EX after 5 weeks of colonization. Open arrows indicate areas of goblet cell (GC) depletion in R-CON vs R-EX mice. Closed arrows indicate areas of inflammatory cell infiltrate. (B) Histological scoring of distal colons from recipient mice after 5 weeks of colonization: i) inflammatory cell infiltration (range 0–4), ii) GC mucus depletion (range 0–4) and, iii) overall histology score (range 0–16); includes aggregate scores of inflammatory cell infiltration, GC mucus depletion, destruction of architecture and crypt abscesses (ranges 0–4; not significant, data not shown). *denotes significant difference at p < 0.05.
Gut microbiota transfer from exercise-trained mice leads to higher body mass in recipient mice and is related to gut SCFA profiles
We observed that recipient mice colonized with the ‘exercised gut microbiota’ were heavier after 5 weeks of colonization than mice colonized with ‘control gut-microbiota’ (R-EX: 24.1 ± 0.63 g vs. R-CON: 22.2 ± 0.36 g, p < 0.05. Fig. 5A.). Of note, absolute body weight differences among recipient mice as result of the ‘exercised gut microbiota colonization’ were observed primarily in male, but not female, mice before and after the DSS treatment (Sex x Exercise-transfer interaction p = 0.10; Table S3). To follow up on possible mechanisms underlying this exercise effect, we determined whether microbial-derived SCFAs, which have been shown to be increased by exercise in conventionally-raised mice and are known contributors to host energy harvest,31 differed in concentration between recipient groups. While there were no statistical differences in concentration of the three most abundant SCFAs, acetate (C2), propionate (C3) and butyrate (C4) (Fig. 5B & C, propionate not shown), the ratio of [C4] to [C2] was significantly elevated in R-EX mice compared to R-CON mice (p < 0.01; Fig. 5D). This ratio is important at it has been previously shown to be elevated in low-dose, antibiotic fed mice that also displayed increased energy harvest and altered body composition profiles compared to their control counterparts.12 In this study, the [C4]:[C2] of recipient mice was strongly associated to host body weight both across and within groups (Pearson r = 0.74; p < 0.001; Fig. 5E). Next, we quantified the genetic potential of the gut microbiota to produce C4 from C2, by measuring the relative abundance of the butyryl coa: acetate CoA transferase (BCoAT) gene; a gene specific to butyrate producers within the gut microbiota and integral in the conversion of acetate to butyrate.32 Interestingly, and in accordance with higher [C4]: [C2] in the cecal contents, the BCoAT gene trended higher in the feces of R-EX compared to the R-CON mice (Fig. 5F, p = 0.09). BCoAT gene abundance was also correlated to cecal content [C4]: [C2] (r = 0.49, p < 0.05; data not shown). Figure 5G is a schematic representing the conversion of acetate and butyryl-CoA to butyrate and acetyl-CoA through the enzyme butyryl:CoA acetate CoA transferase.
Figure 5.
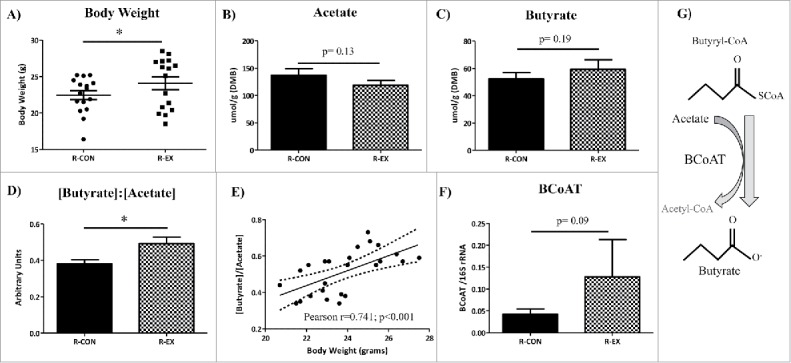
Body weight and cecal SCFA profiles of recipient mice after colonization (Experiment 1). (A) Body weight after 5 weeks of colonization in recipient mice.(B-D) Short chain fatty acid concentrations (μmole/gram) of cecal content dry matter. (E) Relative gene abundance (per 16S rRNA) of the butyrate producing enzyme butyryl CoA: acetate CoA transferase (BCoAT) in the feces of recipient mice.(F) Ratio of [Butyrate]: [Acetate] strongly correlates to body weight. (G) Pathway schematic of acetate to butyrate conversion through BCoAT. *p < 0.05
Experiment 2. Effect of transplant on DSS-induced colitis
Gut microbiota transfer from exercise-trained mice leads to altered gut microbiota composition in recipient mice after colonization and a DSS-colitis insult
In a second experimental cohort of GF mice, we tested whether an ‘exercised-gut microbiota’ colonization prior to treatment with DSS could reduce the clinical symptomology and the inflammatory response to the colitis insult (Recipient-Control-DSS [R-CON-DSS], n = 16 versus Recipient-Exercise-DSS [R-EX-DSS], n = 16). We first confirmed that differences in the community structure (β-diversity) of the gut microbiome observed in the donor mice persisted in the recipient mice after a 28 d colonization period followed by a colitis insult which consisted of five days of DSS (2%) followed by a three-day H2O treatment. Similar to what was observed in Experiment 1, there was a clear disparity between treatments in recipient mice after colonization and DSS treatment (Fig. 6 Unweighted Unifrac and Fig. S1C Weighted UniFrac). Expectedly, the gut microbiota communities after colonization plus DSS treatment were much different than that observed in Experiment 1 with colonization alone, likely due to the large and well-documented effect of DSS treatment on the gut microbiome.33,34 Genus level differences in the gut microbiota of recipient-DSS mouse groups are reported in Table S4.
Figure 6.
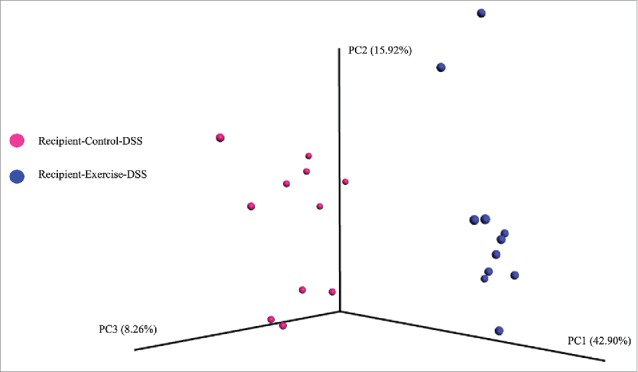
Gut microbiome β-diversity in recipient mice after colonization and an acute, DSS-colitis insult (Experiment 2). Transfer of cecal contents from EX or CON donor groups led to differences in community structure of the fecal microbiota (Unweighted UniFrac) in recipient, gnotobiotic mice after acute DSS colitis insult; dark blue circles = Recipient-Exercise-DSS, pink circles = Recipient-Control-DSS. PERMANOVA, p < 0.01.
