ABSTRACT
The gut microbiota is a contributing factor in obesity-related metabolic disorders. The effect of metformin on the gut microbiota has been reported; however, the relationship between the gut microbiota and the mechanism of action of metformin in elderly individuals is unclear. In this study, the effect of metformin on the gut microbiota was investigated in aged obese mice. The abundance of the genera Akkermansia, Bacteroides, Butyricimonas, and Parabacteroides was significantly increased by metformin in mice fed a high-fat diet. Metformin treatment decreased the expression of IL-1β and IL-6 in epididymal fat, which was correlated with the abundance of various bacterial genera. In addition, both fecal microbiota transplantation from metformin-treated mice and extracellular vesicles of Akkermansia muciniphila improved the body weight and lipid profiles of the mice. Our findings suggest that modulation of the gut microbiota by metformin results in metabolic improvements in aged mice, and that these effects are associated with inflammatory immune responses.
KEYWORDS: aged mice, Akkermansia muciniphila, extracellular vesicles, gut microbiota, metformin
Introduction
Obesity-related metabolic disorders, including type 2 diabetes (T2D) and hyperlipidemia, represent epidemiological and economic burdens on public health worldwide.1 These health problems in aging populations contribute to the early onset of various chronic morbidities.2 The etiology of metabolic disorders includes genetic and environmental factors; dietary pattern is a major contributing factor.3
The gut microbiota also plays important roles in energy metabolism and obesity-related metabolic diseases.4 A high-fat diet (HFD) contributes to dysbiosis of the gut microbiota, leading to metabolic disorders.5 Accordingly, various dietary interventions—including drugs, probiotics and prebiotics—that alter the composition of the gut microbiota are used to improve metabolic parameters.6,7 Moreover, the gut microbiota affects immune homeostasis in the gut, and altered immune responses in adipose tissue are linked with metabolic disorders.8 Chronic inflammation induced by cytokines contributes to metabolic disorders, including insulin resistance and dyslipidemia.9,10
Metformin is primarily used to treat T2D by suppressing glucose production in the liver, increasing insulin sensitivity, and enhancing peripheral glucose uptake in the liver and skeletal muscle.11 The molecular mechanism of action of metformin is not completely understood; however, activation of AMP-activated protein kinase (AMPK) has been proposed to regulate energy balance and glucose metabolism.12
In recent studies, modulation of the gut microbiota by metformin treatment has been suggested to lead to improvement of metabolic parameters, including those of obesity and insulin resistance.6,13 In animal models, metformin significantly modulated the composition of the gut microbiota.6,13 In particular, the increase in numbers of Akkermansia muciniphila caused by metformin was correlated with improved metabolic profiles.14,15 However, the mechanism of the effect of metformin on the gut microbiota is unclear. Also, the effect of metformin in the elderly may differ as elderly individuals have a gut microbiota with a higher proportion of Bacteroidetes than young adults; a significant increase in Akkermansia, and decreases in Bifidobacterium, Bacteriodes, and Clostridium cluster IV abundance have also been reported.16–18 Moreover, Biagi et al. reported that the abundance of specific bacterial genera in the gut is correlated with inflammatory status in elderly individuals.18
The aims of this study were to investigate the effect of metformin on the gut microbiota in elderly subjects. Furthermore, the mechanism underlying this effect was investigated.
Results
Metabolic improvement by metformin
Metformin administration for 16 weeks to mice fed a HFD significantly decreased the body weight and serum glucose level compared to mice fed only a HFD. Metformin also significantly improved glucose tolerance. Moreover, the total cholesterol, and LDL levels were significantly decreased by metformin, probably due to downregulation of ApoA-1 and ApoB (Fig. 1).
Figure 1.
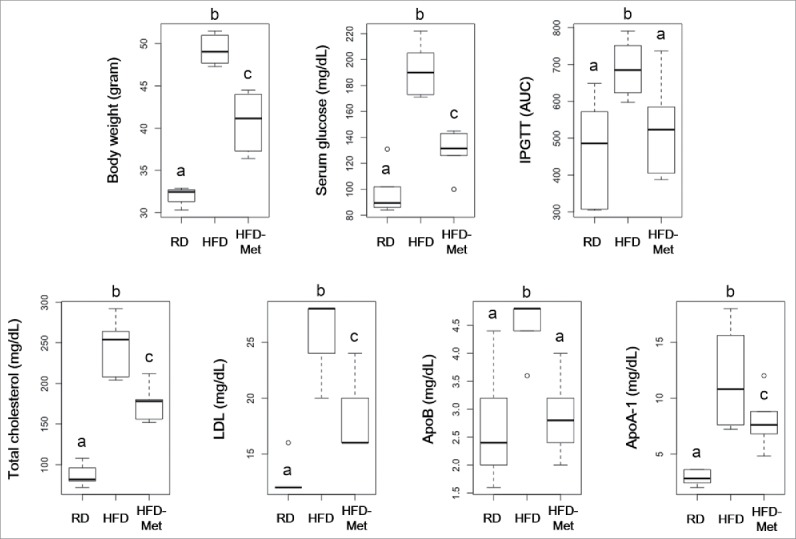
Effect of metformin on body weight, serum glucose, total cholesterol, and LDL. Five-week-old C57BL/6 mice were fed a HFD (45% lipid) for 39 weeks to induce metabolic disorders, then metformin was administered daily for 16 weeks. RD: Regular diet (n = 6); HFD: High-fat diet (n = 6); HFD-Met: Metformin administration during HFD feeding (n = 6). Different superscript letters indicate significant differences (P < 0.05) according to Duncan's post hoc test.
Effect of metformin on the gut microbiota
A total of 288,356 sequences were generated from 18 samples. An average of 11,599 ± 3,380 sequences were recovered per sample and used for comparative analysis. Figure 2a and b shows the differences in microbial diversity among the RD, HFD, and HFD-Met groups. The alpha diversities of the gut microbiota analyzed using Chao1 richness and Shannon index showed no significant differences among the groups (Fig. 2a). PCoA of UniFrac distances showed separation among the RD, HFD, and HFD-Met groups, that was more clearly clustered in unweighted PCoA (Fig. 2b).
