Highlights
-
•
Native rhizobium strains effectively nodulated a popular groundnut variety in Ghana.
-
•
Rhizobium inoculation increased biological N2 fixation in groundnut grown in soil.
-
•
Ghanaian groundnut strains are genetically related to Bradyrhizobium yuanmingense.
-
•
Native isolates are a potential source of strains for local inoculant production.
Keywords: Native isolates, Biological nitrogen fixation, Genetic diversity, Arachis hypogaea L.
Abstract
The identification of locally-adapted rhizobia for effective inoculation of grain legumes in Africa’s semiarid regions is strategic for developing and optimizing cheap nitrogen fixation technologies for smallholder farmers. This study was aimed at selecting and characterising effective native rhizobia, from Ghanaian soils for groundnut (Arachis hypogaea L.) inoculation. From surface-disinfected root nodules of cowpea and groundnut plants grown on farmers’ fields, 150 bacterial isolates were obtained, 30 of which were eventually found to nodulate groundnut plants. After testing the symbiotic potential of these isolates on groundnut on sterilized substrate, seven of them, designated as KNUST 1001–1007, were evaluated in an open field pot experiment using 15N-labelled soil. Although 15N dilution analyses did not indicate differences among treatments in the proportion of nitrogen (N) derived from the atmosphere (%Ndfa), all seven strains increased total N derived from N2 fixation by inoculated groundnut plants as compared to the non-inoculated control. Inoculation with KNUST 1002 led to total N accumulation as high as that of the groundnut reference strain 32H1. Genetic characterisation of the isolates by sequence analysis of 16S rRNA gene, 16S – 23S rRNA intergenic transcribed spacer (ITS) region and nodC gene revealed that isolates KNUST 1003 and 1007 were related to Rhizobium tropici, a common bean symbiont. The other five isolates, including KNUST 1002 belonged to the Bradyrhizobium genus, being closely related to Bradyrhizobium yuanmingense. Therefore, this study revealed novel native Ghanaian rhizobia with potential for the development of groundnut inoculants.
1. Introduction
Groundnut (Arachis hypogaea L.) is a multipurpose grain legume, which is considered a nutritious component in diets and a source of income for smallholder farmers in developing countries (Carlberg, 2012). In Ghana, about 94% of the groundnut production is concentrated in the northern region; a place considered as one of West Africa’s main groundnut production areas (Tsigbey et al., 2003). In terms of symbiotic nitrogen fixation, groundnut has been found to form effective association with both fast and slow growing ‘rhizobia’ of the Rhizobium and Bradyrhizobium genera, respectively (Taurian et al., 2002). Among the Bradyrhizobium strains identified to nodulate groundnut are: Bradyrhizobium arachidis, Bradyrhizobium japonicum, Bradyrhizobium elkanii, Bradyrhizobium lablabi, Bradyrhizobium yuanmingense and Bradyrhizobium iriomotense (Taurian et al., 2006, El-Akhal et al., 2008, Chang et al., 2011, Muñoz et al., 2011, Wang et al., 2013). Other species that nodulate groundnut include Rhizobium gardinii and Rhizobium tropici (Taurian et al., 2006). Despite the nitrogen-fixing ability of groundnut, yields are often below their maximum potential (Nutsugah et al., 2007). These low yields have been partially attributed to low inherent soil fertility and nutrient deficiencies in N and P mostly limit productivity of this crop (Maheswar and Sathiyavani, 2012, Mohamed and Abdalla, 2013).
Options such as mineral nitrogen application and rhizobium inoculation have been considered as means to supply legumes with N (Mweetwa et al., 2014). Apart from the possible adverse environmental consequences of excessive mineral nitrogen application (Trindade et al., 2001, Flechard et al., 2007), farmers are unable to exploit this option due to financial constraints. Thus, the more feasible alternative is the use of rhizobium inoculants. The practice of inoculation with highly effective rhizobium strains has been identified, among other factors, as an essential means to promote biological nitrogen fixation (BNF) with subsequent increases in grain yields (Unkovich and Pate, 2000). The usefulness of this BNF process is made evident when legumes depending on atmospheric N2 produce increased yields in soils in which non-legume crops would require a substantial amount of mineral nitrogen. In addition, inoculation of groundnut has led to considerable increases in nodulation, growth and productivity (Sajid et al., 2010, Sharma et al., 2011, Mohamed and Abdalla, 2013). Despite the potential benefits of inoculation, farmers rarely apply inoculants to groundnut because of the consensus that the association between groundnut and native soil rhizobia is usually adequate. Another factor that could contribute to the limited use of inoculants by farmers is the low awareness of the higher economic returns from the use of inoculants relative to mineral nitrogen (Ndakidemi et al., 2006). Limited availability of groundnut inoculants (i.e. of exotic origin) and lack of local strains for inoculating the crop particularly in Ghana, further exacerbate the limited use of inoculants.
To improve inoculation response of tropical legumes, Nkot et al. (2008) suggested the use of indigenous rhizobia as inoculants. For example, improvement in nodulation and N2 fixation was reported when groundnut was inoculated with native rhizobia (Bogino et al., 2008). Shishido and Pepper, 1990, Sattar et al., 1995 suggested that strains isolated from a particular region are the most effective for a given crop in that same region. In addition, rhizobia that are moderately to highly effective have been found to be well represented among the native population (Herridge et al., 2008) and could serve as a source of elite strains for local inoculant production. This emphasizes the need to identify elite isolates adapted to the prevailing environmental conditions for improved BNF. In selecting rhizobia strains for use as inoculants, the characteristics competitiveness in nodule formation and effectiveness in nitrogen fixation are considered (Stephens and Rask, 2000).
Conversely, the symbiotic potential and genetic diversity of groundnut-nodulating rhizobia is yet to be investigated, particularly in the context of Ghanaian agriculture. Previous reports based on the analyses of 16S rRNA and RFLP revealed a large diversity within cowpea- and soybean-nodulating strains only at the genus level (Abaidoo et al., 2000, Fening et al., 2004). Therefore, it is imperative to assess the diversity within the native rhizobium populations that nodulate groundnut and to estimate their contribution to N2 fixation in grain legumes. The aim of this study was to characterise rhizobia capable of nodulating groundnut using molecular tools and to identify elite strains for groundnut inoculation. To this end, symbiotic potential and phenotypic tests in addition to sequence analyses of 16S rRNA gene, 16S – 23S rRNA intergenic transcribed spacer (ITS) region and symbiotic genes; nodC and nifH were carried out to reveal the diversity within groundnut nodulating rhizobium and identify elite strains for improved inoculation response.
2. Materials and methods
2.1. Recovery and authentication of Rhizobium isolates
Groundnut and cowpea nodules were collected from farmers’ fields across the three regions in northern Ghana at the flowering stage and sampling points were located using a GPS (Supplementary Fig. S1). Recovered nodules were kept on desiccated silica gel and transported to the microbiology laboratory, Kwame Nkrumah University of Science and technology (KNUST) in Kumasi, Ghana, for isolation. Dried nodules were rehydrated in sterile distilled water overnight. After rehydration, whole nodules were surface sterilised using 95% ethanol for 10 s and transferred into a 3% hydrogen peroxide solution for 3 min. The nodules were then rinsed in several changes of sterilised distilled water to remove the remaining hydrogen peroxide as described by Somasegaran and Hoben (1994). Sterilised nodules were carefully crushed onto YMA (yeast mannitol agar) plates (Fred and Waksman, 1928) under aseptic conditions using heat-sterilised forceps. The resulting plates were incubated at 28 °C and monitored for 10 days. Bacterial colonies were repeatedly streaked on YMA medium to obtain pure cultures.
To authenticate isolates as true rhizobia, a nodulation test was carried out under aseptic and controlled conditions using cowpea (Vigna unguiculata L. Walp, cv. Asontem) as the test host. Cowpea was selected for this initial screening because of its highly promiscuous nodulation pattern and for being easily cultivable in growth pouches. Cowpea seeds were prepared (Section 2.4) and pre-germinated on moist sterile tissue paper in Petri dishes and incubated at 28 °C for three days. Seedlings with equal radicle length (2 cm) were selected and aseptically transferred into plastic growth pouches (Mega International, USA) containing N-free plant nutrient solution (Broughton and Dilworth, 1970). After seeding, the growth pouches were arranged on a wooden rack and placed in the greenhouse at KNUST, Kumasi, Ghana. A week after transplanting, broth cultures of each of the isolates were used to inoculate the cowpea seedlings. At 28 days after inoculation, the seedlings were assessed for nodulation and isolates that induced nodule formation on the test host were considered as true rhizobia. Where no nodules were observed, the isolate was not subjected to further studies. Isolates confirmed as true rhizobia were maintained on agar slants and also in 25% (w/v) glycerol (at −20 °C) for short term and long term (−80 °C) storages, respectively.