Gut microbiota transfer from exercise-trained mice leads to attenuated clinical responses to acute DSS-Colitis in recipient mice
Body weights (BW) and clinical symptomology, including diarrhea/rectal bleeding were observed throughout DSS-colitis, and colon length was measured after the acute colitis insult. In response to DSS, both groups (R-CON-DSS vs. R-EX-DSS) lost significant body weight as measured by percent change (% Δ) versus baseline (time main effect, p < 0.001), verifying that the DSS was effective in inducing colitis symptomology (Fig. 7A). Interestingly, R-EX-DSS mice displayed a trend for attenuated BW loss (% Δ) as compared to R-CON-DSS mice (time x activity p = 0.09, Fig. 7A), indicating some level of protection from the colitis insult irrespective of observed differences in BW. Also, similar to what was observed in Experiment 1, absolute BWs were significantly higher in R-EX-DSS group after four weeks of colonization and throughout the eight days of colitis insult (Fig. S2A & Table S5). Importantly, such differences were not likely a result of altered DSS or caloric intake, as both groups consumed similar amounts of fluid and food throughout the treatment periods (Fig. S2B & C).
Figure 7.
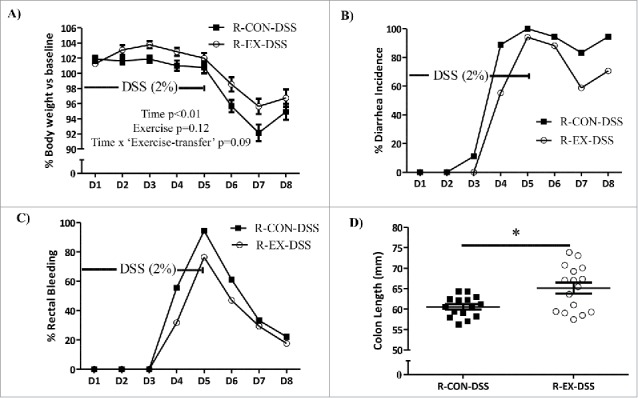
DSS-induced clinical symptoms in recipient mice (Experiment 2). (A) Percent (%) body weight change, (B) percent (%) diarrhea incidence, and (C) percent (%) rectal bleeding incidence of recipient mice during eight days of acute colitis insult. (D) Colon lengths of recipient mice after sacrifice on Day 8 of colitis insult. *denotes significant difference at p < 0.05.
Despite numerical differences, neither the incidence of diarrhea nor the incidence of rectal bleeding was significantly different between physical activity groups throughout the colitis insult (Fig. 7B & C). However, R-EX-DSS mice exhibited longer colons compared to the R-CON-DSS mice at sacrifice on Day 8 (Fig. 7D; p < 0.05), indicating a reduced inflammatory burden in response to colitis as a result of the ‘exercised-gut microbiota’ transplant.
Gut microbiota transfer from exercise-trained mice leads to augmented anti-inflammatory gene expression and attenuated histological parameters in colons of recipient mice after DSS-colitis
To further assess the response to DSS in recipient mice, we analyzed the expression of genes involved in the inflammatory and anti-inflammatory/regenerative response to colitis. Contrary to our hypothesis and the observed attenuation of colon shortening, recipient-DSS groups exhibited similar expression of the pro-inflammatory cytokines IL-1β, IL-6, and TNF-α in the distal colon after the colitis insult on day 8 (Fig. 8A). However, R-EX-DSS mice exhibited augmented colonic expression of cytokines and transcription factors known to be involved in tissue regenerative responses to colitis, including TGFβ, FoxP3, and IL-22, compared to R-CON-DSS mice (Fig. 8B, p < 0.05).
Figure 8.
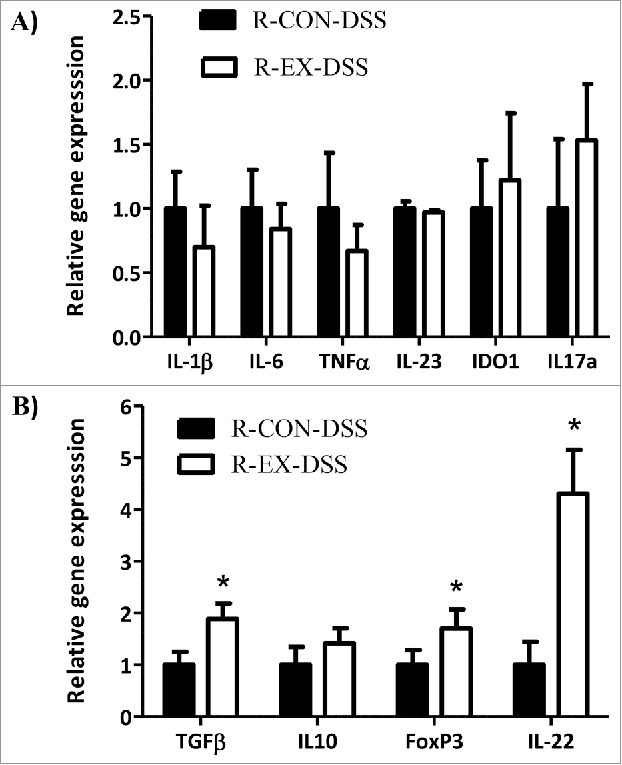
Distal colon gene expression in DSS-treated recipient mice (Experiment 2). Colonic gene expression of (A) pro-inflammatory and (B) anti-inflammatory cytokines in recipient mice that received 5 days of DSS (2%) followed by 3 days of H2O after a 4 week colonization period. *denotes significant difference at p < 0.05.
To examine morphological changes in the gut after DSS-colitis, distal colon histology was quantified by a trained, blinded investigator based on four criteria, as described above. In the colons of R-EX-DSS mice we observed a trend for a reduction in inflammatory cell infiltration (p = 0.11; Fig. 9Ai-ii and 9Bi) along with a significant attenuation of goblet cell mucus depletion versus colons from R-CON-DSS (p < 0.05; Fig. 9Ai-ii and 9Bii), ultimately contributing to an lower overall histological rating compared to colons of R-CON-DSS mice (p < 0.05; Fig. 9Ai-ii and 9Biii).
Figure 9.
