ABSTRACT
Human cell lines are an important resource for research, and are often used as in vitro models of human diseases. In response to the mandate that all cells should be authenticated, we discovered that the MDA-MB-231 cells that were in use in our lab, did not validate based on the alleles of 9 different markers (STR Profile). We had been using this line as a model of triple negative breast cancer (TNBC) that has the ability to form tumors in immuno-compromised mice. Based on marker analysis, these cells most closely resembled the MCF10A line, which are a near diploid and normal mammary epithelial line. Yet, the original cells express carbonic anhydrase IX (CAIX) both constitutively and in response to hypoxia and are features that likely drive the aggressive nature of these cells. Thus, we sought to sub-purify CAIX-expressing cells using Fluorescence Activated Cell Sorting (FACS). These studies have revealed a new line of cells that we have name UFH-001, which have the TNBC phenotype, are positive for CAIX expression, both constitutively and in response to hypoxia, and behave aggressively in vivo. These cells may be useful for exploring mechanisms that underlie progression, migration, and metastasis of this phenotype. In addition, constitutive expression of CAIX allows its evaluation as a therapeutic target, both in vivo and in vitro.
KEYWORDS: breast cancer cells, Breast Biology, Cancer Biology, Cancer Transcriptomes, CAIX, cell sorting, microarray, STR profiling, triple negative model, UFH-001
Introduction
Mammalian carbonic anhydrases (CAs) belong to the α-CA family of metalloenzymes and catalyze the reversible hydration of CO2. There are 13 catalytically active members in this group that differ in their kinetic, inhibitory properties, cell and tissue distribution, and function.1 Carbonic anhydrase IX (CAIX) is one of the membrane-bound CA isoforms.2-4 CAIX is normally expressed in gut epithelial tissue,5 but is upregulated in several forms of cancer, including breast cancer. Indeed, CAIX is a marker for hypoxic regions of breast tumors6,7 and is associated with poor prognosis8-10 and high-grade, ER-negative breast tumors.8 Its role in tumor growth and disease progression has been attributed to its ability to reduce pericellular pH in response to hypoxia,11-13 creating a toxic environment for normal cells.14 Acidic environments also facilitate the breakdown of extracellular matrix15 and enhance metastatic activity.16,17
Triple negative breast cancer (TNBC) is a subtype of breast cancer that is diagnosed by the absence of the estrogen receptor (ER), progesterone receptor (PR), and the human epidermal growth factor receptor 2 (HER2). In the United States, approximately 15 – 20% of all breast cancers are TNBC18,19 which is particularly aggressive,20. Compared to other subtypes of breast cancer, TNBC is more likely to spread beyond the breast,21,22 as well as to recur after surgical23 and/or chemotherapeutic treatment.24 There are no targeted therapies for triple negative breast cancers25 which makes it more difficult to treat. CAIX expression is associated with the TNBC phenotype26,27 and also found in hormone-resistant breast cancers.9 This suggests that CAIX may serve as a therapeutic target for the treatment of TNBC and has attracted the attention of the scientific and pharmaceutical community.
Human cell lines are an important resource for research, and are often used in reverse genetic approaches or as in vitro models of human diseases. Using cell lines in breast cancer research has provided mechanistic insight into the regulation of cell growth, differentiation, tumorigenesis, and metastasis. Due to transcriptional drift in cell culture,28 it is important to continually validate the cell lines that are used in these types of studies. Indeed, many journals and funding agencies now demand this.
In response to this new mandate, we discovered that the MDA-MB-231 cells that we have been using as a cell model for TNBC, and that also show strong expression of CAIX, did not validate based on the alleles of 9 different markers (STR Profile). Because of our interest in CAIX and the strong expression of CAIX in this population, we sought to identify the CAIX-positive cells by flow cytometry. This led to the identification of a new cell line, which derives from MCF10A cells. However, the new line has numerous differences in their transcriptomes when compared against authenticated MCF10A cells. CAIX, specifically, is constitutively expressed (unlike authenticated MCF10A cells) in addition to induction by hypoxia. Further, these cells support tumor growth in a xenograft model. Because these cells lack ER, PR, and HER2 expression, these potentially represent a new TNBC line that we have named UFH-001 (UF Health-001). Herein, we describe its characteristics.
Results
Establishing the UFH-001 cell line
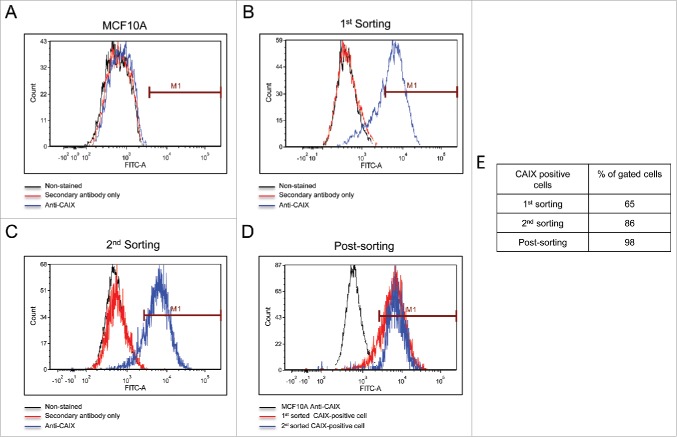
The cells commonly used in the lab include MCF10A (an immortalized breast cancer line), T47D (an ER-positive breast cancer line), and the triple negative MDA-MB-231. We use these to study membrane-bound carbonic anhydrases. We have previously shown that the MCF10A line expresses CAIX only under hypoxic conditions.29 The T47D cells express only carbonic anhydrase XII (CAXII), the expression of which is insensitive to hypoxia.29 In the MDA-MB-231 cell line, CAIX is expressed in a density-dependent manner and induced by hypoxic conditions29. These latter cells also form tumors in SCID mice (Gutwein, Grobmeyer, and Frost, unpublished data). CAIX was originally discovered in HeLa cells30 where it's expression was regulated by cell density31 and later by hypoxia6. Other investigators have shown this same regulation in the MDA-MB-231 cell line.32 That the “MDA-MB-231” cell line in our lab did the same was consistent with these earlier studies. Because of an ongoing collaboration with investigators as the Moffitt Cancer Center in Tampa, FL, we used their Molecular Genomics Core to validate the T47D and the MDA-MB-231 cells. The report revealed that the T47D cells matched with 100% accuracy the unique loci used for STR identification. However, the “MDA-MB-231” cell line did not match the ATCC STR profile for MDA-MB-231 cells, sharing only 25% of the markers. Rather, the presumed MDA-MB-231 cells were a 94% match to the STR profile of MCF10A cells with only a single mis-match. That markers for both lines were identified by this report is somewhat misleading because with a 94% match to the MCF10A line reveals that the presumed MDA-MB-231 cells are from that origin. It is also unlikely that the population is a mixture of MDA-MB-231 cells and MCF10A, because the STR markers that are unique to the MDA-MB-231 cells were not found in the presumed MDA-MB-231 cells (see Fig. 2). Yet, these presumed MDA-MB-231 cells certainly did not express a phenotype that matches the MCF10A cells, because they express CAIX in response to growth, which contrasts to that of MCF10A cells,29 and form tumors in immuno-compromised mice (data not show). Because of the strong expression of CAIX in the presumed MDA-MB-231 cells, we decided to isolate the CAIX-positive cells under normoxic conditions from the CAIX-negative cells using flow cytometry. As a positive control for CAIX-negative cells, we used authenticated MCF10A cells exposed to normoxic conditions. Fig. 1A demonstrates that the normoxic MCF10A cells do not bind the CAIX-specific (M75) monoclonal antibody. In Fig. 1B, our flow cytometry analysis of the “original”, presumed MDA-MB-231 cells, showed that there were two populations: one that was CAIX negative, and one that was CAIX positive. We gated that latter population, 65% of which were CAIX-positive cells (1st sorting). We collected and expanded that gated population and then re-sorted. Fig. 1C shows that we still had two populations of cells: one CAIX negative and the other CAIX positive. Once again, we gated this latter group, 86% of which were now CAIX positive (2nd sorting). That population was collected and expanded for re-evaluation by flow cytometry (Fig. 1D). This population now represented 98% of the CAIX-positive cells. We submitted both the first and second cell-sorted populations for authentication by different companies (Fig. 2). The cells were confirmed to contain only X-chromosomes and were of human origin. But, once again, the genetic profile of the STR markers matched closely that of the 10A line. The sorted cells have one difference compared to the original presumed MDA-MB-231 cells: an additional microsatellite expansion was detected in the vWA gene. This could result from a somatic mutation, trisomy, or gene duplication. Why this was not detected in the original validation protocol is not known. Importantly, the sorted cells appear to be a unique line, with no contamination by another human cell line. From here on, we will refer to these cells as the UFH-001 line.
