Abstract
Patient-derived culture models enable assessment of drug sensitivity and can connect personalized genomics with therapeutic options. However, their clinical translation is constrained by limited fidelity. We outline how the physical microenvironment regulates cell metabolism and describe how engineered culture systems could enhance the predictive power for precision medicine.
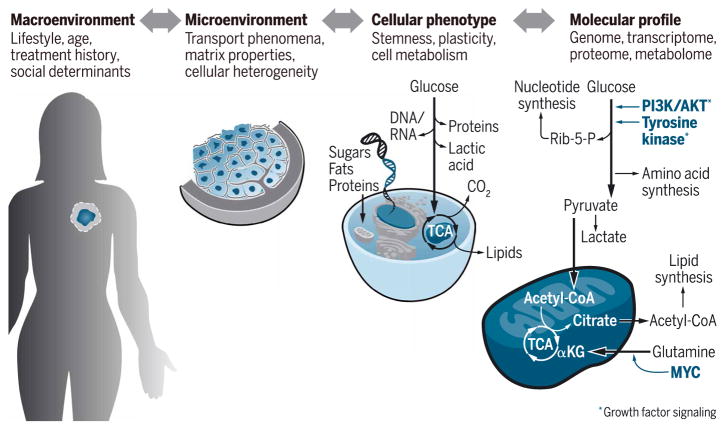
Aberrant cell metabolism is a hallmark of cancer that is driven by interconnected biochemical and biophysical factors acting on cells, tissues, and body fluids. The reciprocal interplay between these factors is controlled at multiple levels, including genetic alterations, cellular composition of the microenvironment (such as immune and stromal cell contributions), lifestyle- and age-dependent variations of organismal metabolism, and environmental exposures (Fig. 1). Moreover, the extent to which these mechanisms affect tumor metabolism is highly dependent on previous treatment history and temporal dynamics that are controlled by the circadian clock (1). As normal cells transform into tumor cells, they must adapt their biochemical networks to coordinate the flux of metabolites through numerous interdependent pathways to meet the new requirements of their oncogenic state (2). These changes govern the conversion of biomass, molecular energy, and redox equivalents required for virtually all biological functions during tumor development (3). The importance of altered metabolism in human cancer is further highlighted by the observation that the most highly recurrent genomic alterations activate oncogenic signaling pathways such as Ras/MAPK (mitogen-activated protein kinase) and PI3K (phosphoinositide 3-kinase)/AKT, which, in turn, are key regulators of cell metabolism (4). Despite these universal mechanisms, metabolic phenotypes vary widely. For example, it has long been thought that aerobic glycolysis is the dominant metabolic phenotype for proliferating cells to generate building blocks for cell division. However, more recent experimental evidence suggests that cancer cells increase flux through both glycolysis and oxidative metabolism to support biomass growth (2). This reprogramming supports tumor growth, allows cells to survive adverse conditions (stresses), and promotes invasive behaviors.
Fig. 1. Determinants of cancer metabolism.
Metabolism is a multiscale process, comprising the full range of biological factors. To understand the interdependent regulatory mechanisms, it is essential that model systems simulate salient features that govern metabolic networks across multiple scales. αKG, α-ketoglutarate; CoA, coenzyme A; Rib-5-P, ribose 5-phosphate.
In addition to altered molecular signaling, metabolic reprogramming is also the cause and consequence of a perturbed microenvironment, which plays a major role in producing the diversity of metabolic phenotypes that can be found not only among tumors but also across different regions of the same tumor (5, 6). Hence, it is perhaps not surprising that nutrient utilization of cancer cells in vitro differs from that of tumors in vivo (7). Although the metabolic phenotype of the tumor cells themselves may be responsible for these observations, it is likely that the interactions with other cell types present in the tumor microenvironment, but lacking in vitro, play a critical role (8). For example, stromal cell metabolism is key to generating a favorable nutrient and energy landscape for tumors, and, more recently, similar observations have also been made for immune cells (9). Our understanding of the role of molecular and cellular changes in the microenvironment is advancing at a rapid pace. However, the direct functional effects that biophysical changes of the microenvironment have on cancer metabolism are largely overlooked. Below, we will highlight mass transport considerations and tumor mechanics as specific examples for physical aberrations intrinsic to the tumor microenvironment and describe model systems to help understand their impact on cancer.
