Abstract
Asthma, hay fever (or allergic rhinitis) and eczema (or atopic dermatitis) often coexist in the same individuals1, partly because of a shared genetic origin2–4. To identify shared risk variants, we performed a genome-wide association study (GWAS, n=360,838) of a broad allergic disease phenotype that considers the presence of any one of these three diseases. We identified 136 independent risk variants (P<3x10-8), including 73 not previously reported, which implicate 132 nearby genes in allergic disease pathophysiology. Disease-specific effects were detected for only six variants, confirming that most represent shared risk factors. Tissue-specific heritability and biological process enrichment analyses suggest that shared risk variants influence lymphocyte-mediated immunity. Six target genes provide an opportunity for drug repositioning, while for 36 genes CpG methylation was found to influence transcription independently of genetic effects. Asthma, hay fever and eczema partly coexist because they share many genetic risk variants that dysregulate the expression of immune-related genes.
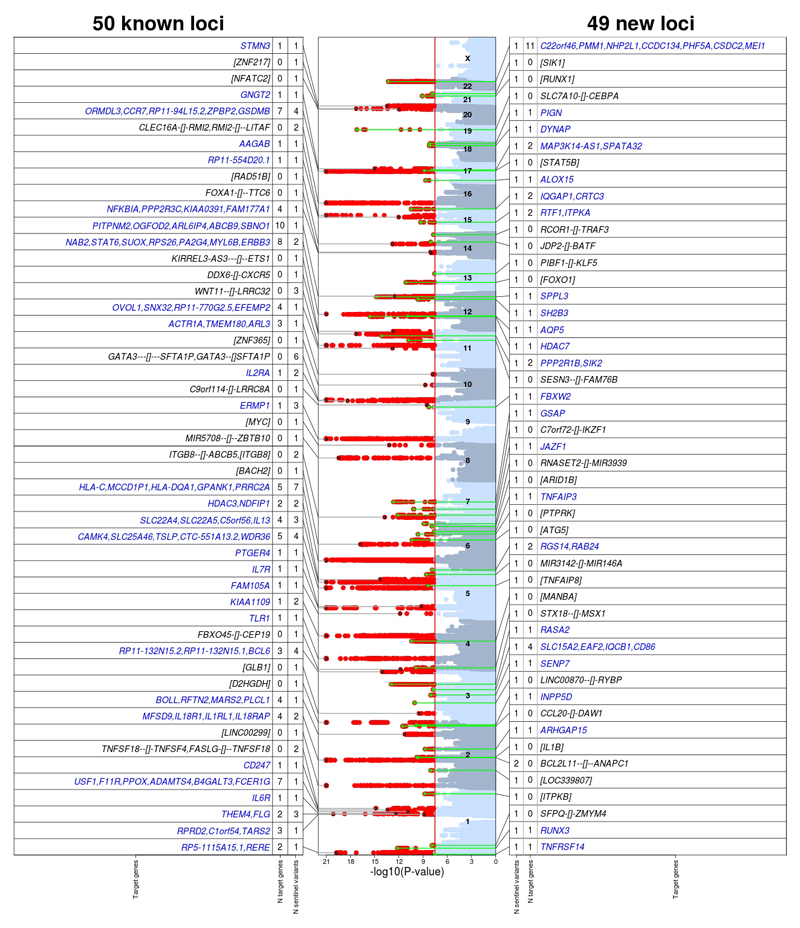
The analytical approach used is summarized in Supplementary Fig. 1. We tested for association with allergic disease 8,307,659 genetic variants that passed quality control filters (Supplementary Table 1), comparing 180,129 cases who reported having suffered from asthma and/or hay fever and/or eczema, and 180,709 controls who reported not suffering from any of these diseases (Supplementary Table 2), all of European ancestry. Meta-analysis of results from the 13 contributing studies (Supplementary Fig. 2) identified 99 genomic regions (i.e. loci) located >1 Mb apart containing at least one genetic variant associated with allergic disease at a genome-wide significance threshold of 3x10-8 (Fig. 1 and Supplementary Table 3). Based on approximate conditional analysis5, 136 genetic variants in these 99 loci had a statistically independent association with disease risk (Supplementary Table 4). Henceforth, we refer to these as “sentinel risk variants”, which either represent, or are in linkage disequilibrium (LD) with, a causal functional variant. These included 86 (in 50 loci) located <1 Mb from risk variants reported in previous GWAS of allergic disease (Supplementary Table 5). Of note, 23/86 sentinel variants were in low linkage disequilibrium (LD, r2<0.05) with the previously reported risk variants, indicating that they represent novel associations in these loci. The remaining 50 sentinel variants (in 49 loci) were located >1Mb from previously reported associations (Supplementary Table 6), of which 17 were in low LD with nearby variants reported for other diseases or traits (Supplementary Table 7). Eighteen loci had multiple independent association signals (Supplementary Table 3). Altogether, we identified 73 (50+23) genetic associations with allergic disease that are new, a substantial increment over the 89 associations reported previously (Supplementary Fig. 3 and Supplementary Table 8).
Figure 1. Loci containing genetic risk variants independently associated with the risk of allergic disease at P<3x10-8.
The 136 sentinel risk variants were located in 50 previously reported (86 variants) and 49 novel (50 variants) risk loci. The numbers of plausible target genes of sentinel risk variants identified for each locus are shown, with target gene names listed in blue font. For loci with many target genes, only a selection is listed. When no target gene was identified (black font), square brackets are used to indicate the location of the sentinel risk variant relative to the nearest gene(s). Specifically, when the risk variant was intergenic (indicated by "gene1--[]--gene2"), the two closest genes (upstream and downstream) are shown; the distance to each gene is proportional to the number of "-" shown. Otherwise, when the risk variant was located within a gene, the respective gene name is shown between square brackets (i.e. [gene]). Red vertical line in Manhattan plot shows genome-wide significance threshold used (P=3x10-8).
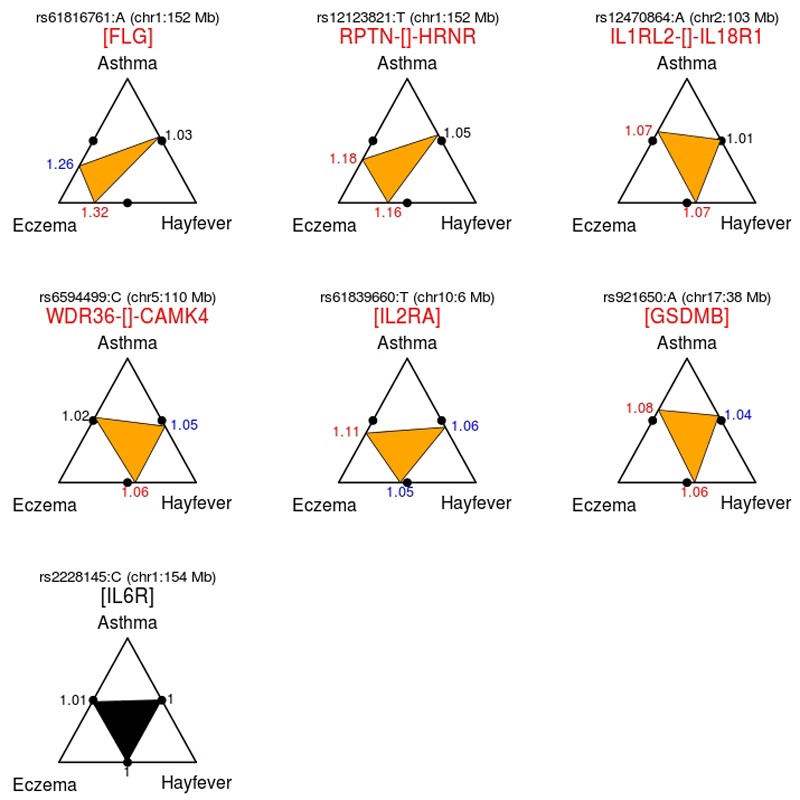
As expected from a study design that maximized power to identify shared risk variants6, we found that 130 of the 136 sentinel variants had similar allele frequencies in case-only association analyses that compared three non-overlapping groups of adults: those who reported suffering from asthma only (n=12,268), hay fever only (n=33,305) or eczema only (n=6,276) (Supplementary Table 9). There was thus no evidence that these 130 variants have differential effects on the three individual diseases. The six variants with evidence for stronger effects in one allergic disease when compared to the other two were located in five known allergy risk loci (e.g. FLG and GSDMB, Fig. 2). On the other hand, many sentinel variants (26 or 19%) were also associated with the age at which symptoms of any allergic disease first developed (n=35,972, Supplementary Table 10), the allele associated with a higher disease risk being always associated with earlier age-of-onset (Supplementary Fig. 4). For 18 of those 26 variants, the effect on age-of-onset was not significantly different between individual diseases (Supplementary Table 10), suggesting that they influence the age at which symptoms first develop for all three diseases.