Figure 2.
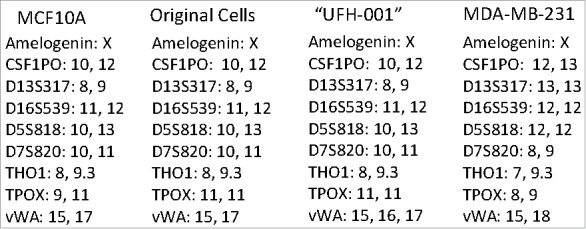
STR analysis of original and UFH-001 cells compared to the ATCC STR marker profile of MCF10A and MDA-MB-231 cells. The Molecular Genomics Core at the Moffitt Cancer Center analyzed the original cells. The UFH-001 cells were analyzed by bioSYNTHESIS (first sorting) and by IDEXX BioResearch (second sorting) with identical results.
Figure 1.
Isolation of CAIX-positive cells using flow cytometry. Panel A. Normoxic MCF10A cells served to identify CAIX-negative cells. The black line represents MCF10A cells that were not exposed to either primary antibody (the M75 monoclonal antibody) or the secondary mouse-specific IgG. The red line represents cells that were exposed to secondary antibody, only. The blue line represents cells that were exposed to both primary and secondary antibodies. Panel B. Separation of CAIX-positive from CAIX-negative cells in the original line. Color coding is the same as described in Panel A. CAIX-positive cells were gated and collected for expansion, representing the 1st sorting. Panel C. The expanded cells (from Panel B) were reanalyzed by flow cytometry for a 2nd sorting. Color coding is the same as described in Panel A. Panel D. Post-sorting analysis. Cells collected at the 2nd sorting were re-analyzed by flow cytometry for a final time and superimposed (blue line) on the gated cells from the 2nd sorting (red line) and CAIX-negative MCF10A cells (black line). Panel E. The table shows the % of gated cells at each step that are CAIX-positive.
Morphology and spheroid formation of the UFH-001 cell line
UFH-001 cells exhibit distinct morphologies from their cell of origin, the MCF10A cells (Fig. 3). At low density, UFH-001 cells display cobblestone morphology and form colonies (Fig. 3A). As they grow, these cells become even more colonized while retaining the cobblestone morphology (Fig. 3B). In contrast, MCF10A cells present as elongated and spindle-like, and do not form colonies at either low (Fig. 3C) or high density (Fig. 3D). Both UFH-001 and MCF10A cells form spheroids (Fig. 3E and 3F). UFH-001 cells form spheroids as early as 24 h after plating (even when the plating density is low) (images not shown). At these lower densities, spheroid size increases every 24 h. At 48 h (Fig. 3E), the size of UFH-001 spheroids depends on the initial cell density. The spheroids appear dense and rounded. In contract, MCF10A cells do not form spheroids at low cell density (40, 200, 1000 cells/well) at any time point (24 h-96 h). At 48 h (Fig. 3F), MCF10A spheroids are in a loose and irregular shape, observed only in wells where the starting cell number was either 5,000 or 10,000 cells.
Figure 3.
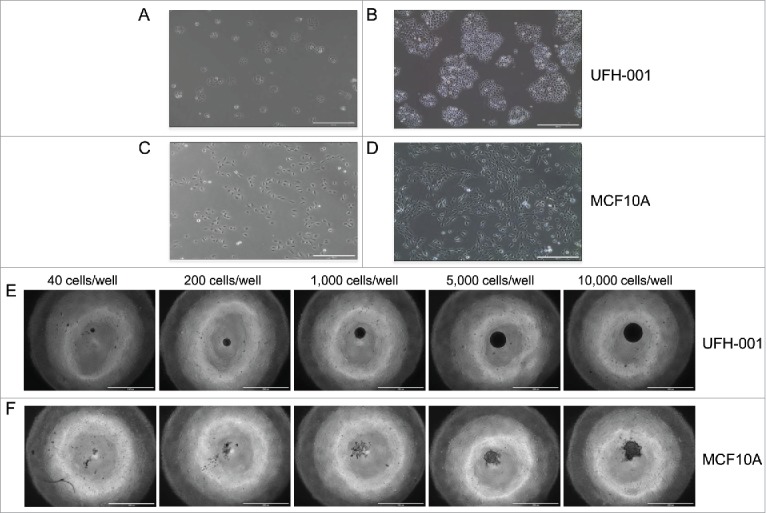
Morphology and spheroid formation of the UFH-001 and MCF10A cells. UFH-001 cells showed a cohesive cobblestone morphology and formed colonies at low (Panel A) and high (Panel B) density (see Methods and Material for details). MCF10A cells displayed an elongated and spindle-like appearance at low (Panel C) and high (Panel D) density. Magnification: 10X, scale bar: 400μM. UFH-001 cells formed circular and dense spheroids across all starting concentrations at 48 h (Panel E). MCF10A cells formed irregular spheroids and only at starting densities of 5,000 and 10,000 cells/well at 48 h (Panel F). The scale bar represents 1 mm. Magnification is 4X.
Microarray data
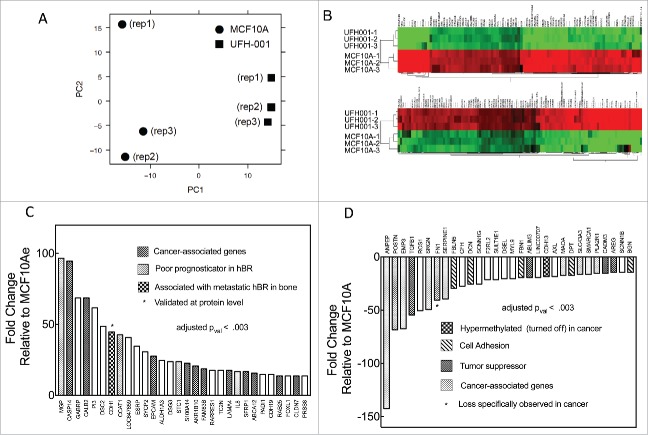
Principal component analysis (PCA) is a dimensional reduction and visualization technique that is used here to project the multivariate data vector of each array into a two-dimensional plot, such that the spatial arrangement of the points in the plot reflects the overall data similarity (or dissimilarity) between arrays. Fig. 4A shows a scatterplot of the triplicate microarray data sets of MCF10A and UFH-001 along the first two principal components. These data show that the UFH-001 transcriptomes clusters away from the MCF10A transcriptomes, predicting that the transcriptomes of UFH-001 and MCF10A cells are unique (Fig. 4A). The heatmap of the top 100 differentially expressed genes (DEG) in UFH-001 vs MCF10A cells show distinct and opposite patterns of regulation (Fig. 4B). Figs 4C and 4D show up- and down-regulated genes, respectively, for the top 30 differentially DEGs. The DEGs were associated with GO terms that suggest extensive extracellular matrix remodeling, in addition to enhancement of exosomal transport and calcium dependent signaling (Table 1). The complete data set for these microarray data has been deposited in the GEO repository (accession number: GSE107209).