Figure 2.
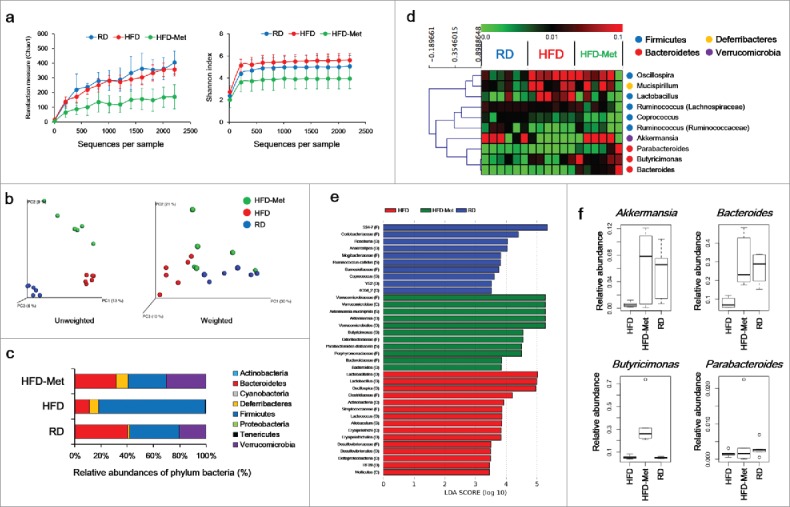
Microbial diversity according to diet and metformin administration. (a) Rarefaction curve and Shannon index of bacterial diversity. (b) PCoA of UniFrac distances. (c) Bacterial classification at the phylum level. (d) Hierarchical clustering of the top 10 most abundant bacterial genera using Spearman's rank correlation. (e) Significant differences in bacterial abundance according to diet and metformin treatment. Significant differences were identified by LEfSe analysis as a P < 0.05 by both the Kruskal–Wallis test (among classes) and Wilcoxon test (between subclasses). The threshold logarithmic LDA score was 3.0. (f) Abundance of Akkermansia, Butyricimonas, Bacteroides, and Parabacteroides according to diet and metformin administration, analyzed by qPCR.
Effect of metformin and diet on the composition of the gut microbiota
In the RD group, the phyla Bacteroidetes and Firmicutes comprised 40.0% ± 20.0% and 38.0% ± 16.0% of the gut microbiota, respectively; the Firmicutes/Bacteroidetes ratio was 1.4 ± 1.3 (Fig. 2c). HFD increased the abundance of Firmicutes (81.4% ± 13.4%), and decreased that of Bacteroidetes (10.8% ± 9.3%), resulting in a ratio of 28.7 ± 29.8 (Fig. 2c). In contrast, in the HFD-Met group, the abundance of Bacteroidetes and Firmicutes was 31.4% ± 35.3% and 29.0% ± 19.4%, respectively; the Firmicutes/Bacteroidetes ratio was 3.1 ± 1.9 (Fig. 2c). Additionally, the abundance of Verrucomicrobia in the HFD-Met group (30.2% ± 25.0%) was greater than that in the RD group (20.3% ± 22.2%) and HFD group (0.2% ± 0.4%) (Fig. 2c).
Among the 10 bacterial genera found in all three groups, Bacteroides, Butyricimonas, and Parabacteroides were of the phylum Bacteroidetes. These three genera clustered separately from the phylum Firmicutes, their abundance was enriched in the HFD-Met group compared to the RD and HFD groups, accounting for 23.1% ± 37.3% of the total identified bacterial OTUs (Fig. 2d). In contrast, five genera (Coprococcus, Lactobacillus, Oscillospira, Ruminococcus [Ruminococcaceae], and Ruminococcus [Lachnospiraceae]) were of the phylum Firmicutes. In LEfSe analysis, the abundance of Akkermansia, Bacteroides, Butyricimonas, and Parabacteroides was significantly greater in the HFD-Met group compared to the RD and HFD groups (Fig. 2e), those abundance was confirmed by qPCR (Fig. 2f).
Immunological changes in epididymal fat
IL-1β and IL-6 expression was significantly decreased in the HFD-Met group compared to the RD and HFD groups (Fig. 3a). Moreover, IL-1β and IL-6 expression was negatively correlated with the abundance of Bacteroides, Butyricimonas, Anaerotruncus, and Akkermansia (Fig. 3b).
Figure 3.
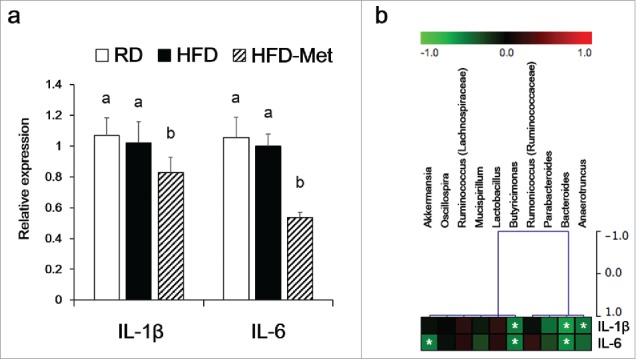
Immunological biomarker levels in epididymal fat pads. Relative mRNA levels compared to the RD group as determined by qPCR. (a) IL-1β and IL-6. Different superscript letters indicate significant differences (P < 0.05) according to Duncan's post hoc test. (b) Correlations between immunological biomarkers and the abundance of bacterial genera in the HFD-Met group (n = 6). *Statistical significance (Spearman's correlation coefficient; P < 0.05).