METABOLIC TRANSPORT: CONVECTION-DIFFUSION REACTION KINETICS
Limited availability of oxygen, local and systemic accumulation of lactic acid, and spatiotemporal variations of metabolic substrates are hallmarks of cancer-associated mass transport that can be attributed to dysfunctional blood vascular and lymphatic systems (10). In tumors, the dysfunction of both systems can be explained by (i) excess amounts of angiogenic factors and (ii) pressure exerted by the expanding tumor mass. Together, these conditions promote the formation of leaky and collapsed blood vessels and lymphatics that negatively affect fluid delivery and drainage. The resulting changes in hydrostatic forces, including increased interstitial pressure, deregulate the spatial and temporal presentation of respiratory gases, metabolites, and soluble factors (10). Abnormal distribution of these factors, in turn, influences metabolic reaction kinetics, further aggravating the emergence of heterogeneous biochemical gradients throughout tumors. Hypoxia and lactic acidosis are principal consequences of aberrant microvascular and lymphatic transport.
Normal cells typically respond to adverse microenvironmental conditions such as hypoxia and acidic pH by exploiting highly conserved regulatory systems of metabolic wiring to support cell survival, growth, and proliferation. These include systems mediated by hypoxia-inducible factor (HIF) (11), mammalian target of rapamycin complex (mTORC) (12), and adenosine 5′-monophosphate–activated protein kinase (AMPK) (13). As cells undergo oncogenic transformation, they co-opt these homeostatic signaling mechanisms and additionally exploit growth factor signaling pathways such as MYC, PI3K, MAPK, phosphatase and tensin homolog (PTEN), p53, and organic cation transporter 1 (OCT1) to support proliferative metabolism at the cellular level. The resulting changes in signaling allow cancer cells to decouple their metabolism from systemic regulation of metabolism by, for example, insulin balance, which supports their hyper-proliferative state under otherwise restrictive conditions (6).
It is the combination of microenvironmental and signaling changes that favors cancer cells exhibiting metabolic plasticity. Metabolic flexibility allows tumor cells to regulate glycolytic and oxidative flux to overcome adverse tissue conditions, including hypoxia, acidosis, increases in reactive oxygen species, and limited nutrient availability at the interior of a tumor. For example, during lactic acidosis, cancer cells can exhibit non-glycolytic metabolism, characterized by reduced glucose uptake and negligible lactate production. This nonglycolytic phenotype provides a protective effect when cells are subsequently exposed to hypoglycemic conditions (12). Metabolic plasticity appears to be at least partially independent of genetic profile, because environmental changes can induce isogenic populations of cancer cells to switch from glycolytic to oxidative phenotypes (14).
The ability of tumor cells to adapt to fluctuating tissue conditions can also result in the emergence of distinct subpopulations that cooperate through complementary modes of metabolism. Adjacent tumor niches interact through asymmetric “supply and demand” for intermediate metabolites, bioactive by-products, and oxygen. For example, some oxygenated cancer cells express monocarboxylate transporter 1 (MCT1), allowing them to consume the energy-rich metabolites produced by Warburg-like cells, which use aerobic glycolysis rather than oxidative phosphorylation (15). This mechanism of nutrient exchange, known as “metabolic symbiosis,” uses lactate from hypoxic cells to fuel oxidative metabolism in aerobic tumor compartments (16). Cancer cells can also manipulate the metabolic properties of stromal fibroblasts to shift toward aerobic glycolysis, thus promoting metabolic efficiency and proliferative capacity. In addition, a variety of other conditions further augment complexity. In particular, metabolic rewiring can directly affect immunity and vice versa (9). This interplay has potentially far-reaching implications not only for the growing field of immunooncology but also for obesity-associated cancers, which tend to be more aggressive and occur in microenvironments that exhibit intrinsically impaired immunity and metabolism (17).
Spatial and temporal variations are key features of the metabolic landscape; tumor cells mutually shape and respond to their soluble environment, driving multiple metabolic phenotypes within individual tumors. Although most of the changes described above occur relatively quickly because of transcriptional changes and/or variations in protein activity, it should also be considered that the periodic flux in metabolic cycles is subject to the 24-hour circadian rhythm (1). To what extent this regulation is dependent on the physical environment remains to be determined. Nevertheless, the full continuum of biochemical niches distributed throughout the tumor across varying time scales produces a mosaic of metabolic phenotypes that ultimately affect a patient’s response to treatment.
