Abstract
Background
The dynamic establishment of the nasal microbiota in early life influences local mucosal immune responses and the susceptibility to childhood respiratory disorders.
Objective
The aim of this case-control study was to monitor, evaluate and compare the development of the nasal microbiota of infants who developed rhinitis and wheeze in the first 18 months of life with those of healthy controls.
Methods
Anterior nasal swabs of 122 subjects belonging to the GUSTO birth cohort were collected longitudinally over seven time points in the first 18 months of life. The nasal microbiota signatures were analyzed using 16S rRNA multiplexed pair-end sequencing from three clinical groups (1) rhinitis alone (n=28), (2) rhinitis with concomitant wheeze (n=34) and (3) healthy controls (n=60).
Results
The maturation of the nasal microbiome followed distinctive patterns in infants from both rhinitis groups compared to controls. Bacterial diversity increased over the period of 18 months of life in control infants, whilst infants with rhinitis showed a decreasing trend (p<0.05). An increase in abundance of Oxalobacteraceae family (Proteobacteria phylum) and Aerococcaceae family (Firmicutes phylum) was associated with rhinitis and concomitant wheeze (adj p<0.01) whilst Corynebacteriaceae family (Actinobacteria phylum) and early colonization with Staphylococcaceae family (Firmicutes phylum) (3 weeks till 9 months) was associated with controls (adj p<0.05). The only difference between the rhinitis group and controls was a reduced abundance of Corynebacteriaceae family (adj p<0.05). Determinants of nasal microbiota succession included gender, mode of delivery, presence of siblings and infant care attendance.
Conclusion
Our results support the hypothesis that nasal microbiome is involved in the development of early onset rhinitis and wheeze in infants.
Keywords: Rhinitis, wheeze, eczema, infant, C-section, nasal microbiota, 16S rRNA gene, Aerococcaceae, Corynebacteriaceae, Oxalobacteraceae, Staphylococcaceae, Moraxellaceae
Introduction
Rhinitis and wheezing disorders in infancy and childhood are common [1, 2]. Birth cohort studies, including our “Growing Up in Singapore Towards healthy Outcomes” (GUSTO) birth cohort, have described infants and toddlers with persistent and recurrent rhinitis. Although the majority do not have allergen sensitization in infancy, early onset rhinitis has been associated with eczema, wheeze and parental history of allergic disorders, suggesting that atopy may play a role in the manifestation of rhinitis. [3–5]
Considerable current evidence supports bacteria-virus interactions as an important factor in lower respiratory tract infections. Respiratory syncytial virus and rhinovirus have been implicated as triggers of wheezing disorders in infants and young children, and as predictors of subsequent asthma at school age [6–10]. It has also been recognized that the resident microbiota of the respiratory tract plays a role as gate keeper that either facilitates or provides resistance to colonization by respiratory virus [10, 11]. The presence of several potentially pathogenic bacteria such as Moraxella catarrhalis in the airways has been associated with asthma and wheeze development [12, 13]. Moreover, farming studies support the concept that environmental microbial exposure has a protective effect on the development of allergic disease [14, 15]. This brings up the concept of “exposome” in which the health status of an individual is determined by his environmental and lifestyle exposure.
Recently, Bosch et al. identified significant differences in the nasal microbiome profiles in infants predisposed to upper respiratory infections in a longitudinal study [16]. Despite these findings, a detailed description of the nasal microbiota in infants with respect to allergic diseases such as rhinitis and wheezing has not been studied longitudinally. In this study, we monitored and evaluated the dynamics of the nasal microbiota in a subcohort of infants belonging to the GUSTO birth cohort over seven time points. The subjects of this subcohort had early onset childhood rhinitis which was associated with parental atopy and atopic comorbidities of atopic dermatitis and wheeze [4]. Given the influential role of the microbiome in the pathogenesis of respiratory tract infections and atopic diseases, the microbiome of the infant’s airways may provide insights into the contributory or inhibitory role of bacteria on the manifestations of atopy.
Methods
Study Population
This case-control study recruited a subset of infants from the GUSTO birth cohort of 1,237 Singaporean newborns [17]. GUSTO is a general population birth cohort and not at risk for atopy cohort. Primary objective of GUSTO study is to identify and evaluate the role of risk factors and determinants influencing body composition and metabolism during early development, and their influence on infants’ health in later childhood. Rhinitis and other atopic outcomes is included as a sub-study of this cohort. Briefly, mothers were recruited antenatally and their children were followed up with scheduled home visits at 3 weeks, 3, 6, 9, 12, 15 months and a clinic visit at 18 months. During these seven visits, caretakers of the subjects were administered a questionnaire on demographic, lifestyle and symptoms of rhinitis and wheeze. Positive cases of rhinitis were then followed up by monthly phone calls to monitor rhinitis progression until a 3-month remission period was over to minimize false-positive cases. This study was approved by the relevant institutions’ ethical review boards (DSRB:B/2009/584 and CIRB:2009/1024/E). Written informed consent was obtained from parents or legal guardians of the subjects.
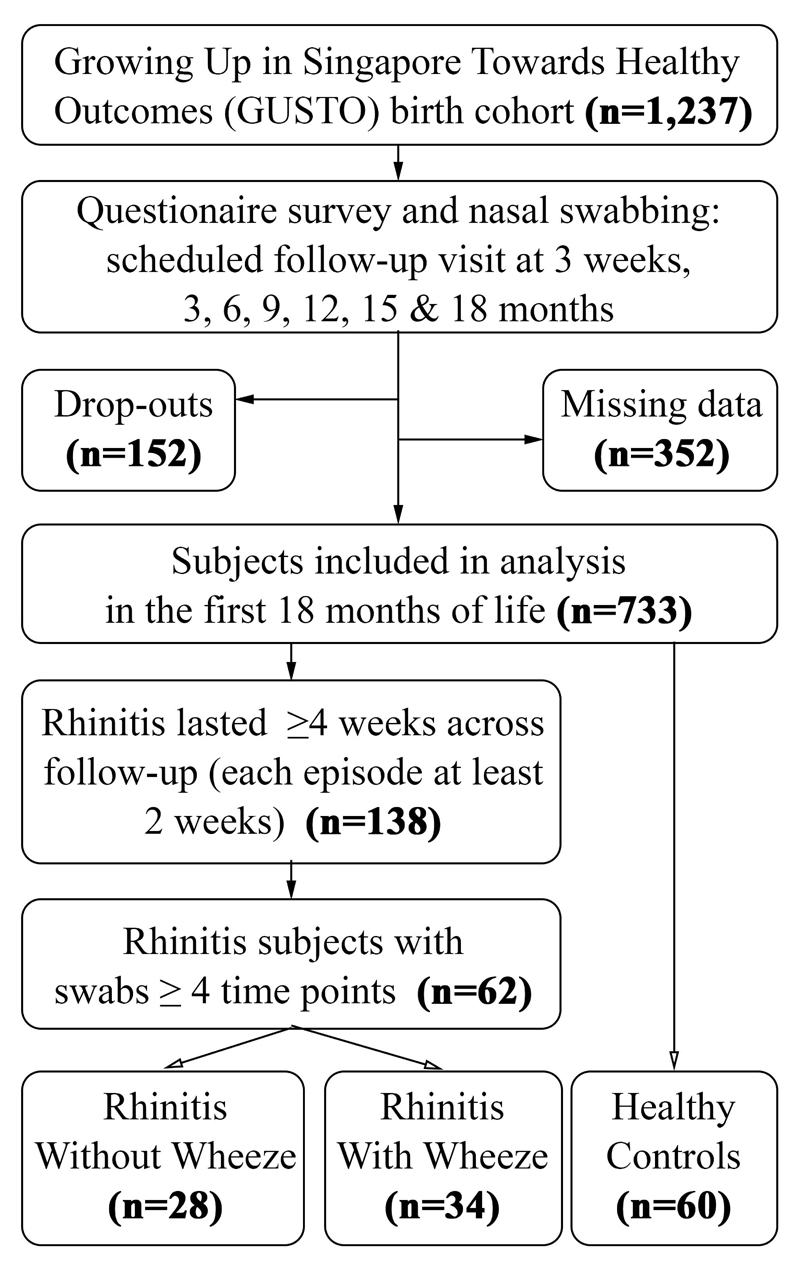
There were 733/1,237 (59.3%) evaluable subjects (excluding drop-outs and those with missing clinical data) who completed follow up till 18 months of age (Fig 1). There were 138 subjects who fulfilled our criteria of rhinitis by 18 months at age. Selection of rhinitis subjects was based on the highest number of available nasal swabs. A total of 62/138 (44.9%) subjects that provided at least four out of the seven nasal swab samples, were then categorized into two groups: rhinitis without wheeze (n=28) and rhinitis with wheeze (n=34). Sixty controls without rhinitis or wheeze were selected with similar characteristics (mode of delivery, age, birth order and day-care attendance) to minimize potential selection bias. Controls had at least four nasal swabs and had negative responses for rhinitis and wheeze (as defined in section below) in the first 18 months of life.