Figure 2. Sentinel variants with significant allele-frequency differences in pairwise case-only association analyses contrasting individuals suffering from a single allergic disease.
For each sentinel variant, we performed three case-only association analyses, comparing asthma-only cases (n=12,268) against hay fever-only cases (n=33,305); asthma-only cases against eczema-only cases (n=6,276); and hay fever-only cases against eczema-only cases. After accounting for multiple testing, significant associations for at least one of these analyses were only observed for six of the 136 sentinel variants, which are shown in the first two rows of the figure. For a given variant, the vertices of the inner triangle point to the position along the edges of the outer triangle that corresponds to the allele frequency difference observed between pairs of single-disease cases. For example, the rs61816761:A allele, which is located in the Fillagrin gene (FLG), was 1.32-fold more common in individuals suffering only from eczema when compared to individuals suffering only from hay fever (P=7.2x10-8), consistent with this SNP being a stronger risk factor for eczema than for hay fever. A similar result (OR = 1.26, P=0.0004) was observed for this variant when contrasting eczema-only cases against asthma-only cases. For comparison, a variant with no allele frequency differences in all three pairwise single-disease association analyses is also shown (rs2228145, in the IL6R gene). In this case, the three estimated odds ratios were approximately equal to 1. The color of the OR font reflects the significance of the association: red for P<1.2x10-4 (correction for multiple testing), blue for P<0.05 and black for P>0.05.
We then used LD-score regression analysis7 (see Methods) to quantify the liability-scale heritability of the three individual diseases that was collectively explained by the 136 top associations in the Nord-Trøndelag Health Study (HUNT, up to n=20,350), which was not part of the discovery meta-analysis. This was found to be 3.2% for asthma, 3.8% for hay fever and 1.2% for eczema, respectively representing about a fifth, a sixth and a tenth of the overall heritability of each disease that is explained by common single nucleotide polymorphisms (SNPs; Supplementary Table 11). Therefore, the inheritance of risk alleles at these loci partly explains why these three conditions coexist.
To understand the biological consequences of allergy risk variants, we then identified plausible target genes of the 136 sentinel variants. There were 5,739 transcripts annotated near (+/- 1 Mb) sentinel variants, including 2,569 protein-coding genes. For 132 of these transcripts, the nearby sentinel variant was in high LD (r2≥0.8) with either a non-synonymous SNP (22 genes; Supplementary Table 12) or a sentinel expression quantitative trait locus (eQTL) identified in relevant tissues or cell types (additional 110 genes; Supplementary Tables 13 and 14). We refer to these 132 transcripts as plausible target genes, which were located in 54 of the 99 risk loci (Fig. 1 and Supplementary Table 15). Studies that confirm the target gene predictions and identify the underlying functional variants are warranted; genes that could be prioritized for functional follow-up include 78 identified using a more conservative LD threshold (r2≥0.95; Supplementary Table 15) or 61 predicted to be the likely targets based on independent evidence from publicly available functional data (Supplementary Tables 16 and 17; see Methods for details). Of note, 79 (60%) of the 132 plausible target genes have not previously been co-cited with allergy-related terms (Supplementary Table 15), and so potentially represent novel key contributors to disease pathophysiology (examples in Table 1).
Table 1. Selected examples of plausible target genes not previously implicated in the pathophysiology of allergic disease.
| Gene | Summary | Possible role(s) in allergic diseasea |
|---|---|---|
| RERE | Nuclear receptor coregulator that positively regulates retinoic acid signaling | Positive regulation of B cell differentiation, eosinophil survival and migration |
| PPP2R3C | Sub-unit of protein phosphatase 2A (PP2A) that regulates immune cell function | Th2 differentiation, Treg function, response to viral infection |
| RASA2 | GTPase-activating protein of Ras that regulates receptor signal transduction | Unknown. RASA3: hematopoiesis. RASA4: macrophage phagocytosis. |
| SIK2 | Salt-inducible kinase | Regulation of macrophage inflammatory phenotype, metabolic homeostasis |
| RTF1 | Component of the PAF complex, that is involved in transcriptional regulation | Anti-viral response, regulation of TNF expression |
| SMARCE1 | Sub-unit of the BAF chromatin remodeling complex | Repressor of CD4 differentiation |
| DYNAP | Dynactin-associated protein that activates protein kinase B | Cytokine signaling, T cell function |
| THEM4 | Mithocondrial thioesterase that is a negative regulator of protein kinase B | Vitamin D-dependent macrophage-mediated inflammation |
| ARHGAP15 | Rho GTPase activating protein that down-regulates RAC1 | Rac1-dependent inflammatory response |
| SENP7 | Sentrin/small ubiquitin-like modifier (SUMO)-specific protease | Susceptibility to viral infection |
References that support the possible role(s) listed are cited in the Supplementary Note.
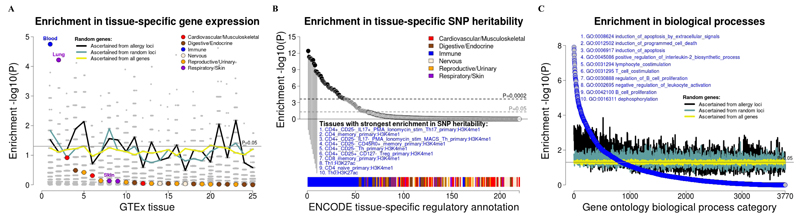
Next, based on data from the GTEx consortium8, we identified broad tissue types in which the plausible target genes were disproportionally expressed, using the Tissue Specific Expression Analysis (TSEA) approach described previously9. We excluded genes located in the major histocompatibility complex (MHC) or not present in the TSEA GTEx database, leaving 112 plausible target genes for analysis. When compared to the remaining 17,671 non-MHC genes in the genome, we found that the list of plausible targets was enriched for genes specifically expressed in whole-blood and lung (Fig. 3A). Both associations remained significant (Supplementary Fig. 5) after restricting the background gene list to the subset of 12,804 non-MHC genes with eQTLs reported in the same studies used to identify the plausible target genes (Supplementary Table 13). These results indicate that the plausible targets are enriched for genes preferentially expressed in whole-blood and lung, and that this is unlikely to arise because the plausible targets were also enriched for genes with eQTLs in those tissues.
Figure 3. Tissues and biological processes influenced by allergy risk variants.