Figure 4.
Microarray data. Panel A. Principal component analysis (PCA) of the microarray data. Panel B. Heat-map comparing microarray data of differentially expressed genes between UFH-001 and MCF10A cells (top 100 differentially- expressed genes are shown). Panel C. Top 30 up-regulated mRNA species (fold-change) in UFH-001 cells relative to MCF10A. Panel D. Top 30 down-regulated mRNA species (fold-change) in UFH-001 cells relative to MCF10A.
Table 1.
Microarray pathway analysis in UFH-001 cells relative to MCF10A cells.
| Accession # | Pathway Identifier | Number | Enrichment | p-value | |
|---|---|---|---|---|---|
| Up-Regulated | GO:0098742 | cell adhesion via PM | 7/30 | 22.6 | 1.72 × 10–4 |
| GO:0005509 | calcium ion binding | 9/30 | 9.3 | 8.63 × 10–4 | |
| GO:0009888 | tissue development | 13/30 | 5.7 | 8.05 × 10–4 | |
| GO:0070062 | exosome and vesicular transport | 16/30 | 4.2 | 1.28 × 10–4 | |
| Down-Regulated | GO:0030198 | extracellular matrix organization | 9/30 | 21.1 | 2.44 × 10–6 |
| GO:0005539 | glycosaminoglycan binding | 6/30 | 20.4 | 1.33 × 10–3 | |
| GO:0031012 | extracellular matrix | 10/30 | 13.3 | 2.41 × 10–6 | |
| GO:0007155 | cell adhesion | 10/30 | 8.3 | 1.23 × 10–3 |
TNBC characterization
CAIX is a marker for the triple negative phenotype in breast cancer ([26, 27]). Fig. 5A demonstrates that the UFH-001 line expresses CAIX both constitutively and in response to hypoxia. MCF10A cells also expressed CAIX but only in response to hypoxia. It is thought that the MCF10A line is of basal cell origin even though it does not support tumor growth in nude mice.33 Microarray data collected from normoxic UFH-001 and MCF10A cells (Fig. 5B) show elevated expression of CAIX (3.7 fold) and CAII (7.5 fold) in UFH-001 cells relative to MCF10A cells. Interestingly, CAXII mRNA levels are reasonably high in both MCF10A and UFH-001 but no CAXII protein was detected in either cell type (Fig. 5A). T47D cells were used as a positive control.
Figure 5.
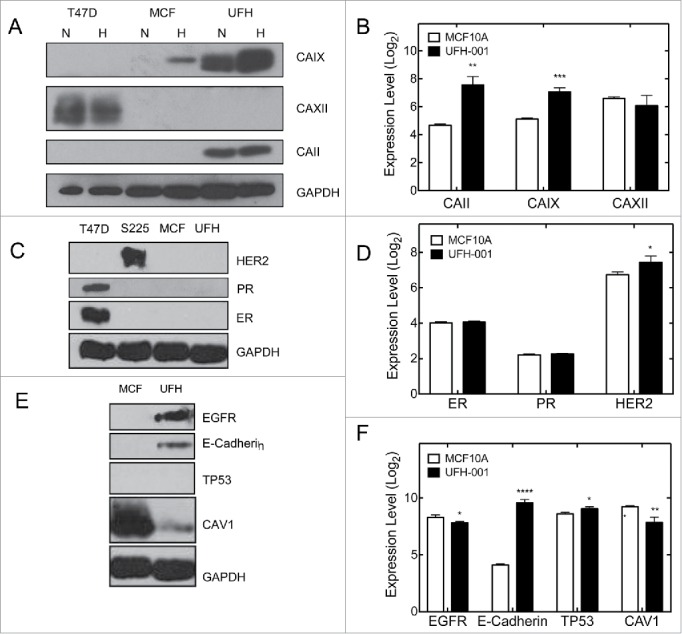
Selected markers for the TNBC phenotype. Panels A, C, and E are representative western blots. Panels B, D, and F show comparative mRNA expression based on Log2 scores of microarray data. T47D and Sum225 cells were used as positive controls in the western blots, where appropriate. The number of asterisks denotes the statistical significance between mRNA levels in UFH-001 and MCF10A cells: * p < 0.05, ** p<0.01, *** p < 0.001, **** p < 0.0001)
TNBC breast cancers express neither the estrogen or progesterone receptors (ER, PR), nor overexpress the human isoform of ERBB2 (HER2). Our microarray data (Fig. 5D) show that UFH-001 cells have the same ER and PR expression as MCF10A cells, and a small but significant increase in HER2 expression when compared with MCF10A cells. Despite this, neither cell type expressed ER, PR nor HER2 protein (Fig. 5C). T47D and SUM225 cells served as the positive control for ER and PR, and HER2 expression, respectively.
Epidermal growth factor receptor (EGFR), E-cadherin, TP53, and caveolin 1 (CAV1) expression are associated with TNBC breast cancer.34-36 In UFH-001 cells, we observed both EGFR and E-cadherin expression (Fig. 5E). Neither of these proteins was expressed in MCF10A cells. No TP53 was observed in UFH-001 cells (nor in MCF10A cells), and CAV1 was significantly decreased when compared with MCF10A cells. At the mRNA level (Fig. 5F), EGFR was lower in UFH-001 cells relative to MCF10A, as was CAV1. E-cadherin was increased by 42-fold relative to MCF10A cells while TP53 was only slightly elevated.
Proliferation, migration, and invasion of UFH-001 cells
The malignant cell is characterized by its rapid proliferative activity. In order to determine the proliferative ability of UFH-001 cells, we examined cell growth in culture (Fig. 6A). Equal cell number (15,000 cells) was plated for both UFH-001 and MCF10A cells. By day 3, there was a clear difference in proliferative capacity of the UFH-001 cells relative to the MCF10A cells, which became significantly different at later time points. The UFH-001 cells divide in less that 24 h, while the MCF10A cells divide once during a 48 h period.
Figure 6.
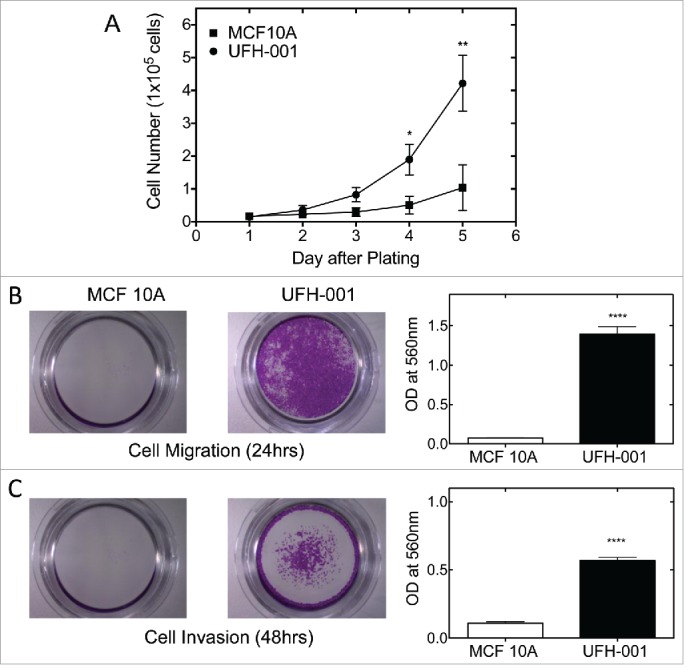
Proliferation, migration, and invasion of UFH-001 and MCF10A cells. Panel A. Cell growth curves of UFH-001 and MCF10A, plated at 15,000 cells/plate in 35 mm plates, were cultured over 5 days. Panel B. Representative images show the migration of UFH-001 and MCF10A cells in transwell plates. Panel C. Representative images show the invasive capacity of UFH-001 and MCF10A cells in transwell plates. * p < 0.05, ** p < 0.01, **** p < 0.0001).