Figure 1.
Flowchart of Subject Selection.
Case Definitions
Rhinitis was defined as having symptoms of sneezing, runny and/or blocked nose that lasted for at least four weeks in single or multiple episodes (each episode lasting at least two weeks) This definition was based on the Allergic Rhinitis and its Impact on Asthma (ARIA) guidelines [4]. Wheeze was defined as presence of wheeze symptoms (noisy breathing with a high-pitched, whistling sound heard from the chest, not the mouth). Eczema was defined as physician-diagnosed eczema in the first 18 months. At the 18 month visit, subjects were assessed via skin prick test (SPT) for the presence of allergic sensitization to a panel of food (milk, peanut, egg) and house dust mites allergens (Dermatophagoides pteronyssinus, Dermatophagoides farinae) from Greer Laboratories (Lenoir, NC, USA) and Blomia tropicalis (extract obtained from in-house laboratory) [18]. Wheal size of at least 3mm larger than the negative control (saline) was inferred as a positive reaction.
Sample Collection
Anterior nasal swabbing was carried out on infants during the visits at seven time points (3 weeks, 3, 6, 9, 12, 15 and 18 months) using dry flocked swabs and stored in Universal Transport Medium (Copan Diagnostics, USA) tubes at -80°C on the same day of collection until analysis.
Nucleic Acid Extraction, 16S rRNA Gene Amplification and Sequencing
DNA was extracted using QIAGEN EZ1 DNA Extraction kit and QIAGEN EZ1 Advanced XL automated purification machine (Qiagen, Germany) according to the manufacturer’s instruction. Bacterial 16S rRNA gene sequences (V3-V6) were amplified and purified as described in Ong et al. [19]. Purified amplicons were fragmented using E220 Focused-ultrasonicator (Covaris, USA) to yield product of an average length of 180 bases. QIAGEN GeneRead DNA Library Prep I Kit (QIAGEN, Germany) was used for library preparation. Multiplexing Sample Preparation Oligonucleotide Kit (Illumina, USA) was used to label DNA sequencing libraries. Multiplexed pair-end sequencing (2 x 76bp reads) was performed using Illumina HISEQ RAPID platform.
16S rRNA Sequencing Analysis
Samples with less than 100,000 reads were removed. Reconstruction of the original amplicon (V3-V6) sequences and classification of amplicon sequences followed the workflow described in Ong et al. [19], which is based on “Expectation Maximization Iterative Reconstruction of Genes from the Environment” (EMIRGE) [20]. EMIRGE-reconstructed sequences were trimmed to the primer-amplified section and searched using BLAST against the complete Greengenes database (dated May, 2013; greengenes/13_5/99_otus.fasta). Hits below predefined percent identity (97% at the species level, 94.5% at the genus level, 86.5% at the family level and 75% at the phylum level) were not considered for classification and removed. EMIRGE assigns abundance estimates to reconstructed sequences. Those quality-passed sequences were clustered into operational taxonomy units (OTUs). The relative abundance of OTUs and Shannon diversity index (SDI) were calculated for each sample at each taxonomic level.
Bioinformatics Analysis
All bioinformatics analyses were performed in the R version 3.4.1 within R studio version 1.0.136. Package “vegan” was used to plot rarefaction curves and Principal Coordinates Analysis (PCoA) based on Bray-Curtis distance to visualize the separation of microbiota profiles between groups. “Envfit” function was used to fit bacterial abundance onto ordination space (p<0.001, 999 permutations) to determine the microbial determinants that shape the bacterial community composition [21]. The Sankey plots was constructed by a custom C++ script (https://github.com/BetaCollins/Sankey). Spearman correlation analysis was done using CoNet plugin in Cytoscape as described by Hannigan et al. [22, 23]. Correlation heatmap and network were visualized in Graphpad Prism 7.0 and Cytoscape 3.4.0 respectively.
Statistical Analysis
All statistical analyses were carried out using IBM Statistical Package for the Social Sciences (SPSS) Statistics version 22 (IBM Cooperation, New York). Univariate analysis (chi-square test) was used to study the demographic, lifestyle and clinical factors between groups. Linear mixed model and linear regression were used to evaluate the bacterial abundance longitudinally and at specific time points respectively, as well as to explore the effect of demographic factors on abundance of main bacterial groups, while adjusting for potential confounders. All statistical significance tests and confidence intervals were two-sided and set at p<0.05. Geometric means were used to represent bacterial abundance and SDI due to non-parametric data distribution.
Results
Demographic, Lifestyle and Clinical Characteristics
The demographic, lifestyle and clinical characteristics of the population were summarized in Table 1. Eczema symptoms and allergen sensitization to any of the 6 tested allergens were more common in rhinitis groups than controls (p<0.05). Maternal history of eczema, and paternal history of rhinitis and eczema was also higher in rhinitis without wheeze group than controls (p<0.05). The use of postnatal antibiotics, Chinese/Malay ethnicity, maternal history of rhinitis, infant-care attendance in the first year were significantly associated with rhinitis with concomitant wheeze compared to control infants (p<0.05). Other demographic and clinical factors were not significantly different between groups.
Table 1.