2.2. Symbiotic potential of native isolates on groundnut in sterilised sand in Ghana
The sixty-five isolates, that were considered true rhizobia based on the authentication test on cowpea, were evaluated for their symbiotic potential together with recommended/commercial strains namely: Bradyrhizobium diazoefficiens USDA 110 (soybean strain from Florida, USA) (Delamuta et al., 2013), and two Brazilian elite-strains, Bradyrhizobium pachyrhizi strain BR 3262 and Bradyrhizobium yuanmingense strain BR 3267 (Leite et al., 2017). USDA 110 is a strain widely used in commercial inoculants for soybean in Africa and in characterising newly cultured isolates.
The groundnut variety ‘Chinese’ (an early maturing variety preferred by most farmers in Ghana) was used. For the first experiment, four-litre capacity pots were filled with 3 kg of sterilised river sand and arranged in the greenhouse at KNUST, Kumasi, Ghana. Prior to filling the pots, the sand was sterilised in an autoclave at 121 °C for 1 h (Lupwayi and Haque, 1994). Broughton and Dilworth (Broughton and Dilworth, 1970) N-free nutrient solution was used to irrigate the plants weekly. The strains were classified by a symbiotic effectiveness index (SEI) that was calculated from the shoot dry matter (SDM) of the groundnut plants inoculated with a specific isolate divided by the SDM of groundnut plants inoculated with the reference strain BR 3267, expressed as a percentage (Yates et al., 2016).
2.3. Nitrogen fixation contribution of isolates on groundnut in 15N labelled soil in Brazil
The second experiment was conducted in pots in the open field at Embrapa Agrobiologia, Seropédica, Brazil. The planting medium used was soil classified as an Alfisol (US Soil Taxonomy Classification) obtained from Piracicaba, São Paulo State, Brazil, with a history of 15N enrichment since the 1980s through the application of 15N labelled organic matter (Tsai, Siu Mui, CENA, Piraicicaba, personal communication). Due to the compacted nature of the soil, it was mixed with 50% sand to improve drainage. Prior to the experiment, the chemical properties of the soil were analysed using the methods of Souza and Nogueira (2005): pH in H2O, 5.3; exchangeable Al, Ca and Mg: 0.04, 0.96 and 0.18 cmolcd−1, respectively; and P and K were 16.2 and 21.7 mgL−1, respectively. The soil had sand, silt and clay fractions of 14%, 22% and 64% respectively and fell within the clay textural class. Seven effective isolates identified from the first experiment alongside the reference strains BR 3267 and three other effective/recommended groundnut strains, Bradyrhizobium sp. strain BR 10254 (Torres-J et al., 2014), Bradyrhizobium sp. strain 32H1 (Urtz and Elkan, 1996) and SEMIA 6144 (Menna et al., 2009) were used as the treatments. Also included were three non-N2-fixing reference plants: non-nodulating (NN) soybean (Glycine max), NN common bean (Phaseolus vulgaris) and sorghum (Sorghum bicolor, cv. BR 305). Each experimental unit consisted of five-litre capacity pot filled with 4 kg of soil, amended with 732 mg P2O5, 241 mg K2O and the specific treatment. Clean tap water was used to irrigate the experiment every week. In both experiments, two un-inoculated controls; (1) with nitrogen (70 ppm in the form of 0.05% KNO3 for the first experiment and 100 mg N in the form of NH4NO3 for the second experiment) and (2) without nitrogen (-N) were included.
2.4. Bacterial culture, experiment management and experimental design
Broth cultures of each of the isolates and reference strains used in this study were prepared by inoculating a loop-full of pure culture in yeast mannitol broth (YMB). The cultures were then incubated in an orbital incubator at 125 rpm and 28 °C until the late logarithmic growth phrase where an O.D.600 nm of 1.0 was achieved. Groundnut seeds were surfaced sterilised with 95% ethanol for 30 s and 3% hydrogen peroxide solution for 3 min followed by several rinses in sterilised distilled water (Somasegaran and Hoben, 1994). Five seeds were planted per pot with the help of a pair of sterile forceps and thinned to two plants one week after planting. One mL culture of each of the test isolates or reference strain was used to inoculate the seeds at planting. Unless otherwise stated, all bacteria cultures, seed preparation and planting in this study followed the procedure outlined in this section. Both trials were arranged in a randomized complete block design (RCBD) with four replicates.
2.5. Data collection
Plants were harvested 45 days after planting for the experiments conducted in sand and soil/sand mixture (i.e. in Ghana and Brazil, respectively). Groundnut shoots were separated from the roots at the soil surface level. Nodulated roots and detached nodules were collected and stored in polythene bags. Samples were transported to the microbiology laboratory, KNUST in Ghana and Embrapa-Agrobiologia in Brazil, respectively, for processing. Nodules were separated from roots by gently washing the root system under running tap water to remove all debris and adhering sand or soil after which the nodules were detached, counted and oven dried together with shoots at 65 °C for 72 h to estimate dry biomass.
For the second experiment, dried plant shoots were ground to fine powder using a roller miller similar to that described by Arnold and Schepers (2004). Total N contents of all plant samples and seeds were analysed using the semi-micro Kjeldahl procedure as described by Urquiaga et al. (1992). The 15N enrichment of aliquots of sub-samples containing between 35 and 70 µg of N were weighed into tin capsules and determined using an automated continuous-flow isotope-ratio mass spectrometer consisting of a Costech EA Model ECS 4010 automatic C and N analyzer coupled to a Thermo Delta V Advantage mass spectrometer (Costech Analytical, Valencia, CA, USA). Since the 15N dilution technique was used for this study, the 15N enrichment of the N derived from the soil was estimated by discounting the N derived from seed and its excess 15N content using the formula proposed by Boddey et al. (1995):
where SC indicates corrected for seed N, at.% xs is the atom% 15N excess and Ps is the proportion of the seed N that was assimilated by the plant tissue. Ps was assumed to be 50% since the correction was done for shoot tissue only (Okito et al., 2004).
For the proportion of N derived from the air (%Ndfa) via BNF, the equation of (Chalk, 1985) was used:
where at.% 15Nxs is the atom% 15N excess of the shoot tissue of the plants. As three different non-N2-fixing reference crops were included, individual as well as combined estimates of the %Ndfa for each isolate could be calculated using both the total N difference method and 15N isotope dilution method.
2.6. Statistical analysis
Data measured for the various experiments were subjected to analyses of variance using SISVAR (Ferreira, 2008). Where overall probability was significant (p < 0.05), means were separated using Scott Knott at 5% probability. Relationships among isolates and sampling sites were explored with principal component analysis using the software STATGRAPHICS Centurion V16.1.11 (StatPoint Technologies Inc, Warrenton, VA)
2.7. Morpho - cultural characteristics of effective isolates
Characterisation of effective isolates was carried out when at least three isolated colonies were observed after streaking on YMA with bromothymol blue as pH indicator. Characteristics analysed were: pH reaction of culture medium, number of days to form colonies, colony elevation and form and mucus production. Bacterial cultures were given identification numbers (Table 1) and deposited in the culture collections of Johanna Döbereiner Biological Resources Center (CRB-JD), Embrapa Agrobiologia, Brazil and the KNUST Microbiology Laboratory, Kumasi, Ghana.
Table 1.
GenBank accession numbers of sequences obtained in this study.
|
Bradyrhizobium isolatesa |
Genbank accession number |
||||
|---|---|---|---|---|---|
| KNUST ID | CRB-JD ID | 16S rRNA | ITS | nodC | nifH |
| KNUST 1001 | BR 10839 | KY229769 | MF108830 | KY040459 | KY040464 |
| KNUST 1002 | BR 10840 | KY229770 | MF108831 | KY040460 | KY040465 |
| KNUST 1004 | BR 10824 | KY229772 | MF108832 | KY040461 | KY040466 |
| KNUST 1005 | BR 10841 | KY229773 | MF108833 | KY040462 | KY040467 |
| KNUST 1006 | BR 10842 | KY229774 | MF108834 | KY040463 | KY040468 |
| Rhizobium isolates | |||||
| KNUST 1003 | BR 10837 | KY229771 | – | – | – |
| KNUST 1007 | BR 10838 | KY229775 | – | – | – |
KNUST ID: Kwame Nkrumah University Technology culture collection identification, CRB-JD: Johanna Döbereiner Biological Resource Center culture collection identification.
2.8. DNA extraction, PCR amplification and gene sequencing
Bacterial genomic DNA was extracted using the Wizard genomic DNA purification kit (Promega, USA). Extracted DNA was submitted to amplification of 16S rRNA gene (Lane, 1991) and the intergenic transcribed spacer (ITS) region between the 16S and 23S rRNA genes (Cardinale et al., 2004). Additionally, the symbiotic gene nodC (Sarita et al., 2005) and the nitrogenase reductase gene nifH (Poly et al., 2001) were amplified. For each of the genes amplified, conditions for the PCR specified by the cited references were employed. PCR amplicons were subjected to bi-directional sequencing using the BigDye® Terminator v3.1 Cycle Sequencing Kit (ThermoFisher). Sequencing reaction products were subjected to post-reaction clean-up and analysed using an ABI 3500 Genetic Analyzer (Thermo Fisher). Quality control and sequence assembly were performed using BioNumerics 7 (Applied Maths, Belgium).