Effect of fecal material from metformin-treated mice on metabolic parameters
The body weights of the HFD- and fHFD-fed groups increased by 1.9 ± 1.9 and 3.3 ± 0.6 g after 4 weeks, respectively (Fig. 4a). The body weights of the fRD- and fMet-fed groups decreased by 2.9 ± 0.7 and 0.3 ± 1.6 g, respectively (Fig. 4a). Total cholesterol and LDL levels were lower in the fMet group than the fHFD group, albeit not significantly so (Fig. 4c). Serum glucose level was not significantly different between fMet group and fHFD group (Fig. 4b). In addition, the abundance of Bacteroidetes was significantly increased in the fMet group, and that of the genus Bacteroides was increased markedly, compared to the fHFD-fed group (Fig. 4d and e). FMT did not changed the expression level of IL-1β and IL-6 in epididymal fat (data now shown).
Figure 4.
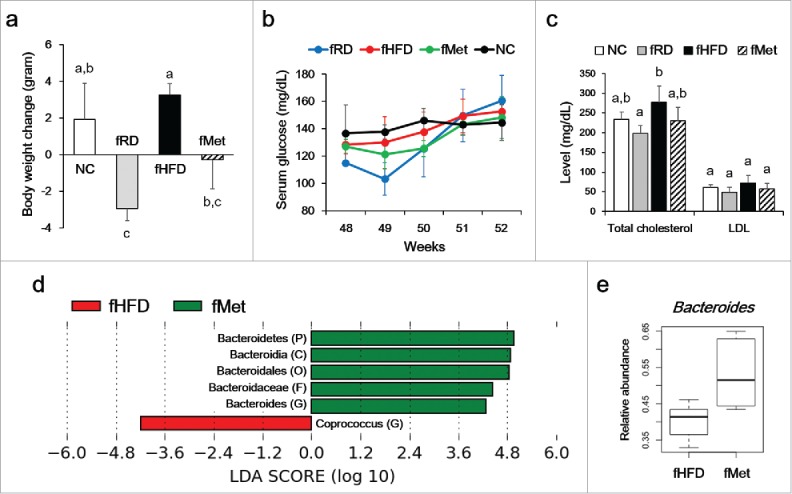
Effect of fecal microbiota transplantation (FMT) on body weight (a), serum glucose level (b), and lipid profile (c). Six-week-old mice were fed a HFD for 43 weeks, then inoculated orally with pooled fecal material from RD (fRD, n = 3)- metformin (fMet, n = 3)-, and HFD (fHFD, n = 3)-treated mice for 4 weeks. Different superscript letters indicate significant differences (P < 0.05) by Duncan's post hoc test. (d) Differences in bacterial communities after FMT. The threshold logarithmic LDA score was 3.0. (e) Abundance of Bacteroides was analyzed by qPCR.
Effect of extracellular vesicles of Akkermansia muciniphila (AkkEV) on metabolic parameters
After 5 weeks, weight gain in the AkkEV-fed group (0.5 ± 1.5 g) was significantly less than that in the HFD-fed group (3.0 ± 1.2 g) (Fig. 5a). The total cholesterol level in the AkkEV-fed group was lower than that in the HFD-fed group (Fig. 5c). The epididymal fat pad weight in the AkkEV-fed group was lower than that in the NC group, albeit not significantly so (P = 0.057) (Fig. 5d). There was no significant change in serum glucose level between HFD-fed group and AkkEV-fed group (Fig. 5b). AkkEV did not significantly change the composition of the gut microbiota (data not shown).
Figure 5.
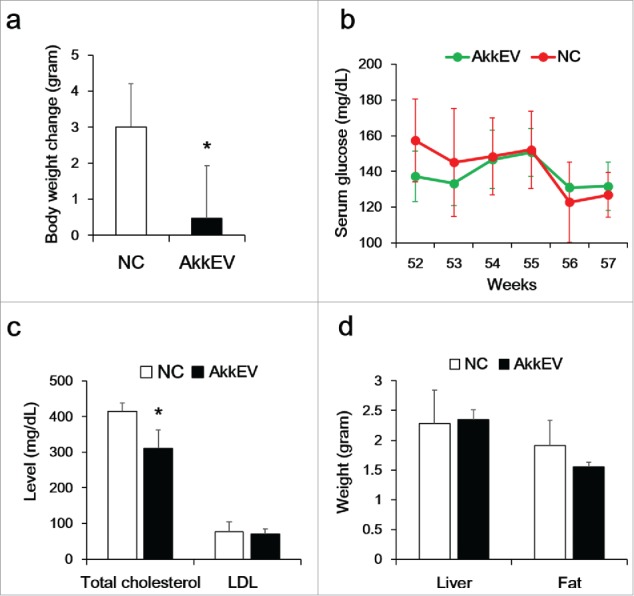
Effect of EVs of Akkermansia muciniphila (AkkEV) on body weight (a), serum glucose (b), and lipid profile (c). Six-week-old mice were fed a HFD for 48 weeks, and then orally administered AkkEV daily for 9 weeks (n = 3). (d) Weight of liver and epididymal fat. *Statistical significance (Wilcoxon rank-sum test; P < 0.05) compared to the NC group (n = 4).
Discussion
Metformin treatment affects the gut microbiota, leading to improvements in metabolic parameters, including obesity and insulin resistance.6 The metformin-induced increase in the abundance of A. muciniphila was associated with improvements in metabolic parameters. However, these effects were detected in young adult mice. Our findings indicate an age-specific effect of metformin on the gut microbiota and improvements in metabolic parameters in mice with HFD-induced obesity.