Demographic, Lifestyle and Clinical Characteristics of the Three Clinical Groups.
| All Rhinitis† | ||||
|---|---|---|---|---|
| Control† | Rhinitis Without Wheeze† | Rhinitis With Wheeze† | ||
| n = 60 | n = 62 | n = 28 | n = 34 | |
| Male gender | 31 (51.67) | 42 (67.74) | 19 (67.86) | 23 (67.65) |
| Presence of siblings | 37 (61.67) | 38 (61.29) | 17 (60.71) | 21 (61.76) |
| Caesarean delivery | 10 (16.67) | 14 (22.58) | 5 (17.86) | 9 (26.47) |
| Pre-term <37 weeks gestation | 4 (6.67) | 4 (6.45) | 2 (7.14) | 2 (5.88) |
| Intrapartum antibiotic prophylaxis | 18 (30.51) | 22 (35.48) | 5 (17.86) | 17 (50.00) |
| Postnatal antibiotics within first year | 19 (31.67) | 35 (58.33) ** | 14 (51.85) | 21 (63.64)** |
| Ethnicity | ||||
| Chinese | 36 (60.00) | 30 (48.39)* | 17 (60.71) | 13 (38.24)** |
| Malay | 10 (16.67) | 21 (33.87)* | 8 (28.57) | 13 (38.24)* |
| Indian | 8 (13.33) | 11 (17.74) | 3 (10.71) | 8 (23.53) |
| Maternal history | ||||
| Any of three atopic diseases | 12 (20.34) | 23 (39.66)* | 11 (40.74) | 12 (38.71)* |
| Rhinitis | 6 (10.17) | 13 (22.41) | 4 (14.81) | 9 (29.03)* |
| Eczema | 3 (5.08) | 11 (18.97)* | 7 (25.93)* | 4 (12.90) |
| Asthma | 7 (11.86) | 9 (15.52) | 2 (7.41) | 7 (22.58) |
| Paternal History | ||||
| Any of three atopic diseases | 18 (30.51) | 25 (43.10) | 16 (59.26)* | 9 (29.03) |
| Rhinitis | 11 (18.64) | 13 (22.41) | 10 (37.04)* | 3 (9.68) |
| Eczema | 3 (5.08) | 7 (12.07) | 5 (18.52)* | 2 (6.45) |
| Asthma | 8 (13.56) | 10 (17.24) | 4 (14.81) | 6 (19.35) |
| Infant care attendance within first year | 7 (15.56) | 6 (17.65) | 2 (10.53) | 4 (26.67)* |
| Exclusive breastfeeding till month 6 | 10 (16.67) | 8 (12.90) | 7 (25.00) | 1 (2.94) |
| Eczema symptoms | 6 (10.00) | 19 (30.65)** | 9 (32.14)** | 10 (29.41)* |
| Allergen sensitization | ||||
| Any of six tested allergens | 4 (7.02) | 13 (23.21)* | 7 (28.00)* | 6 (19.35) |
| Egg | 1 (1.75) | 4 (7.14) | 3 (12.00) | 1 (3.23) |
| Milk | 0 (0.00) | 3 (5.36) | 1 (4.00) | 2 (6.45) |
| Peanut | 0 (0.00) | 4 (7.14) | 3 (12.00)* | 1 (3.23) |
| Dermatophagoides pteronyssinus | 3 (5.26) | 10 (17.86) | 6 (24.00)* | 4 (12.90) |
| Dermatophagoides farina | 3 (5.26) | 5 (8.93) | 3 (12.00) | 2 (6.45) |
| Blomia tropicalis | 0 (0.00) | 2 (3.57) | 2 (8.00) | 0 (0.00) |
significant at p<0.05 compared to healthy group in univariate analysis.
significant at p<0.01 compared to healthy group in univariate analysis.
Some variables had subjects with missing data.
In the clinical groups with rhinitis, the mean (± SD) onset of rhinitis symptoms was 7.1 ± 4.8 months (range 3 weeks to 18 months) and wheeze symptoms onset was 8.2 ± 4.6 months (range 3 to 18 months).
16S rRNA Sequencing Summary
There was a total of 670 nasal swab samples collected from 122 subjects (62 cases and 60 controls). Excluding samples with low yield of DNA concentration and low reads count, this yielded 620 nasal swab samples. The sequencing summary is tabulated in Table E1. The rarefaction curve for number of observed OTUs at family level were generated (Fig E1).
Dynamics of Bacterial Diversity
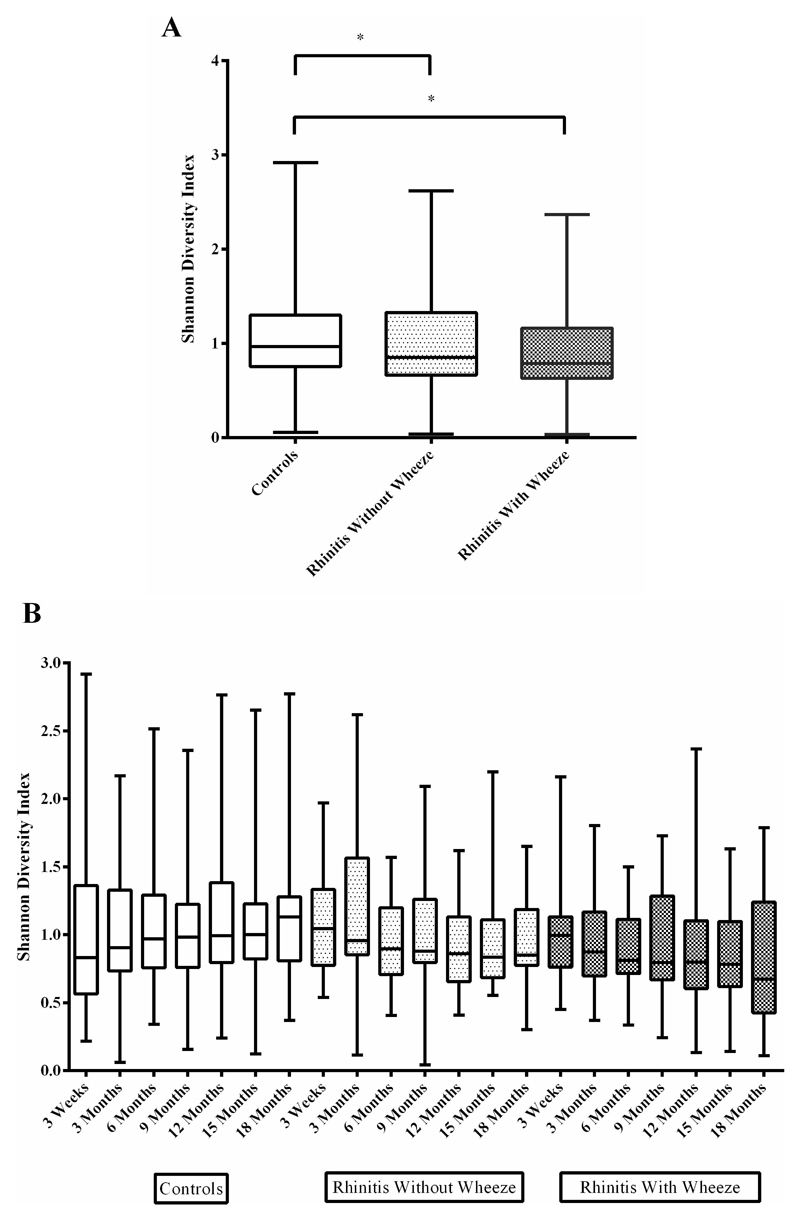
Overall SDI was significantly lower in both rhinitis groups (without wheeze: geometric mean SDI=0.85; with wheeze: geometric mean SDI=0.83) compared to controls (geometric mean SDI=0.97, p=0.03 and 0.01 respectively) (Fig 2A). Diversity in controls increased (p=0.019) while diversity in infants from both rhinitis groups declined over time (without wheeze, p=0.028; with wheeze, p=0.025) (Fig 2B).
Figure 2.
Microbial diversity of the three clinical groups. (A) Overall Shannon Diversity combining all seven time-points from 3 weeks till 18 months; (B) Shannon diversity of the three clinical groups by time points. Max, min, 25th percentile, 75th percentile and geometric mean of relative abundance were used for box and whisker plot.
* denotes significance at p<0.05 between groups.