BIOPHYSICAL FORCES: THE MECHANICS OF TUMOR METABOLISM
In addition to the soluble microenvironment, metabolic pathways are also sensitive to variations in tumor mechanical properties. Increased tumor stiffness can be attributed to a variety of parameters. In particular, increased cell density as well as deposition and contraction of fibrillar extracellular matrix (ECM) proteins, resulting from the formation of an activated tumor stroma, play important roles. Furthermore, increased concentrations of glycosylated matrix components control tissue hydration and interstitial osmotic pressure (18). Water absorption, in turn, causes swelling, which further contributes to the palpable stiffness of solid tumors and the transport limitations highlighted above (19).
The resulting changes in tissue architecture, as well as matrix structure and mechanics, can affect tumor metabolism through altered mechanotransduction, a process by which cells convert extracellular mechanical cues into biochemical outputs. ECM structural elements that were once considered to be static architectural features are now known to regulate an intricate network of force-sensitive signaling functions that play a critical role in the development of disease (20). In particular, integrin receptors, which are transmembrane heterodimers that physically link the ECM and the actin cytoskeleton, are primary mediators of mechanotransduction (21). Activation of integrin-mediated mechanosignaling can be induced by external forces applied to the cells (for example, due to dynamic mechanical loading in the skeleton) or by the cells themselves. More specifically, cells generate forces in response to tumor stiffness by increasing contractility after phosphorylation of myosin light chain (MLC) by Rho kinase (ROCK) (20). Under mechanical tension, actin filaments bundle into stress fibers, and integrins cluster to form focal adhesions, which both play active roles in regulating multiple aspects of cell metabolism (22).
Focal adhesions are important elements of mechanical signal transduction, and multiple signaling pathways that regulate metabolic processes also converge at this interface. For example, focal adhesions are canonically associated with activation of focal adhesion kinase (FAK) and c-Src tyrosine kinase (CTK), which activate Ras and the MAPK pathway (23). In addition, the PI3K-AKT-mTOR pathway is a core element of focal adhesion signaling by operating as a transducer for growth factor receptors and Ras. Through receptor clustering, focal adhesions directly enhance growth factor–dependent PI3K signaling and a host of downstream functions (24). By exerting mechanical control over tyrosine kinase effector networks, focal adhesion proteins directly contribute to malignant invasion, as demonstrated by the loss of tissue organization and concomitant changes in metabolic activity on stiffer substrates (25). Inhibiting the adhesion apparatus, or restoring appropriate matrix mechanics, results in cell quiescence and tissue homeostasis (26). Nevertheless, adhesion-dependent signaling may vary as a function of the specific ECM context in which cells are located, with potential consequences for metabolism. For example, changes in the spatial presentation of ECM components can activate focal adhesion protein signaling in the absence of detectable focal adhesion formation, and these differences may regulate processes such as tumor cell invasion and interactions with the surrounding stroma (27).
Remodeling of the actin cytoskeleton not only is a key component of mechanosignaling but also had already been described as a potential regulator of glycolytic enzymes in 1968 (28). Recent evidence helps explain why. Actin polymerization involves the rapid assembly and disassembly of fiber bundles that provide the structural framework of the cell. When actin fibers disassemble, they release the metabolic enzyme aldolase, which catalyzes the conversion of 6-carbon fructose molecules to the 3-carbon molecules glyceraldehyde and dihydroxyacetone, ultimately accelerating glycolysis (29). This mechanism sensitizes glucose catabolism to external mechanical forces, thus establishing a potential direct link between matrix mechanical properties and cell metabolism.
Reciprocally, metabolic changes also affect the way cells interpret their mechanical environment. For example, glutamine metabolism appears to play a key role in mechanosignaling (30). A recent study reported that glutamine regulation partly controls the activity of RhoA, an oncogenic guanosine triphosphatase (GTPase) involved in focal adhesions and actin stress fiber assembly. Hyperactive RhoA signaling typically causes oncogenic transformation, but this effect could be reversed using a small-molecule inhibitor of glutaminase (GLS1), the amidohydrolase enzyme that converts glutamine to glutamate for entry into the tricarboxylic acid (TCA) cycle (22). Another study corroborated this observation by showing that stiffness-induced changes in glutamine flux were mediated in part by transcriptional regulation of GLS1 (31), suggesting reciprocal feedback between mechanosignaling and glutamine metabolism.