2.9. Phylogenetic analyses
Multiple nucleotide sequence alignments were generated using CLUSTAL W (Thompson et al., 1994) and phylogenetically analysed using MEGA7 software (Kumar et al., 2016). Concatenated sequence analyses of the 16S rRNA and the ITS region for the isolates in this study as well as type-strains of recognized Bradyrhizobium species were aligned and trimmed to the same length. Aligned sequences of the different genes were then concatenated using the Seaview program (Galtier et al., 1996). The maximum likelihood reconstruction method was used in calculating the phylogenetic trees of individual and concatenated genes. The most suitable models for generating phylogenetic trees of individual and concatenated genes were determined for each alignment using the integrated model selection tool of MEGA7. The strength of the phylogenetic tree topologies was evaluated using the bootstrap method by applying 500 pseudo replicates (Felsenstein, 1985). DNA sequences obtained for the various isolates after sequencing have been deposited in the GenBank database under the accession numbers in Table 1.
3. Results
3.1. Symbiotic potential of Ghanaian rhizobium isolates on groundnut in sterilised sand
A total of 65 bacterial isolates were authenticated after inducing root nodules when inoculated individually on cowpea plants grown in sterile growth pouches in a greenhouse.
These 65 authenticated strains were then tested for their performance by inoculating them on groundnut grown in pots with sterilised sand medium. The negative control (groundnut plants without inoculation or nitrogen fertilizer) did not form any nodules and showed nitrogen deficiency symptoms. Among the 65 authenticated rhizobia, only 30 induced nodulation in groundnut, demonstrating a difference in micro-symbiont specificity between groundnut and cowpea. Significant treatment effects on nodule number were observed following analysis of variance with three of the isolates producing statistically more (p < 0.05) nodules than all the reference strains (Table 2). Isolate KNUST 1006 produced nodule numbers similar to that of the reference strain BR 3267. With the exception of KNUST 1002, all the isolates that induced increased nodule numbers also resulted in increased nodule dry weights that were significantly (p < 0.05) higher than that observed for the reference strain, BR 3267. Isolate KNUST 1007 also caused a significant increase in nodule dry weight. Treatment with isolates KNUST 1001 and 1002 caused the greatest increase (p < 0.05) in shoot dry weight (Table 2). Symbiotic effectiveness of isolates also varied significantly (p < 0.05) among the isolates with isolates KNUST 1001 and 1002 performing better than the reference strain BR 3267 (Table 2). Twelve of the isolates had significantly lower symbiotic effectiveness indices (SEI) than all the reference strains. The lowest SEI was recorded for the control treatment without nitrogen and isolate KNUST 1020. Profiling the symbiotic effectiveness of isolates placed 23% into the effective group (i.e. SEI > 75%). The remaining isolates were considered as partially effective (Table 2).
Table 2.
Nodulation and shoot dry weight of inoculated groundnut and symbiotic effectiveness of isolates in sterilised sand.
| Isolate/strain |
Source | Nodule number | Nodule dry weight (mg pot−1) | Shoot dry weight (g pot−1) | Symbiotic effectiveness index (%) | |
|---|---|---|---|---|---|---|
| FIELD ID | KNUST ID | |||||
| 2NAG 52b1 | KNUST 1001† | Konta | 137.3 a | 150.0c | 7.04 a | 132.95 a |
| 2NAG 53e | KNUST 1002† | Yipaani | 105.0 b | 100.0 d | 7.20 a | 136.00 a |
| 2NAG 9d | KNUST 1003† | Punyoro kb | 99.3 b | 290.3 a | 4.94 c | 93.34 c |
| 2NAG 8a | KNUST 1004† | Kandiga 2 | 24.0 e | 70.0 e | 3.89 e | 73.56 e |
| 2NAG 75b | KNUST 1005† | Akuokayili | 18.0 e | 44.0 f | 4.23 d | 79.98 d |
| 2NAG 08e | KNUST 1006† | Kandiga 2 | 84.7c | 183.3 b | 5.84 b | 110.48 b |
| 2NAG 87c | KNUST 1007† | Boro | 12.3 f | 180.0 b | 5.02 c | 94.86 c |
| 2NAG 01e | KNUST 1008 | Tamale | 37.3 d | 47.7 f | 2.34 h | 44.23 h |
| 2NAG 08d | KNUST 1009 | Kandiga 2 | 10.3 f | 45.7 f | 3.25 f | 61.35 f |
| 2NAG 09b | KNUST 1010 | Punyoro kb | 5.0 f | 24.7 f | 2.54 h | 47.95 h |
| 2NAG 11d | KNUST 1011 | Kandiga | 6.3 f | 34.0 f | 2.93 g | 55.33 g |
| 2NAG 11f | KNUST 1012 | Kandiga | 13.3 f | 23.0 f | 2.39 h | 45.19 h |
| 2NAG 11 g | KNUST 1013 | Kandiga | 15.3 f | 86.3 d | 3.88 e | 73.32 e |
| 2NAG 13e | KNUST 1014 | Naaga | 7.0 f | 24.7 f | 2.41 h | 45.62 h |
| 2NAG 19d | KNUST 1015 | Akuokayili 1 | 9.3 f | 24.3 f | 2.36 h | 44.53 h |
| 2NAG 20a | KNUST 1016 | Pishigu | 6.3 f | 28.7 f | 2.45 h | 46.23 h |
| 2NAG 70 g | KNUST 1017 | Kuncheni | 5.3 f | 28.3 f | 3.17 f | 59.96 f |
| 2NAG 71b | KNUST 1018 | Zaguo deryiri | 5.0 f | 36.3 f | 3.73 e | 70.36 e |
| 2NAG 72a | KNUST 1019 | Zaguo deryiri | 13.0 f | 46.0 f | 3.70 e | 69.99 e |
| 2NAG 73e | KNUST 1020 | Gbare | 6.7 f | 44.0 f | 2.01 i | 37.91 i |
| 2NAG 75b | KNUST 1021 | Saawie | 12.3 f | 46.0 f | 3.79 e | 71.65 e |
| 2NAG 80d | KNUST 1022 | Varimpere | 12.0 f | 21.3 f | 3.13 f | 59.13 f |
| 2NAG 81b | KNUST 1023 | Varimpere | 8.3 f | 57.7 e | 2.65 h | 50.03 h |
| 2NAG 84e | KNUST 1024 | Chiatanga | 8.7 f | 19.7 f | 2.48 h | 46.86 h |
| 2NAG 85c | KNUST 1025 | Dorima | 7.0 f | 34.3 f | 2.57 h | 48.59 h |
| 2NAG 87a | KNUST 1026 | Boro | 9.3 f | 56.0 e | 3.32 f | 62.75 f |
| 2NAG 87d | KNUST 1027 | Boro | 12.3 f | 40.7 f | 3.47 f | 65.66 f |
| 2NAG 92b | KNUST 1028 | Tabiasi 1 | 5.3 f | 19.0 f | 2.94 g | 55.49 g |
| 2NAG 93e | KNUST 1029 | Tabiasi 2 | 15.3 f | 41.3 f | 2.84 g | 53.59 g |
| 2NAG 97a | KNUST 1030 | Kpalga | 22.7 e | 130.0 c | 4.18 d | 78.90 d |
| Non-Inoculated | – | – | 1.81 i | 34.21 i | ||
| Reference strains | ||||||
| USDA 110 | USA | 10.0 f | 31.7 f | 3.39 f | 64.1 f | |
| BR 3262 | Brazil | 21.3 e | 60.3 e | 3.54 f | 66.84 f | |
| BR 3267 | Brazil | 80.7 c | 78.3 d | 5.29 c | 100.00 c | |
| CV (%) | 27.93 | 23.35 | 7.17 | 7.34 | ||
Means in the same column followed by the same letter are not significantly different at P < 0.05 (Scott Knott Test). 2NAG = Phase2N2Africa, † Isolates selected for second experiment.
Principal component analysis gave a better understanding of the symbiotic potential of the isolates and their biogeographic distribution across sampling sites. Two principal components explained 94.5% of the variation in symbiotic potential of test isolates and the three reference strains. The first principal component explained 84.1% of the variation, which was dominated by shoot dry weight (Supplementary Fig. S2). The second component explained 10.5% of the variation with nodule dry weight being the main contributing variable. Shoot dry weight and symbiotic effectiveness index pointed towards the same direction and were close to each other demonstrating a correlation between these two variables (Supplementary Table S1). Isolates that clustered in the direction of the shoot dry weight and symbiotic effectiveness variables, together with the reference strain BR 3267, recorded high values. On the other hand, isolates that clustered on the opposite side (i.e. to the left) produced lower values for the variables considered. A large proportion of the sampling sites harboured strains with SEI of between 25 and 75% and clustered on the left side.