Maturation of the Nasal Microbiota In the First 18 Months of Life
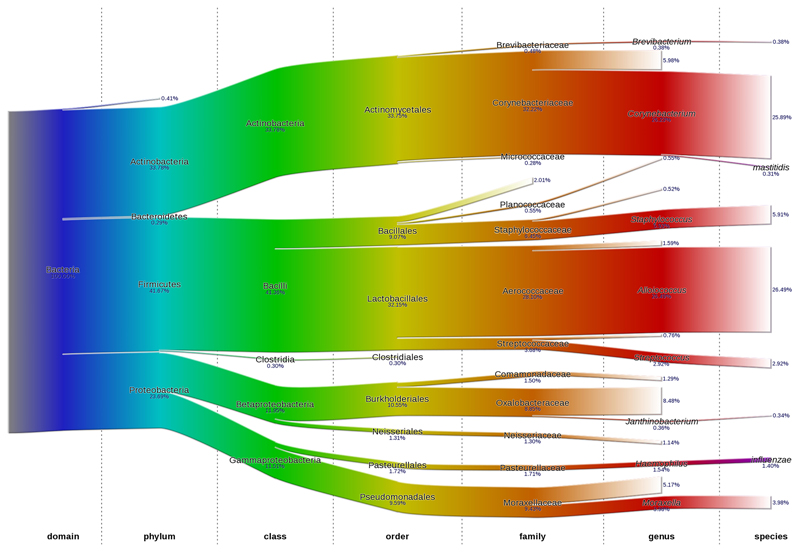
The overall distribution of nasal microbiota down to species level is shown using Sankey plots (Fig 3). The Sankey plots for each clinical group are shown in Figure E2. The abundance of predominant phyla, family and genus are summarized in Figure 4 and Table E2. Abundance data down to species level were not analyzed further due to low bacterial identification rate.
Figure 3.
Sankey plot representing the overall nasal bacterial composition and their corresponding average abundance for three clinical groups. Each colour represents different taxonomy classification levels. The arithmetic mean of relative abundance is inserted below each microbial group.
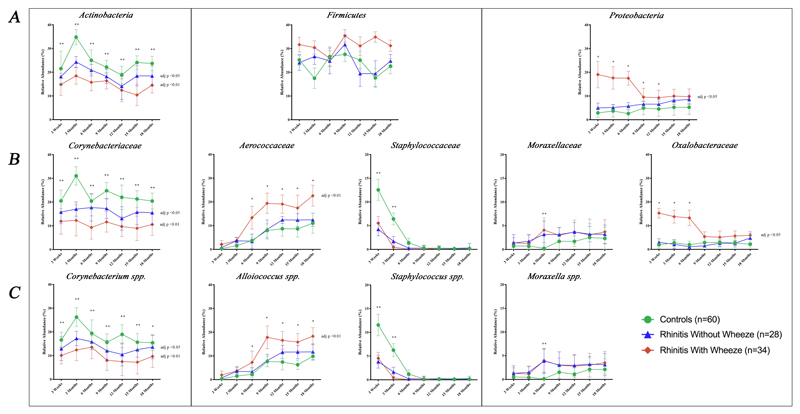
Figure 4.
Bacterial abundance of major (A) Phyla, (B) Family and (C) Genus of the three clinical groups by time points. Data presented as geometric mean of relative abundance. Adjusted p values denotes significant difference of bacterial relative abundance between rhinitis and control groups using linear mixed model analysis and linear regression after adjusting for confounders (gender, family history of respiratory disease, presence of siblings, mode of delivery, use of intrapartum antibiotic prophylaxis and postnatal antibiotics and breastfeeding pattern).
* denotes significant difference at p<0.05 between rhinitis with wheeze and control group at specific time point.
** denotes significant difference p<0.05 between both rhinitis groups and control group at specific time point.
Phyla and Family
For all the three clinical groups, the predominant phyla were Actinobacteria, Proteobacteria and Firmicutes, which accounted for more than 97% of the total nasal microbiota. Other minor phyla made up less than 2% of the whole bacterial community.
Higher abundance of Actinobacteria was associated with control group compared to both rhinitis groups till the age of 18 months (without wheeze: adj p=0.034; with wheeze: adj p<0.01). Corynebacteriaceae family comprised 95.1% of this phylum. Correspondingly, the abundance of Corynebacteriaceae was higher in controls than both rhinitis groups (without wheeze: adj p=0.037; with wheeze: adj p<0.01).
In contrast, higher abundance of Proteobacteria phylum was observed for the rhinitis groups compared to controls, but statistical significance was seen only for rhinitis with wheeze (adj p <0.01). At family level, Oxalobacteraceae and Moraxellaceae comprised 36.9% and 39.3% of this phylum respectively. However, only Oxalobacteraceae was significantly higher in rhinitis with wheeze than controls over time (adj p<0.01).
Firmicutes phylum did not show distinct trends longitudinally for all clinical groups. However, Aerococcaceae family, which comprised 67.6% of this phylum, was higher in rhinitis with wheeze group compared to controls (adj p <0.01). Staphylococcaceae made up 14.8% of the Firmicute phylum and was significantly decreased in both rhinitis groups till 9 month (adj p<0.05).
Genera
The Corynebacteriaceae, Aerococcaceae, Moraxellaceae and Staphylococcaceae families were made up mainly of Corynebacterium spp. (82.0%), Alloiococcus spp. (94.3%), Moraxella spp. (43.7%) and Staphylococcus spp. (91.1%) respectively. The trends of these genera in the first 18 months were similar to their corresponding families (Fig 4C). Janthinobacterium spp. was the only genus of Oxalobacteraceae family that was identified with the sequencing data processing method employed. However, the average relative abundance of this genus was only 0.3%.
In summary, the relative abundance of bacteria between the three clinical groups showed distinct trends throughout 18 months of life. Although differences in bacteria composition were observed for both groups of rhinitis (with and without wheeze) compared to controls, the differences were more marked in the rhinitis with concomitant wheeze group. Furthermore, correlation analysis showed that Oxalobacteraceae family (Proteobacteria phylum) had direct negative interactions with Aerococcaceae family (Firmicutes phylum) and Corynebacteriaceae family (Actinobacteria phylum) in all three clinical groups throughout the first 18 months. This reinforces the results of our longitudinal analysis (Fig 4) that a distinct pattern of microbial abundance of the major bacterial groups was associated with the clinical outcomes; where high abundance of Corynebacteriaceae and Staphylococcaceae, and low abundance of Oxalobacteraceae and Aerococcaceae were associated with controls and the reverse pattern of abundance for rhinitis and wheeze (Fig E3). There was lower abundance of the Corynebacteriaceae and showed little fluctuation throughout the 18 month period in both rhinitis groups (with and without wheeze) compared to controls.