The ability to migrate and invade are key factors responsible for metastasis. To metastasize, cancer cells must migrate from the original growth site, invade surrounding tissues, and relocate to distal sites through the circulation or lymphatic system. To examine the migration and invasion abilities of UFH-001 cells, we used the trans-well assay to quantify the migratory and invasive potential of UFH-001 cell (Fig. 6B, C). After 24 hours of incubation, about 19 times more UFH-001 cells migrated as compared to MCF10A cells (Fig. 6B). For invasion, we increased the time frame to 48 h because the cells were less invasive than migratory. At that time point, UFH-001 cells had a greater invasion rate (5x) than MCF10A cells (Fig. 6C).
Tumorigenicity of UFH-001 cells
Finally, we assessed the ability of the UFH-001 cells to form tumors in NOD/SCID mice. To monitor xenograft tumor growth, we planned to use Luciferase-transfected UFH-001 cells (UFH-Luc). First, we compared the growth of non-transfected UFH-001 cells and UFH-Luc cells in cell culture. These data show that there is no difference in the rate of cell growth between the parental and transfected lines (Fig. 7A). Luciferase-transfected UFH-001 cells (3 × 105) then were injected into the 4th mammary gland of replicate mice. Fig. 7B shows tumor formation over time. At each time point, luciferase activity was detected after injection of luciferin (150 mg/kg body weight) into the peritoneum. As early as 1.5 weeks after injection, small tumors could be observed. Growth increased substantially over the 4 weeks that we continued to observe the animals. At the end-point, the center of the tumors showed significant necrosis (data not shown). Western blot analysis showed that xenograft tumors retain the expression pattern of UFH-001 cells with respect to carbonic anhydrases and E-cadherin.
Figure 7.
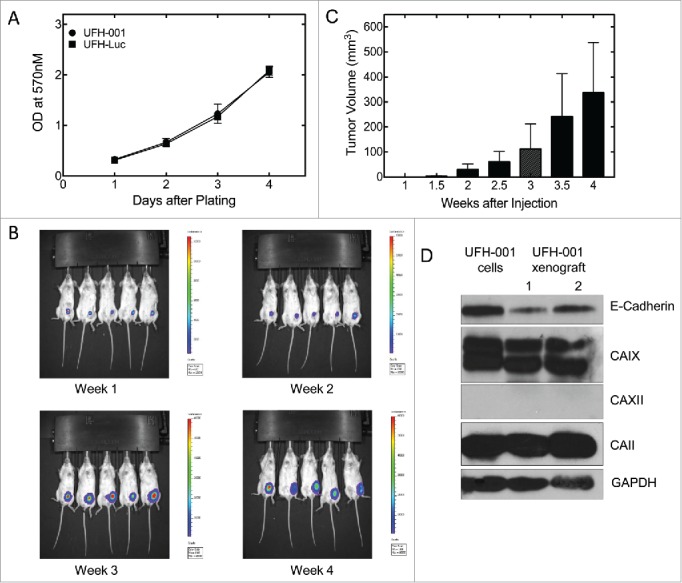
Tumorigenicity of UFH-001 cells in NOD/SCID mice. Panel A. UFH-001 and UFH-Luc cells were grown in culture for four days. At each day an MTT assay was performed to evaluate cell proliferation. Panel B. UF-Luc cells were orthotopically implanted (3 × 105 cells) into the first mammary gland of NOD/SCID mice, and evaluated by in vivo (see Materials and Methods). Panel C. Tumor size was quantified by calculating tumor volume twice a week after the injection of UFH-Luc cells. Luminescence followed similar trends as tumor volume except at 4 weeks where there was central necrosis that reduced luminescent readings (data not shown). Panel D. Samples of UFH-001 cells and tissue from UFH-Luc generated tumors were compared by western blotting.
Discussion
The cancer research community relies heavily on cells isolated from original tumors and metastatic sites, to understand alterations in signal transduction, to define differential gene expression, and to identify drug targets for therapeutic intervention. These cells often undergo minor changes in genotype and phenotype, especially across different labs. There is also the risk that lines get mixed or mislabeled when shared across labs. One striking example of this was the misidentification of the MDA-MB-435 cells as breast cancer cells.37 These cells originally may have been breast cancer cells, but some time before 1982 they became contaminated with M14 (melanoma cells) that essentially overgrew and replaced the breast cancer cells.
For over a decade, our lab has been using a line identified originally as MDA-MB-231 breast cancer cells. They exhibited all of the typical attributes described in the literature: they displayed the triple negative phenotype, were responsive to hypoxia, expressed CAIX, and grew orthotopic tumors in nude mice. Yet, upon subjecting these cells to microsatellite screening (STR), it was revealed that they exhibit an STR profile that was strikingly more similar to the MCF10A line than MDA MB 231 cells. Because our lab has a strong interest in understanding the role of CAIX in cancer cell progression, we pursued the isolation of the CAIX-positive cells from the original line, thinking that these cells may have been a mixed population. When authenticated, the purified cells exhibited an STR profile identical to that of the original line, with the exception of an additional marker in the vWA microsatellite. We have interpreted this to mean that neither the original cells nor the CAIX-purified cells were/are a mixed population of cells. Rather, they represent a new line. Overall, the STR profile of the CAIX-purified line is more than 80% identical to MCF10A cells, which strongly implies that the MCF10A is the line of origin of the purified cells. We have named this new line, UFH-001, to distinguish them from the parental line (MCF10A). There are multiple lines that have arisen from MCF10A line, either by oncogenic overexpression (pre-malignant MCF-10AT cells) or repeated selection from the MCF10AT line for increasingly tumorigenic behavior (MCF-10CA1a cells).38 Because the expression of H-ras does not differ between MCF10A and the UFH-001 cells, it is unlikely that the UFH-001 cells originated from the MCF10AT or MCF-10CA1a lines.
Despite the near identity in microsatellite markers for MCF10A and UFH-001 cells, principle component analysis (PCA) of the comparative microarray data demonstrates a significant difference in the RNA population of the UFH-001 cells compared to that of the MCF10A cells. This includes changes in the expression of 1272 coding RNA species and 458 non-coding RNAs. UFH-001 cells also grow faster than the MCF10A cells, show in vitro migratory and metastatic behavior and exhibit a tumorigenic phenotype when inoculated orthotopically in the mammary fat pad, clearly different from the MCF10A line. While genetic drift is often blamed for changes to cells in extended culture, the transforming event must have been significant to alter such a large population of the RNA pool.