The above connections suggest that matrix mechanics and mechanosignaling directly regulate metabolic functions. Still, it has to be kept in mind that ECM is not a static element of the tumor microenvironment but undergoes spatiotemporal variations, which, in turn, can be influenced by varied metabolic transport. For example, an increase in hypoxia causes up-regulation of lysyl oxidase (32), which can induce structural and mechanical changes in collagen type I that activate mechanosignaling (33). Hence, tumor-associated transport and mechanics are functionally linked and should be carefully considered when studying tumor cell metabolism. By incorporating these factors into models of patient disease, the predictive value of such models will be improved, as will their ability to identify effective therapeutic strategies.
EXPERIMENTAL PLATFORMS: ENGINEERING MODEL SYSTEMS
Although in vitro models can never capture a complete representation of the tumor microenvironment, they can be useful for examining the effect of its most salient features on cellular metabolism, including physical properties of the matrix and the impact of fluid transport. One of the greatest challenges in studying metabolism is navigating between levels of space, time, and complexity, from molecular details to whole-body physiology. This is one area in which new tissue-engineered model systems are proving especially useful.
Conventional in vitro assays are largely based on monotypic populations of cells cultured on plastic or glass substrates. Monolayer cultures of homogeneous cells, or mouse xenografts derived from commercial cell lines, diverge from the original tumor, most notably through the dramatic loss of heterogeneity and tissue structure. Microarray-based comparisons of cancer cells in two-dimensional (2D) versus 3D culture revealed broad changes in hypoxia response and proinflammatory pathways (34). These observations reflect a growing consensus that the culture environment is a critical determinant of cell behavior and that 3D models may be able to recapitulate changes in cellular metabolism that cannot be studied using conventional in vitro approaches.
Varying degrees of complexity can be accomplished with 3D culture techniques. Simply embedding cells within 3D substrates can restore a mélange of important functions, including 3D morphogenesis, assembly of multiprotein adhesions, secretory functions, and tissue homeostasis. However, “3D culture” has become an umbrella term that obscures a more nuanced reality. There are many differences between 2D, 3D, and physiological settings; dimensionality is complicated by the numerous parameters that define each model system. For example, “matrix mechanics” encompasses fiber architecture and conformation, matrix composition and porosity, covalent and ionic cross-linking, cell density, polarity, and contractility. Similarly, “mass transport” not only is a question of vascular proximity but also involves osmotic and hydrostatic pressure gradients, fluid viscosity and shear stress, cell density and metabolic activity, concentrations of ions and dissolved gases, and matrix binding kinetics. Functional consequences emerge from the integrative effects of these manifold variables, rather than from a perfunctory switch from 2D to 3D. 3D assays provide demonstrable evidence that context is important, but we must carefully consider how various features of the experimental system provide instructive cues that alter cell behavior, especially with regard to metabolic programming. Because many physical factors can affect tumor metabolism by altering nutrient availability, it is also critical to consider that media composition alone can have profound effects on metabolic wiring and regulation. This was highlighted by a recent study showing that culturing cells with human plasma-like medium affected their metabolism, including the metabolome, redox state, and glucose utilization (35).
Emerging tissue-engineering technologies now provide attractive tools to control the physical microenvironment for studies of cancer cell metabolism. Hydrogel-based biomaterials are frequently the basis for 3D culture, and a catalog of natural and synthetic materials is available to support cell adhesion, viability, and remodeling; nutrient and waste exchange; and appropriate mechanical properties. Natural materials have inherent biological functions, such as adhesive ligands and cleavage sites, but it can be difficult to precisely define and manipulate the composition and/or structural properties. On the other hand, synthetic gels afford greater control over materials properties, and can be readily manipulated with biological moieties, but lack the biological complexity of native tumor-associated ECM. Nevertheless, it should be kept in mind that cells remodel their ECM environment over time. Hence, the ECM that exists at any given time will differ from its initial state, which, in turn, will influence the type of ECM that will be deposited.