(A) Enrichment of tissue-specific gene expression in 25 broad tissues studied by the GTEx consortium. We used the TSEA approach9 to test if genes specifically expressed in a given tissue were enriched amongst the list of plausible target genes when compared to other genes in the genome. The enrichment (y-axis) is shown as the -log10 of the Fisher’s exact test P-value. For comparison, we analyzed 1,000 lists of random genes instead of the plausible target genes. We selected genes at random using three strategies (see Methods for details). First, genes were randomly drawn from the 98 non-MHC allergy risk loci identified in our GWAS, matching on the number selected per locus and in total. The enrichment P-value for each of the 1,000 lists of random genes is shown by a grey circle. The black-solid line shows the P-value for the 50th most significant random list (i.e. corresponding to the 5th percentile): under the null hypothesis of no enrichment, this P-value should be close to 0.05 (horizontal grey line). Second, genes were drawn at random from 2 Mb loci selected at random from the genome, matching on the number of genes selected (and available for selection) per locus and in total. Third, genes were drawn at random from all 18,300 genes available for analysis. For the latter two strategies, the P-value for the 50th most significant random gene list is shown by the blue and yellow lines, respectively; enrichment results for each individual random dataset are not shown. Similar results were obtained after restricting the random genes and the background gene list to the subset of genes with eQTLs (Supplementary Fig. 5). Genes in the MHC were excluded from these analyses.
(B) Enrichment of SNP-based heritability in 220 individual cell type-specific regulatory annotations. We used stratified LD score regression analysis 10 to quantify the contribution of SNPs that overlap cell type-specific regulatory annotations to the SNP-based disease heritability. Annotations with an enrichment in SNP heritability (-log10 of the P-value of the regression coefficient, y-axis) that was significant after correcting for multiple testing (P<0.0002) are shown in black circles (top 10 listed in blue font; all results in Supplementary Table 19). SNPs in the MHC were excluded from these analyses.
(C) Biological processes enriched amongst the list of plausible target genes. We used GeneNetwork12 to test if the plausible target genes as a group were more likely to be part of a specific biological process category when compared to the rest of the genes in the genome. The enrichment (y-axis) is shown as the –log10 of the Wilcoxon rank-sum test P-value (see Methods for details). The top 10 pathways are listed in blue font. For comparison, we analyzed 1,000 lists of random genes generated using the same three strategies described above. For each of these strategies, the P-value for the 50th most significant random gene list is shown by the black (random genes from allergy loci), blue (random genes from random loci) and yellow (random genes selected from all available genes) lines. Similar results were obtained after restricting the random genes and the background gene list to the subset of genes with eQTLs (not shown). Genes in the MHC were excluded from these analyses.
The enrichment in whole-blood and lung expression could be a general feature of arbitrary genes located near the sentinel risk variants. To address this possibility, we determined how often the enrichment observed with the plausible target genes was exceeded when analyzing 1,000 lists of random genes. When genes were randomly selected from the same 98 non-MHC allergy risk loci identified in the meta-analysis, matching on the number of plausible target genes identified per locus (range 0 to 11) and in total (i.e. 112), the enrichment observed in whole-blood was not exceeded in any of the 1,000 random lists when considering results for all 25 tissues tested (Fig. 3A and Supplementary Table 18). Similar results were observed for lung. For comparison, arbitrary genes were also selected from 2 Mb loci drawn at random from the genome, or simply from all genes in the genome, and results were very similar (Fig. 3A and Supplementary Table 18). Randomly selecting genes from the subset with eQTLs also had no impact on the results (Supplementary Fig. 5). Therefore, we conclude that the enrichment in expression observed in whole-blood and lung was specific to the genes identified as plausible targets of sentinel risk variants.
To identify specific cell types that were likely to contribute to the enrichment in whole-blood, we used an orthogonal approach10 that quantifies tissue-specific enrichments in SNP heritability rather than in gene expression. Specifically, this approach quantifies the trait heritability that is explained by SNPs that overlap cell type-specific regulatory annotations measured by the ENCODE project in 100 different cell types. In this analysis, the strongest enrichment in SNP heritability was observed for regulatory annotations measured in helper T cells (including Th17, Th1 and Th2), regulatory T cells, CD4+ and CD8+ memory T cells, CD56+ NK cells and CD19+ B cells (Fig. 3B and Supplementary Table 19). These results are consistent with previous findings11 and the widely documented contribution of these T cell subsets to allergic responses. Similar results were obtained after removing the 136 top associations from our GWAS results (Supplementary Fig. 6 and Supplementary Table 19), indicating that the observed enrichments extend beyond genome-wide significant SNPs. These results demonstrate that genetic risk variants shared between asthma, hay fever and eczema, including but not limited to the ones that reached genome-wide significance, operate to a large extent by modulating gene expression in cells of the immune system.
To help understand how the sentinel variants might influence immune cell function, we then identified biological processes over-represented amongst the plausible target genes when compared to the rest of the genes in the genome (MHC excluded), using GeneNetwork12. As for the analysis of tissue-specific enrichment in gene expression, for each specific biological process, we compared the enrichment observed with the list of plausible target genes with that observed with 1,000 lists of genes randomly drawn from the same allergy risk loci. After correcting for the 3,770 biological processes tested, we found 35 pathways for which the enrichment observed with the plausible target genes was exceeded in <5% of the random gene lists (Fig. 3C and Supplementary Table 20). These included biological processes related to T and B cell activation, B cell proliferation and isotype switching, interleukin (IL-) 2 and IL-4 production, confirming a key role for the sentinel variants and the likely target genes on lymphocyte-mediated immunity. Other noteworthy enrichments were observed for pathways related to induction of cell death, lipid phosphorylation and NK cell differentiation.
Consistent with a widespread effect of allergy risk variants on immune cell function, many sentinel risk variants have been reported to associate with other immune-related traits, notably blood cell counts (Supplementary Table 21) and auto-immune diseases (Supplementary Table 22). The genetic overlap with auto-immune diseases was not restricted to sentinel variants, as evidenced by significant positive genetic correlations with celiac disease, Crohn's disease and inflammatory bowel disease obtained after excluding the 136 top associations from our GWAS results (Supplementary Table 23). Other significant genetic correlations were observed for obesity- and depression-related traits, both previously suggested by twin studies13. The former provides support for a role of allergy risk variants in the regulation of metabolic homeostasis.
We then investigated whether any of the plausible target genes identified could potentially represent a new opportunity for drug repositioning, as shown by others14. We found that 29 genes have been or are being considered as drug targets, including nine for the treatment of allergic diseases (Supplementary Table 24), four for auto-immune diseases (Supplementary Table 25) and 16 for other diseases (Supplementary Table 26), mostly cancer. Therefore, for 20 genes, drugs currently in development for other indications might influence biological mechanisms underlying allergic disease. For six of these genes, the effect on gene expression of the allergy protective allele (Supplementary Table 27) and the existing drug matched (Table 2), suggesting that the latter might attenuate (and not exacerbate) allergy symptoms, and so could be prioritized for pre-clinical testing.