3.2. Nitrogen fixation potential of selected isolates on groundnut grown in soil in Brazil
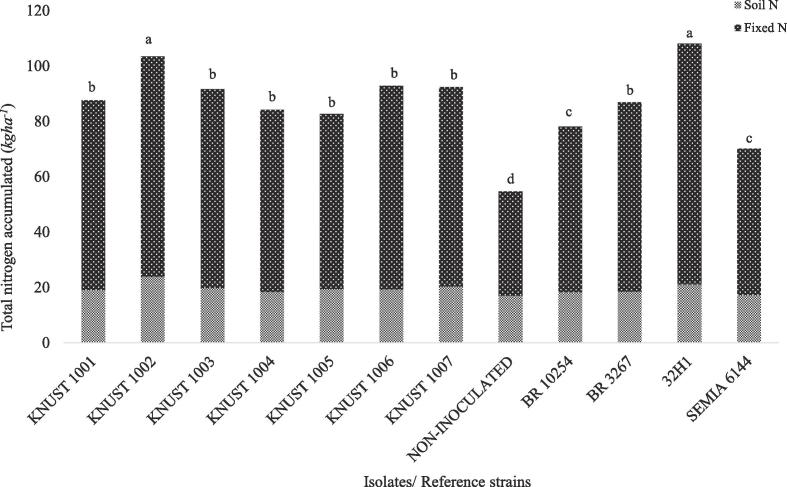
Seven best-performing isolates from the first experiment were selected for a second test using 15N labelled soil as the growth medium. All the selected isolates, except KNUST 1004, were ranked effective (with SEI > 75%) in the previous experiment. Although isolate KNUST 1030 had a SEI > 75% in the first experiment, it was not selected because re-isolation after the first experiment failed, thus the next best performing isolate from the partially effective group (i.e. KNUST 1004 with SEI = 73.6%) was selected. Significant (p < 0.05) differences were observed among isolates in terms of nodulation, shoot dry weight and nitrogen accumulated in the shoot. Nodulation in all the treatments was significantly (p < 0.05) higher than the un-inoculated control (–N). The non-nodulating (NN) soybean and common bean on the other hand did not show any signs of nodulation. The symbiotic association between three of the isolates and the test host resulted in increased nodule dry weight comparable to the control treatment inoculated with the 32H1 reference strain. Isolates that induced high nodule dry weights did not necessarily produce higher nodule numbers and vice versa (Table 3). Among the reference strains used, 32H1 was the most effective in terms of shoot dry weight while isolate KNUST 1002 produced the highest (p < 0.05) shoot dry weight among the test isolates. Considering the effect of inoculation on shoot N accumulation, isolate KNUST 1002 promoted a significant (p < 0.05) increase in N accumulation of groundnut shoot when compared to all the other test isolates (Fig. 1). The performance of isolate KNUST 1002 was not significantly different from that of the reference strain 32H1. Generally, the shoot dry weight and N accumulation of inoculated groundnut plants were superior to the reference plants (Table 3 and Fig. 1).
Table 3.
Nodulation and shoot dry weight of inoculated groundnut and reference plants grown in 15N labelled soil.
| Isolate/strain/reference plant | Nodule number | Nodule dry weight |
Shoot dry weight |
|---|---|---|---|
| kg ha−1 | |||
| KNUST 1001 | 341.0 e | 146.6 a | 2672.5 c |
| KNUST 1002 | 411.5 d | 147.5 a | 2867.5 b |
| KNUST 1003 | 564.8 a | 132.9 a | 2745.0 c |
| KNUST 1004 | 337.5 e | 146.5 a | 2556.3 d |
| KNUST 1005 | 456.3 c | 114.8 b | 2331.3 e |
| KNUST 1006 | 495.8 c | 120.4 b | 2687.5 c |
| KNUST 1007 | 488.3c | 120.1 b | 2596.3 d |
| NON-INOCULATED | 416.0 d | 87.0 c | 1677.5 g |
| BR 10254 | 523.0 b | 147.8 a | 2340.0 e |
| BR 3267 | 499.5 c | 126.4 b | 2565.0 d |
| 32H1 | 498.3 c | 151.3 a | 3146.3 a |
| SEMIA 6144 | 476.3 c | 137.1 a | 2193.8 f |
| NN common bean | – | – | 466.3 i |
| NN Soybean | – | – | 712.5 h |
| Sorghum | – | – | 300.0 j |
| CV (%) | 5.9 | 8.6 | 2.9 |
Means in the same column followed by the same letter are not significantly different at P < 0.05 (Scott Knott Test). NN: non-nodulating.
Fig. 1.
Total nitrogen accumulation and estimates of N derived from the air. Bars followed by the same letter are not significantly different at P < 0.05 (Scott Knott Test).
In general, the 15N enrichment values of inoculated plants were lower than all the reference plants. The 15N enrichment data showed that there were very small amounts of N derived from the soil. The level of 15N enrichment suggests that, for the N-rich legume seeds of NN common bean (9.3 mg N seed−1) and NN soybean (13.0 mg N seed−1), the large differences in seed N content (which were not enriched with 15N) were responsible for much of the isotope dilution. For this reason, only the sorghum plant (seed N content 0.62 mg N seed−1) was used as a reference for estimating the 15N enrichment of the N derived from the soil with the assumption that 50% of the shoot N was derived from the seed. The values for the percentage of nitrogen derived from the atmosphere (%Ndfa) ranged from 88 to 93% with no significant (p < 0.05) difference between treatments (Supplementary Table S2). However, there were large and significant differences between the values for the total N derived from BNF (Fig. 1). Isolate KNUST 1002 accumulated more N from BNF than any other strain except the reference strain 32H1 and most of the other strains isolated in Ghana were statistically similar to that of the reference strain BR 3267. The lowest amount of N fixed was recorded for the non-inoculated treatment.
3.3. Morpho – cultural and genetic characterisation of effective isolates
The seven isolates selected as most effective on groundnut grown on sterilised sand were characterised based on morpho-cultural and molecular genetic characteristics. Five isolates (KNUST 1001, 1002, 1004, 1005 and 1006) formed isolated colonies within six to seven days, alkalinized the culture medium and formed opaque-white colonies; traits that are consistent with the genus Bradyrhizobium. Isolates KNUST 1003 and KNUST 1007 acidified the culture medium, forming circular and elevated colonies. These fast-growing isolates, the colonies of which formed within three days, produced abundant mucus that was shiny in appearance while slow-growing isolates produced colonies with reduced mucus and were more opaque in appearance (Supplementary Table S3).
The phylogeny of the selected strains was studied by analysing the near-complete sequence of their 16S rRNA gene and ITS region. Basic Local Alignment Search Tool (BLAST) analysis of the 16S rRNA sequences confirmed that isolate KNUST 1001, 1002, 1004, 1005 and 1006 belonged to the genus Bradyrhizobium. The fast-growing isolates KNUST 1003 and 1007 were highly similar to members of the genus Rhizobium (Supplementary Fig. S3a and b).
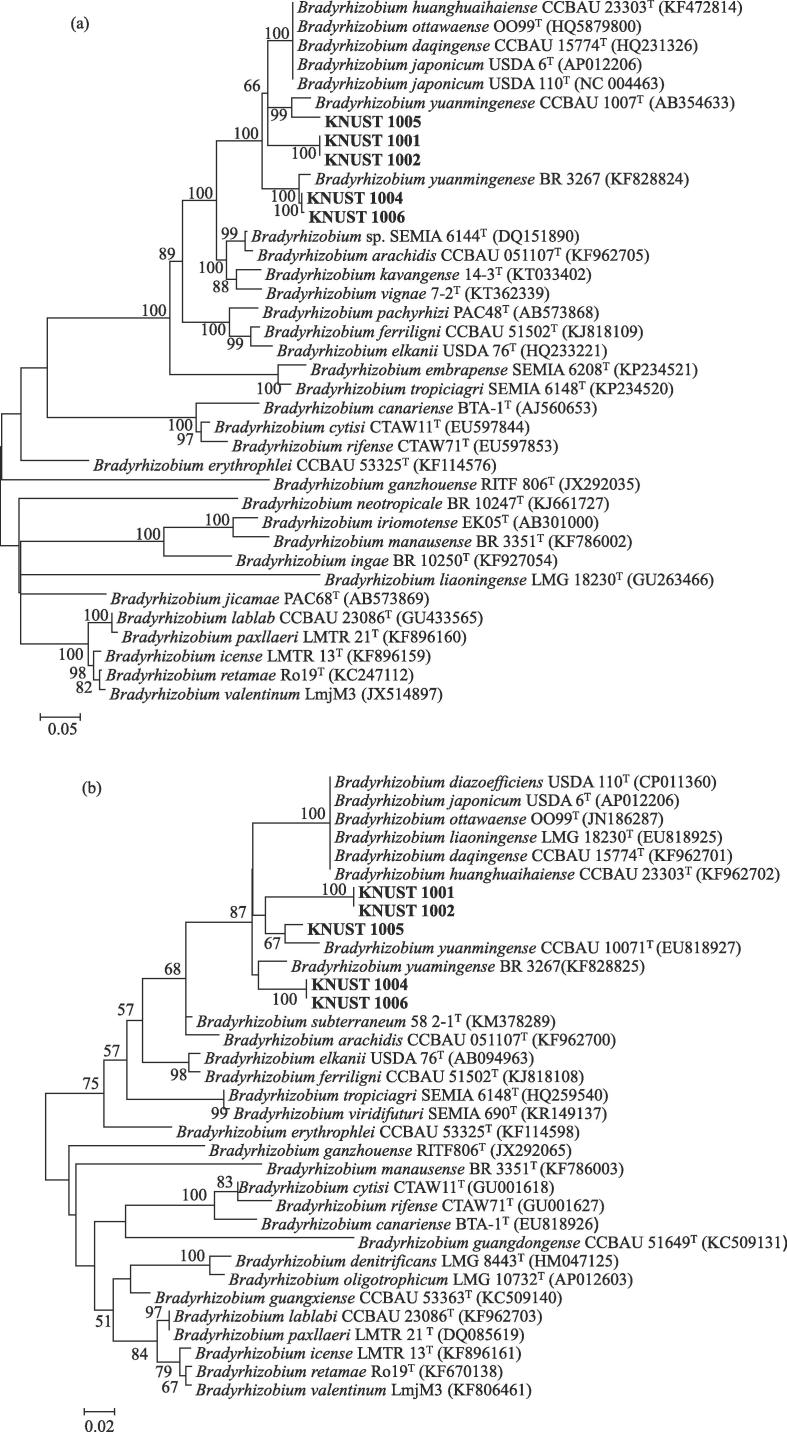
Within the genus Bradyrhizobium, 16S rRNA sequences are too conserved to permit for a more detailed phylogenetic classification at the species level. The ITS sequence between the 16S and 23S rRNA genes can be used to improve this phylogenetic resolution. Therefore, in this study, phylogenetic analysis was performed on the concatenated sequences of the 16S rRNA gene (1243 nt) and the ITS region (1027 nt) giving a total of 2270 nt. In this analysis, the five Bradyrhizobium isolates from this study clustered together on the same branch with close relation to B. yuanmingense CCBAU 10071T, Bradyrhizobium daqingense CCBAU 15774T and Bradyrhizobium subterraneum 48 2-1T with bootstrap support of at least 71% and with nucleotide sequence similarity values between 99.1 and 99.3% (Fig. 2, Supplementary Table S4). Generally, the concatenated analyses of 16S rRNA gene and ITS region revealed a clearer relationship between clustering of Bradyrhizobium strains or isolates compared to their individual analyses (Supplementary Fig. S3a and S4).