The gut microbiota of patients with T2D has been characterized, but recent studies have revealed that the results could have been biased by the confounding effect of metformin treatment.19–21 This confounding effect may have been due to imbalances in amino acid metabolism and SCFA production caused by the gut microbiota.20,21 As its effect on the gut microbiota contributes to the antidiabetic effect of metformin, we evaluated the characteristics of the gut microbiota in patients treated with metformin with the aim of identifying new therapeutic targets for T2D.22
A decrease in the Firmicutes/Bacteroidetes ratio is considered predictive of metabolic improvement by metformin treatment. Obese individuals have a high Firmicutes/Bacteroidetes ratio, particularly in the presence of metabolic comorbidities.23,24 Although various health conditions—such as antibiotic-associated diarrhea, Crohn's disease, and ulcerative colitis—could be confounding factors in the relationship between the Firmicutes/Bacteroidetes ratio and obesity.25,26, the difference in the Firmicutes/Bacteroidetes ratio between the HFD (28.7 ± 29.8) and HFD-Met (3.1 ± 1.9) groups was consistently significant in aged and adult mice (HFD, 2.6 ± 0.9; HFD-Met, 0.2 ± 0.2) in a previous study.6
The abundance of A. muciniphila (phylum Verrucomicrobia) was increased by metformin, irrespective of age, and was related to improvements in metabolic parameters. Metformin treatment significantly increased the abundance of A. muciniphila in obese mice.6,13 Moreover, the abundance of A. muciniphila was significantly higher in patients with T2D treated with metformin.27 Cross-talk between A. muciniphila and the host was reported recently.14,28 An increased abundance of A. muciniphila was associated with improved metabolic parameters, including obesity and insulin resistance, and regulation of IFN-γ expression in the small intestine was mediated by the effect of A. muciniphila on glucose metabolism.14,15
In addition, the abundance of Bacteroides, Butyricimonas, and Parabacteroides was significantly increased by metformin treatment; moreover, these three genera were not abundant in adult mice or RD mice.6 Therefore, an increased abundance of these genera is characteristic of the gut microbiota of aged obese mice fed a HFD. The increase in Bacteroides and Butyricimonas abundance was associated with improved metabolic parameters in mice treated with metformin. Short-chain fatty acids (SCFAs) produced by the gut microbiota ameliorate insulin resistance and inflammation.29 Butyrate, which is produced by Butyricimonas spp., protects against diet-induced obesity.30,31 Moreover, butyrate increased mucin production, which was also regulated by A. muciniphila, and mucus layer thickness highly related with metabolic improvement.14,32 Bacteroides spp. are abundant in non-obese individuals.33 Several strains of Bacteroides produce propionate and succinate, which protect against various metabolic disorders such as insulin resistance and diet-induced obesity.34,35 Further studies of the influence of Parabacteroides on metabolic parameters are warranted.
The improvements in metabolic parameters induced by metformin treatment were mediated by inflammatory immune responses in epididymal fat, and possibly associated with the altered gut microbiota. Various inflammatory responses in adipose tissue were related with metabolic disorders.36 IL-6 is a pleiotropic cytokine involved not only in inflammation but also in metabolic homeostasis.37,38 IL-6 is produced in the liver, adipose tissue, and muscle and has context-dependent pro- and anti-inflammatory effects, which influence metabolic disorders, including insulin resistance.37 IL-6 levels in adipose tissue reportedly increase with age,39 and IL-6 attenuates insulin signaling in adipocytes.40–42 In this study, a metformin-induced decrease in IL-6 expression by metformin treatment was detected in epididymal fat, and IL-6 expression was significantly negatively correlated with the abundance of Bacteroides and Butyricimonas. Therefore, the effect of the metformin-induced increase in the abundance of Bacteroides and Butyricimonas on insulin resistance in aged mice may be related to downregulation of IL-6. Moreover, IL-6 expression was not affected by metformin treatment in HFD-fed adult mice in a previous study,6 and downregulation of IL-6 was suggested to be related to the effect of modulation of the gut microbiota on insulin signaling in aged individuals. IL-1β, a major pro-inflammatory cytokine, plays an important role in obesity-related inflammation and insulin resistance.40-42 IL-1β signaling crosstalks with the IL-6 signaling cascade via activation of signal transducer and activator of transcription 3 (STAT3).37 IL-1β expression and the abundance of Bacteroides and Butyricimonas were negatively correlated in this study; therefore, IL-1β, as well as IL-6, was involved in the improvements of metabolic parameters.
FMT did not significantly modulate the bacterial community composition, except for Bacteroides, and did not downregulate IL-1β or IL-6. Therefore, the increase in the abundance of Akkermansia and Butyricimonas caused by metformin may play a key role in the downregulation of IL-1β and IL-6, which are reportedly related to the abundance of Akkermansia and Butyricimonas, respectively.13,44
The effect of A. muciniphila on metabolic parameters, such as body weight and lipid profiles, has been reported.13,14 Plovier et al. demonstrated that a membrane protein of A. muciniphila improved metabolism in obese and diabetic mice,43 which is in agreement with our data. Moreover, the abundance of A. muciniphila was negatively correlated with IL-6 expression. Therefore, the downregulation of IL-6 caused by the increased A. muciniphila abundance was involved in the improvement of metabolic parameters by metformin treatment. The active components of AkkEV have been reported to be 100–200 nm proteins.41 Further studies are required to better understand the characteristics of AkkEV.
The effect of metformin on the gut microbiota might be involved in its antidiabetic activity. Based on Koch's postulates, the mechanism of action of metformin involving the gut microbiota could be validated by FMT, which is used to assess the effect of alterations in the gut microbiota, from metformin-treated mice.45 FMT from metformin-treated donors improved the glucose tolerance of germ-free mice.22 This suggested the potential of FMT in the treatment of various metabolic disorders. However, the clinical application of FMT is limited, because an aseptic environment cannot be achieved in the gut. Moreover, daily feeding of fecal materials from HFD-Met mice decreased both body weight gain and lipid levels. In addition, although FMT did not alter the gut microbiota, FMT significantly increased the abundance of Bacteroides spp. This result suggests that FMT may exert a therapeutic effect without altering the pre-existing gut microbiota. Thus, further study of FMT using antibiotic-treated animal models is warranted. However, the mechanism underlying the effect of FMT on metabolic parameters is unclear, and should be the focus of further research.