Table 2. Plausible target genes with drugs in development for indications other than allergic diseases, for which the effect on gene expression of the allergy protective allele and the existing drug matched.
| Plausible target gene | Effect of allergy protective allele on gene expression | Drug Action | Drug Status | Drug Name | Originator Company | Active Indications |
|---|---|---|---|---|---|---|
| CD86 | Increased | Agonist | Discovery | BR-02001 | Boryung_Pharm_Co_Ltd | Autoimmune_disease |
| CCR7 | Decreased | Antagonist | Discovery | anti-CCR7_chimeric_IgG1_antibodies | North_Coast_Biologics_LLC | Unidentified_indication |
| CCR7 | Decreased | Antagonist | Discovery | anti-CCR7_monoclonal_antibody | Pepscan_Systems_BV | Cancer |
| CCR7 | Decreased | Antagonist | Discovery | CCR7-targeting_antibody | Abilita_Bio_Inc | Metastatic_breast_cancer |
| CCR7 | Decreased | Antagonist | NA | chemokine_antagonists | Neurocrine_Biosciences_Inc | NA |
| CCR7 | Decreased | Antagonist | NA | chemokine_receptor_inhibitors | Sosei_Group_Corp | NA |
| F11R | Decreased | Antagonist | Discovery | F11R_inhibitors | Provid_Pharmaceuticals_Inc | Cardiovascular_disease |
| F11R | Decreased | Antagonist | Discovery | F-50073 | Pierre_Fabre_SA | Cancer |
| PHF5A | Decreased | Antagonist | Discovery | PHF5A_inhibitors | Fred_Hutchinson_Cancer_Research_Center | Glioblastoma |
| RGS14 | Decreased | Antagonist | NA | regulator_of_G-protein_signaling_14_inhibitor | University_of_Malaga | Memory loss |
| TARS2 | Decreased | Antagonist | Discovery | borrelidin | Scripps_Research_Institute | Infectious_disease |
Finally, based on data from the BIOS consortium15 (n=2,101), we found that a substantial fraction of target genes (36 or 27%) had a nearby CpG site for which methylation levels were significantly correlated with mRNA levels in blood, independently of SNP effects (Supplementary Table 28). This observation raises the possibility that environmental effects on the methylation state of these CpGs might influence target gene expression and, by extension, allergic disease risk. Well powered studies that address this possibility are warranted. In exploratory analyses, we tested the association between five established risk factors for allergic disease (see Methods) and the methylation state of expression-associated CpGs for those 36 genes (largest n=1,221). We observed only one significant association, between smoking and the methylation state of PITPNM2 (Supplementary Table 29), which was reported in a previous study16. These results indicate that smoking might influence the risk of allergic disease partly by modulating the methylation state of expression-associated CpGs for PITPNM2, a PYK2-binding protein17 potentially involved in neutrophil function18,19.
In conclusion, we substantially increased the number of known risk variants for allergic disease through a large GWAS of a multi-disease phenotype defined based on information from three genetically correlated diseases, asthma, hay fever and eczema. With a few exceptions, the variants identified had similar effects on the individual disease entities. The risk variants, and their likely target genes, are predicted to influence overwhelmingly the function of immune cells. Novel drugs for allergy are proposed based on genomics-guided drug repositioning. Finally, our results raise the possibility that environmental factors such as smoking might influence allergic disease risk through modulation of target gene methylation.
Online Methods
Meta-analysis of allergic disease GWAS results conducted in 13 studies (n=360,838)
In each of 13 participating studies (Supplementary Tables 1 and 2), a GWAS was performed using an additive genetic model in individuals of European descent that reported suffering from asthma and/or hay fever and/or eczema (case-group, total n=180,129), against those who never reported suffering from any of these three conditions (control group, total n=180,709). A detailed description of the procedures used to identify cases and controls, as well as for SNP genotyping, imputation and association testing, is provided for each study in the Supplementary Note.
Prior to the meta-analysis, standard quality control (QC) filters were applied to results from individual studies (Supplementary Table 1). After QC, and restricting the analysis to SNPs present in at least the two largest studies (UK Biobank and 23andMe, Inc., combined n=256,623), results were available for 8,307,659 variants, of which most (89%) were available in >95% of the overall sample size. Intercept estimates from LD score regression analysis 7, which reflect inflation of test statistics that are likely due to technical biases, ranged between 1.00 and 1.16 (Supplementary Table 1). Results from individual studies were adjusted for the observed inflation by multiplying the square of the standard error of each genetic effect estimate by the respective LD score regression intercept. We then used METAL 20 to combine association results across studies using an inverse-variance-weighted, fixed-effects meta-analysis. P-values from the meta-analysis were further adjusted for the meta-analysis LD score regression intercept of 1.04. The genome-wide significance threshold was set at 3x10-8, as suggested previously for GWAS analyzing variants with MAF≥1% 21.
Identification of independent associations through approximate conditional analyses
For each chromosome, we identified all SNPs with a P≤3x10-8, sorted these based on base pair position, and then grouped variants into the same locus if the distance between consecutive variants was <1Mb. Variants located >1 Mb from the previous genome-wide significant variant were assigned to a new locus. Next, for each of these loci, we identified statistically independent associations using approximate conditional analyses, as implemented in GCTA 5. We refer to these as sentinel risk variants. In these analyses, LD calculations were based on a subset of 5,000 individuals from the UKBiobank study. Briefly, for each locus, we (1) identified the most significantly-associated SNP [i]; (2) adjusted the summary statistics of all SNPs in that locus by the effect of that top SNP; (3) identified the most significantly-associated SNP [j] that remained genome-wide significant in that locus; (4) adjusted the summary statistics of all SNPs in that locus by the effects of SNPs i and j. We repeated this process until there were no SNPs associated with allergic disease at P≤3x10-8 after adjusting for the effect of other, more strongly independently associated variants in that locus. Lastly, we estimated the LD between sentinel variants located in different risk loci (i.e. >1 Mb apart) and confirmed that the r2 was always close to 0 (no pairs of sentinel variants with r2>0.02).
Determining the novelty status of independent SNP associations with allergic disease
Previous GWAS identified 185 SNPs associated with the risk of various allergic conditions, which we grouped into 89 independent associations based on the LD between variants (see Supplementary Note). We used that information to classify each of our independent SNP associations into two major groups: located in known (<1Mb from any of those 185 previously reported associations; “KnownLocus”) or new (>1Mb from those variants; “NewLocus”) allergy risk loci. For the first group, we then estimated the LD between each sentinel variant identified in our study and all variant(s) reported in previous GWAS. If all reported variants had an r2<0.05 with our sentinel variant, then our association was considered to represent a new risk variant in a known risk locus (“KnownLocus-NewVariant”). Alternatively, when at least one reported variant had an r2≥0.05, our association was considered to be a known risk variant in a known risk locus (“KnownLocus-KnownVariant”). The second major group was composed of variants located in new allergy risk loci. Within this group, we used the same approach just described to determine if our associations were novel when considering any disease or trait with genome-wide significant associations reported in the NHGRI-EBI GWAS catalog.
Comparison of risk allele frequencies between individuals suffering from a single allergic disease
By combining information from asthma, hay fever and eczema in the case-control definition used in our GWAS, we expected our study design to improve power to identify risk variants shared between, but not specific to any of, the three diseases 6. To understand if the associations discovered in our GWAS were indeed likely to represent risk factors shared across allergic diseases, we took advantage of the observation that not all affected individuals report allergic co-morbidities 1,22,23, and compared allele frequencies between three groups of adults: asthma-only cases (n=12,268), hay fever-only cases (n=33,305) and eczema-only cases (n=6,276). The studies that contributed to this analysis are indicated in Supplementary Table 1 and described in detail in the Supplementary Note. We performed three sets of association analyses contrasting three non-overlapping groups of individuals: asthma-only (g1) vs. hay fever-only (g2); asthma-only (g1) vs. eczema-only (g3); and hay fever-only (g2) vs. eczema-only (g3). These analyses are statistically independent from the case-control analysis carried out as part of the GWAS, which facilitates interpretation of the results. For a given sentinel SNP, results from these analyses indicate if the risk allele is more (odds ratio [OR] >1) or less (OR<1) common in e.g. group 1 (g1) when compared to group 2 (g2). For example, if a SNP contributed similarly to the risks of asthma and hay fever but not eczema, then one would expect an OR~1 in the asthma-only vs. hay fever-only comparison, but an OR>1 in the asthma vs. eczema and hay fever vs. eczema analyses. The significance threshold for these analyses was set at 1.2x10-4, which corresponds to a Bonferroni correction for the 136 SNPs and three sets of analyses performed (i.e. P<0.05/(136x3)).