Fig. 2.
Unrooted maximum likelihood phylogenetic tree based on concatenated 16S rRNA gene and ITS sequences showing relationships among isolates and type-strains (T) of the genus Bradyrhizobium. Bootstrap values were inferred from 500 replicates and are indicated at the tree nodes when ≥50%. GenBank accession numbers are provided in parentheses. The bar represents two estimated substitutions per 100 nucleotide positions.
3.4. Analyses of symbiotic genes
The phylogenetic relationship of the nodC gene from the novel Bradyrhizobium isolates in relation to validly described species was studied. The nodC phylogenetic analyses placed two of the isolates, KNUST 1004 and 1006, on a branch together with B. yuanmingense BR 3267T with 100% bootstrap support (Fig. 3a). The isolates KNUST 1001, 1002 and 1005 were together on a branch with their sequences most closely related to B. yuanmingense CCBAU 1007T, Bradyrhizobium ottawaense OO99T, Bradyrhizobium japonicum USDA 6T, Bradyrhizobium huanghuaihaiense CCBAU 23303T and Bradyrhizobium denitrificans LMG 8443T, with 65% bootstrap support. In agreement with the nodC phylogeny, phylogenetic analyses of the nifH gene also placed the isolate KNUST 1005 in a branch together with B. yuanmingense CCBAU 10071T followed by the inclusion of isolates KNUST 1001 and 1002. Isolates KNUST 1004 and 1006 also shared close relation to B. yuanmingense BR 3267T (Fig. 3b). Therefore, in general, the nodC and nifH phylogenies were congruent with the phylogeny estimated based on the concatenated 16S rRNA and ITS sequences.
Fig. 3.
Unrooted maximum likelihood phylogenetic tree based on nodC (a) and nifH (b) genes showing relationships among isolates and type-strains (T) of the genus Bradyrhizobium. Bootstrap values were inferred from 500 replicates and are indicated at the tree nodes when ≥50%. GenBank accession numbers are provided in the parenthesis. The bar represents five or two estimated substitutions per 100 nucleotide positions.
4. Discussion
4.1. Authentication and symbiotic potential of isolates
In tropical soils, there is an enormous diversity of rhizobia with different nodulation capacities, which forms a natural reserve of germplasm for the selection of strains with desired characteristics (Dilworth et al., 2001). When assessing the relationship between rhizobia and their host, infectivity and symbiotic effectiveness are the two essential features commonly considered (Brockwell, 1998). The symbioses between legumes and rhizobium must be effective for enhanced BNF and subsequent yield improvement to be realized. In this study, a preliminary screening for authentic rhizobia was performed using cowpea in growth pouches because this species is easier to grow under such conditions than the target species groundnut. The variation in numbers of infective isolates between cowpea and groundnut reflects differences in host-range among bacteria and host plant species and also confirms earlier studies that report that groundnut has a more restrictive nodulation pattern than cowpea (Thies et al., 1991). Nevertheless, the inoculation experiments described herein demonstrate that it was possible to obtain several effective rhizobia for groundnut inoculation, notably Bradyrhizobium sp. KNUST 1002, which caused increments in nodulation, shoot dry weight and amount of N fixed in both experiments. The increase in nodulation observed following inoculation may be attributed to the favorable chemical properties of the study soil (such as pH). The possibility of achieving good nodulation and N2 fixation above a pH of 5.2 has been reported by Hungria and Vargas (2000) and which was the case in this study. The outstanding performance of Bradyrhizobium sp. KNUST 1002 could imply that it was more compatible with the legume host than the other test isolates.
The effectiveness of the symbiotic association between the infective isolates and their host in this study revealed varying effectiveness classes with some test isolates resulting in significantly higher SEI than the reference strains used. Useful variations in characteristics required in inoculant strains such as symbiotic effectiveness have also been observed within the natural pool of soil rhizobia (O’Hara et al., 2002). Nitrogen fixation efficiency has been found to be diverse, ranging from symbiotic interactions leading to little or no nitrogen fixation, to those that obtain nitrogen in levels equivalent to or even greater than plants treated with mineral N (Terpolilli et al., 2008). Similar results have been reported in other studies carried out to evaluate rhizobium cultures of various tropical legumes for their symbiotic capacity (Florentino et al., 2010, Marra et al., 2012). Additionally, the uneven distribution of effective isolates demonstrated in the principal component analysis (Supplementary Fig. S2) highlights a wide variation in terms of geographic distribution and symbiotic performance (Abaidoo et al., 2007). In order to overcome sub-optimal N2 fixation, the need arises to acquire rhizobium with high N2 fixing ability that are also well adapted to the prevailing environment (Yates et al., 2016). Estimation of the contribution of strains to nitrogen fixation has been based on methods such as N2 balance, N2 difference, 15N natural abundance, ureide analyses, acetylene reductase assay, hydrogen evolution and 15N isotope dilution method (Unkovich et al., 2008). The latter method was employed in this study to quantify the amount of N2 fixed for the selected effective isolates (experiment 2). Three reference plants were included in this experiment to estimate % Ndfa, because of the difficulty in directly determining which reference crop would accumulate N with the same 15N enrichment as the legume crop. Boddey et al. (1995) thus recommended that several reference plants should be utilized to produce individual estimates of BNF contribution with the range of these estimates considered as an index of their accuracy. The lower 15N enrichment recorded by inoculated plants implies that they received contributions of unlabeled N through BNF. On the other hand, the estimates of 15N enrichments of the NN legume reference plants were extremely high and which could be due to their large seed N contents. Naegle et al. (2005), indicated that the response of seedlings’ (of soybean) to available N is strongly related to available seed N resources. Since the seed N content of the NN reference plants were so high, and the availability of labelled soil N was so low, the results suggest that the N derived from the seed was considerably higher than 50%. The estimated 15N enrichments for these two NN legume crops of approximately 0.19–0.20 atom% excess were far higher than that of the small-seeded sorghum and as such, it is thought impossible that these NN legumes obtained only half of their N from seed reserves. Therefore, to avoid over estimation of BNF, the proportion of N derived from the atmosphere was estimated using sorghum as the reference crop.
The insignificant differences observed for the various treatments in terms of the 15N enrichment was not surprising since all the isolates selected for this experiment were potentially effective. The un-inoculated treatment resulted in the proportion of N derived from the atmosphere that was similar to all other treatments, but the amount of BNF contributed by the former was significantly lower. The results also show that, the proportion of plant accumulated N derived from BNF by inoculated plants ranged between 88 and 93%. The large differences between the different strains was apparent only in the total accumulated N and the total N derived from BNF. The Bradyrhizobium strain 32H1 was the best and most consistent reference strain in all parameters measured supporting the claim that it is an effective strain on groundnut (Urtz and Elkan, 1996). All isolates except KNUST 1005 performed similar to the reference strain BR 3267. The results from this study indicate the presence of indigenous rhizobia strains with highly effective symbiotic capacities that can be used as inoculants.