Multivariate Analysis of Nasal Microbial Community Composition
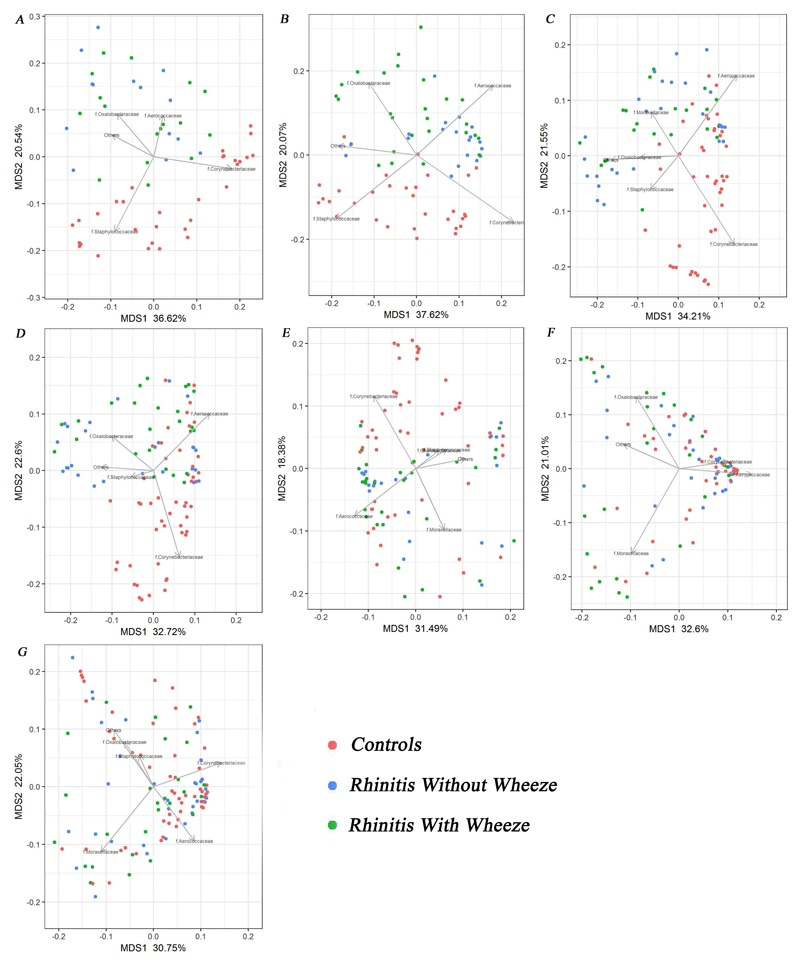
To investigate the differences in community composition between clinical groups, PCoA was plotted at seven time points (Fig 5). The five bacterial groups (Corynebacteriaceae, Oxalobacteraceae, Moraxellaceae, Aerococcaceae and Staphylococcaceae families) together explained an average of 54.6% (range: 49.87%-57.69%) of the overall variability in the microbiota composition. Distinct bacterial community composition was seen between two groups of rhinitis and controls at 3 weeks and 3 months. At these time points, Corynebacteriaceae and Staphylococcaceae significantly influenced controls whilst Oxalobacteraceae and Aerococcaceae shaped the rhinitis groups (p<0.001). The clusters between groups became less distinct from 6 months onwards. Moraxellaceae drove bacterial composition of rhinitis groups at this time point (p<0.001). The microbial communities after 9 months showed an obvious shift and disappearing of distinct clusters between clinical groups. Of note, Corynebacteriaceae remained the key bacterial drivers of bacterial community in controls till 12 months (p<0.001).
Figure 5.
Principal coordinates analysis (PCoA) based on Bray–Curtis dissimilarity between nasal microbiome profiles (Family level) for the three clinical groups at various time-points. (A) 3 week; (B) 3 month; (C) 6 month; (D) 9 month; (E) 12 month; (F) 15 month and (G) 18 month. Arrow shows the directions from the origin for which clusters have significant abundances for the bacterial groups (p<0.001, 999 permutations) and its length is proportional to the correlation between ordination and the bacterial groups.
Influence of Demographic and Clinical Determinants on Establishment of the Main Nasal Bacterial Groups
Linear regression was employed to evaluate the effect of demographic and clinical factors on the overall dynamic establishment of the five main bacterial families (Fig E4). Caesarean-delivery and male gender were associated with higher abundance of Aerococcaceae and lower abundance of Staphylococcaceae (adj p<0.05). Presence of siblings and infant care attendance were associated with lower colonization of Corynebacteriaceae, and higher carriage of Oxalobacteraceae and Moraxellaceae (adj p<0.05). Other factors (pre-term, use of intrapartum antibiotic prophylaxis and postnatal antibiotics, family history of atopic diseases and exclusive breastfeeding pattern) did not show significant associations.
Discussion
The longitudinal design of this study with serial sampling over seven time points describes the dynamic changes and provides valuable snapshots of the succession of the nasal microbiota in early childhood. We observed distinct differences in the profiles of microbiota of infants who developed rhinitis, especially in those who also had concomitant wheeze in the first 18 months of life compared to healthy controls. As these differences were present from the earliest time points (3 weeks and 3 months of age), and before the onset of clinical symptoms in the majority of subjects, our findings strongly suggest a role of the nasal microbiome in the development of respiratory disease.
Nasal Microbial Diversity
The nasal microbial diversity increased with age in the controls, whilst the rhinitis groups (with or without concomitant wheeze) was reversed with decreasing trends with age. Other cross-sectional studies in infants and young children with respiratory disorders such as asthma, allergic rhinitis and acute respiratory infection, have described similar findings [24–26].
Distinct Bacterial Profiles Between Disease Groups and Healthy
The three dominating phyla of the infant nasal microbiome were Actinobacteria, Proteobacteria and Firmicutes which is consistent with other longitudinal studies in children [13, 16].
Both the rhinitis (without and with wheeze) groups had decreased abundance of Corynebacteriaceae family (Actinobacteria phylum) compared to controls. Other studies have also shown reduced abundance of Corynebacteriaceae with other respiratory conditions such as acute otitis media and wheezing [13, 27]. Of interest is the finding that Corynebacterium accolens from the human nasal mucosa releases antibacterial free fatty acid from human triacylglycerols [28]. Taken together, these findings suggest that a high abundance of Corynebacteriaceae confers protection by inhibiting other potentially pathogenic bacteria against respiratory diseases in infants.
In contrast to Corynebacteriaceae, there was higher abundance of Proteobacteria in those with rhinitis and concomitant wheeze compared to controls. This observation was consistent with other studies that reported its association with wheezing respiratory disorders [29]. Although there have not been other reports of Oxalobacteriaceae family in association with wheeze in infants specifically, its relation with asthma in adults has been reported [30]. Our current study in infants strengthens the possibility that Oxalobacteraceae may contribute to wheezing disorders.
The evidence that Moraxella spp. (Moraxellaceae family) influences subsequent respiratory illness appears to vary between studies. Several studies have shown that Moraxella spp. was associated with an increased risk of pneumonia in children in their first 3 years of life [31]. On the other hand, it has also been shown to be a part of a commensal bacterial community in the first 2 years of life and protects against subsequent respiratory infection [32]. In our study, there was no difference in abundance of Moraxella spp. between clinical groups longitudinally but it was significantly increased in both rhinitis groups at the 6 month time point.
Similar to Oxalobacteraceae family, the abundance of Aerococcaceae family (Firmicutes phylum) was significantly higher with the rhinitis and concomitant wheeze group compared to controls. As in all clinical groups, Aerococcaceae colonization appears later in age (6 months and beyond) compared to Oxalobacteraceae and Corynebacteriaceae. This observation is consistent with previous reports in children studied longitudinally from 1 till 24 months [32, 33]. The microbials belonging to the Aerococcaceae were further identified to belong to the Alloiococcus spp. Although this genus appeared to be strongly associated with rhinitis and wheeze in our study, other studies have shown the contrary, in that lower abundance of Alloiococcus spp. during infancy was associated with higher numbers of acute respiratory infections [13]. Little is known of its role in early nasal microbial colonization. Alloiococcus is a Gram-positive genus with only one known species, Alloiococcus otitidis (A. otitidis) [34]. This species is frequently detected in the outer/middle ear canal and nasopharyngeal of children with acute otitis media. Studies from Japan found that nasopharyngeal colonization of A. otitidis is increased in otitis-prone children compared to that in non-otitis-prone children [35, 36]. This organism is also shown to induce local immune response in the middle ear as a pathogen [37, 38].