In summary, metformin treatment significantly altered the gut microbiota in aged mice with HFD-induced obesity. In particular, the changes in Bacteroides and Butyricimonas abundance were age-specific. The abundance of A. muciniphila increased markedly; in previous studies, AkkEV were associated with the metformin-induced improvements in metabolic parameters in adult animals irrespective of age. Moreover, downregulation of IL-1β and IL-6 by metformin treatment in fat was negatively correlated with the abundance of several bacterial genera involved in the metformin-induced improvements in metabolic parameters. These results suggest modulation of the gut microbiota by metformin has a therapeutic effect on metabolic disorders in elderly individuals.
Materials and methods
Animal model
Male C57BL/6N mice were purchased from Samtako Co. Ltd (Osan, Republic of Korea) and were housed with free access to water and food in a temperature-humidity controlled animal facility under a 12 h light-dark cycle at 22 ± 2°C and 55 ± 5% humidity. Six-week-old mice were fed a HFD (45% kcal fat; FeedLab Inc.) for 39 weeks to induce metabolic disorders, including obesity and T2D. Metformin (HFD-Met: 250 mg/kg body weight, n = 6) was administered daily for the final 16 weeks of HFD feeding. Mice fed a regular diet (RD; 10% kcal fat, Purina Korea Inc., n = 6) and a HFD without metformin (HFD, n = 6) were used as the negative controls. All experimental protocols were approved by the Institutional Animal Care and Use Committee (IACUC) of Sahmyook University (No. 2015001).
Metabolic measurements
Body weight, serum glucose level, and food intake were measured once every other week. Serum glucose level was measured using an Accu-Chec Performa system (Roche) after starvation for 12 h. Intraperitoneal glucose tolerance testing (IPGTT) was performed at 16 weeks after metformin administration. Mice were interaperitoneally injected with glucose solution (2 g/kg body weight, in phosphate-buffered saline [PBS]), and glucose levels were measured 30, 60, 90, and 120 min after injection. Total cholesterol, low-density lipoprotein (LDL), apolipoprotein A-1 (ApoA-1), and apolipoprotein B (ApoB) levels were measured using a biochemical analyzer (AU480, Beckman Coulter).
Immunological biomarkers
Interleukin-1β (IL-1β; forward primer: 5′-CAGGATGAGGACATGACACC-5′, reverse primer: 5′-CTCTGCAGACTCAAACTCCAC-5′), and interleukin-6 (IL-6; forward primer: 5′-GTA CTC CAG AAG ACC AGA GC-5′, reverse primer: 5′-TGC TGG TGA CAA CCA CGG CC-5′) expression was determined in epididymal fat pads. Total RNA was extracted using a RiboEx™ (GeneAll, Korea), cDNA synthesis was performed using a HyperScript™ RT premix (GeneAll, Korea) according to the manufacturer's instructions. To quantify mRNA levels, SYBR® Green PCR Master Mix (Applied Biosystems) and a StepOnePlus™ real-time PCR system (Applied Biosystems) were used. β-Actin was used as an internal control.
Gut microbiota analysis
Total DNA was extracted using a PowerSoil DNA Isolation Kit (MO BIO Laboratories Inc.) from cecum samples including fecal materials. Amplification of partial sequences of 16S rRNA genes was performed based on a 16S rRNA amplification protocol from the Earth Microbiome Project.46 16S rRNA genes were amplified using the 515F/806R primer set, which includes an adapter sequence for amplification of the V4 region (515F forward primer: 5′-TCG TCG GCA GCG TCA GAT GTG TAT AAG AGA CAG GTG CCA GCM GCC GCG GTA A- 5′; 806R reverse primer: 5′-GTC TCG TGG GCT CGG AGA TGT GTA TAA GAG ACA GGG ACT ACH VGG GTW TCT AAT). To attach the dual indices and adapter to amplified polymerase chain reaction (PCR) products, index PCR was performed using an AmpONE™ α-Pfu DNA polymerase (GeneAll, Korea) and Nextera® XT Index Kit v2 (Illumina). PCR products after amplification and attach were purified using the Expin™ PCR SV (GeneAll, Korea). Sequencing of partial bacterial 16S rRNA genes was performed using the MiSeq Reagent Kit V3 (600 cycles) and MiSeq platform (Illumina) at KoBioLabs Inc.
Prior to analysis of 16S rRNA sequences, BCL files were converted into raw FASTQ files including read1, index and read2 sequences using CASAVA-1.8.2. After preprocessing (quality filtering and trimming steps using FASTX-Toolkit), sequences were assigned to operational taxonomic units (OTUs, 97% identity), and representative sequences were selected using QIIME 1.7.0 software.47 Next, taxonomic composition, alpha diversity and beta diversity were analyzed. LDA Effect Size (LEfSe) was used to estimate taxonomic abundance and characterize differences between groups.48 A heat map of functional gene abundance was generated using MultiExperiment Viewer (MEV) software (v4.8.1).
The relative abundance of four bacterial genera was confirmed using SYBR® Green PCR Master Mix (Applied Biosystems) and a StepOnePlus™ real-time PCR system (Applied Biosystems). The genus specific primer sets for universal total bacterial (UniF340; forward primer: 5′-ACTCCTACGGGAGGCAGCAGT-5′, UniR514; reverse primer: 5′-ATTACCGCGGCTGCTGGC-5′), Akkermansia (AM1; forward primer: 5′-CAGCACGTGAAGGTGGGGAC-5′, AM2; reverse primer: 5′-CCTTGCGGTTGGCTTCAGAT-5′) Bacteroides (AllBac296f; forward primer: 5′-GAGAGGAAGGTCCCCCAC-5′, AllBac412r; reverse primer: 5′-CGCTACTTGGCTGGTTCAG-5′), Butyricimonas (Buty1f; forward primer: 5′-GGTGAGTAACACGTGTGCAAC-5′, Buty1r; reverse primer: 5′-TACCCCGCCAACTACCTAATG-5′), and Parabacteroides (ASF519f; forward primer: 5′-TTGCCGTTGAAACTGGTTGA-5′, ASF519r; reverse primer: 5′-GGAGTTCTGCGTGATATCTATGCA-5′) were used for amplification.49–52
Fecal microbiota transplantation (FMT)
Fecal material was collected daily for 16 weeks and stored at −70°C. Fecal material from RD (fRD, n = 3)-, HFD-metformin (fMet, n = 3)-, and HFD (fHFD, n = 3)-treated mice was pooled in PBS, and mice fed a HFD for 48 weeks were orally administered 20 mg of fecal material for 4 weeks.