4.2. Genetic characterisation of effective isolates
Morpho-cultural characterisation of isolates used in this study indicated that they belong to the genera Bradyrhizobium and Rhizobium and this was confirmed by BLAST analysis of 16S rRNA gene sequences. Groundnut has been found previously to form associations with strains from the Rhizobium genus in addition to Bradyrhizobium symbionts (Van Rossum et al., 1995, Urtz and Elkan, 1996, Yang et al., 2005). Five out of seven isolates in this study belong to the Bradyrhizobium genus affirming the observation by several authors that bacteria associated with peanut are predominantly Bradyrhizobium (Van Rossum et al., 1995, Zhang et al., 1999, Yang et al., 2005). The classification of novel species has been based on a polyphasic approach, which considers phenotypic and genetic characteristics (Vandamme et al., 1996). This approach employs the sequencing of 16S rRNA gene as the backbone of genetic classification (Garrity and Holt, 2001). However, this gene has been found to be limited in delineating the diversity within Bradyrhizobium at the species level (Wang and Martínez-Romero, 2000) corroborating the findings in this study where diversity within Bradyrhizobium isolates was not clearly defined. The Rhizobium isolates characterised in this study showed close relation to the Rhizobium tropici group (Dall’Agnol et al., 2013). Strains belonging to this group are characterised with broad-host-range, high tolerance to environmental stress and genetic stability (Hungria et al., 2000, Hungria et al., 2003). This observation is interesting since the isolates in this study were obtained from areas with harsh environmental conditions. The analysis of concatenated 16S rRNA and ITS regions in this study revealed that the isolates shared more than 95.5% similarity to the B. yuanmingense strain CCBAU 1007T; implying that these isolates belong to this species (Willems et al., 2001). Willems et al. (2003) reported that the ITS gene sequencing and DNA-DNA hybridization shared a high correlation such that a sequence similarity of more than 95.5% shared by strains indicates that they belong to the same genospecies and have more than 60% DNA-DNA hybridization.
Symbiotic genes, on the other hand, have been found useful in the determination of host range, nodulation capacity and symbiovars between rhizobia and legumes (Rogel et al., 2011). The observation from the analyses of symbiotic genes of isolates in this study revealed that some areas in northern Ghana harbour strains related to B. yuanmingense in addition to the already identified geographical origins of this species (So et al., 1994, Vinuesa et al., 2005, Ormeno-Orrillo et al., 2006, Gu et al., 2007, Steenkamp et al., 2008, Leite et al., 2017). This suggests that strains of this species may be widely distributed in nature. Although nifH phylogeny is characterised by lateral gene transfer related to the host (Vinuesa et al., 2005), isolates that effectively nodulated groundnut in this study did not necessarily show any close relation with typical groundnut symbionts such as Bradyrhizobium arachidis (Wang et al., 2013), Bradyrhizobium subterraneum (Grönemeyer et al., 2015) and Bradyrhizobium vignae (Grönemeyer et al., 2016). This observation maybe because effective isolates used in this study were originally obtained from nodules of cowpea plants.
To this end, B. yuanmingense has been confirmed to be an important micro-symbiont of groundnut as reported previously in other studies (Gu et al., 2007, Steenkamp et al., 2008, Leite et al., 2017).
5. Conclusion
Increased nodulation, biomass production and N accumulation in soil-grown groundnut were achieved after inoculation with native rhizobium strains of northern Ghana. Among the isolates tested, KNUST 1002 was highly effective, performing similar to the groundnut reference strain 32H1. Apart from two Rhizobium isolates (KNUST 1003 and 1007), all the strains selected in this study were closely related to B. yuanmingense, confirming this species as a major micro-symbiont of groundnut.
Acknowledgments
Acknowledgements
This work was supported by the Bill & Melinda Gates Foundation through the Soil Health Project (Grant Number 2013 SHP025), the Africa-Brazil Agricultural Innovation Marketplace project (ID 1705) and the M-BoSs (MarketPlace-Building on Success) program (ID 5095). The authors express their appreciation to Embrapa Agrobiologia and Kwame Nkrumah University of Science and Technology for the technical and infrastructural assistance. We are grateful to Dr Tsai Sui Mui of CENA, Piracicaba, SP, for the provision of the 15N-labelled soil. We also express our appreciation to Mr. Ivan de A. J. Menezes, Dr. Janaina R. C. Rouws and Dr. Jerri E. Zilli for their support during the pot experiment, statistical analyses and recommendation of control rhizobium strains, respectively. The author RMB gratefully acknowledges research fellowships from the Brazilian National Research Council (CNPq) and the Rio de Janeiro State Research Foundation (FAPERJ).
Footnotes
Supplementary data associated with this article can be found, in the online version, at https://doi.org/10.1016/j.apsoil.2018.03.003.
Appendix A. Supplementary data
References
- Abaidoo R., Keyser H., Singleton P., Dashiell K., Sanginga N. Population size, distribution, and symbiotic characteristics of indigenous Bradyrhizobium spp. that nodulate TGx soybean genotypes in Africa. Appl. Soil Ecol. 2007;35:57–67. [Google Scholar]
- Abaidoo R.C., Keyser H.H., Singleton P.W., Borthakur D. Bradyrhizobium spp. (TGx) isolates nodulating the new soybean cultivars in Africa are diverse and distinct from bradyrhizobia that nodulate North American soybeans. Int. J. Syst Evol. Microbiol. 2000;50:225–234. doi: 10.1099/00207713-50-1-225. [DOI] [PubMed] [Google Scholar]
- Arnold S., Schepers J. A simple roller-mill grinding procedure for plant and soil samples. Commun. Soil Sci. Plan. 2004;35:537–545. [Google Scholar]
- Boddey R.M., de Oliveira O.C., Alves B.J., Urquiaga S. Field application of the15N isotope dilution technique for the reliable quantification of plant-associated biological nitrogen fixation. Fertil. Res. 1995;42:77–87. [Google Scholar]
- Bogino P., Banchio E., Bonfiglio C., Giordano W. Competitiveness of a Bradyrhizobium sp. strain in soils containing indigenous rhizobia. Curr. Microbiol. 2008;56:66–72. doi: 10.1007/s00284-007-9041-4. [DOI] [PubMed] [Google Scholar]
- Brockwell J. Matching rhizobia and temperate species of Acacia. ACIAR Proc. ACIAR. 1998:264–273. [Google Scholar]
- Broughton W.J., Dilworth M.J. Methods in legume-rhizobium technology: plant nutrient solutions. Handbook Rhizobia. 1970:245–249. [Google Scholar]
- Cardinale M., Brusetti L., Quatrini P., Borin S., Puglia A.M., Rizzi A., Zanardini E., Sorlini C., Corselli C., Daffonchio D. Comparison of different primer sets for use in automated ribosomal intergenic spacer analysis of complex bacterial communities. Appl. Environ. Microbiol. 2004;70:6147–6156. doi: 10.1128/AEM.70.10.6147-6156.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlberg, E.J., 2012. An economic evaluation of groundnut research in Uganda and Ghana. The University of North Carolina at Chapel Hill.