Staphylococcaceae family (Firmicutes phylum) differ between clinical groups longitudinally till 9 months, and its abundance was significantly higher at early time points (3 week and 3 month) in controls compared to the rhinitis groups. This was strengthened by PCoA at 3 week and 3 month, Staphylococcaceae is, therefore, a feature of early colonization in healthy individuals before its declines in later infancy, and this is supported by other studies in healthy children aged below 18 months of age [13, 33]. Although cutaneous colonization of Staphylococcus spp., particularly Staphylococcus aureus is associated with atopic eczema [39], our longitudinal analysis did not show any association between eczema and colonization of nasal Staphylococcus spp. (data not shown).
Our data indicate that the succession of nasal bacterial over the first 18 months of life influence the susceptibility to wheeze. We observed that high carriage of Oxalobacteraceae and low abundance of Corynebacterium spp. (Corynebacteriacecae family) and Staphylococcaceae in the early time points (till 9 months) appeared to encourage the early colonization and growth of Alloiococcus spp. (Aerococcaceae family) at an accelerated rate in subjects with wheeze compared to controls. Thus, the initial establishment of Oxalobacteraceae rather than Corynebacterium spp. might be a predictive marker of transition to the subsequent increase in abundance of Alloiococcus spp. Of note is the ‘graded’ degree of dysbiosis between the rhinitis alone and rhinitis with concomitant wheeze groups, with the rhinitis with wheeze group showing the more extreme difference from controls. It is tempting to speculate that larger deviation from ‘normal’ microbial establishment in the airway is required to predispose wheeze in infancy. Our findings are supported by Bosch et al. who reported an accelerated maturation of microbiota in children with more than two respiratory tract infection episodes per year compared to controls with less than two episodes [16].
Determinants of Establishment of Nasal Microbiota
This study found that male gender, presence of siblings, Caesarean delivery and infant-care attendance influenced the establishment and maturation of the nasal microbiome. These factors could be the determinants for dysbiotic nasal microbiota that could predispose to rhinitis and wheeze later in life.
The presence of siblings and daycare attendance was associated with lower abundance of Corynebacteriaceae and higher abundance of Oxalobacteraceae and Moraxellaceae. This interaction with other young children has also been shown to be related to frequent wheezing at the age of 2 years compared to children with little or no exposure [40]. An Australian study by Teo et al., showed that significantly higher relative abundances of Moraxella spp. and lower relative abundances of Corynebacterium spp. in infants attending day care compared to those at home [13]. A cross-sectional American study showed that infants with siblings were more likely be dominated by Moraxella than a Corynebacterium while opposite pattern was observed in those without siblings [41]. Whether the increased exposure to respiratory viruses or its influence on the establishment of the nasal microbiome through the interaction with other young children plays a more important role in the development of wheezing illness in early life will require further study.
Our results also showed that male gender and delivery by Caesarean section was associated with a more ‘dysbiotic’ nasal microbial profile with higher abundance of Aerococcaceae and lower abundance of Staphylococcaceae. Although not observed in other studies, the mode of delivery did impact on the succession of nasal microbiome in early life [16]. A birth cohort study up to 6 months led by Bosch et al. showed that children born by Caesarean section had a higher abundance of Staphylococcus aureus than vaginally born children, even though this trend was most prominent at the early time points. In relation to gender, male gender has been shown to be a universal risk factor for respiratory disorders such as rhinitis, wheeze and asthma in early childhood in most populations published and not confined to this cohort [42, 43]. This may arise from gender differences in lung airway anatomy and maturation [44]. The airways in males have been found to be structurally narrower than females and this anatomical difference may contribute to the higher prevalence of wheezing disorders in the male [45, 46]. However, there are other gender related differences that may explain the gender differences in nasal bacterial colonization that was observed in our cohort. Firstly, the delay in pulmonary surfactant in the male fetal lung compared to females [47, 48] may contribute to the pattern of early microbial colonization. Surfactant proteins have the ability to aggregate and control the clearance of bacteria through binding and neutralization by acting as opsonin [49–51]. Secondly, the innate and adaptive immune responses of male infants are also weaker than females, and this difference may influence differential colonization of the nasal mucosa [52–54].
Our study did not show a significant impact of intrapartum antibiotic prophylaxis or post-natal antibiotic on the nasal microbiome. Other studies in infants with respiratory tract infection and acute otitis media have shown a negative association between antibiotic usage and Corynebacteriaceae carriage [16, 27]. These differences between studies may in part be explained by the different collection sites of our study (anterior nares) and other studies (nasopharyngeal).
Limitations and Conclusion
The limitation in this study is that the sampling area is the anterior nares. Several studies have highlighted the variation in bacterial composition between different sites of respiratory tract [55–58]. Moreover, the impact of virus was not analyzed in our study which deserves further investigation. We were also unable to analyze all the bacteria groups down to species level. Future studies involving metagenomics sequencing may provide more precise identification of bacteria and may also provide information on viruses.
Despite these limitations, our longitudinal birth cohort with dense serial sampling support the hypothesis that distinct profiles of the nasal microbiota in the first 18 months of life impact on the development of rhinitis and wheezing in later infancy. Our GUSTO follow up data showed that 20% of infants with rhinitis persisted to have rhinitis at the age 5 years, whilst controls remained rhinitis free (data not shown), indicating that rhinitis status at 18 months is a predictor for rhinitis status at 5 years. Further work to verify these findings and its impact on subsequent allergic rhinitis and asthma in later childhood will provide important insights into the role of the nasal microbiome in these disorders. These are important steps towards developing novel strategies for the treatment and management of these disorders.
Supplementary Material
Capsule Summary.
The nasal microbiome in the first 18 months of life is involved in the development of rhinitis and wheeze in early life. A decrease in Corynebacteriaceae and Staphylococcaceae, and increase in Oxalobacteraceae and Aerococcaceae of the anterior nares are associated with early onset rhinitis and wheeze.
Clinical Implications.
Research into the mechanisms behind longitudinal variation in the respiratory microbiome may pave the way for the prevention and treatment of early onset rhinitis and wheezing disorders.
Acknowledgements
We would like to express our gratitude to members of the GUSTO group for their assistance which includes Allan Sheppard, Amutha Chinnadurai, Anne Rifkin-Graboi, Anqi Qiu, Arijit Biswas, Birit F.P. Broekman, Boon Long Quah, Borys Shuter, Chai Kiat Chng, Cheryl Ngo, Choon Looi Bong, Christiani Jeyakumar Henry, Cornelia Yin Ing Chee, Doris Fok, Fabian Yap, George Seow Heong Yeo, Helen Chen, Iliana Magiati, Inez Bik Yun Wong, Ivy Yee-Man Lau, Jeevesh Kapur, Jenny L. Richmond, Jerry Kok Yen Chan, Joanna D. Holbrook, Joshua J. Gooley, Kok Hian Tan, Krishnamoorthy Niduvaje, Leher Singh, Lin Lin Su, Lourdes Mary Daniel, Marielle V. Fortier, Mark Hanson, Mary Foong-Fong Chong, Mary Rauff, Mei Chien Chua, Michael Meaney, Mya Thway Tint, Neerja Karnani, Ngee Lek, P. C. Wong, Pratibha Agarwal, Rob M. van Dam, Salome A. Rebello, Shang Chee Chong, Shirong Cai, Sok Bee Lim, Chin-Ying Stephen Hsu, Victor Samuel Rajadurai, Walter Stunkel, Wee Meng Han, Wei Wei Pang, Yin Bun Cheung and Yung Seng Lee. We would especially like to thank the GIS platforms for their support during the project in the areas of scientific and research computing (led by Chih Chuan Shih), research pipeline development (led by Andreas Wilm) and the next generation sequencing platform (led by Wendy Soon). We also thank Professor Lee Yuan Kun for his intellectual input.