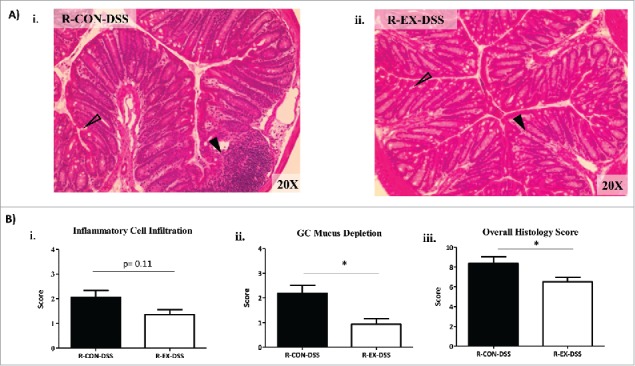
Distal colon histology in DSS-treated recipient mice (Experiment 2). (A) Representative H&E stains of distal colons from i) R-CON-DSS (20x) and ii) R-EX-DSS (20x) after 4 weeks of colonization followed by DSS colitis insult. Open arrows indicate areas of goblet cell depletion. Closed arrows indicate areas of inflammatory cell infiltrate. (B) Histological scoring of distal colons: i) inflammatory cell infiltration (range 0–4), ii) GC mucus depletion (range 0–4), and iii) overall histology score (range 0–16); includes aggregate scores of inflammatory cell infiltration, GC mucus depletion, destruction of architecture and crypt abscesses (ranges 0–4; not significant, data not shown). *denotes significant difference at p < 0.05.
Discussion
In this study, we show that exercise training-induced changes in the gut microbiota of donor mice can persist and cause biologically relevant perturbations in recipient, gnotobiotic mice after a gut microbiota transplant and colonization. This is evidenced by distinct inflammatory responses to microbial colonization, altered luminal concentrations of microbial-derived metabolites, and attenuated responses to an acute colitis insult in recipient mice that received a gut microbiota transplant from donor mice that were exercise trained versus those mice that remained without access to a running wheel.
Gut microbiota composition profiles in recipient mice were highly dependent on the physical activity status of their respective donor origins. We speculate that such disparity may be shaped by a physiologically relevant consortium of bacteria that were observed to be sensitive to exercise training. Of these exercise-altered taxa, Akkermansia spp. is a mucus colonizing bacteria shown to reduce colonic inflammation and protect against diet-induced metabolic syndrome.35 In addition, the Lachnospiraceae family contains bacterial species with well-documented butyrate producing capabilities.36 Lastly, species from Ruminococcus spp. appear to be involved in early life growth, as cultured strains were able to rescue a blunted growth phenotype in GF mice.37
The mucosal tissue of the gut responds to a microbiota colonization with a complex array of pro- and anti-inflammatory processes.38 Here, we observed R-EX mice to have attenuated colonic expression of IL-1β and IL-23, two cytokines involved in the innate immune response to commensal and pathogenic bacteria within the gut.14,39 IL-1β, in particular, is expressed in a multitude of innate immune cells and its release is sensitive to cell surface binding of bacterial associated components, including lipopolysaccharide (LPS) of gram-negative bacteria, to host toll-like receptors (TLRs).40 R-EX mice also exhibited lower colonic expression of IDO1; a highly conserved enzyme that mediates the conversion of amino acid tryptophan towards the bioactive metabolite kynurenine.41 Importantly, IDO1 expression is highly regulated by IL-1β, reflects mucosal inflammation, and is responsive to commensal load on the gut epithelial layer.42,43 In the current study, we postulate that the reduced expression of these pro-inflammatory factors alongside the attenuation of immune cell infiltration observed through histology, indicate a potential reduction in overall bacterial adherence to the mucus/epithelial layer within the gut of R-EX mice.
SCFAs have been shown to be upregulated by exercise training in the murine gut6 and contribute to numerous metabolic and inflammatory processes within the host, including energy harvest and reduced gut inflammation.16,44 Here, we show that R-EX mice that displayed higher body weights after the colonization period also displayed a significantly higher ratio of butyrate [C4] to acetate [C2] compared to R-CON mice. Intriguingly, this ratio was strongly correlated to body weight across and within both recipient groups, providing evidence that [C4]:[C2] may be an important indicator of metabolic potential of the gut microbiota.
Probing further, we found that the abundance of butyryl CoA: Acetate CoA transferase (BCoAT), a prevalent and obligatory feature of a large portion of butyrate-producing bacteria and a gene necessary for the conversion of C2 to C4, trended higher in R-EX mice. These data are similar to previous data from Cho et al. (2012), who observed higher cecal [C4]:[C2] and BCoAT levels in mice that received low-dose antibiotics, which was reported in accordance with differences in body composition between control and antibiotic-exposed animals.12
These data support the hypothesis that augmented capacity of the gut microbiota to produce SCFA may contribute to enhanced energy harvest within a physically active host. Such an adaptation, we propose, may be evolutionarily advantageous during periods of high energy expenditure and caloric deficit, such as during foraging periods of rodents. Future studies will be needed to investigate the mechanisms behind such phenomena.
In a second cohort of colonized mice, we demonstrated that exercise-induced gut microbiota changes modulated the host response during and after an acute, dextran-sodium-sulfate (DSS)-induced colitis insult. Most notably, R-EX-DSS mice exhibited augmented colonic expression of anti-inflammatory cytokines and transcription factors involved in the regenerative response to acute DSS-colitis. TGF-β, in particular, is required for protection against the two models of T-cell mediated colitis,45,46 and directly mediates differentiation and responsiveness of subsets of suppressive myeloid and lymphoid cells.47 Of these cell types regulated by TGF-β, regulatory T-cells constitutively express the anti-inflammatory cytokine IL-10, and are required for maintaining epithelial barrier and regenerative response to colitis.48 IL-22, meanwhile, is a cytokine released primarily from innate lymphoid cells (ILCs) that has been shown to mitigate colitis symptomology in mice and humans.49,50 In a mouse model, IL-22 gene delivery led to rapid amelioration of local intestinal inflammation,51 while IL-22 binding protein administration (which blocks action of IL-22) led to suppressed goblet cell restitution during the recovery phase in mice that received DSS.52 Here, we demonstrate that alongside a higher expression of IL-22, mice that received the ‘exercised-gut microbiota’ had higher abundance of mucus-containing goblet cells at day 8 of the colitis insult than mice that received the ‘control-gut microbiota’. This is important, as DSS-colitis is known to cause mucus layer degradation and goblet cell depletion, ultimately leading to exposure of the epithelial layer to both commensal and pathogenic microbes.53 Therefore, enhanced goblet cell mucus repletion observed in R-EX-DSS mice may be an important feature in maintaining a barrier between commensal microbes and host gut epithelial cells, ultimately limiting bacterial adherence and ameliorating the inflammatory insult to DSS. Together, we surmise that the differences in cytokine expression profiles and measures of histology between treatment groups indicate augmented anti-inflammatory and regenerative responses to DSS-colitis as a result of the ‘exercised-gut microbiota’ colonization.