Extracellular vesicles of Akkermansia muciniphila (AkkEV)
Akkermansia muciniphila was purchased from the American Type Culture Collection (ATCC; BAA-835) and cultured in brain heart infusion broth (BHI; 0.025% resazurin, 0.05% L-cysteine) under anaerobic conditions at 37°C for 7 days. Culture medium was separated from A. muciniphila by centrifugation and filtration. Extracellular vesicles of A. muciniphila (AkkEVs) were isolated by ultracentrifugation at 150,000 g for 3 h at 4°C. AkkEVs were resuspended in PBS, and mice fed HFD for 52 weeks were orally administrated 20 µg AkkEVs daily for 5 weeks (n = 3). HFD-fed mice were used as negative controls (n = 4).
Statistical analysis
Data are expressed as means ± standard deviation (SD). In a relative abundance analysis using LEfSe based on the Kruskal–Wallis and Wilcoxon tests, significance was defined as a value of P < 0.05. The logarithmic LDA score threshold was set at 3.0. To quantify in vivo mRNA levels relative to an internal control (β-actin), the 2−ΔΔCt relative quantification method (ΔΔCt = (Ct.Target – Ct.β-actin)Group1 – (Ct.Target – Ct. β-actin)Group2) was used. Statistical significance was assessed by one-way analysis of variance (ANOVA), followed by Duncan's post hoc test. All statistical analyses were performed using RStudio. A P value < 0.05 was considered to indicate statistical significance.
Funding Statement
This work was supported by Korea Institute of Planning & Evaluation for Technology in Food, Agriculture, Forestry and Fisheries (iPET) (314044-3).
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
This paper was supported by the Sahmyook University research fund.
References
- 1.Mortality GBD, Causes of Death C Global, regional, and national life expectancy, all-cause mortality, and cause-specific mortality for 249 causes of death, 1980–2015: a systematic analysis for the Global Burden of Disease Study 2015. Lancet. 2016;388:1459–544. doi: 10.1016/S0140-6736(16)31012-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stuckler D. Population causes and consequences of leading chronic diseases: a comparative analysis of prevailing explanations. Milbank Quarterly. 2008;86:273–326. doi: 10.1111/j.1468-0009.2008.00522.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carnethon MR, Loria CM, Hill JO, Sidney S, Savage PJ, Liu K, Coronary Artery Risk Development in Young Adults study . Risk factors for the metabolic syndrome: the Coronary Artery Risk Development in Young Adults (CARDIA) study, 1985–2001. Diabetes Care. 2004;27:2707–15. doi:https://doi.org/ 10.2337/diacare.27.11.2707. [DOI] [PubMed] [Google Scholar]
- 4.Cani PD, Delzenne NM. The role of the gut microbiota in energy metabolism and metabolic disease. Curr Pharm Des. 2009;15:1546–58. doi: 10.2174/138161209788168164. [DOI] [PubMed] [Google Scholar]
- 5.Brown K, DeCoffe D, Molcan E, Gibson DL. Diet-induced dysbiosis of the intestinal microbiota and the effects on immunity and disease. Nutrients. 2012;4:1095–119. doi: 10.3390/nu4081095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee H, Ko G. Effect of metformin on metabolic improvement and gut microbiota. Applied Environmental Microbiol. 2014;80:5935–43. doi: 10.1128/AEM.01357-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xiao S, Fei N, Pang X, Shen J, Wang L, Zhang B, Zhang M, Zhang X, Zhang C, Li M, et al.. A gut microbiota-targeted dietary intervention for amelioration of chronic inflammation underlying metabolic syndrome. FEMS Microbiol Ecol. 2014;87:357–67. doi: 10.1111/1574-6941.12228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation. Cell. 2014;157:121–41. doi: 10.1016/j.cell.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gregor MF, Hotamisligil GS. Inflammatory mechanisms in obesity. Ann Rev Immunol. 2011;29:415–45. doi: 10.1146/annurev-immunol-031210-101322. [DOI] [PubMed] [Google Scholar]
- 10.Cai D, Yuan M, Frantz DF, Melendez PA, Hansen L, Lee J, Shoelson SE. Local and systemic insulin resistance resulting from hepatic activation of IKK-beta and NF-kappaB. Nat Med. 2005;11:183–90. doi: 10.1038/nm1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rotella CM, Monami M, Mannucci E. Metformin beyond diabetes: new life for an old drug. Curr Diabetes Rev. 2006;2:307–15. doi: 10.2174/157339906777950651. [DOI] [PubMed] [Google Scholar]
- 12.Hardie DG, Ross FA, Hawley SA. AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nat Rev Mol Cell Biol. 2012;13:251–62. doi: 10.1038/nrm3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shin NR, Lee JC, Lee HY, Kim MS, Whon TW, Lee MS, Bae JW. An increase in the Akkermansia spp. population induced by metformin treatment improves glucose homeostasis in diet-induced obese mice. Gut. 2013;63(5):727–35. doi: 10.1136/gutjnl-2012-303839. [DOI] [PubMed] [Google Scholar]