- Chalk P.M. Estimation of N2 fixation by isotope dilution: an appraisal of techniques involving 15N enrichment and their application. Soil Biol. Biochem. 1985;17:389–410. [Google Scholar]
- Chang Y.L., Wang J.Y., Wang E.T., Liu H.C., Sui X.H., Chen W.X. Bradyrhizobium lablabi sp. nov., isolated from effective nodules of Lablab purpureus and Arachis hypogaea. Int. J. Syst. Evol. Microbiol. 2011;61:2496–2502. doi: 10.1099/ijs.0.027110-0. [DOI] [PubMed] [Google Scholar]
- Dall’Agnol R.F., Ribeiro R.A., Ormeño-Orrillo E., Rogel M.A., Delamuta J.R.M., Andrade D.S., Martínez-Romero E., Hungria M. Rhizobium freirei sp. nov., a symbiont of Phaseolus vulgaris that is very effective at fixing nitrogen. Int. J. Syst. Evol. Microbiol. 2013;63:4167–4173. doi: 10.1099/ijs.0.052928-0. [DOI] [PubMed] [Google Scholar]
- Delamuta J.R.M., Ribeiro R.A., Ormeño-Orrillo E., Melo I.S., Martínez-Romero E., Hungria M. Polyphasic evidence supporting the reclassification of Bradyrhizobium japonicum group Ia strains as Bradyrhizobium diazoefficiens sp. nov. Int. J. Syst. Evol. Microbiol. 2013;63:3342–3351. doi: 10.1099/ijs.0.049130-0. [DOI] [PubMed] [Google Scholar]
- Dilworth M., Howieson J., Reeve W., Tiwari R., Glenn A. Acid tolerance in legume root nodule bacteria and selecting for it. Anim. Prod. Sci. 2001;41:435–446. [Google Scholar]
- El-Akhal M.R., Rincón A., Arenal F., Lucas M.M., El Mourabit N., Barrijal S., Pueyo J.J. Genetic diversity and symbiotic efficiency of rhizobial isolates obtained from nodules of Arachis hypogaea in Northwestern Morocco. Soil Biol. Biochem. 2008;40:2911–2914. [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Fening J., Sessitsch A., Offei S., Danso S. Genomic heterogeneity within cowpea bradyrhizobia isolated from Ghanaian soils. WAJAE. 2004;6 [Google Scholar]
- Ferreira D.F. SISVAR: um programa para análises e ensino de estatística. Revista Symposium Lavras. 2008:36–41. [Google Scholar]
- Flechard C., Ambus P., Skiba U., Rees R., Hensen A., Van Amstel A., Van Den Pol-Van Dasselaar A., Soussana J.-F., Jones M., Clifton-Brown J. Effects of climate and management intensity on nitrous oxide emissions in grassland systems across Europe. Agric. Ecosyst. Environ. 2007;121:135–152. [Google Scholar]
- Florentino L.A., Sousa P.M.D., Silva J.S., Silva K.B., Moreira F.M.d.S. Diversity and efficiency of Bradyrhizobium strains isolated from soil samples collected from around Sesbania virgata roots using cowpea as trap species. Revista Brasileira de Ciência do Solo. 2010;34:1113–1123. [Google Scholar]
- Fred E.B., Waksman S.A. 1928. Laboratory manual of general microbiology. [Google Scholar]
- Galtier N., Gouy M., Gautier C. SEAVIEW and PHYLO_WIN: two graphic tools for sequence alignment and molecular phylogeny. Comput. Appl. Biosci.: CABIOS. 1996;12:543–548. doi: 10.1093/bioinformatics/12.6.543. [DOI] [PubMed] [Google Scholar]
- Garrity G.M., Holt J.G. The road map to the manual. Bergey’s Manual® of Systematic Bacteriology. Springer; 2001. pp. 119–166. [Google Scholar]
- Grönemeyer J.L., Chimwamurombe P., Reinhold-Hurek B. Bradyrhizobium subterraneum sp. nov., a symbiotic nitrogen-fixing bacterium from root nodules of groundnuts. Int. J. Syst. Evol. Microbiol. 2015;65:3241–3247. doi: 10.1099/ijsem.0.000403. [DOI] [PubMed] [Google Scholar]
- Grönemeyer J.L., Hurek T., Bünger W., Reinhold-Hurek B. Bradyrhizobium vignae sp. nov., a nitrogen-fixing symbiont isolated from effective nodules of Vigna and Arachis. Int. J. Syst. Evol. Microbiol. 2016;66:62–69. doi: 10.1099/ijsem.0.000674. [DOI] [PubMed] [Google Scholar]
- Gu J., Wang E., Chen W.X. Genetic diversity of rhizobia associated with Desmodium species grown in China. Lett. Appl. Microbiol. 2007;44:286–292. doi: 10.1111/j.1472-765X.2006.02071.x. [DOI] [PubMed] [Google Scholar]
- Herridge D.F., Peoples M.B., Boddey R.M. Global inputs of biological nitrogen fixation in agricultural systems. Plant Soil. 2008;311:1–18. [Google Scholar]
- Hungria M., Campo R.J., Mendes I.C. Benefits of inoculation of the common bean (Phaseolus vulgaris) crop with efficient and competitive Rhizobium tropici strains. Biol. Fert. Soils. 2003;39:88–93. [Google Scholar]
- Hungria M., de S Andrade D., de O Chueire L.M., Probanza A., Guttierrez-Mañero F.J., Megi’as M. Isolation and characterisation of new efficient and competitive bean (Phaseolus vulgaris L.) rhizobia from Brazil. Soil Biol. Biochem. 2000;32:1515–1528. [Google Scholar]
- Hungria M., Vargas M.A. Environmental factors affecting N2 fixation in grain legumes in the tropics, with an emphasis on Brazil. Field Crops Res. 2000;65:151–164. [Google Scholar]
- Kumar S., Stecher G., Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016 doi: 10.1093/molbev/msw054. msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane D. 16S/23S rRNA sequencing. Nucl. Acid Techniq. Bacterial Systematics. 1991:125–175. [Google Scholar]
- Leite J., Passos S.R., Simões-Araújo J.L., Rumjanek N.G., Xavier G.R., Zilli J.É. Genomic identification and characterization of the elite strains Bradyrhizobium yuanmingense BR 3267 and Bradyrhizobium pachyrhizi BR 3262 recommended for cowpea inoculation in Brazil. Braz. J. Microbiol. 2017 doi: 10.1016/j.bjm.2017.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupwayi, N., Haque, I., 1994. Legume-Rhizobium technology manual. ILCA Environmental Sciences Working Document (ILCA). no. 29.
- Maheswar N.U., Sathiyavani G. Solubilization of phosphate by Bacillus Sps, from groundnut rhizosphere (Arachis hypogaea L) J. Chem. Pharm. Res. 2012;4:4007–4011. [Google Scholar]
- Marra L.M., Soares C.R.F.S., de Oliveira S.M., Ferreira P.A.A., Soares B.L., de Fráguas Carvalho R., de Lima J.M., de Souza Moreira F.M. Biological nitrogen fixation and phosphate solubilization by bacteria isolated from tropical soils. Plant Soil. 2012;357:289–307. [Google Scholar]
- Menna P., Barcellos F.G., Hungria M. Phylogeny and taxonomy of a diverse collection of Bradyrhizobium strains based on multilocus sequence analysis of the 16S rRNA gene, ITS region and glnII, recA, atpD and dnaK genes. Int. J. Syst. Evol. Microbiol. 2009;59:2934–2950. doi: 10.1099/ijs.0.009779-0. [DOI] [PubMed] [Google Scholar]
- Mohamed S., Abdalla A. Growth and yield response of groundnut (Arachis hypogaea L.) to microbial and phosphorus fertilizers. J. Agri-Food Appl. Sci. 2013;1:78–85. [Google Scholar]
- Muñoz V., Ibanez F., Tonelli M.L., Valetti L., Anzuay M.S., Fabra A. Phenotypic and phylogenetic characterization of native peanut Bradyrhizobium isolates obtained from Córdoba, Argentina. Syst. Appl. Microbiol. 2011;34:446–452. doi: 10.1016/j.syapm.2011.04.007. [DOI] [PubMed] [Google Scholar]
- Mweetwa A.M., Mulenga M., Mulilo X., Ngulube M., Banda J.S., Kapulu N., N'gandu S.H. Response of cowpea, soya beans and groundnuts to non-indigenous legume inoculants. Sustain. Agric. Res. 2014;3:84. [Google Scholar]
- Naegle E., Burton J., Carter T., Rufty T. Influence of seed nitrogen content on seedling growth and recovery from nitrogen stress. Plant Soil. 2005;271:329–340. [Google Scholar]
- Ndakidemi P., Dakora F., Nkonya E., Ringo D., Mansoor H. Yield and economic benefits of common bean (Phaseolus vulgaris) and soybean (Glycine max) inoculation in Northern Tanzania. Anim. Prod. Sci. 2006;46:571–577. [Google Scholar]
- Nkot L.N., Krasova-Wade T., Etoa F., Sylla S., Nwaga D. Genetic diversity of rhizobia nodulating Arachis hypogaea L. in diverse land use systems of humid forest zone in Cameroon. Appl. Soil Ecol. 2008;40:411–416. [Google Scholar]
- Nutsugah S., Oti-Boateng C., Tsigbey F., Brandenburg R. Assessment of yield losses due to early and late leaf spots of groundnut (Arachis hypogaea L.). Ghana. J. Agric. Sci. 2007;40:21–27. [Google Scholar]
- O’Hara G., Yates R., Howieson J. Selection of strains of root nodule bacteria to improve inoculant performance and increase legume productivity in stressful environments. Inoculants and nitrogen fixation of legumes in Vietnam. ACIAR Proc. 2002 [Google Scholar]
- Okito A., Alves B., Urquiaga S., Boddey R. Isotopic fractionation during N2 fixation by four tropical legumes. Soil Biol. Biochem. 2004;36:1179–1190. [Google Scholar]
- Ormeno-Orrillo E., Vinuesa P., Zuniga-Davila D., Martínez-Romero E. Molecular diversity of native bradyrhizobia isolated from Lima bean (Phaseolus lunatus L.) in Peru. Syst. Appl. Microbiol. 2006;29:253–262. doi: 10.1016/j.syapm.2005.09.002. [DOI] [PubMed] [Google Scholar]
- Poly F., Monrozier L.J., Bally R. Improvement in the RFLP procedure for studying the diversity of nifH genes in communities of nitrogen fixers in soil. Res. Microbiol. 2001;152:95–103. doi: 10.1016/s0923-2508(00)01172-4. [DOI] [PubMed] [Google Scholar]
- Rogel M.A., Ormeno-Orrillo E., Romero E.M. Symbiovars in rhizobia reflect bacterial adaptation to legumes. Syst. Appl. Microbiol. 2011;34:96–104. doi: 10.1016/j.syapm.2010.11.015. [DOI] [PubMed] [Google Scholar]
- Sajid M., Rab A., Wahid F., Shah S., Jan I., Khan M., Hussain S., Khan M., Iqbal Z. Influence of rhizobium inoculation on growth and yield of groundnut cultivars. Sarhad J. Agric. 2010;27:573–576. [Google Scholar]
- Sarita S., Sharma P.K., Priefer U.B., Prell J. Direct amplification of rhizobial nodC sequences from soil total DNA and comparison to nodC diversity of root nodule isolates. FEMS Microbiol. Ecol. 2005;54:1–11. doi: 10.1016/j.femsec.2005.02.015. [DOI] [PubMed] [Google Scholar]
- Sattar M., Quader M., Danso S. Nodulation, N2 fixation and yield of chickpea as influenced by host cultivar and Bradyrhizobium strain differences. Soil Biol Biochem. 1995;27:725–727. [Google Scholar]
- Sharma P., Sardana V., Kandola S. Response of groundnut (Arachis hypogaea L.) to Rhizobium Inoculation. Libyan Agri. Res. Centre J. Int. 2011;2:101–104. [Google Scholar]
- Shishido M., Pepper I. Identification of dominant indigenous Rhizobium meliloti by plasmid profiles and intrinsic antibiotic resistance. Soil Biol. Biochem. 1990;22:11–16. [Google Scholar]
- So R.B., Ladha J.K., Young J.P.W. Photosynthetic symbionts of Aeschynomene spp. form a cluster with bradyrhizobia on the basis of fatty acid and rRNA analyses. Int. J. Syst. Evol. Microbiol. 1994;44:392–403. doi: 10.1099/00207713-44-3-392. [DOI] [PubMed] [Google Scholar]
- Somasegaran P., Hoben H.J. Handbook for Rhizobia. Springer; 1994. Quantifying the growth of rhizobia; pp. 47–57. [Google Scholar]
- Souza, G., Nogueira, A., 2005. Manual de laboratório: solo, água, nutrição vegetal, nutrição animal e alimentos. São Carlos, Embrapa Pecuária Sudeste, p. 334.