The exercise-induced changes in the microbiome and host phenotype reported here coincide with data reported in previous studies examining the effects of exercise on the gut microbiome in conventionally-raised mice. For instance, Matsumoto et al. provided evidence that voluntary exercise increased the level of cecal butyrate concentrations6 while Mika et al. showed that exercise training in juvenile rats increased butyrate-producing bacterial genera, which was associated with enhanced early life lean mass gains.4 Moreover, others have reported exercise-induced changes in the gut microbiome were associated reduced inflammation and altered responses to inflammatory stimuli, including a high-fat diet.5,8,9 At the taxa level, comparison of our data to previous studies is difficult as species and strain of animal, vendor origin, and sequencing technologies differed widely between experiments. Nevertheless, the observation in this study that exercise alters gut microbiome communities, increases butyrate-producing capabilities, and reduces host colon inflammation confirms reports by others and should be investigated further to understand the mechanisms behind such a response.
There are some limitations to this study. For instance, despite the fact that a large percentage of taxa effectively transferred from donor to recipient mice (Table S2), the compositional profile of the gut microbiota (as measured by relative taxa abundance) shifted as a result of the transfer and colonization. This sensitivity of the gut microbiota to colonization is also relevant when investigating host responses, as the mucus layer and immune system are in constant flux during early time periods of colonization.14 In addition, we also observed significant effects of sex in recipient mice of both experiments. Beyond the obvious differences in body weight, we observed that recipient male mice were significantly more inflamed than female mice in both experiments, as evidenced by higher expression of inflammatory cytokines in the distal colon. We are unsure of the mechanisms behind these observations. Nevertheless, we observed no statistical sex x ‘exercise-transfer’ interactions amongst major outcomes studied. Considering this, we deemed it appropriate to collapse groups based on sex before outcome analysis. Future studies should examine both the effects of sex and the temporal dynamics of gut microbiota colonization to confirm the results presented in this study.
Conclusions
Overall, this study provides seminal evidence that exercise training can alter the gut microbiota in a fashion that directly alters host response during states of health and disease. Of primary significance, gut microbiota modifications induced by exercise appear be important for inhibiting gut inflammation, altering gut morphology and potentially increasing energy harvest within colonized, previously germ-free host. Since host response to microbiota colonization in gnotobiotic mice is important for development for gut architecture and the immune system, we surmise that exercise in conventionally-raised animals may be a particularly strong modulator of gut microbiota-host interactions during periods where these systems are most malleable, including during states of disease. To this end, we observed an enhanced regenerative response to a colitis insult in recipient, gnotobiotic mice that received a gut microbiota from exercised donors. In conclusion, exercise-induced modifications in the gut microbiota can directly modify host-microbial interactions with potentially beneficial outcomes for the host. Future studies aimed at understanding the mechanisms responsible for these effects may highlight potential targets to improve health.
Methods
Experimental design
Figure 1 depicts the research design of two independent experiments intended to: 1) determine the effects of gut microbiota transplantation from exercise-trained (EX) or untrained (CON) donor (D) mice on the host response to colonization in recipient, previously GF mice (Experiment 1) and, 2) determine the effects of gut microbiota transplantation from EX or CON donor mice on the response to acute experimental colitis in previously GF recipient mice (Experiment 2). All experiments were completed in accordance with the guidelines of the Institutional Animal Care and Use Committees of the University of Illinois at Urbana-Champaign and the Mayo Clinic (Rochester, MN).
Donor mice, exercise protocols and tissue collection
Adult male, 6-week old, C57Bl/6N mice were purchased from Taconic Farms (Germantown, NY) and were singly housed in an AAALAC-accredited facility at the University of Illinois at Urbana-Champaign (UIUC). All mice were maintained on Lab Diet 5K67 and fed ad libitum (St. Louis, MO). The diet consisted of 22.37 % protein, 15.6 % fat, 61.98 % carbohydrate and 4.4 % of fiber (crude). After a one-week acclimation, mice were randomly assigned to two activity groups: exercise (Donor-Exercise, D-EX, n = 10), or sedentary-control (Donor-Control, D-CON, n = 10). The D-EX mice were housed in cages with free access to telemetered running wheels (Respironics Bend, OR) while D-CON remained without access to a wheel for 42 days. D-EX ran 5.1 ± 0.7 km.day−1. One mouse died unexpectedly in within the first week of beginning the experiment leaving n = 9 within the D-CON group. After 42 days, mice were euthanized via rapid CO2 asphyxiation followed by cervical dislocation. Ceca, colon contents and fecal samples were snap frozen by liquid N2 and stored at −80°C.
Gut microbiota transplant and colonization
Cecal contents from donor mice were pooled together by activity group (i.e. control or exercise). Contents were diluted in pre-reduced phosphate buffered saline (PBS) (1:2 volume [contents]/volume [PBS]) and orally gavaged into two cohorts (Exp1 or Exp2) of equally distributed GF male and female, 4–5 week old C57Bl6/N mice (n = 15-16/activity group; n = 9 female/group and n = 6-7 males/group). All recipient mice were maintained in gnotobiotic isolators at the Mayo Clinic (Rochester, MN) prior to and after the gut microbiota transplant. The germ-free status of all mice prior to transplant was confirmed by aerobic and anaerobic culture, gram stain and PCR using universal 16S rRNA gene primers. For Experiment 1 (colonization effect alone), mice were sacrificed 35 days after initial colonization (Recipient-Control [R-CON] n = 15; n = 9 female, n = 6 male); Recipient-Exercise [R-EX] n = 16; (n = 10 female, n = 6 male) (Fig. 1). For Experiment 2, a second cohort of gnotobiotic mice were colonized for 28 days and then subjected to an acute five-day dextran-sodium sulfate colitis (DSS) insult followed by three days of regular drinking water (Recipient-Control-DSS [R-CON-DSS] n = 16; n = 8 male, n = 8 female and Recipient-Exercise-DSS [R-EX-DSS]; n = 16; n = 8 male, n = 8 female) (Fig. 1). All recipient mice were co-housed by sex (4-5 per cage), remained without access to a running wheel during the colonization (Exp1 and Exp2) and DSS treatment (Exp2). All recipient mice were maintained on an autoclavable Lab Diet 5K67 standard chow (same as donor mice) and fed ad libitum throughout the experimental protocols.