While there does not seem to be a single alteration that has caused this change from a non-tumorigenic to a tumorigenic line, there are a few changes in RNA expression that could significantly alter phenotype. Recently, it has been shown that CAIX ablation blocks hypoxia-induced expression of STC1 (stanniocalcin-1).39 This protein has been linked to poor survival in breast cancer patients with the basal B phenotype, and shown to stimulate invasiveness and tumor progression in TNBC.40,41 The expression of STC1 mRNA is 32 times higher in UFH-001 compared to the MCF parental line. E-cadherin, which is normally lost in the epithelial to mesenchymal transition. One anomaly that we noticed is the lack of expression of E-cadherin in the MCF10A line. This line is of basal origin42 but several investigators have shown that E-cadherin is expressed in this line33,42,43). One difference that we noted was the medium in which the MCF10A line is maintained. In other labs, cholera toxin is included, among other reagents, in the DMEM/Ham's F-12 medium. We found that the MCF10A line did not require this for growth, so for lab safety we removed the toxin, but because this toxin perpetually activates adenylate cyclase this could have an effect on E-cadherin expression. Qu et al. showed that E-cadherin was expressed in only 70% of MCF10A spheroids.42 So another possibility is that the MCF10A line we are using selected against it's expression. Also of note is that our line was specifically validated. In the other studies, they were not although purchased directly from ATCC. The other anomaly is the overexpression of E-cadherin in UFH-001 cells compared to the MCF10A line. One explanation for this phenomenon comes from a study in which E-cadherin protein re-appears in clumps of metastatic breast cancer cells associated with bone.44 This reversion to an epithelial state (MET) is not uncommon during the growth of carcinoma metastases.45,46 The high frequency of E-cadherin expression at metastatic sites may play a role in the intercellular adhesion of metastatic cells at these sites. UFH-001 cells may represent such cells, supported by their epithelial phenotype.
TNBC accounts for approximately 10–15% of breast cancers and has a poorer prognosis relative to ER-positive patients.47 The former type of breast cancer also shows aggressiveness, invasiveness, and early metastases in vivo. CAIX is a transmembrane zinc metalloenzyme functioning as the catalyst for the reversible hydration of carbon dioxide to bicarbonate with the release of a proton.48 CAIX expression is a marker for hypoxia, poor prognosis, and the triple negative phenotype.8-10 It is also a therapeutic target because loss of function leads to reduced cell growth in culture and tumor growth in animal models.49,50 That the UFH-001 cells constitutively express CAIX both in vitro and in established tumors, along with their ability for form spheroids, make this line useful for further investigating the role of CAIX in breast cancer progression.
Materials and methods
Cell culture
Cells originally designated as MDA-MB-231 were a gift from Dr. Kevin Brown (University of Florida). MCF10A and T47D cells were gifts from Dr. Brian Law (University of Florida). The presumed MDA-MB-231 cells and the new line isolated from these cells, UFH-001, were maintained in Dulbecco's Modified Eagle's medium (DMEM), supplemented with 10% fetal bovine serum (FBS) (Sigma Aldrich #F2442). These cells did not grow well in medium designed for MCF10A cell growth (data not shown) despite the fact that they are derived from this line. Thus, UFH-001 cells appear to be significantly less hormone-dependent than the MCF10A cells, other than that which is naturally in FBS. MCF10A cells were cultivated in DMEM/Ham's F12 medium (Fisher #MT10080CM), supplemented with 5% horse serum (Sigma Aldrich #H1138-6X), 10 μg/ml insulin (Gibco, A1138211), 20ng/mL epidermal growth factor (EGF) (Upstate Biochem) and 100 ng/mL dexamethasone (Sigma, D4902). T47D cells were validated and cultivated in McCoys 5A medium (Fisher #16-600-108), supplemented with 10% FBS and insulin (10 μg/mL). HER2-positive SUM225 cells were purchased from Asterand, and cultivated in Ham's F-12 medium containing 10 mM HEPES, supplemented with 5% FBS, dexamethasone (100 ng/mL) and insulin (5 μg/mL). HEK293T cells (ThermoFisher Scientific), used for the packaging lentivirus with luciferase expression, were cultured in DMEM supplemented with 10% FBS and 1X antibiotics/antimycotic solution (Corning, #30-004). All cell lines were maintained at 37°C in humidified air with 5% CO2. For hypoxia, cells were placed in humidified Billups Rothenberg Metabolic Chambers and exposed to 1% O2, 5% CO2 and balanced N2 for 16 h at 37°C.
Sorting UFH-001 cells by flow cytometry
The original MDA-MB-231 cell mixture was detached with cell dissociation buffer (Life technologies) and re-suspended in flow cytometry staining buffer (PBS, 2%BSA, 2 mM EDTA). The “mixed” population of cells were labeled with anti-CAIX antibody (M75) (a gift from Dr. Egbert Oosterwijk, but originally made by Dr. Sylvia Pastorekova51) for 1 h on ice. After washing with the staining buffer, the mixed cells were incubated with anti-mouse FITC (Sigma Aldrich) for 30 minutes on ice and washed again with the staining buffer. Cell sorting was performed by the Cytometry Core in the Institutional Center for Biotechnology (ICBR) on a BD FACSAria-IIu (BD Biosciences) using Diva 8 software. For this assay, a 50 milliwatt laser emitting at 488 nanometers was used. Cells were gated by forward light scatter (FSC) vs side light scatter (SSC) and cell aggregates removed using SSC pulse-area vs SSC pulse-width dot plots. FITC fluorescence (through a 530nm +/-15nm bandpass filter) was plotted as a histogram. A sorting gate was set at the 99.5 percentile value based on a negative control sample (MCF10A) and applied to the positive sample (original MDA-MB-231 cell mixture). Data files for 10,000 events were recorded and exported for further plotting and analysis using FCS Express, Version 5 (Denovo Software). The CAIX-positive cells were renamed UFH-001.
The details for culture conditions during the process of flow cytometry are as follows
For the first sorting, cells were plated at 10,000 cells/mL and cultured for 4 days. The cells from one 10 cm plate were used for sorting. After the first sorting, the cells were transferred into a single 10 cm plate. Cells were grown for four days. The culture was split at a 1:10 ratio. One sample was used for authentication and the other samples were frozen at −80°C. A frozen sample from the first sorting was thawed and grown for four days and split at a 1:10 ratio. After growing for four days in culture, the cells from one plate were used for the second sorting. The gated cells were transferred to a single 10 cm plate and cultured for four days. Cells were split at a 1:10 ratio and grown for another four days. Samples were taken for authentication, while other samples were cryopreserved. During this authentication, mycoplasma was detected. Cells were treated to eliminate the infection, expanded, and frozen for storage. These treated cells are those in present use in the lab.
Morphological appearance of cells
UFH-001 and MCF10A cells were plated at a density of 10,000 and 20,000 cells/mL, respectively, in 10 cm plates. UFH-001 and MCF10A cells were photographed at day 1 (low density) and day 4 (high density) at 10x magnification by light microscope (EVOS, Thermo Fisher Scientific).
Spheroid formation assay
UFH-001 and MCF10A cells (total volume of 100 μL in appropriate medium) at 40, 200, 1,000, 5,000, and 10,000 cells/well into 96 well low attachment microplates (Corning, #4515). Cells were incubated up to 96 h in 5% CO2 at 37°C. Spheroids were photographed at specific intervals using the EVOS microscope system (Thermo Fisher Scientific). The 48 h time points are shown in Fig. 3.