Association between sentinel risk variants and variation in allergy age-of-onset
There is considerable variation in the age allergic diseases are first reported, and this has been shown to be influenced by genetic risk factors 24. We therefore studied the association between the sentinel variants identified in our GWAS and age-of-onset observed in the UK Biobank study (n=35,972). For each individual, we first considered the earliest age of any allergic disease (asthma or hay fever/eczema; the latter two were covered by the same question, and so could not be differentiated) being reported. SNPs were tested for association with this phenotype, with sex and a SNP array variable included as covariates. The significance threshold used for this analysis was 3.6x10-4 (i.e. P<0.05/136). Because significant SNP associations with this broad age-of-onset phenotype could be driven by different risk allele frequencies amongst cases suffering from different individual conditions (for example, a FLG variant might be associated with earliest age-of-onset because it is more prevalent in eczema cases, which tends to precede the development of asthma and hay fever 25), we repeated the analysis by considering individuals who had reported suffering only from a single disease: asthma-only (n=7,445), hay fever-only (n=4,232) and eczema-only (n=1,225). For a given SNP, differences in effect size (beta) between groups were quantified using the formula z = sigma / SE_sigma, where sigma = beta_groupA – beta_groupB, and SE_sigma = sqrt(SE_beta_groupA^2 + SE_beta_groupB^2), which follows a normal distribution.
Estimating the contribution of the sentinel variants to the heritability of asthma, hay fever and eczema
Five steps were involved. First, we performed a GWAS of the individual diseases in the HUNT study, which was not included in the discovery meta-analysis. The HUNT study is described in greater detail in the Supplementary Note. Briefly, based on self-reported questionnaire information, we identified 1,875 cases and 16,463 controls for the asthma GWAS; 6,939 cases and 12,844 controls for the hay fever GWAS; and 2,630 cases and 16,131 controls for the eczema GWAS. After quality control filters, we analyzed 7.6 million common variants (genotyped and imputed) for association with each individual phenotype. The genomic inflation factor (i.e. lambda) for these analyses were 1.049 for asthma, 1.078 for hay fever, and 1.041 for eczema. Second, for each of the three diseases, we quantified the overall SNP-based heritabilities with LD score regression 7 using a subset of 1.2 million HapMap SNPs. To obtain a heritability estimate on the liability scale, we set the population prevalence to be the same as the sample prevalence, given that this was a population-based study. Third, we removed the 136 sentinel variants (and all correlated variants, r2>0.05) from the individual disease GWAS results. Fourth, we re-estimated SNP-based heritabilities as described for step two, but now using the GWAS results without the 136 top associations. In the fifth and final step, the contribution of the 136 sentinel variants towards the heritability of each disease was calculated as the difference between the SNP-based heritability estimated in steps two (all SNPs) and four (without 136 top associations).
Identification of plausible target genes of sentinel risk variants
Two independent strategies were used to identify plausible target genes underlying the observed associations. By 'target gene' we mean a gene for which protein sequence and/or variation in transcription is associated with a sentinel risk variant or one of its proxies (r2>0.8).
First, we used wANNOVAR 26 to identify genes containing non-synonymous SNPs amongst all variants in LD (r2>0.8) with any sentinel risk variant. SNPs in LD with sentinel risk variants were identified using genotype data from individuals of European descent from the 1000 Genomes Project 27 (n=294, release 20130502_v5a).
Second, to identify genes with transcription levels associated with a sentinel risk variant or one of its proxies (r2>0.8), we queried publicly available results from 39 published expression quantitative trait loci (eQTL) studies conducted in 19 tissues or cell types relevant to allergic disease (Supplementary Table 13). We used a conservative significance threshold to identify significant SNP-gene expression associations, specifically a P<2.3x10-9 for cis effects (<1 Mb). We selected this threshold based on a Bonferroni correction that considers the total number of protein-coding genes (G) and the number of SNPs likely to have been tested per gene (M): P<0.05/(GxM). G was set at 21,742, based on the GeneCards database28, queried on October 19th, 2016. We approximate M to be 1,000, as indicated by others 29–31, and so the threshold becomes P=0.05/(21,472 genes x 1,000 SNPs per gene)=2.3x10-9. We did not use information from trans eQTLs to identify plausible target genes of sentinel risk variants, because often these are thought to involve indirect effects32 (e.g. sentinel SNP influences the expression of a transcript in cis, which in turn affects the expression of many other genes in trans).
For each eQTL study, and within each study for each tissue, we created a list of SNPs associated with gene expression in cis at a P<2.3x10-9. Then, for each gene in that study-tissue dataset, we used the --clump procedure in PLINK to reduced the list of expression-associated SNPs (which often included many correlated SNPs) to a set of ‘sentinel eQTLs’, defined as the SNPs with strongest association with gene expression and in low LD (r2<0.05, LD window of 2 Mb) with each other. This procedure was repeated for each of the 94 study-tissue datasets listed in Supplementary Table 13. Finally, we identified as a likely target of a sentinel allergy risk variant any gene for which a sentinel eQTL in any of the 94 study-tissue datasets had an LD r2>0.8 with the sentinel risk variant. That is, we only considered genes for which there was strong LD between a sentinel variant and a sentinel eQTL, which reduces the chance of spurious co-localization. We did not use statistical approaches developed to distinguish co-localization from shared genetic effects because these have very limited resolution at high LD levels (r2>0.8) 33.
To help prioritize plausible target genes for functional validation in subsequent studies, we identified genes for which publicly available functional data supported not just the presence of chromatin interactions between an enhancer and a gene promoter (based on 5C34, promoter capture Hi-C35, ChIA-PET36 or in situ Hi-C37 data), but also an association between variation in enhancer epigenetic marks and variation in gene transcription levels (based on PreSTIGE38, H3K27ac enhancer and super-enhancer annotation 39, IM-PET40 or FANTOM541 analyses). We considered data from immune cell types, lung and skin (Supplementary Table 16) and putative enhancers that overlapped a sentinel risk variant (or one of its strongly correlated proxies, r2>0.95).
Genes that were unlikely to have been previously implicated in the pathophysiology of allergic disease were identified using the procedure described in the Supplementary Note.
Enrichment in tissue-specific gene expression
We used the TSEA approach 9 to identify tissues that were likely to be affected functionally by the biological effects of the sentinel risk variants. We implemented this approach locally using custom scripts. Specifically, for each of 25 broad tissue types studied by the GTEx consortium, we tested if genes with tissue-specific expression (based on a Specificity Index threshold 9 [pSI] of 0.05; listed in file TableS3_NAR_Dougherty_Tissue_gene_pSI_v3-1.txt, downloaded from http://genetics.wustl.edu/jdlab/psi_package/) were enriched amongst the list of plausible target genes, when compared to the rest of the genes in the genome. After excluding genes without a pSI value and in the MHC, there were 112 plausible target genes and 17,671 background genes available for analysis. To test if the plausible target genes were enriched for genes with specific expression in a given tissue, we used Fisher’s exact test (one-sided). To rule out the possibility that a significant enrichment could arise because the list of plausible targets was enriched for genes with eQTLs, we repeated the analysis after restricting the background gene list to a subset of 12,804 genes that were found to have eQTLs in the same eQTL studies that were used to identify plausible target genes of sentinel variants.