Dextran sodium sulfate-induced colitis
Regular drinking water was replaced by 2% DSS (w/v, MP Biomedical, 36,000-50,000 MW) and administered for 5 days, followed by a return to regular drinking water for 3 days before sacrifice. Daily measurements of body weight and food and fluid intake were made during and after DSS administration. In addition, mouse feces were evaluated for consistency/diarrhea (e.g. formed pellet vs. semi-formed/soft), and mice were observed daily for the presence of rectal bleeding.
Tissue collection
All recipient mice were euthanized via rapid CO2 asphyxiation followed by cervical dislocation. The colon was removed and colon length was assessed using digital calipers to the nearest millimeter. Small distal colon tissue sections (∼ 2 mm) were excised and then placed in Carnoy's fixative (60% ethanol, 20% chloroform and 10% acetic acid) for 4 hours and then placed in 100% ethanol until tissue embedding. The remaining colon tissue and feces were excised and stored at −80°C for gene expression and sequencing analysis, respectively. For SCFA analysis, whole ceca were excised and aliquots of fresh of cecal contents were weighed for dry matter (DM) or placed in acid (see below for complete SCFA protocol).
Bacterial DNA isolation and 16S rRNA gene sequencing
Fecal bacterial DNA was extracted from all donor (n = 9-10/group) and a subset of recipient mice (n = 11/group) by using the PowerLyzer PowerSoil DNA Isolation Kit (MOBIO Laboratories, Inc.). After extraction and DNA quality assurance through gel electrophoresis, library construction was completed using a Fluidigm Access Array system in the Functional Genomics Unit of the Roy J. Carver Biotechnology Center at the University of Illinois at Urbana. After library construction, 300 bp of the V4 region of the 16SrRNA gene were amplified according to Caporaso et al.54 and sequenced at the WM Keck Center for Biotechnology at the University of Illinois using an Illumina MiSeq2000 with the use of V3 reagents (Illumina Inc.). High-quality (> 25) sequence data (FASTQ) were analyzed with QIIME 1.8.0.54 For quality control, sequences were depleted of barcodes and primers, and short sequences (< 237 bp), sequences with ambiguous base calls, and sequences with homopolymer runs exceeding 6 bp were removed. After removal of singletons, OTUs were then classified using closed reference picking with the Ribosomal Database Project (RDP) at 97% similarity. Weighted and unweighted UniFrac distances were computed at an even sampling depth of 11,058 sequences per sample, a value chosen based off of alpha-diversity rarefaction curves (data not shown).
Distal colon gene expression
Quantitative real-time PCR was used to assess differences in gene expression in colon tissues of recipient mice (n = 15/16 per group). RNA from ∼20–30 mg of homogenized colon tissue was extracted using a modified RNeasy mini kit protocol (Qiagen) and stored at −80°C. Complementary DNA was synthesized by reverse transcription reaction using commercial kits [Life Technologies Co. (Applied Biosystems)] and random primers. The RNA sample concentrations and purities were analyzed by a spectrophotometer. Next, qRT-PCR was completed on with the 7900HT Fast Real-Time PCR system (SDS Enterprise Database; Life Technologies; Carlsbad, CA) with use of Taqman® master mix (Applied Biosystems, Carlsbad, CA). Target genes were expressed relative to Gapdh (Mm99999915_g1) using the ΔΔ-Ct method as fold change, with the R-CON or R-CON-DSS serving as calibrator control for experiments 1 and 2, respectively. All primers used for colon gene expression are referenced in Table S6.
Distal colon histology
Carnoy's fixed distal colon samples were paraffin embedded, and 3-μm sections were stained with hematoxylin and eosin (H&E). Histology was scored by a trained, blinded observer on a modified version of a published scoring55 system that considers four criteria: inflammatory cell infiltration (0–4), goblet cell mucus depletion (0–4); destruction of architecture (0–4) and crypt abscesses (0–4). Overall histology score was quantified using an aggregate of these scores.
Short chain fatty acid analysis
SCFAs were analyzed as described previously by Panasevich et al (2015).56 Briefly, cecal contents were weighed and acidified in 6.25% meta-phosphoric acid solution and stored at −20°C until analysis. Cecal SCFA concentrations were determined by gas chromatography by using a gas chromatograph (Hewlett-Packard 5890A Series II) and a glass column (180 cm × 4 mm i.d.), packed with 10% SP-1200/1% H3PO4 on 80/100 + mesh Chromosorb WAW (Supelco, Inc.). Nitrogen was the carrier gas with a flow rate of 75 mL/min. Oven, detector, and injector temperatures were 125°, 175°, and 180°C, respectively. Acetic, n-butyric, and propionic acid solutions (Sigma-Aldrich) were used as standards.
Butyryl CoA:Acetate CoA transferase (BcoAT) gene quantification
Isolated bacterial DNA (as described above) was assessed for the relative abundance of butyryl CoA: acetate CoA transferase gene content by qPCR as described by Louis and Flint (2007).32 Briefly, real-time PCR experiments were performed with SYBR Green Master Mix (Applied Biosystems) as a probe in a total volume of 20ul. BCoAT gene quantification was analyzed in parallel with the 16S rRNA (total bacteria). The amplification cycle used was 1 cycle of 95°C for 3 min; 40 cycles of 95°C, 53°C, and 72°C for 30 s each with data acquisition at 72°C. BCoAT and Universal 16S rRNA primers are referenced in Table S6.
Statistical analysis
Gut microbiome β-diversity was analyzed based on unweighted UniFrac distance metrics generated from QIIME, visualized using EMPeror and analyzed by permutation multivariate analysis of variance (PERMANOVA).31,45 Taxa level comparisons between treatment groups were analyzed by independent Student's t-test (or non-parametric Mann-Whitney U test) and corrected by false discovery rate (FDR). Comparisons of inflammatory gene expression, SCFA concentrations and BCoAT gene abundance between groups were analyzed by independent Student's t-tests. For Exp2, the effects of DSS on body weight loss was analyzed by a 2-way mixed model analysis of variance (ANOVA) with time (8 days) and activity group (R-EX-DSS and R-EX-DSS) as independent variables. Incidence rates of diarrhea and rectal bleeding were analyzed by Pearson Chi-Square on each day after the beginning of DSS administration. Correlations between bacterial taxa, SCFA concentrations and host phenotype measures were analyzed by either Pearson r or Spearman rank correlations depending on normality. Only one sex x ‘exercise-transfer’ interaction was observed for the major outcome variables (i.e., body weight) in either experiment. Therefore, analysis of major outcome variables in recipient mice were collapsed by sex. The effects of sex on outcome variables are reported for reference in Tables S3 and S5, as well as Figure S3. Significance was set a priori at p < 0.05. In cases of multiple comparisons, FDR p < 0.05 was deemed significant.