RNA isolation and microarray analysis
UFH-001 cells and MCF10A cells were plated at a density of 10,000 and 20,000 cells/ mL, respectively in 10cm plates. RNA extraction was performed when the cells were at 75–80% confluence using an RNase Easy Plus Mini kit (Qiagen #74134). All steps were followed as recommended by the manufacturer. RNA concentration was calculated at 260nm and the ratio of the absorbance values at 260/280 was used as a monitor for purity and stability. Total RNA was converted to cRNA, and microarrays (Human Transcriptome Array) were performed by the Gene Expression and Genotyping Core in the ICBR at the University of Florida. The analysis was performed in triplicate, biological replicates. Expression values were deduced from RMA normalized intensities. CEL files were processed with the limma package from R/Bioconductor
Western blot analysis
Western blot analysis was performed as previously described.29 In brief, cells were washed with ice-cold PBS and lysed in RIPA buffer at 4°C containing a cocktail of protease inhibitors (Sigma-Aldrich, P8340). Lysates were clarified by centrifugation. The supernatants were collected and total protein concentration was determined by the Markwell assay.52 Western blot analysis was performed after SDS-PAGE (10% gel; equal protein concentration per lane) and transferred onto nitrocellulose membranes. Membranes were incubated with specific antibodies: CAIX (M75 monoclonal, originally made by Pastorekova,51 but a gift from Dr. Egbert Oosterwijk, used at a dilution of 1:4000), CAII (Novus #NB600-919, used at a dilution of 1:4000), CAXII (R&D # AF2190, used at a dilution of 1:1000), estrogen receptor (Santa Cruz # sc-8002, used at a dilution of 1:200), HER2 (Cell Signaling #2165S, used at a dilution of 1:1000), E-cadherin (BD biosciences #610181, used at a dilution of 1:1000), EGFR (Cell Signaling #4267, used at a dilution of 1:1000), TP53 (Santa Cruz #sc-126, used at a dilution of 1:200), CAV1 (Santa Cruz # sc07875, used at a dilution of 1:1000), and GAPDH (Cell Signaling #5174, used at a dilution of 1:4000), PR (Santa Cruz #SC-810 used at a dilution of 1:200) followed by incubation with horseradish peroxidase–conjugated secondary antibodies (Sigma-Aldrich). The secondary antibodies were detected by enhanced chemiluminescence (GE Healthcare Biosciences, #RPN2232, or RPN2235).
Cell number determination
UFH-001 cells and MCF10A cells were plated at a density of 15,000 cells/well in 35 mm plates. Cell number was determined on a Coulter Counter ZM (Beckman/Coulter) over a 5-day period. The medium was replaced every other day.
Cell migration and invasion assays
UFH-001 and MCF10A cells were exposed to serum-free medium for 24 h. Cells were released from plates with cell dissociation buffer (Gibco #13151-014), which lacks trypsin. They were then plated in serum-free medium (specific for cell type) at a density of 50,000 cells/300μL/insert in 24 well cell migration and invasion plates (Cell Biolabs CBA-100-C-5). The cells were allowed to migrate (24 h) or invade (48 h) across the insert towards the well that contained 10% FBS. The assay was terminated after the appropriate time point. Cells were fixed, stained, and photographed for image analysis. All cell lines were maintained at 37°C in humidified air with 5% CO2 for the duration of the experiment.
Transfection of luciferase
Luciferase expressing lentivirus was packed into HEK293T cells by transfection with plasmids pLenti-PGK V5-LUC Neo, psPAX2, and pMD2.G (Addgene, MA, USA). Culture medium with virus was collected at 48 and 72 h after transfection, passed through a 0.45μm filter and frozen at -80°C. UFH-001 cells were cultured with 1:1 virus /DMEM medium (v/v) with 8 μg/mL polybrene (Sigma-Aldrich Corp) for 4 h, which was then exchanges for fresh DMEM (10%FBS). After selection with 400 μg/mL G418 (Gibco, #11811031) for 2 weeks, the transfected cells (UFH-Luc) were validated by the detection of luciferin-dependent luciferase activity.
MTT assay
UFH-001 parental cells and UFH-Luc cells were plated at a density of 1500 cells/well into 96-well plates. Cell growth was determined by using a standard tetrazolium bromide (MTT) assay on sequential days after plating.
Animal models
All procedures were conducted in accordance with the National Institutes of Health regulations and approved by the University of Florida Institutional Animal Care and Use Committee (IACUC protocol # 201603567). For mouse xenografts, a total of 3 × 105 UFH-Luc cells suspended in culture medium was injected into the fourth left mammary fat pad of NOD/SCID mice (Jackson Laboratory, ME, USA), aged around 10–12 weeks old. Tumor sizes were measured with a digital caliper (Fisher Scientific, MA, USA), and the volume was calculated using the formula: 0.5 x L x W2 (L- length, W-width). The Xenogen IVIS Spectrum system (Caliper Lifesciences, MA, USA) was used for in vivo imaging to track tumor growth and metastasis.
Statistical evaluation
Differences in gene expression between UFH-001 and MCF10A cells were evaluated with Student's t test. Student's t test was also used to study cell growth, migration, invasion and tumor growth. All p values were calculated based on two-sided testing and p < 0.05 was considered as statistically significant. Statistical analysis was performed using Prism software.
Funding Statement
NIH, CA165284.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- 1.Hilvo M, Salzano AM, Innocenti A, Kulomaa MS, Scozzafava A, Scaloni A, Parkkila S, Supuran CT. Cloning, Expression, Post-Translational Modifications and Inhibition Studies on the Latest Mammalian Carbonic Anhydrase Isoform, CA XV. J Med Chem. 2009;52:646–54. doi: 10.1021/jm801267c. PMID:19193158. [DOI] [PubMed] [Google Scholar]
- 2.Opavsky R, Pastorekova S, Zeln¡k V, Gibadulinov A, Stanbridge EJ, Z vada J, Kettmann R, Pastorek J. Human MN/CA9 Gene, A Novel Member of the Carbonic Anhydrase Family: Structure and Exon to Protein Domain Relationships. Genomics. 1996;33:480–7. doi: 10.1006/geno.1996.0223. PMID:8661007. [DOI] [PubMed] [Google Scholar]
- 3.Li Y, Wang H, Tu C, Shiverick KT, Silverman DN, Frost SC. Role of Hypoxia and EGF on Expression, Activity, Localization, and Phosphorylation of Carbonic Anhydrase IX in MDA-MB-231 Breast Cancer Cells. Biochim Biophy Acta. 2011;1813:159–67. doi: 10.1016/j.bbamcr.2010.09.018. PMID:20920536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hilvo M, Baranauskiene L, Salzano AM, Scaloni A, Matulis D, Innocenti A, Scozzafava A, Monti SM, Di Fiore A, De Simone G, et al.. Biochemical Characterization of CA IX, One of the Most Active Carbonic Anhydrase Isozymes. J Biol Chem. 2008;283:27799–809. doi: 10.1074/jbc.M800938200. PMID:18703501. [DOI] [PubMed] [Google Scholar]