We also tested if a significant enrichment in tissue-specific expression could be a general feature of genes near sentinel risk variants, and not specific to the list of genes identified as plausible targets. To address this possibility, we generated 1,000 arbitrary gene lists, each containing 112 random genes instead of the plausible target genes. We selected genes at random from the 17,783 with an available pSI value and not in the MHC, using three strategies. First, genes were randomly drawn from allergy risk loci (+/- 1 Mb of a sentinel variant). To generate each list of random genes, for each non-MHC allergy risk locus L, we randomly selected a locus R from the subset of non-MHC allergy risk loci for which the number of genes available for selection was the same or greater than the actual number of plausible target genes (T) selected for that locus L. Then, for that locus R, we selected T genes at random from the available genes in that locus. This procedure was repeated for all non-MHC allergy risk loci, ensuring that the same locus was not selected twice in a given random dataset.
In the second strategy, genes were randomly drawn from 2 Mb loci selected at random from the genome. In this case, to generate each list of random genes, we first partitioned the autosomes (excluding the MHC) into 1,430 consecutive 2 Mb loci, and counted how many genes with an available pSI value were present in each of these loci. Then, for each non-MHC allergy risk locus L, we randomly selected a locus R from the subset of 2 Mb loci for which the number of genes available for selection satisfied the following criteria: (1) was the same or greater than the actual number of plausible target genes (T) selected for that locus L; and (2) matched (within 10%) the number of genes available for selection for that locus L. This was important to ensure that the randomly selected locus R was comparable to the allergy risk locus L in terms of the number of genes available for selection. Then, for that locus R, we selected T genes at random from the available genes in that locus.
In the third and final strategy, we simply selected genes at random from all 17,783 non-MHC genes with an available pSI value, ignoring where the genes were located in the genome. As a result, for a given random list, the genes selected could only be in close proximity to other genes in that same list by chance alone.
The same approach used to test the enrichment in tissue-specific expression for the plausible target genes was then used to analyze each of the 1,000 lists of random genes. For each of these lists, the smallest P-value observed across all 25 tissues tested was retained (Pmin). The proportion of random gene lists (out of 1,000) with a Pmin that was the same or lower than the enrichment P-value observed with the plausible target genes (Pobs) was then calculated. This corresponds to the probability of exceeding that enrichment when analyzing the random gene lists, after correcting for the 25 tissues tested. As we did for the analysis of the plausible target genes, we repeated the generation and analysis of random gene lists after restricting the genes available for selection (and the background gene list) to the subset of genes with a known eQTL.
Enrichment in tissue-specific SNP heritability
Finucane et al. 10 developed an approach to identify tissues likely affected by the functional effects of disease risk variants, called stratified LD score regression. This approach quantifies the contribution of SNPs located in tissue-specific regulatory annotations to the overall disease heritability. As such, it does not require the identification of likely target genes of allergy risk variant and considers all SNPs in the genome, not just those with a genome-wide significant association with disease risk. Specifically, up to four histone marks (H3K4e1, H3K4me3, H3K9ac and H3K27ac) measured by the ENCODE project are used to define regulatory annotations (e.g. enhancers) in 100 different cell types. SNPs that overlap these regulatory annotations are then identified and their contribution as a group to the disease heritability quantified. As recommended by Finucane et al. 10, we ranked cell types based on the P-value of the regression coefficient, rather than the P-value of total enrichment. To ensure that significant SNP heritability enrichments were not explained by the effects of sentinel variants, we removed the top SNPs (and any variants with r2>0.05 with these) from the meta-analysis GWAS results and repeated the LD score regression analysis.
Enrichment of biological processes
To identify biological processes enriched amongst the non-MHC target genes, we used GeneNetwork 12. With this approach, gene sets originally included in a given GO biological process (BP) were expanded to include other genes based on a 'guilt-by-association' procedure 12. After excluding BPs with <10 or >500 genes, 3,770 BPs were available for analysis. For each BP, we tested its enrichment amongst the list of plausible target genes as follows. First, we downloaded a gene set file containing z-scores for each of 19,976 unique genes in the genome from http://129.125.135.180:8080/GeneNetwork/resources/ontology?ontology=GO_BP&term=[pathway]], where ‘pathway’ was replaced with the actual name of the BP being tested (e.g. “GO:0000002”). The z-score for gene X in that file reflects the probability that gene X is part of that BP. Second, we compared the distribution of z-scores between the list of plausible target genes (107 non-MHC genes were in the GeneNetwork gene set files, and so were available for analysis) and a background gene list of 18,193 genes (obtained after excluding MHC genes, the 107 plausible target genes and genes not listed in GENCODE release 19), using a one-sided Wilcoxon rank-sum test. The P-value from this test represents the probability that genes in that BP are enriched amongst the list of plausible target genes, when compared to the background gene list.
As for the enrichment analysis of tissue-specific expression, we estimated how often a BP enrichment observed with the list of plausible target genes would be expected had we sampled genes at random from the allergy risk loci or from random loci. This analysis addresses the possibility that an observed enrichment might not be a specific feature of the plausible target genes identified but instead a general feature of genes located near sentinel allergy risk variants, or simply in close proximity to each other. We used the same three strategies described above to generate 1,000 lists of random genes, sampling from the 18,300 non-MHC with an available z-score and in GENCODE release 19. To determine if using eQTL information to identify plausible target genes could have biased the enrichment analysis, we generated and analysed random gene lists after restricting the genes available for selection to the subset with known eQTLs (12,913), but found very similar results (not shown).
Common traits and diseases associated with allergic disease risk variants
We first identified all variants in LD (r2>0.8) with a sentinel risk variant using data from Europeans of the 1000 Genomes Project 27 (n=294, release 20130502_v5a), and extracted any associations with these reported in the NHGRI-EBI GWAS catalog database 42 (queried on December 13, 2016) or by Astle et al. 43, a large GWAS of blood cell counts (n=173,480). To complement this analysis, we estimated the SNP-based genetic correlation between our GWAS and results reported for 229 common traits or diseases, using LD Hub 44. In these analyses, results from our meta-analysis were not corrected for the LD score intercept, either at the study level or after the meta-analysis.
Identification of target genes considered as drug targets for human diseases
To identify genes that encode transcripts that are targets of drugs considered for clinical development, we queried the Thomson Reuters Cortellis™ Drug database between November 7 and 15, 2016, which included 63,417 drugs. The drug search was carried out individually for each gene. First, a search query was built based on the following format: HGNC approved gene name OR alias_1 OR … OR alias_N. Gene name aliases were obtained from the Bioconductor annotation package org.Hs.eg.db. For example, to find drugs that target IL6R, the search query used was: "CD126" OR "IL-6R-1" OR "IL-6RA" OR "IL6Q" OR "IL6RA" OR "IL6RQ" OR "gp80" OR "IL6R" OR "interleukin 6 receptor". Second, after running the search query, results were filtered based on the ascribed “Target-based Actions”, keeping only entries that corresponded to the gene name or an alias. For example, of the 65 results obtained with the IL6R query above, only for 20 did the target-based action mention IL6R or an alias. Third, drug results were downloaded, and the gene and respective drug allocated to one of three groups: (1) gene with at least one drug considered for the treatment of allergic diseases; (2) gene considered for the treatment of immune-related conditions, but not allergic diseases specifically; and (3) gene considered for the treatment of other conditions.
Directional effect of the allergy protective allele on target gene expression
In an attempt to predict if existing drugs would be expected to attenuate or exacerbate allergic symptoms, we compared the effect on gene expression between the allergy protective allele and the existing drug. We acknowledge that this is a simplistic comparison, because it assumes that the effect of the protective allele is not tissue- or context-dependent, which is true for most but not all expression-associated SNPs 45–47, and extends to protein levels.