Ethics approval and consent to participate
All experiments were completed in accordance with the guidelines of the Institutional Animal Care and Use Committees of the University of Illinois and the Mayo Clinic (Rochester, MN).
Availability of data and materials
The remaining datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Supplementary Material
Abbreviations
- BCoAT
Butyryl-CoA:Acetate-CoA tranferase
- C2
Acetate
- C4
Butyrate
- DSS
Dextran sodium sulfate
- GC
goblet cell
- GF
germ-free
- GM
gut microbiota
- IDO
Indolamine 2,3 dioxygenase
- IL
Interleukin
- PCoA
principle coordinates analysis
- TGF
Transforming growth factor
- SCFAs
Short chain fatty acids
- TNF
Tumor necrosis factor
Disclosure of potential conflicts of interest
The authors declare that they have no competing interests
Acknowledgments
The authors would like to thank Mark Band, Alvaro Hernandez and Chris Wright at the Roy J. Carver Biotechnology Center at the University of Illinois for their assistance in 16S rRNA sequencing.
Funding
This work supported by a grant from the Mayo Clinic-University of Illinois Alliance for Technology-based Healthcare and National Institute of Diabetes and Digestive Kidney Diseases Grant K08 DK-100638 (to PCK).
References
- [1].Walsh NP, Gleeson M, Shephard RJ, Gleeson M, Woods JA, Bishop NC, Fleshner M, Green C, Pedersen BK, Hoffman-Goetz L, et al.. Position statement. Part one: Immune function and exercise. Exerc Immunol Rev. 2011;17:6-63. PMID:21446352 [PubMed] [Google Scholar]
- [2].Walsh NP, Gleeson M, Pyne DB, Nieman DC, Dhabhar FS, Shephard RJ, Oliver SJ, Bermon S, Kajeniene A. Position statement. Part two: Maintaining immune health. Exerc Immunol Rev. 2011;17:64-103. PMID:21446353 [PubMed] [Google Scholar]
- [3].Allen JM, Berg Miller ME, Pence BD, Whitlock K, Nehra V, Gaskins HR, White BA, Fryer JD, Woods JA. Voluntary and forced exercise differentially alters the gut microbiome in C57BL/6J mice. J Appl Physiol (1985). 2015;118:1059-66. doi: 10.1152/japplphysiol.01077.2014. PMID:25678701 [DOI] [PubMed] [Google Scholar]
- [4].Mika A, Van Treuren W Gonzalez A, Herrera JJ, Knight R, Fleshner M. Exercise is More Effective at Altering Gut Microbial Composition and Producing Stable Changes in Lean Mass in Juvenile versus Adult Male F344 Rats. PLoS One. 2015;10:e0125889. doi: 10.1371/journal.pone.0125889. PMID:26016739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Choi JJ, Eum SY, Rampersaud E, Daunert S, Abreu MT, Toborek M. Exercise attenuates PCB-induced changes in the mouse gut microbiome. Environ Health Perspect. 2013;121:725-30. doi: 10.1289/ehp.1306534. PMID:23632211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Matsumoto M, Inoue R, Tsukahara T, Ushida K, Chiji H, Matsubara N, Hara H. Voluntary running exercise alters microbiota composition and increases n-butyrate concentration in the rat cecum. Biosci Biotechnol Biochem. 2008;72:572-6. doi: 10.1271/bbb.70474. PMID:18256465 [DOI] [PubMed] [Google Scholar]
- [7].Queipo-Ortuno MI, Seoane LM, Murri M, Pardo M, Gomez-Zumaquero JM, Cardona F, Casanueva F, Tinahones FJ. Gut Microbiota Composition in Male Rat Models under Different Nutritional Status and Physical Activity and Its Association with Serum Leptin and Ghrelin Levels. PloS one. 2013;8:e65465. doi: 10.1371/journal.pone.0065465. PMID:23724144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Kang SS, Jeraldo PR, Kurti A, Miller ME, Cook MD, Whitlock K, Goldenfeld N, Woods JA, White BA, Chia N, et al.. Diet and exercise orthogonally alter the gut microbiome and reveal independent associations with anxiety and cognition. Mol Neurodegener. 2014;9:36. doi: 10.1186/1750-1326-9-36. PMID:25217888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Evans CC, Lepard KJ, Kwak JW, Stancukas MC, Laskowski S, Dougherty J, Moulton L, Glawe A, Wang Y, Leone V, et al.. Exercise prevents weight gain and alters the gut microbiota in a mouse model of high fat diet-induced obesity. PLoS One. 2014;9:e92193. doi: 10.1371/journal.pone.0092193. PMID:24670791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Lambert JE, Myslicki JP, Bomhof MR, Belke DD, Shearer J, Reimer RA. Exercise training modifies gut microbiota in normal and diabetic mice. Appl Physiol Nutr Metab. 2015;40:749-52. doi: 10.1139/apnm-2014-0452. PMID:25962839 [DOI] [PubMed] [Google Scholar]
- [11].Turnbaugh PJ, Backhed F, Fulton L, Gordon JI. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe. 2008;3:213-23. doi: 10.1016/j.chom.2008.02.015. PMID:18407065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Cho I, Yamanishi S, Cox L, Methé BA, Zavadil J, Li K, Gao Z, Mahana D, Raju K, Teitler I, et al.. Antibiotics in early life alter the murine colonic microbiome and adiposity. Nature. 2012;488:621-6. doi: 10.1038/nature11400. PMID:22914093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Rokhsefat S, Lin A, Comelli EM. Mucin-Microbiota Interaction During Postnatal Maturation of the Intestinal Ecosystem: Clinical Implications. Dig Dis Sci. 2016. doi: 10.1007/s10620-016-4032-6. PMID:26792279 [DOI] [PubMed] [Google Scholar]
- [14].Earle KA, Billings G, Sigal M, Lichtman JS, Hansson GC, Elias JE, Amieva MR, Huang KC, Sonnenburg JL. Quantitative Imaging of Gut Microbiota Spatial Organization. Cell host & microbe. 2015;18:478-88. doi: 10.1016/j.chom.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Willemsen LE, Koetsier MA, van Deventer SJ, van Tol EA. Short chain fatty acids stimulate epithelial mucin 2 expression through differential effects on prostaglandin E(1) and E(2) production by intestinal myofibroblasts. Gut. 2003;52:1442-7. doi: 10.1136/gut.52.10.1442. PMID:12970137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027-31. doi: 10.1038/nature05414. PMID:17183312 [DOI] [PubMed] [Google Scholar]