- 5.Saarnio J, Parkkila S, Parkkila AK, Waheed A, Casey MC, Zhou XY, Pastorekova S, Pastorek J, Karttunen T, Haukipuro K, et al.. Immunohistochemistry of Carbonic Anhydrase Isozyme IX (MN/CA IX) in Human Gut Reveals Polarized Expression in Epithelial Cells with the Highest Proliferative Capacity. J Histochem Cytochem. 1998;46:497–504. doi: 10.1177/002215549804600409. PMID:9524195. [DOI] [PubMed] [Google Scholar]
- 6.Wykoff CC, Beasley NJP, Watson PH, Turner KJ, Pastorek J, Sibtain A, Wilson GD, Turley H, Talks KL, Maxwell PH, et al.. Hypoxia-inducible Expression of Tumor-associated Carbonic Anhydrase. Cancer Res. 2000;60:7075–83. PMID:11156414. [PubMed] [Google Scholar]
- 7.Tafreshi NK, Lloyd MC, Proemsey JB, Bui MM, Kim J, Gillies RJ, Morse DL. Evaluation of CAIX and CAXII Expression in Breast Cancer at Varied O2 Levels: CAIX is the Superior Surrogate Imaging Biomarker of Tumor Hypoxia. Mol Imaging Biol. 2016;18(2):219–31. doi: 10.1007/s11307-015-0885-x. PMID:26276155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Span PM, Bussink J, Manders P, Beex LVAM, Sweep CGJ. Carbonic Anhydrase-9 Expression Levels and Prognosis in Human Breast Cancer: Association with Treatment Outcome. Br J Cancer. 2003;89:271–6. doi: 10.1038/sj.bjc.6601122. PMID:12865916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Generali D, Fox SB, Berruti A, Brizzi MP, Campo L, Bonardi S, Wigfield SM, Bruzzi P, Bersiga A, Allevi G, et al.. Role of Carbonic Anhydrase IX Expression in Prediction of the Efficacy and Outcome of Primary Epirubicin/Tamoxifen Therapy for Breast Cancer. Endocrine-Related Cancer. 2006;13:921–30. doi: 10.1677/erc.1.01216. PMID:16954440. [DOI] [PubMed] [Google Scholar]
- 10.Chia SK, Wykoff CC, Watson PH, Han C, Leek RD, Pastorek J, Gatter KC, Ratcliffe P, Harris AL. Prognostic Significance of a Novel Hypoxia-Regulated Marker, Carbonic Anhydrase IX, in Invasive Breast Cancer. J Clin Oncol. 2001;19:3660–8. doi: 10.1200/JCO.2001.19.16.3660. [DOI] [PubMed] [Google Scholar]
- 11.Helmlinger G, Sckell A, Dellian M, Forbes NS, Jain RK. Acid Production in Glycolysis-impaired Tumors Provides New Insights into Tumor Metabolism. Clin Cancer Res. 2002;8:1284–91. [PubMed] [Google Scholar]
- 12.Swietach P, Vaughan-Jones RD, Harris AL. Regulation of Tumor pH and the Role of Carbonic Anhydrase 9. Cancer Metastasis Rev. 2007;26:299–310. doi: 10.1007/s10555-007-9064-0. PMID:17415526. [DOI] [PubMed] [Google Scholar]
- 13.Swietach P, Wigfield S, Cobden P, Supuran CT, Harris AL, Vaughan-Jones RD. Tumor-Associated Carbonic Anhydrase 9 Spatially Coordinates Intracellular pH in Three-Dimensional Multicellular Growths. J Biol Chem. 2008;283:20473–83. doi: 10.1074/jbc.M801330200. PMID:18482982. [DOI] [PubMed] [Google Scholar]
- 14.Gottlieb RA, Giesing HA, Zhu JY, Engler RL, Babior BM. Cell Acidification in Apoptosis: Granulocyte Cology-stimulating Factor Delays Programed Cell Death in Neutrophils by Up-regulating the Vaculolar H + -ATPase. Proc Natl Acad Sci USA. 1995;92:5965–8. doi: 10.1073/pnas.92.13.5965.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gatenby RA, Gawlinski ET, Gmitro AF, Kaylor B, Gillies RJ. Acid-mediated Tumor Invasion: A Multidisplinary Study. Cancer Res. 2006;66:5216–23. doi: 10.1158/0008-5472.CAN-05-4193. PMID:16707446. [DOI] [PubMed] [Google Scholar]
- 16.Goetze K, Walenta S, Ksiazkiewicz M, Kunz-Schughart LA, Mueller-Klieser W. Lactate enhances motility of tumor cells and inhibits monocyte migration and cytokine release. Int J Oncol. 2011;39(2):453–63. PMID:21617859. [DOI] [PubMed] [Google Scholar]
- 17.Estrella V, Chen T, Lloyd M, Wojtkowiak J, Cornnell HH, Ibrahim-Hashim A, Bailey K, Balagurunathan Y, Rothberg JM, Sloane BF, et al.. Acidity generated by the tumor microenvironment drives local invasion. Cancer Res. 2013;73(5):1524–35. doi: 10.1158/0008-5472.CAN-12-2796. PMID:23288510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bertucci F, Finetti P, Cervera N, Esterni B, Hermitte F, Viens P, Birnbaum D. How basal are triple-negative breast cancers? Int J Cancer. 2008;123(1):236–40. doi: 10.1002/ijc.23518. PMID:18398844. [DOI] [PubMed] [Google Scholar]
- 19.Rakha EA, Elsheikh SE, Aleskandarany MA, Habashi HO, Green AR, Powe DG, El-Sayed ME, Benhasouna A, Brunet J, Akslen LA, et al.. Triple-Negative Breast Cancer: Distinguishing between Basal and Nonbasal Subtypes. Clin Cancer Res. 2009;15:2302–10. [DOI] [PubMed] [Google Scholar]
- 20.Peddi PF, Ellis MJ, Ma C. Molecular Basis of Triple Negative Breast Cancer and Implications for Therapy. Internat J Breast Cancer. 2012; 2012, Article ID 217185, 7 pages. doi: 10.1155/2012/217185:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dent R, Hanna WM, Trudeau M, Rawlinson E, Sun P, Narod SA. Pattern of metastatic spread in triple-negative breast cancer. Breast Cancer Res Treat. 2009;115(2):423–8. doi: 10.1007/s10549-008-0086-2. PMID:18543098. [DOI] [PubMed] [Google Scholar]
- 22.Sorlie T, Perou CM, Tibshirani R, Aas R, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, et al.. Gene Expression Patterns of Breast Carcinomas Distinguish Tumor Subclasses with Clinical Implications. Proc Natl Acad Sci USA. 2001;98:10869–74. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eiermann W, Vallis KA. Locoregional treatments for triple-negative breast cancer. Ann Oncol. 2012;23 Suppl 6:vi30−34. doi: 10.1093/annonc/mds192. PMID:23012299. [DOI] [PubMed] [Google Scholar]
- 24.Silver DP, Richardson AL, Eklund AC, Wang ZC, Szallasi Z, Li Q, Juul N, Leong CO, Calogrias D, Buraimoh A, et al.. Efficacy of neoadjuvant Cisplatin in triple-negative breast cancer. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2010;28(7):1145–53. doi: 10.1200/JCO.2009.22.4725. PMID:20100965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wahba HA, El-Hadaad HA. Current approaches in treatment of triple-negative breast cancer. Cancer Biol Med. 2015;12(2):106–16. PMID:26175926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tan EY, Yan M, Campo L, Han C, Takano E, Turley H, Candiloro I, Pezzella F, Gatter KC, Millar EKA, et al.. The Key Hypoxia Regulated Gene CAIX is Upregulated in Basal-like Breast Tumours and is Associated with Resistance to Chemotherapy. British Journal of Cancer. 2009;100:405–11. doi: 10.1038/sj.bjc.6604844. PMID:19165203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trastour C, Benizri E, Ettore F, Ramaioli A, Chamorey E, Pouyssegur J, Berra E. HIF-1 alpha and CA IX Staining in Invasive Breast Carcinomas: Prognosis and Treatment Outcome. Int J Cancer. 2007;120:1451–8. [DOI] [PubMed] [Google Scholar]
- 28.Hughes P, Marshall D, Reid Y, Parkes H, Gelber C. The costs of using unauthenticated, over-passaged cell lines: how much more data do we need? Biotechniques. 2007;43(5):575, 577−578, 581-572 passim. doi: 10.2144/000112598. [DOI] [PubMed] [Google Scholar]