To determine if the allergy protective allele of a sentinel variant was associated with higher or lower target gene expression, we focused on the subset of target genes identified via an eQTL (see above). This was straightforward to assess when the sentinel SNP and the expression-associated SNP were the same variant: for example, if the allergy-protective allele had a negative effect (e.g. beta or z-score) on gene expression in the published eQTL study, then that allele was associated with lower gene expression. On the other hand, when the two SNPs did not correspond to the same variant, but were in high LD (r2>0.8) with each other, we first determined which allele of the expression-associated SNP was on the same haplotype as the allergy-risk allele. Then we used that allele to infer the direction of effect of the allergy-risk allele on gene expression.
Modulation of target gene methylation by environmental risk factors
We first tested if variation in DNA CpG methylation was associated with variation in target gene expression, independently of SNP effects, using data from the Biobank-based Integrative Omics Study (BIOS) consortium that is described in detail elsewhere 15,48. Methylation and expression levels in whole blood samples (n=2,101) were quantified respectively with Illumina Infinium HumanMethylation450 BeadChip Kit arrays and RNA-seq (2x50bp paired-end, Hiseq2000, >15M read pairs per sample). For each target gene, we identified CpG sites in cis (<250 Kb from gene) for which methylation levels were significantly associated with gene expression levels (FDR<5%), after adjusting the methylation levels for methylation-associated SNPs and expression levels for expression-associated SNPs. Such CpG sites, called cis-eQTMs, were identified in a previous study 15 and downloaded from http://genenetwork.nl/biosqtlbrowser. For most genes, there were multiple cis-eQTMs, and so we selected the CpG site most strongly associated with variation in gene expression for downstream analyses.
Next, we tested the association between methylation levels at these sentinel CpGs with five established risk factors for allergic disease using data from unrelated individuals of the Netherlands Twin Register (NTR) study, which was included in the BIOS consortium studies 15,48. The risk factors tested were current smoking (n=1,221), maternal smoking (n=637), BMI (n=1,214), birth weight (n=1,015) and number of older siblings (n=775). Information on BMI and current smoking was collected as part of the NTR biobank project 49 at blood draw. Birth weight was obtained in multiple NTR surveys as previously described 50. Maternal smoking during pregnancy was measured in NTR Survey 10 (data collection in 2013) with the following question: Did your mother ever smoke during pregnancy? with answer categories: no, yes, I don’t know. Information on the number of older siblings was obtained through self-report in NTR surveys 2, 3 and 6. For twin pairs, the answers were checked for consistency and missing data for one twin were supplemented with data from the co-twin where possible. Linear or logistic regression was used to test the association between methylation (β-value) and individual risk factors, with the following variables included as covariates: sex, age at blood sampling, methylation array row, bisulphite plate and white blood cell percentages (% neutrophils, % monocytes, and % eosinophils). The association with maternal smoking was tested while also adjusting for smoking status.
Data availability
Summary statistics of the meta-analysis without the 23andMe study are available at https://genepi.qimr.edu.au/staff/manuelF/gwas_results/main.html. The full GWAS summary statistics for the 23andMe discovery data set will be made available through 23andMe to qualified researchers under an agreement with 23andMe that protects the privacy of the 23andMe participants. Please contact David Hinds (dhinds@23andme.com) for more information and to apply to access the 23andMe data.
Supplementary Material
Acknowledgments
This research was conducted using the UK Biobank resource under Application Number 10074. Detailed acknowledgments and funding details are provided for each contributing study in the Supplementary Note.
Footnotes
Author Contributions
Data collection and analysis in the contributing studies. AAGC study: M.A.F., M.C.M., S.C.D., L.M.B., P.J.T., N.G.M., D.L.D.; LifeLines study: J.M.V., G.H.K.; GENEVA study: H.B., E.R., M.H., A.F., N.N., H.S.,S.K., C.G., K.S., S.W.; GENUFAD studies: I.M., F.R., J.E-G., S.G., A.A., G.H., C.O.S., N.H., Y-A,L.; 23andMe study: C.T., D.A.H.; GERA study: J.D.H., J.S.W., R.B.M, E.J.; NTR study: Q.H., J-J,H., G.W., D.I.B.; CATSS, TWINGENE and SALTY studies: A.T., V.U., Y.L., P.K.E.M., C.A., R.K.; ALSPAC study: L.P.; HUNT study: B.M.B., L.F., M.E.G., J.B.N., W.Z., K.H., A.L., O.L.H., M.L., G.A., C.W.; UK Biobank study: L.P., M.A.F.
Methylation analysis: J.vD., D.I.B., R.J.
Biological and drug annotation: M.A.F., C.W.M., E.M., K.B., O.H., J.Z., J.A.R., J.B., B.B.
Quality control, meta-analysis, tables and figures: M.A.F.
Writing group: M.A.F., J.M.V., I.M., C.T., J.D.H., Q.H., A.T., V.U., J.vD., Y.L., J.E-G., B.M.B., J.B., S.C.D., S.W., P.K.E.M., R.J., E.J., Y-A.L., D.I.B., C.A., R.K., G.H.K., L.P.
Study design and management: M.A.F., D.A.H., B.M.B., S.W., P.K.E.M., R.J., E.J., Y-A.L., D.I.B., C.A., R.K., G.H. K., L.P.
Competing Financial Interests
The authors declare no competing financial interests.