- [17].Tolhurst G, Heffron H, Lam YS, Parker HE, Habib AM, Diakogiannaki E, Cameron J, Grosse J, Reimann F, Gribble FM. Short-chain fatty acids stimulate glucagon-like peptide-1 secretion via the G-protein-coupled receptor FFAR2. Diabetes. 2012;61:364-71. doi: 10.2337/db11-1019. PMID:22190648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, deRoos P, Liu H, Cross JR, Pfeffer K, Coffer PJ, et al.. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504:451-5. doi: 10.1038/nature12726. PMID:24226773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Kaplan GG, Ng SC. Understanding and Preventing the Global Increase of Inflammatory Bowel Disease. Gastroenterology. 2017;152(2):313-21. [DOI] [PubMed] [Google Scholar]
- [20].Kanauchi O, Mitsuyama K, Andoh A. The therapeutic impact of manipulating microbiota in inflammatory bowel disease. Curr Pharm Des. 2009;15:2074-86. doi: 10.2174/138161209788489195. PMID:19519445 [DOI] [PubMed] [Google Scholar]
- [21].Cario E. Microbiota and innate immunity in intestinal inflammation and neoplasia. Curr Opin Gastroenterol. 2013;29:85-91. doi: 10.1097/MOG.0b013e32835a670e. PMID:23207600 [DOI] [PubMed] [Google Scholar]
- [22].Moehle C, Ackermann N, Langmann T, Aslanidis C, Kel A, Kel-Margoulis O, Schmitz-Madry A, Zahn A, Stremmel W, Schmitz G. Aberrant intestinal expression and allelic variants of mucin genes associated with inflammatory bowel disease. J Mol Med (Berl). 2006;84:1055-66. doi: 10.1007/s00109-006-0100-2. PMID:17058067 [DOI] [PubMed] [Google Scholar]
- [23].McGuckin MA, Eri R, Simms LA, Florin TH, Radford-Smith G. Intestinal barrier dysfunction in inflammatory bowel diseases. Inflamm Bowel Dis. 2009;15:100-13. doi: 10.1002/ibd.20539. PMID:18623167 [DOI] [PubMed] [Google Scholar]
- [24].Cleynen I, Vermeire S. Paradoxical inflammation induced by anti-TNF agents in patients with IBD. Nat Rev Gastroenterol Hepatol. 2012; 9: 496-503. doi: 10.1038/nrgastro.2012.125. PMID:22751454 [DOI] [PubMed] [Google Scholar]
- [25].George LA, Gadani A, Cross RK, Jambaulikar G, Ghazi LJ. Psoriasiform Skin Lesions Are Caused by Anti-TNF Agents Used for the Treatment of Inflammatory Bowel Disease. Dig Dis Sci. 2015;60:3424-30. doi: 10.1007/s10620-015-3763-0. PMID:26115749 [DOI] [PubMed] [Google Scholar]
- [26].Lemann M. Treatment of chronic inflammatory bowel diseases. Bull Acad Natl Med. 2007;191:1125-41; discussion 41. PMID:18402168 [PubMed] [Google Scholar]
- [27].Bilski J, Mazur-Bialy A, Brzozowski B, Magierowski M, Zahradnik-Bilska J, Wójcik D, Magierowska K, Kwiecien S, Mach T, Brzozowski T. Can exercise affect the course of inflammatory bowel disease? Experimental and clinical evidence. Pharmacological reports : PR. 2016;68:827-36. doi: 10.1016/j.pharep.2016.04.009. PMID:27255494 [DOI] [PubMed] [Google Scholar]
- [28].Trojian TH, Mody K, Chain P. Exercise and colon cancer: primary and secondary prevention. Curr Sports Med Rep. 2007;6:120-4. PMID:17376341 [DOI] [PubMed] [Google Scholar]
- [29].Cook MD, Martin SA, Williams C, Whitlock K, Wallig MA, Pence BD, Woods JA. Forced treadmill exercise training exacerbates inflammation and causes mortality while voluntary wheel training is protective in a mouse model of colitis. Brain Behav Immun. 2013;33:46-56. doi: 10.1016/j.bbi.2013.05.005. PMID:23707215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Bilski J, Mazur-Bialy AI, Brzozowski B, Magierowski M, Jasnos K, Krzysiek-Maczka G, Urbanczyk K, Ptak-Belowska A, Zwolinska-Wcislo M, Mach T, et al.. Moderate exercise training attenuates the severity of experimental rodent colitis: the importance of crosstalk between adipose tissue and skeletal muscles. Mediators of inflammation. 2015;2015:605071.v 10.1155/2015/605071. PMID:25684862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Murphy EF, Cotter PD, Healy S, Marques TM, O'Sullivan O, Fouhy F, Clarke SF, O'Toole PW, Quigley EM, Stanton C, et al.. Composition and energy harvesting capacity of the gut microbiota: relationship to diet, obesity and time in mouse models. Gut. 2010;59:1635-42. doi: 10.1136/gut.2010.215665. PMID:20926643 [DOI] [PubMed] [Google Scholar]
- [32].Louis P, Flint HJ. Development of a semiquantitative degenerate real-time pcr-based assay for estimation of numbers of butyryl-coenzyme A (CoA) CoA transferase genes in complex bacterial samples. Appl Environ Microbiol. 2007;73:2009-12. doi: 10.1128/AEM.02561-06. PMID:17259367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Brinkman BM, Becker A, Ayiseh RB, Hildebrand F, Raes J, Huys G, Vandenabeele P. Gut microbiota affects sensitivity to acute DSS-induced colitis independently of host genotype. Inflamm Bowel Dis. 2013;19:2560-7. doi: 10.1097/MIB.0b013e3182a8759a. PMID:24105395 [DOI] [PubMed] [Google Scholar]
- [34].Liang X, Li H, Tian G, Li S. Dynamic microbe and molecule networks in a mouse model of colitis-associated colorectal cancer. Sci Rep. 2014;4:4985. doi: 10.1038/srep04985. PMID:24828543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Everard A, Belzer C, Geurts L, Ouwerkerk JP, Druart C, Bindels LB, Guiot Y, Derrien M, Muccioli GG, Delzenne NM, et al.. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc Natl Acad Sci U S A. 2013;110:9066-71. doi: 10.1073/pnas.1219451110. PMID:23671105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Vital M, Howe AC, Tiedje JM. Revealing the bacterial butyrate synthesis pathways by analyzing (meta)genomic data. mBio. 2014;5:e00889. doi: 10.1128/mBio.00889-14. PMID:24757212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Blanton LV, Charbonneau MR, Salih T, Barratt MJ, Venkatesh S, Ilkaveya O, Subramanian S, Manary MJ, Trehan I, Jorgensen JM, et al.. Gut bacteria that prevent growth impairments transmitted by microbiota from malnourished children. Science. 2016;351:830-7. doi: 10.1126/science.aad3311. PMID:26912898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Gensollen T, Iyer SS, Kasper DL, Blumberg RS. How colonization by microbiota in early life shapes the immune system. Science. 2016;352:539-44. doi: 10.1126/science.aad9378. PMID:27126036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Abraham C, Cho JH. IL-23 and autoimmunity: new insights into the pathogenesis of inflammatory bowel disease. Annu Rev Med. 2009;60:97-110. doi: 10.1146/annurev.med.60.051407.123757. PMID:18976050 [DOI] [PubMed] [Google Scholar]