This research is supported by the Singapore National Research Foundation under its Translational and Clinical Research (TCR) Flagship Programme and administered by the Singapore Ministry of Health’s National Medical Research Council (NMRC), Singapore- NMRC/TCR/004-NUS/2008, NMRC/CSA/022/2010 and NMRC/CIRG/1344/2012. Additional funding is provided by the NRF CREATE Programme (NRF370062-HUJ-NUS - PROJECT 10) and NUHS Cross-Department Grant 2011. KMG is supported by the National Institute for Health Research through the NIHR Southampton Biomedical Research Centre.
Abbreviations Used
- ARIA
Allergic Rhinitis and its Impact on Asthma
- DNA
Deoxyribonucleic Acid
- EMIRGE
Expectation Maximization Iterative Reconstruction of Genes from the Environment
- GERMS
Genome Institute of Singapore Efficient Rapid Microbial Sequencing
- GUSTO
Growing Up in Singapore Towards healthy Outcomes
- OTUs
Operational Taxonomic Units
- PCoA
Principal Coordinates Analysis
- PCR
Polymerase Chain Reaction
- SDI
Shannon Diversity Index
- SPT
Skin Prick Test
- spp.
species
- 16S rRNA
16S Ribosomal Ribonucleic Acid
Footnotes
Disclosure
All authors declare no conflict of interest. Christophe Lay is an employee of Danone Nutricia Research.
References
- 1.Tan TN, et al. Prevalence of allergy-related symptoms in Singaporean children in the second year of life. Pediatr Allergy Immunol. 2005;16(2):151–6. doi: 10.1111/j.1399-3038.2005.00242.x. [DOI] [PubMed] [Google Scholar]
- 2.Tan TN, et al. Prevalence of asthma and comorbid allergy symptoms in Singaporean preschoolers. Asian Pac J Allergy Immunol. 2006;24(4):175–82. [PubMed] [Google Scholar]
- 3.Grabenhenrich LB, et al. Prediction and prevention of allergic rhinitis: A birth cohort study of 20 years. J Allergy Clin Immunol. 2015;136(4):932–40 e12. doi: 10.1016/j.jaci.2015.03.040. [DOI] [PubMed] [Google Scholar]
- 4.Hardjojo A, et al. Rhinitis in the first 18 months of life: exploring the role of respiratory viruses. Pediatr Allergy Immunol. 2015;26(1):25–33. doi: 10.1111/pai.12330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Riedler J, et al. Exposure to farming in early life and development of asthma and allergy: a cross-sectional survey. Lancet. 2001;358(9288):1129–33. doi: 10.1016/S0140-6736(01)06252-3. [DOI] [PubMed] [Google Scholar]
- 6.Lukkarinen M, et al. Rhinovirus-induced first wheezing episode predicts atopic but not nonatopic asthma at school age. J Allergy Clin Immunol. 2017 doi: 10.1016/j.jaci.2016.12.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu S, et al. Predictors of asthma following severe respiratory syncytial virus (RSV) bronchiolitis in early childhood. Pediatr Pulmonol. 2016;51(12):1382–1392. doi: 10.1002/ppul.23461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Del Rosal T, et al. Recurrent wheezing and asthma after bocavirus bronchiolitis. Allergol Immunopathol (Madr) 2016;44(5):410–4. doi: 10.1016/j.aller.2015.07.004. [DOI] [PubMed] [Google Scholar]
- 9.Sun H, et al. Prevalence of rhinovirus in wheezing children: a comparison with respiratory syncytial virus wheezing. Braz J Infect Dis. 2016;20(2):179–83. doi: 10.1016/j.bjid.2015.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lynch JP, et al. The Influence of the Microbiome on Early-Life Severe Viral Lower Respiratory Infections and Asthma-Food for Thought? Front Immunol. 2017;8:156. doi: 10.3389/fimmu.2017.00156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beigelman A, Bacharier LB. Early-life respiratory infections and asthma development: role in disease pathogenesis and potential targets for disease prevention. Curr Opin Allergy Clin Immunol. 2016;16(2):172–8. doi: 10.1097/ACI.0000000000000244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mansbach JM, et al. Respiratory syncytial virus and rhinovirus severe bronchiolitis are associated with distinct nasopharyngeal microbiota. J Allergy Clin Immunol. 2016;137(6):1909–1913 e4. doi: 10.1016/j.jaci.2016.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Teo SM, et al. The infant nasopharyngeal microbiome impacts severity of lower respiratory infection and risk of asthma development. Cell Host Microbe. 2015;17(5):704–15. doi: 10.1016/j.chom.2015.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schuijs MJ, et al. Farm dust and endotoxin protect against allergy through A20 induction in lung epithelial cells. Science. 2015;349(6252):1106–10. doi: 10.1126/science.aac6623. [DOI] [PubMed] [Google Scholar]
- 15.Stein MM, et al. Innate Immunity and Asthma Risk in Amish and Hutterite Farm Children. N Engl J Med. 2016;375(5):411–21. doi: 10.1056/NEJMoa1508749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bosch AA, et al. Maturation of the Infant Respiratory Microbiota, Environmental Drivers and Health Consequences: A Prospective Cohort Study. Am J Respir Crit Care Med. 2017 doi: 10.1164/rccm.201703-0554OC. [DOI] [PubMed] [Google Scholar]
- 17.Soh SE, et al. Cohort profile: Growing Up in Singapore Towards healthy Outcomes (GUSTO) birth cohort study. Int J Epidemiol. 2014;43(5):1401–9. doi: 10.1093/ije/dyt125. [DOI] [PubMed] [Google Scholar]
- 18.Yi FC, et al. Culture of Blomia tropicalis and IgE immunoblot characterization of its allergenicity. Asian Pac J Allergy Immunol. 1999;17(3):189–94. [PubMed] [Google Scholar]
- 19.Ong SH, et al. Species identification and profiling of complex microbial communities using shotgun Illumina sequencing of 16S rRNA amplicon sequences. PLoS One. 2013;8(4):e60811. doi: 10.1371/journal.pone.0060811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller CS, et al. EMIRGE: reconstruction of full-length ribosomal genes from microbial community short read sequencing data. Genome Biol. 2011;12(5):R44. doi: 10.1186/gb-2011-12-5-r44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oksanen JKR, Blanchet FG, Legendre P, Minchin PR, O’Hara RB, Simpson GL, Solymos P, Stevens MHH, Wagner H. vegan: Community Ecology, Package. R Package Version 2.2– 1. 2013. [Google Scholar]
- 22.Faust K, Raes J. CoNet app: inference of biological association networks using Cytoscape. F1000Res. 2016;5:1519. doi: 10.12688/f1000research.9050.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hannigan GD, et al. The human skin double-stranded DNA virome: topographical and temporal diversity, genetic enrichment, and dynamic associations with the host microbiome. MBio. 2015;6(5):e01578–15. doi: 10.1128/mBio.01578-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chiu CY, et al. Airway Microbial Diversity is Inversely Associated with Mite-Sensitized Rhinitis and Asthma in Early Childhood. Sci Rep. 2017;7(1):1820. doi: 10.1038/s41598-017-02067-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pettigrew MM, et al. Upper respiratory tract microbial communities, acute otitis media pathogens, and antibiotic use in healthy and sick children. Appl Environ Microbiol. 2012;78(17):6262–70. doi: 10.1128/AEM.01051-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sakwinska O, et al. Nasopharyngeal microbiota in healthy children and pneumonia patients. J Clin Microbiol. 2014;52(5):1590–4. doi: 10.1128/JCM.03280-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hilty M, et al. Nasopharyngeal microbiota in infants with acute otitis media. J Infect Dis. 2012;205(7):1048–55. doi: 10.1093/infdis/jis024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bomar L, et al. Corynebacterium accolens Releases Antipneumococcal Free Fatty Acids from Human Nostril and Skin Surface Triacylglycerols. MBio. 2016;7(1):e01725–15. doi: 10.1128/mBio.01725-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hyde ER, et al. Nasopharyngeal Proteobacteria are associated with viral etiology and acute wheezing in children with severe bronchiolitis. J Allergy Clin Immunol. 2014;133(4):1220–2. doi: 10.1016/j.jaci.2013.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bisgaard H, et al. Childhood asthma after bacterial colonization of the airway in neonates. N Engl J Med. 2007;357(15):1487–95. doi: 10.1056/NEJMoa052632. [DOI] [PubMed] [Google Scholar]