- 29.Li Y, Wang H, Oosterwijk E, Tu C, Shiverick KT, Silverman DN, Frost SC. Expression and Activity of Carbonic Anhydrase IX Is Associated with Metabolic Dysfunction in MDA-MB-231 Breast Cancer Cells. Cancer Investigation. 2009;27:613–23. doi: 10.1080/07357900802653464. PMID:19367501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pastorek J, Pastorekova S, Callebaut I, Mornon JP, Zeln¡k V, Opavsky R, Zat'ovicov M, Liao S, Portetelle D, Stanbridge EJ, et al.. Cloning and Characterization of MN, a Human Tumor-associated Protein with a Domain Homologous to Carbonic Anhydrase and a Putative Helix-Loop-Helix DNA Binding Segment. Oncogene. 1994;9:2877–88. PMID:8084592. [PubMed] [Google Scholar]
- 31.Zavada J, Zavadova Z, Pastorekova S, Ciampor F, Pastorek J, Zelnik V. Expression of MaTu-MN protein in human tumor cultures and in clinical specimens. Int J Cancer. 1993;54(2):268–74. doi: 10.1002/ijc.2910540218. PMID:8486430. [DOI] [PubMed] [Google Scholar]
- 32.Robertson N, Potter C, Harris AL. Role of Carbonic Anhydrase IX in Human Tumor Cell Growth, Survival, and Invasion. Cancer Res. 2004;64:6160–5. doi: 10.1158/0008-5472.CAN-03-2224. PMID:15342400. [DOI] [PubMed] [Google Scholar]
- 33.Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, Clark L, Bayani N, Coppe J, Tong F, et al.. A Collection of Breast Cancer Cell Lines for the Study of Functionally Distinct Cancer Subtypes. Cancer Cell. 2006;10:515–27. doi: 10.1016/j.ccr.2006.10.008. PMID:17157791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Elsheikh SE, Green AR, Rakha EA, Samaka RM, Ammar AA, Powe D, Reis-Filho JS, Ellis IO. Caveolin 1 and Caveolin 2 are associated with breast cancer basal-like and triple-negative immunophenotype. Br J Cancer. 2008;99(2):327–34. doi: 10.1038/sj.bjc.6604463. PMID:18612310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nakagawa M, Bando Y, Nagao T, Morimoto M, Takai C, Ohnishi T, Honda J, Moriya T, Izumi K, Takahashi M, et al.. Expression of p53, Ki-67, E-cadherin, N-cadherin and TOP2A in triple-negative breast cancer. Anticancer research. 2011;31(6):2389–93. PMID:21737670. [PubMed] [Google Scholar]
- 36.Changavi AA, Shashikala A, Ramji AS. Epidermal Growth Factor Receptor Expression in Triple Negative and Nontriple Negative Breast Carcinomas. J Lab Physicians. 2015;7(2):79–83. doi: 10.4103/0974-2727.163129. PMID:26417156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rae JM, Creighton CJ, Meck JM, Haddad BR, Johnson MD. MDA-MB-435 Cells Are Derived from M14 Melanoma Cells – a Loss for Breast Cancer, but a Boon for Melanoma Reseach. Breast Cancer Research and Treatment. 2007;104:13–19. doi: 10.1007/s10549-006-9392-8. PMID:17004106. [DOI] [PubMed] [Google Scholar]
- 38.Imbalzano KM, Tatarkova I, Imbalzano AN, Nickerson JA. Increasingly transformed MCF-10A cells have a progressively tumor-like phenotype in three-dimensional basement membrane culture. Cancer Cell Int. 2009;9:7. doi: 10.1186/1475-2867-9-7. PMID:19291318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zandberga E, Zayakin P, Abols A, Pupola D, Trapencieris P, Line A. Depletion of carbonic anhydrase IX abrogates hypoxia-induced overexpression of stanniocalcin-1 in triple negative breast cancer cells. Cancer Biol Ther. 2017:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jeon M, Han J, Nam SJ, Lee JE, Kim S. STC-1 expression is upregulated through an Akt/NF-kappaB-dependent pathway in triple-negative breast cancer cells. Oncol Rep. 2016;36(3):1717–22. doi: 10.3892/or.2016.4972. PMID:27461417. [DOI] [PubMed] [Google Scholar]
- 41.Chang AC, Doherty J, Huschtscha LI, Redvers R, Restall C, Reddel RR, Anderson RL. STC1 expression is associated with tumor growth and metastasis in breast cancer. Clin Exp Metastasis. 2015;32(1):15–27. doi: 10.1007/s10585-014-9687-9. PMID:25391215. [DOI] [PubMed] [Google Scholar]
- 42.Qu Y, Han B, Yu Y, Yao W, Bose S, Karlan BY, Giuliano AE, Cui X. Evaluation of MCF10A as a Reliable Model for Normal Human Mammary Epithelial Cells. PLoS One. 2015;10(7):e0131285. doi: 10.1371/journal.pone.0131285. PMID:26147507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen A, Beetham H, Black MA, Priya R, Telford BJ, Guest J, Wiggins GA, Godwin TD, Yap AS, Guilford PJ. E-cadherin loss alters cytoskeletal organization and adhesion in non-malignant breast cells but is insufficient to induce an epithelial-mesenchymal transition. BMC Cancer. 2014;14:552. doi: 10.1186/1471-2407-14-552. PMID:25079037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saha B, Chaiwun B, Imam SS, Tsao-Wei DD, Groshen S, Naritoku WY, Imam SA. Overexpression of E-cadherin protein in metastatic breast cancer cells in bone. Anticancer Res. 2007;27(6B):3903–8. PMID:18225549. [PubMed] [Google Scholar]
- 45.Chaffer CL, Thompson EW, Williams ED. Mesenchymal to epithelial transition in development and disease. Cells Tissues Organs. 2007;185(1-3):7–19. doi: 10.1159/000101298. PMID:17587803. [DOI] [PubMed] [Google Scholar]
- 46.Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer. 2009;9(4):265–73. doi: 10.1038/nrc2620. PMID:19262571. [DOI] [PubMed] [Google Scholar]
- 47.Schneider BP, Winer EP, Foulkes WD, Garber J, Perou jM, Richardson A, Sledge GW, Carey LA. Triple-Negative Breast Cancer: Risk Factors to Potential Targets. Clin Cancer Res. 2008;14:8010–8. [DOI] [PubMed] [Google Scholar]
- 48.Silverman DN, Lindskog S. The Catalytic Mechanism of Carbonic Anhydrase – Implication of a Rate Limiting Protolysis of Water. Acc Chem Res. 1988;21:30–36. doi: 10.1021/ar00145a005. [DOI] [Google Scholar]
- 49.Pacchiano F, Carta F, McDonald PC, Lou Y, Vullo D, Scozzafava A, Dedhar S, Supuran CT. Ureido-substituted benzenesulfonamides potently inhibit carbonic anhydrase IX and show antimetastatic activity in a model of breast cancer metastasis. J Med Chem. 2011;54(6):1896–902. doi: 10.1021/jm101541x. PMID:21361354. [DOI] [PubMed] [Google Scholar]
- 50.Lou Y, McDonald PC, Oloumi A, Chia S, Ostlund C, Ahmadi A, Kyle A, auf dem Keller U, Leung S, Huntsman D, et al.. Targeting Tumor Hypoxia: Suppression of Breast Tumor Growth and Metastasis by Novel Carbonic Anhydrase IX Inhibitors. Cancer Res. 2011;71:3364–76. doi: 10.1158/0008-5472.CAN-10-4261. PMID:21415165. [DOI] [PubMed] [Google Scholar]
- 51.Pastorekova S, Zavadova Z, Kostal M, Babusikova O, Zavada J. A Novel Quasi-viral Agent, MaTu, is a Two-Component System. Virology. 1992;187:620–6. doi: 10.1016/0042-6822(92)90464-Z. PMID:1312272. [DOI] [PubMed] [Google Scholar]
- 52.Markwell MAK, Haas SM, Lieber LL, Tolbert NE. A Modification of the Lowry Procedure to Simplify Protein Determination in Membrane and Lipoprotein Samples. Anal Biochem. 1978;87:206–10. doi: 10.1016/0003-2697(78)90586-9. PMID:98070. [DOI] [PubMed] [Google Scholar]