References
- 1.Pinart M, et al. Comorbidity of eczema, rhinitis, and asthma in IgE-sensitised and non-IgE-sensitised children in MeDALL: a population-based cohort study. The Lancet Respiratory medicine. 2014;2:131–140. doi: 10.1016/S2213-2600(13)70277-7. [DOI] [PubMed] [Google Scholar]
- 2.Thomsen SF, et al. Findings on the atopic triad from a Danish twin registry. The international journal of tuberculosis and lung disease : the official journal of the International Union against Tuberculosis and Lung Disease. 2006;10:1268–1272. [PubMed] [Google Scholar]
- 3.van Beijsterveldt CE, Boomsma DI. Genetics of parentally reported asthma, eczema and rhinitis in 5-yr-old twins. Eur Respir J. 2007;29:516–521. doi: 10.1183/09031936.00065706. [DOI] [PubMed] [Google Scholar]
- 4.Loh PR, et al. Contrasting genetic architectures of schizophrenia and other complex diseases using fast variance-components analysis. Nat Genet. 2015;47:1385–1392. doi: 10.1038/ng.3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang J, et al. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat Genet. 2012;44:369–375. doi: 10.1038/ng.2213. S361–363, ng.2213 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ferreira MA. Improving the power to detect risk variants for allergic disease by defining case-control status based on both asthma and hay fever. Twin Res Hum Genet. 2014;17:505–511. doi: 10.1017/thg.2014.59. [DOI] [PubMed] [Google Scholar]
- 7.Bulik-Sullivan BK, et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015 doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Consortium GT. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wells A, et al. The anatomical distribution of genetic associations. Nucleic Acids Res. 2015;43:10804–10820. doi: 10.1093/nar/gkv1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Finucane HK, et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat Genet. 2015;47:1228–1235. doi: 10.1038/ng.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Farh KK, et al. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature. 2015;518:337–343. doi: 10.1038/nature13835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fehrmann RS, et al. Gene expression analysis identifies global gene dosage sensitivity in cancer. Nat Genet. 2015;47:115–125. doi: 10.1038/ng.3173. [DOI] [PubMed] [Google Scholar]
- 13.Thomsen SF, Kyvik KO, Backer V. Etiological relationships in atopy: a review of twin studies. Twin Res Hum Genet. 2008;11:112–120. doi: 10.1375/twin.11.2.112. [DOI] [PubMed] [Google Scholar]
- 14.Sanseau P, et al. Use of genome-wide association studies for drug repositioning. Nature biotechnology. 2012;30:317–320. doi: 10.1038/nbt.2151. [DOI] [PubMed] [Google Scholar]
- 15.Bonder MJ, et al. Disease variants alter transcription factor levels and methylation of their binding sites. Nat Genet. 2017;49:131–138. doi: 10.1038/ng.3721. [DOI] [PubMed] [Google Scholar]
- 16.Joehanes R, et al. Epigenetic Signatures of Cigarette Smoking. Circ Cardiovasc Genet. 2016;9:436–447. doi: 10.1161/CIRCGENETICS.116.001506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lev S, et al. Identification of a novel family of targets of PYK2 related to Drosophila retinal degeneration B (rdgB) protein. Mol Cell Biol. 1999;19:2278–2288. doi: 10.1128/mcb.19.3.2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yan SR, Novak MJ. Beta2 integrin-dependent phosphorylation of protein-tyrosine kinase Pyk2 stimulated by tumor necrosis factor alpha and fMLP in human neutrophils adherent to fibrinogen. FEBS Lett. 1999;451(1):33–38. doi: 10.1016/s0014-5793(99)00539-6. [DOI] [PubMed] [Google Scholar]
- 19.Kamen LA, Schlessinger J, Lowell CA. Pyk2 is required for neutrophil degranulation and host defense responses to bacterial infection. J Immunol. 2011;186:1656–1665. doi: 10.4049/jimmunol.1002093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. doi:btq340 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fadista J, Manning AK, Florez JC, Groop L. The (in)famous GWAS P-value threshold revisited and updated for low-frequency variants. Eur J Hum Genet. 2016;24:1202–1205. doi: 10.1038/ejhg.2015.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gough H, et al. Allergic multimorbidity of asthma, rhinitis and eczema over 20 years in the German birth cohort MAS. Pediatr Allergy Immunol. 2015;26:431–437. doi: 10.1111/pai.12410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mortz CG, Andersen KE, Dellgren C, Barington T, Bindslev-Jensen C. Atopic dermatitis from adolescence to adulthood in the TOACS cohort: prevalence, persistence and comorbidities. Allergy. 2015;70:836–845. doi: 10.1111/all.12619. [DOI] [PubMed] [Google Scholar]
- 24.Sarnowski C, et al. Identification of a new locus at 16q12 associated with time to asthma onset. J Allergy Clin Immunol. 2016;138:1071–1080. doi: 10.1016/j.jaci.2016.03.018. [DOI] [PubMed] [Google Scholar]
- 25.Dharmage SC, et al. Atopic dermatitis and the atopic march revisited. Allergy. 2014;69:17–27. doi: 10.1111/all.12268. [DOI] [PubMed] [Google Scholar]
- 26.Chang X, Wang K. wANNOVAR: annotating genetic variants for personal genomes via the web. J Med Genet. 2012;49:433–436. doi: 10.1136/jmedgenet-2012-100918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Genomes Project C, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rebhan M, Chalifa-Caspi V, Prilusky J, Lancet D. GeneCards: integrating information about genes, proteins and diseases. Trends in genetics: TIG. 1997;13:163. doi: 10.1016/s0168-9525(97)01103-7. [DOI] [PubMed] [Google Scholar]
- 29.Davis JR, et al. An Efficient Multiple-Testing Adjustment for eQTL Studies that Accounts for Linkage Disequilibrium between Variants. Am J Hum Genet. 2016;98:216–224. doi: 10.1016/j.ajhg.2015.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Montgomery SB, et al. Transcriptome genetics using second generation sequencing in a Caucasian population. Nature. 2010;464:773–777. doi: 10.1038/nature08903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lappalainen T, et al. Transcriptome and genome sequencing uncovers functional variation in humans. Nature. 2013;501:506–511. doi: 10.1038/nature12531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Westra HJ, et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat Genet. 2013;45:1238–1243. doi: 10.1038/ng.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chun S, et al. Limited statistical evidence for shared genetic effects of eQTLs and autoimmune-disease-associated loci in three major immune-cell types. Nat Genet. 2017;49:600–605. doi: 10.1038/ng.3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sanyal A, Lajoie BR, Jain G, Dekker J. The long-range interaction landscape of gene promoters. Nature. 2012;489:109–113. doi: 10.1038/nature11279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mifsud B, et al. Mapping long-range promoter contacts in human cells with high-resolution capture Hi-C. Nat Genet. 2015;47:598–606. doi: 10.1038/ng.3286. [DOI] [PubMed] [Google Scholar]
- 36.Li G, et al. Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell. 2012;148:84–98. doi: 10.1016/j.cell.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rao SS, et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Corradin O, et al. Combinatorial effects of multiple enhancer variants in linkage disequilibrium dictate levels of gene expression to confer susceptibility to common traits. Genome Res. 2014;24:1–13. doi: 10.1101/gr.164079.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hnisz D, et al. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.He B, Chen C, Teng L, Tan K. Global view of enhancer-promoter interactome in human cells. Proc Natl Acad Sci U S A. 2014;111:E2191–2199. doi: 10.1073/pnas.1320308111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Andersson R, et al. An atlas of active enhancers across human cell types and tissues. Nature. 2014;507:455–461. doi: 10.1038/nature12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Welter D, et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42:D1001–1006. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Astle WJ, et al. The Allelic Landscape of Human Blood Cell Trait Variation and Links to Common Complex Disease. Cell. 2016;167:1415–1429 e1419. doi: 10.1016/j.cell.2016.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zheng J, et al. LD Hub: a centralized database and web interface to perform LD score regression that maximizes the potential of summary level GWAS data for SNP heritability and genetic correlation analysis. Bioinformatics. 2017;33:272–279. doi: 10.1093/bioinformatics/btw613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fairfax BP, et al. Genetics of gene expression in primary immune cells identifies cell type-specific master regulators and roles of HLA alleles. Nat Genet. 2012;44:502–510. doi: 10.1038/ng.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fairfax BP, et al. Innate immune activity conditions the effect of regulatory variants upon monocyte gene expression. Science. 2014;343:1246949. doi: 10.1126/science.1246949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fu J, et al. Unraveling the regulatory mechanisms underlying tissue-dependent genetic variation of gene expression. PLoS Genet. 2012;8:e1002431. doi: 10.1371/journal.pgen.1002431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhernakova DV, et al. Identification of context-dependent expression quantitative trait loci in whole blood. Nat Genet. 2017;49:139–145. doi: 10.1038/ng.3737. [DOI] [PubMed] [Google Scholar]
- 49.Willemsen G, et al. The Netherlands Twin Register biobank: a resource for genetic epidemiological studies. Twin Res Hum Genet. 2010;13:231–245. doi: 10.1375/twin.13.3.231. [DOI] [PubMed] [Google Scholar]
- 50.Tsai PC, et al. DNA Methylation Changes in the IGF1R Gene in Birth Weight Discordant Adult Monozygotic Twins. Twin Res Hum Genet. 2015;18:635–646. doi: 10.1017/thg.2015.76. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Summary statistics of the meta-analysis without the 23andMe study are available at https://genepi.qimr.edu.au/staff/manuelF/gwas_results/main.html. The full GWAS summary statistics for the 23andMe discovery data set will be made available through 23andMe to qualified researchers under an agreement with 23andMe that protects the privacy of the 23andMe participants. Please contact David Hinds (dhinds@23andme.com) for more information and to apply to access the 23andMe data.