- Steenkamp E.T., Stępkowski T., Przymusiak A., Botha W.J., Law I.J. Cowpea and peanut in southern Africa are nodulated by diverse Bradyrhizobium strains harboring nodulation genes that belong to the large pantropical clade common in Africa. Mol. Phylogenet. Evol. 2008;48:1131–1144. doi: 10.1016/j.ympev.2008.04.032. [DOI] [PubMed] [Google Scholar]
- Stephens J., Rask H. Inoculant production and formulation. Field Crops Res. 2000;65:249–258. [Google Scholar]
- Taurian T., Aguilar O.M., Fabra A. Characterization of nodulating peanut rhizobia isolated from a native soil population in Córdobar Argentina. Symbiosis. 2002;33:59–72. [Google Scholar]
- Taurian T., Ibañez F., Fabra A., Aguilar O.M. Genetic diversity of rhizobia nodulating Arachis hypogaea L. in central Argentinean soils. Plant Soil. 2006;282:41–52. [Google Scholar]
- Terpolilli J.J., O’Hara G.W., Tiwari R.P., Dilworth M.J., Howieson J.G. The model legume Medicago truncatula A17 is poorly matched for N2 fixation with the sequenced microsymbiont Sinorhizobium meliloti 1021. New Phytol. 2008;179:62–66. doi: 10.1111/j.1469-8137.2008.02464.x. [DOI] [PubMed] [Google Scholar]
- Thies J.E., Bohlool B.B., Singleton P.W. Subgroups of the cowpea miscellany: symbiotic specificity within Bradyrhizobium spp. for Vigna unguiculata, Phaseolus lunatus, Arachis hypogaea, and Macroptilium atropurpureum. Appl. Environ. Microbiol. 1991;57:1540–1545. doi: 10.1128/aem.57.5.1540-1545.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.D., Higgins D.G., Gibson T.J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucl. Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-J, unior C.V., Leite J., Ros, d., alia C.E., Santos S., Fernandes-J, unior P.I., Edson Zilli J., Rumjanek N.G., Xavier G.R. Diversity and symbiotic performance of peanut rhizobia from Southeast region of Brazil. Afr. J. Microbiol. Res. 2014;8:566–577. [Google Scholar]
- Trindade H., Coutinho J., Jarvis S., Moreira N. Nitrogen mineralization in sandy loam soils under an intensive double-cropping forage system with dairy-cattle slurry applications. Eur. J. Agron. 2001;15:281–293. [Google Scholar]
- Tsigbey, F., Brandenburg, R., Clottey, V., 2003. Peanut production methods in Northern Ghana and some disease perspectives. World Geography of the Peanut Knowledge Base Website 9.
- Unkovich, M., Herridge, D., Peoples, M., Cadisch, G., Boddey, B., Giller, K., Alves, B., Chalk, P., 2008. Measuring plant-associated nitrogen fixation in agricultural systems. Australian Centre for International Agricultural Research (ACIAR).
- Unkovich M.J., Pate J.S. An appraisal of recent field measurements of symbiotic N2 fixation by annual legumes. Field Crops Res. 2000;65:211–228. [Google Scholar]
- Urquiaga S., Cruz K.H., Boddey R.M. Contribution of nitrogen fixation to sugar cane: nitrogen-15 and nitrogen-balance estimates. Soil Sci. Soc. Am. J. 1992;56:105–114. [Google Scholar]
- Urtz B.E., Elkan G.H. Genetic diversity among Bradyrhizobium isolates that effectively nodulate peanut (Arachis hypogaea) Can. J. Microbiol. 1996;42:1121–1130. doi: 10.1139/m96-144. [DOI] [PubMed] [Google Scholar]
- Van Rossum D., Schuurmans F.P., Gillis M., Muyotcha A., Van Verseveld H.W., Stouthamer A.H., Boogerd F.C. Genetic and phenetic analyses of Bradyrhizobium strains nodulating peanut (Arachis hypogaea L.) roots. Appl. Environ. Microbiol. 1995;61:1599–1609. doi: 10.1128/aem.61.4.1599-1609.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandamme P., Pot B., Gillis M., De Vos P., Kersters K., Swings J. Polyphasic taxonomy, a consensus approach to bacterial systematics. Microbiol. Rev. 1996;60:407–438. doi: 10.1128/mr.60.2.407-438.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinuesa P., Silva C., Werner D., Martínez-Romero E. Population genetics and phylogenetic inference in bacterial molecular systematics: the roles of migration and recombination in Bradyrhizobium species cohesion and delineation. Mol. Phylogenet. Evol. 2005;34:29–54. doi: 10.1016/j.ympev.2004.08.020. [DOI] [PubMed] [Google Scholar]
- Wang R., Chang Y.L., Zheng W.T., Zhang D., Zhang X.X., Sui X.H., Wang E.T., Hu J.Q., Zhang L.Y., Chen W.X. Bradyrhizobium arachidis sp. nov., isolated from effective nodules of Arachis hypogaea grown in China. Syst. Appl. Microbiol. 2013;36:101–105. doi: 10.1016/j.syapm.2012.10.009. [DOI] [PubMed] [Google Scholar]
- Wang E., Martínez-Romero E. Prokaryotic nitrogen fixation: a model system for the analysis of a biological process. 2000. Phylogeny of root-and stem-nodule bacteria associated with legumes; pp. 177–186. [Google Scholar]
- Willems A., Doignon-Bourcier F., Goris J., Coopman R., de Lajudie P., De Vos P., Gillis M. DNA-DNA hybridization study of Bradyrhizobium strains. Int. J. Syst. Evol. Microbiol. 2001;51:1315–1322. doi: 10.1099/00207713-51-4-1315. [DOI] [PubMed] [Google Scholar]
- Willems A., Munive A., de Lajudie P., Gillis M. In most Bradyrhizobium groups sequence comparison of 16S–23S rDNA internal transcribed spacer regions corroborates DNA-DNA hybridizations. Syst. Appl. Microbiol. 2003;26:203–210. doi: 10.1078/072320203322346056. [DOI] [PubMed] [Google Scholar]
- Yang J.K., Xie F.L., Zou J., Zhou Q., Zhou J.C. Polyphasic characteristics of bradyrhizobia isolated from nodules of peanut (Arachis hypogaea) in China. Soil Biol. Biochem. 2005;37:141–153. [Google Scholar]
- Yates, R., Howieson, J., Hungria, M., Bala, A., O’Hara, G., Terpolilli, J., 2016. Authentication of rhizobia and assessment of the legume symbiosis in controlled plant growth systems, p 73–108. Working with rhizobia. ACIAR, Canberra, Australia.
- Zhang X., Nick G., Kaijalainen S., Terefework Z., Paulin L., Tighe S.W., Graham P.H., Lindström K. Phylogeny and diversity of Bradyrhizobium strains isolated from the root nodules of peanut (Arachis hypogaea) in Sichuan, China. Syst. Appl. Microbiol. 1999;22:378–386. doi: 10.1016/S0723-2020(99)80046-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.