- 31.Vissing NH, Chawes BL, Bisgaard H. Increased risk of pneumonia and bronchiolitis after bacterial colonization of the airways as neonates. Am J Respir Crit Care Med. 2013;188(10):1246–52. doi: 10.1164/rccm.201302-0215OC. [DOI] [PubMed] [Google Scholar]
- 32.Biesbroek G, et al. Early respiratory microbiota composition determines bacterial succession patterns and respiratory health in children. Am J Respir Crit Care Med. 2014;190(11):1283–92. doi: 10.1164/rccm.201407-1240OC. [DOI] [PubMed] [Google Scholar]
- 33.Bogaert D, et al. Variability and diversity of nasopharyngeal microbiota in children: a metagenomic analysis. PLoS One. 2011;6(2):e17035. doi: 10.1371/journal.pone.0017035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tano K, et al. Alloiococcus otitidis--otitis media pathogen or normal bacterial flora? APMIS. 2008;116(9):785–90. doi: 10.1111/j.1600-0463.2008.01003.x. [DOI] [PubMed] [Google Scholar]
- 35.Durmaz R, et al. Detection of Alloiococcus otitidis in the nasopharynx and in the outer ear canal. New Microbiol. 2002;25(2):265–8. [PubMed] [Google Scholar]
- 36.Harimaya A, et al. High frequency of Alloiococcus otitidis in the nasopharynx and in the middle ear cavity of otitis-prone children. Int J Pediatr Otorhinolaryngol. 2006;70(6):1009–14. doi: 10.1016/j.ijporl.2005.10.012. [DOI] [PubMed] [Google Scholar]
- 37.Harimaya A, et al. Evidence of local antibody response against Alloiococcus otitidis in the middle ear cavity of children with otitis media. FEMS Immunol Med Microbiol. 2007;49(1):41–5. doi: 10.1111/j.1574-695X.2006.00166.x. [DOI] [PubMed] [Google Scholar]
- 38.Harimaya A, et al. Induction of CD69 expression and Th1 cytokines release from human peripheral blood lymphocytes after in vitro stimulation with Alloiococcus otitidis and three middle ear pathogens. FEMS Immunol Med Microbiol. 2005;43(3):385–92. doi: 10.1016/j.femsim.2004.10.014. [DOI] [PubMed] [Google Scholar]
- 39.Lee JH, Son SW, Cho SH. A Comprehensive Review of the Treatment of Atopic Eczema. Allergy Asthma Immunol Res. 2016;8(3):181–90. doi: 10.4168/aair.2016.8.3.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ball TM, et al. Siblings, day-care attendance, and the risk of asthma and wheezing during childhood. N Engl J Med. 2000;343(8):538–43. doi: 10.1056/NEJM200008243430803. [DOI] [PubMed] [Google Scholar]
- 41.Hasegawa K, et al. Household siblings and nasal and fecal microbiota in infants. Pediatr Int. 2017;59(4):473–481. doi: 10.1111/ped.13168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Almqvist C, et al. Impact of gender on asthma in childhood and adolescence: a GA2LEN review. Allergy. 2008;63(1):47–57. doi: 10.1111/j.1398-9995.2007.01524.x. [DOI] [PubMed] [Google Scholar]
- 43.Kurukulaaratchy RJ, et al. The influence of gender and atopy on the natural history of rhinitis in the first 18 years of life. Clin Exp Allergy. 2011;41(6):851–9. doi: 10.1111/j.1365-2222.2011.03765.x. [DOI] [PubMed] [Google Scholar]
- 44.Becklake MR, Kauffmann F. Gender differences in airway behaviour over the human life span. Thorax. 1999;54(12):1119–38. doi: 10.1136/thx.54.12.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Doershuk CF, Fisher BJ, Matthews LW. Specific airway resistance from the perinatal period into adulthood. Alterations in childhood pulmonary disease. Am Rev Respir Dis. 1974;109(4):452–7. doi: 10.1164/arrd.1974.109.4.452. [DOI] [PubMed] [Google Scholar]
- 46.Fleisher B, et al. Lung profile: sex differences in normal pregnancy. Obstet Gynecol. 1985;66(3):327–30. [PubMed] [Google Scholar]
- 47.Nielsen HC. Testosterone regulation of sex differences in fetal lung development. Proc Soc Exp Biol Med. 1992;199(4):446–52. doi: 10.3181/00379727-199-43379. [DOI] [PubMed] [Google Scholar]
- 48.Nielsen HC. Androgen receptors influence the production of pulmonary surfactant in the testicular feminization mouse fetus. J Clin Invest. 1985;76(1):177–81. doi: 10.1172/JCI111943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hohlfeld JM. The role of surfactant in asthma. Respir Res. 2002;3:4. doi: 10.1186/rr176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Han S, Mallampalli RK. The Role of Surfactant in Lung Disease and Host Defense against Pulmonary Infections. Ann Am Thorac Soc. 2015;12(5):765–74. doi: 10.1513/AnnalsATS.201411-507FR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nkadi PO, Merritt TA, Pillers DA. An overview of pulmonary surfactant in the neonate: genetics, metabolism, and the role of surfactant in health and disease. Mol Genet Metab. 2009;97(2):95–101. doi: 10.1016/j.ymgme.2009.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Klein SL, Flanagan KL. Sex differences in immune responses. Nat Rev Immunol. 2016;16(10):626–38. doi: 10.1038/nri.2016.90. [DOI] [PubMed] [Google Scholar]
- 53.Fish EN. The X-files in immunity: sex-based differences predispose immune responses. Nat Rev Immunol. 2008;8(9):737–44. doi: 10.1038/nri2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Muenchhoff M, Goulder PJ. Sex differences in pediatric infectious diseases. J Infect Dis. 2014;209(Suppl 3):S120–6. doi: 10.1093/infdis/jiu232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Costello EK, et al. Bacterial community variation in human body habitats across space and time. Science. 2009;326(5960):1694–7. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Biesbroek G, et al. Deep sequencing analyses of low density microbial communities: working at the boundary of accurate microbiota detection. PLoS One. 2012;7(3):e32942. doi: 10.1371/journal.pone.0032942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lemon KP, et al. Comparative analyses of the bacterial microbiota of the human nostril and oropharynx. MBio. 2010;1(3) doi: 10.1128/mBio.00129-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Akmatov MK, et al. Determination of nasal and oropharyngeal microbiomes in a multicenter population-based study - findings from Pretest 1 of the German National Cohort. Sci Rep. 2017;7(1):1855. doi: 10.1038/s41598-017-01212-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.