- 14.Everard A, Belzer C, Geurts L, Ouwerkerk JP, Druart C, Bindels LB, Guiot Y, Derrien M, Muccioli GG, Delzenne NM, et al.. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc Natl Acad Sci U S A. 2013;110:9066–71. doi: 10.1073/pnas.1219451110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Greer RL, Dong X, Moraes AC, Zielke RA, Fernandes GR, Peremyslova E, Vasquez-Perez S, Schoenborn AA, Gomes EP, Pereira AC, et al.. Akkermansia muciniphila mediates negative effects of IFNgamma on glucose metabolism. Nat Commun. 2016;7:13329. doi: 10.1038/ncomms13329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zwielehner J, Liszt K, Handschur M, Lassl C, Lapin A, Haslberger AG. Combined PCR-DGGE fingerprinting and quantitative-PCR indicates shifts in fecal population sizes and diversity of Bacteroides, bifidobacteria and Clostridium cluster IV in institutionalized elderly. Exp Gerontol. 2009;44:440–6. doi: 10.1016/j.exger.2009.04.002. [DOI] [PubMed] [Google Scholar]
- 17.Mariat D, Firmesse O, Levenez F, Guimaraes V, Sokol H, Dore J, Corthier G, Furet JP. The Firmicutes/Bacteroidetes ratio of the human microbiota changes with age. BMC Microbiol. 2009;9:123. doi: 10.1186/1471-2180-9-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Biagi E, Nylund L, Candela M, Ostan R, Bucci L, Pini E, Nikkïla J, Monti D, Satokari R, Franceschi C, et al.. Through ageing, and beyond: gut microbiota and inflammatory status in seniors and centenarians. PloS One. 2010;5:e10667. doi: 10.1371/journal.pone.0010667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F, Guan Y, Shen D, Peng Y, Zhang D, et al.. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490:55–60. doi: 10.1038/nature11450. [DOI] [PubMed] [Google Scholar]
- 20.Mardinoglu A, Boren J, Smith U. Confounding Effects of Metformin on the human gut microbiome in type 2 diabetes. Cell Metab. 2016;23:10–2. doi: 10.1016/j.cmet.2015.12.012. [DOI] [PubMed] [Google Scholar]
- 21.Forslund K, Hildebrand F, Nielsen T, Falony G, Le Chatelier E, Sunagawa S, Prifti E, Vieira-Silva S, Gudmundsdottir V, Pedersen HK, et al.. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature. 2015;528:262–6. doi: 10.1038/nature15766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu H, Esteve E, Tremaroli V, Khan MT, Caesar R, Manneras-Holm L, Ståhlman M, Olsson LM, Serino M, Planas-Fèlix M, et al.. Metformin alters the gut microbiome of individuals with treatment-naive type 2 diabetes, contributing to the therapeutic effects of the drug. Nat Medic. 2017;23(7):850–858. doi: 10.1038/nm.4345. [DOI] [PubMed] [Google Scholar]
- 23.Clarke SF, Murphy EF, Nilaweera K, Ross PR, Shanahan F, O'Toole PW, Cotter PD. The gut microbiota and its relationship to diet and obesity: new insights. Gut Microbes. 2012;3:186–202. doi: 10.4161/gmic.20168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Louis S, Tappu RM, Damms-Machado A, Huson DH, Bischoff SC. Characterization of the gut microbial community of obese patients following a weight-loss intervention using whole metagenome shotgun sequencing. PloS One. 2016;11:e0149564. doi: 10.1371/journal.pone.0149564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carding S, Verbeke K, Vipond DT, Corfe BM, Owen LJ. Dysbiosis of the gut microbiota in disease. Microbial Ecol Health Dis. 2015;26:26191. doi: 10.3402/mehd.v26.26191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ott SJ, Musfeldt M, Wenderoth DF, Hampe J, Brant O, Folsch UR, Timmis KN, Schreiber S. Reduction in diversity of the colonic mucosa associated bacterial microflora in patients with active inflammatory bowel disease. Gut. 2004;53:685–93. doi: 10.1136/gut.2003.025403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de la Cuesta-Zuluaga J, Mueller NT, Corrales-Agudelo V, Velasquez-Mejia EP, Carmona JA, Abad JM, Escobar J. Metformin is associated with higher relative abundance of mucin-degrading akkermansia muciniphila and several short-chain fatty acid-producing microbiota in the gut. Diabetes Care. 2017;40:54–62. doi: 10.2337/dc16-1324. [DOI] [PubMed] [Google Scholar]
- 28.Derrien M, Vaughan EE, Plugge CM, de Vos WM. Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int J Systematic Evolutionary Microbiol. 2004;54:1469–76. doi: 10.1099/ijs.0.02873-0. [DOI] [PubMed] [Google Scholar]
- 29.den Besten G, van Eunen K, Groen AK, Venema K, Reijngoud DJ, Bakker BM. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J Lipid Res. 2013;54:2325–40. doi: 10.1194/jlr.R036012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gao Z, Yin J, Zhang J, Ward RE, Martin RJ, Lefevre M, Cefalu WT, Ye J. Butyrate improves insulin sensitivity and increases energy expenditure in mice. Diabetes. 2009;58:1509–17. doi: 10.2337/db08-1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin HV, Frassetto A, Kowalik EJ Jr, Nawrocki AR, Lu MM, Kosinski JR, Hubert JA, Szeto D, Yao X, Forrest G, et al.. Butyrate and propionate protect against diet-induced obesity and regulate gut hormones via free fatty acid receptor 3-independent mechanisms. PloS One. 2012;7:e35240. doi: 10.1371/journal.pone.0035240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hatayama H, Iwashita J, Kuwajima A, Abe T. The short chain fatty acid, butyrate, stimulates MUC2 mucin production in the human colon cancer cell line, LS174T. Biochem Biophys Res Commun. 2007;356:599–603. doi: 10.1016/j.bbrc.2007.03.025. [DOI] [PubMed] [Google Scholar]