- [40].Mortha A, Chudnovskiy A, Hashimoto D, Bogunovic M, Spencer SP, Belkaid Y, Merad M. Microbiota-dependent crosstalk between macrophages and ILC3 promotes intestinal homeostasis. Science. 2014;343:1249288. doi: 10.1126/science.1249288. PMID:24625929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Cherayil BJ. Indoleamine 2,3-dioxygenase in intestinal immunity and inflammation. Inflamm Bowel Dis. 2009;15:1391-6. doi: 10.1002/ibd.20910. PMID:19322906 [DOI] [PubMed] [Google Scholar]
- [42].Wolf AM, Wolf D, Rumpold H, Moschen AR, Kaser A, Obrist P, Fuchs D, Brandacher G, Winkler C, Geboes K, et al.. Overexpression of indoleamine 2,3-dioxygenase in human inflammatory bowel disease. Clin Immunol. 2004;113:47-55. doi: 10.1016/j.clim.2004.05.004. PMID:15380529 [DOI] [PubMed] [Google Scholar]
- [43].Clarke G, McKernan DP, Gaszner G, Quigley EM, Cryan JF, Dinan TG. A Distinct Profile of Tryptophan Metabolism along the Kynurenine Pathway Downstream of Toll-Like Receptor Activation in Irritable Bowel Syndrome. Front Pharmacol. 2012;3:90. doi: 10.3389/fphar.2012.00090. PMID:22661947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Machiels K, Joossens M, Sabino J, De Preter V Arijs I, Eeckhaut V, Ballet V, Claes K, Van Immerseel F Verbeke K, et al.. A decrease of the butyrate-producing species Roseburia hominis and Faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut. 2014;63:1275-83. doi: 10.1136/gutjnl-2013-304833. PMID:24021287 [DOI] [PubMed] [Google Scholar]
- [45].Groux H, O'Garra A, Bigler M, Rouleau M, Antonenko S, de Vries JE, Roncarolo MG. A CD4+ T-cell subset inhibits antigen-specific T-cell responses and prevents colitis. Nature. 1997;389:737-42. doi: 10.1038/39614. PMID:9338786 [DOI] [PubMed] [Google Scholar]
- [46].Powrie F, Carlino J, Leach MW, Mauze S, Coffman RL. A critical role for transforming growth factor-beta but not interleukin 4 in the suppression of T helper type 1-mediated colitis by CD45RB(low) CD4+ T cells. J Exp Med. 1996;183:2669-74. doi: 10.1084/jem.183.6.2669. PMID:8676088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Becker C, Fantini MC, Neurath MF. TGF-beta as a T cell regulator in colitis and colon cancer. Cytokine Growth Factor Rev. 2006;17:97-106. doi: 10.1016/j.cytogfr.2005.09.004. PMID:16298544 [DOI] [PubMed] [Google Scholar]
- [48].Chen J, Xie L, Toyama S, Hünig T, Takahara S, Li XK, Zhong L. The effects of Foxp3-expressing regulatory T cells expanded with CD28 superagonist antibody in DSS-induced mice colitis. Int Immunopharmacol. 2011;11:610-7. doi: 10.1016/j.intimp.2010.11.034. PMID:21163250 [DOI] [PubMed] [Google Scholar]
- [49].Lamas B, Richard ML, Leducq V, Pham HP, Michel ML, Da Costa G, Bridonneau C, Jegou S, Hoffmann TW, Natividad JM, et al.. CARD9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nat Med.. 2016;22:598-605. doi: 10.1038/nm.4102. PMID:27158904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Martin JC, Beriou G, Heslan M, Bossard C, Jarry A, Abidi A, Hulin P, Ménoret S, Thinard R, Anegon I, et al.. IL-22BP is produced by eosinophils in human gut and blocks IL-22 protective actions during colitis. Mucosal Immunol. 2016;9:539-49. doi: 10.1038/mi.2015.83. PMID:26329427 [DOI] [PubMed] [Google Scholar]
- [51].Sugimoto K, Ogawa A, Mizoguchi E, Shimomura Y, Andoh A, Bhan AK, Blumberg RS, Xavier RJ, Mizoguchi A. IL-22 ameliorates intestinal inflammation in a mouse model of ulcerative colitis. J Clin Invest. 2008;118:534-44. PMID:18172556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Bergstrom KS, Morampudi V, Chan JM, Bhinder G, Lau J, Yang H, Ma C, Huang T, Ryz N, Sham HP, et al.. Goblet Cell Derived RELM-beta Recruits CD4+ T Cells during Infectious Colitis to Promote Protective Intestinal Epithelial Cell Proliferation. PLoS Pathog. 2015;11:e1005108. doi: 10.1371/journal.ppat.1005108. PMID:26285214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Johansson ME, Gustafsson JK, Sjoberg KE, Petersson J, Holm L, Sjövall H, Hansson GC. Bacteria penetrate the inner mucus layer before inflammation in the dextran sulfate colitis model. PloS One. 2010;5:e12238. doi: 10.1371/journal.pone.0012238. PMID:20805871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4516-22. doi: 10.1073/pnas.1000080107. PMID:20534432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Erben U, Loddenkemper C, Doerfel K, Spieckermann S, Haller D, Heimesaat MM, Zeitz M, Siegmund B, Kühl AA. A guide to histomorphological evaluation of intestinal inflammation in mouse models. Int J Clin Exp Pathol. 2014;7:4557-76. PMID:25197329 [PMC free article] [PubMed] [Google Scholar]
- [56].Panasevich MR, Allen JM, Wallig MA, Woods JA, Dilger RN. Moderately Fermentable Potato Fiber Attenuates Signs and Inflammation Associated with Experimental Colitis in Mice. J Nutr. 2015;145:2781-8. doi: 10.3945/jn.115.218578. PMID:26491118 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The remaining datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.