- 33.Kasai C, Sugimoto K, Moritani I, Tanaka J, Oya Y, Inoue H, Tameda M, Shiraki K, Ito M, Takei Y, et al.. Comparison of the gut microbiota composition between obese and non-obese individuals in a Japanese population, as analyzed by terminal restriction fragment length polymorphism and next-generation sequencing. BMC Gastroenterol. 2015;15:100. doi: 10.1186/s12876-015-0330-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chakraborti CK. New-found link between microbiota and obesity. World J Gastrointestinal Pathophysiol. 2015;6:110–9. doi: 10.4291/wjgp.v6.i4.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rios-Covian D, Arboleya S, Hernandez-Barranco AM, Alvarez-Buylla JR, Ruas-Madiedo P, Gueimonde M, et al.. Interactions between Bifidobacterium and Bacteroides species in cofermentations are affected by carbon sources, including exopolysaccharides produced by bifidobacteria. Applied Environmental Microbiol. 2013;79:7518–24. doi: 10.1128/AEM.02545-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lackey DE, Olefsky JM. Regulation of metabolism by the innate immune system. Nat Rev Endocrinol. 2016;12:15–28. doi: 10.1038/nrendo.2015.189. [DOI] [PubMed] [Google Scholar]
- 37.Mauer J, Denson JL, Bruning JC. Versatile functions for IL-6 in metabolism and cancer. Trends Immunol. 2015;36:92–101. doi: 10.1016/j.it.2014.12.008. [DOI] [PubMed] [Google Scholar]
- 38.Scheller J, Chalaris A, Schmidt-Arras D, Rose-John S. The pro- and anti-inflammatory properties of the cytokine interleukin-6. Biochim Et Biophys Acta. 2011;1813:878–88. doi: 10.1016/j.bbamcr.2011.01.034. [DOI] [PubMed] [Google Scholar]
- 39.Starr ME, Saito M, Evers BM, Saito H. Age-associated increase in cytokine production during systemic inflammation-II: The role of IL-1beta in age-dependent IL-6 upregulation in adipose tissue. J Gerontol Series A, Biol Sci Med Sci. 2015;70:1508–15. doi: 10.1093/gerona/glu197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rotter V, Nagaev I, Smith U. Interleukin-6 (IL-6) induces insulin resistance in 3T3-L1 adipocytes and is, like IL-8 and tumor necrosis factor-alpha, overexpressed in human fat cells from insulin-resistant subjects. J Biol Chem. 2003;278:45777–84. doi: 10.1074/jbc.M301977200. [DOI] [PubMed] [Google Scholar]
- 41.Kang CS, Ban M, Choi EJ, Moon HG, Jeon JS, Kim DK, Park SK, Jeon SG, Roh TY, Myung SJ, et al.. Extracellular vesicles derived from gut microbiota, especially Akkermansia muciniphila, protect the progression of dextran sulfate sodium-induced colitis. PloS One. 2013;8:e76520. doi: 10.1371/journal.pone.0076520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weigert C, Hennige AM, Brodbeck K, Haring HU, Schleicher ED. Interleukin-6 acts as insulin sensitizer on glycogen synthesis in human skeletal muscle cells by phosphorylation of Ser473 of Akt. Am J Physiol Endocrinol Metab. 2005;289:E251–7. doi: 10.1152/ajpendo.00448.2004. [DOI] [PubMed] [Google Scholar]
- 43.Plovier H, Everard A, Druart C, Depommier C, Van Hul M, Geurts L, Chilloux J, Ottman N, Duparc T, Lichtenstein L, et al.. A purified membrane protein from Akkermansia muciniphila or the pasteurized bacterium improves metabolism in obese and diabetic mice. Nat Med. 2017;23:107–13. doi: 10.1038/nm.4236. [DOI] [PubMed] [Google Scholar]
- 44.Jangi S, Gandhi R, Cox LM, Li N, von Glehn F, Yan R, Patel B, Mazzola MA, Liu S, Glanz BL, et al.. Alterations of the human gut microbiome in multiple sclerosis. Nat Commun. 2016;7:12015. doi: 10.1038/ncomms12015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lundberg R, Toft MF, August B, Hansen AK, Hansen CH. Antibiotic-treated versus germ-free rodents for microbiota transplantation studies. Gut Microbes. 2016;7:68–74. doi: 10.1080/19490976.2015.1127463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gilbert JA, Meyer F, Antonopoulos D, Balaji P, Brown CT, Brown CT, Desai N, Eisen JA, Evers D, Field D, et al.. Meeting report: the terabase metagenomics workshop and the vision of an Earth microbiome project. Stand Genomic Sci. 2010;3:243–8. doi: 10.4056/sigs.1433550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, et al.. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–6. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Collado MC, Derrien M, Isolauri E, de Vos WM, Salminen S. Intestinal integrity and Akkermansia muciniphila, a mucin-degrading member of the intestinal microbiota present in infants, adults, and the elderly. Applied Environmental Microbiol. 2007;73:7767–70. doi: 10.1128/AEM.01477-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Croswell A, Amir E, Teggatz P, Barman M, Salzman NH. Prolonged impact of antibiotics on intestinal microbial ecology and susceptibility to enteric Salmonella infection. Infect Immunity. 2009;77:2741–53. doi: 10.1128/IAI.00006-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Layton A, McKay L, Williams D, Garrett V, Gentry R, Sayler G. Development of Bacteroides 16S rRNA gene TaqMan-based real-time PCR assays for estimation of total, human, and bovine fecal pollution in water. Applied Environmental Microbiol. 2006;72:4214–24. doi: 10.1128/AEM.01036-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cahenzli J, Koller Y, Wyss M, Geuking MB, McCoy KD. Intestinal microbial diversity during early-life colonization shapes long-term IgE levels. Cell Host Microbe. 2013;14:559–70. doi: 10.1016/j.chom.2013.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]