Both natural and synthetic materials are readily integrated with microfluidic technologies to recapitulate matrix mechanics and fluid transport processes that affect tumor metabolism. For example, microfluidic biomaterials can be generated using a confined gel, whereby microchannels are patterned within a transparent silicone mold (36). Alternatively, dense hydrogels allow imprinting of microfluidic conduits directly within the scaffold to control the spatial and temporal gradients of exogenous factors or drugs, reminiscent of microvascular function (37, 38). In addition to forming predefined vascular structures, endothelial cells can also be mixed into natural or synthetic ECMs to allow them to assemble into microvascular networks that model those of primary tumors (39). These strategies have been used to construct biomimetic vascularized tissue constructs to recapitulate the individual and combinatorial effects of matrix structure, solute transport, and cellular composition that define the metabolic environment.
Microfluidic biomaterials can also be integrated with live cell imaging techniques to acquire spatial and temporal information about the complex interdependencies between the microenvironment and cell metabolism. These tools can be used to readily manipulate and measure real-time, single-cell dynamics by using endogenous or genetically encoded fluorescent sensors. For example, a vascularized microtumor (VMT) model was used to map metabolic activity within different regions of hybrid microfluidic organoids via fluorescence lifetime imaging of NAD+ (nicotinamide adenine dinucleotide) and NADH (reduced form of NAD+) (40). The VMT platform mimicked stromal composition, matrix structure, and vascular function, and it simulated metabolic responses to pharmacologic agents (40).
Advances in biomaterials and microfabrication technologies (like the VMT model described above) afford new methods to integrate vascular, stromal, and epithelial compartments with precise arrangements of parenchymal and interstitial elements (41). In addition, emerging tissue culture techniques have produced a new generation of micro-physiological devices (“tissue chips”) that capture increasingly accurate representations of whole organs, including liver, kidney, heart, lungs, brain, gastrointestinal (GI) system, blood vessels, skin, adipose, cervix, uterus, and ovaries (41). Originally designed for preclinical drug screening, organ-on-a-chip models have been used to simulate first-pass metabolism; activation of anticancer prodrugs; synergistic actions of drug combinations; modulation of tissue bioavailability; membrane barrier function; off-target toxicity; and mechanisms of drug adsorption, distribution, metabolism, and excretion (42). Tumor metabolism is a multi-scale phenomenon, and these platforms make it possible to simulate higher-order metabolic regulation in vitro.
The next milestone for microphysiological platforms involves the serial integration of multiple organs within a single device. A complete “body-on-a-chip” would simulate interactions between organs, such as drug adsorption through the GI tract, metabolism in the liver, clearance in the kidneys, and cytotoxic effects in the heart or other tissues (42). Already, pioneering systems have been strategically validated as physical analogs for pharmacokinetic/pharmacodynamic modeling (43). By carefully controlling tissue volume and fluid residence time, integrated micro-physiological systems can mimic drug distribution, uptake, and activity in surrogate organs (44). One such platform predicted nontarget drug retention in adipose tissue and nephrotoxicity. Similarly, a commercial model called Hurel was instrumental for identifying drug metabolites that were not present in traditional, monotypic cell culture (45).
Collectively, microphysiological devices present a promising opportunity to navigate across cell, tissue, and organ systems when investigating cancer metabolism and drug response. Microtissue devices are not delicate “artisan” products but increasingly robust platforms for broad application in the laboratory and the clinic. Several platforms are commercially available, with high simplicity, reliability, and throughput, making these technologies suitable for implementation in non-engineering laboratories.
CLINICAL TRANSLATION: METABOLISM IN PRECISION ONCOLOGY
The manifold variables that influence metabolism and drug response make it challenging to infer susceptibility to targeted pharmaceutical agents. Factors related to metabolism and energetics (diet, physical activity, weight control, and vascular health) interact with genetic, physiological, and environmental variables to influence cancer risk, prognosis, and treatment outcomes in ways that are often impossible to predict. Even the time of day and its relationship to fed/fast states and circadian rhythm have been demonstrated to affect drug efficacy (1, 46). Every patient manifests a unique and ever-changing gene expression, metabolic profile, and tissue microenvironment.
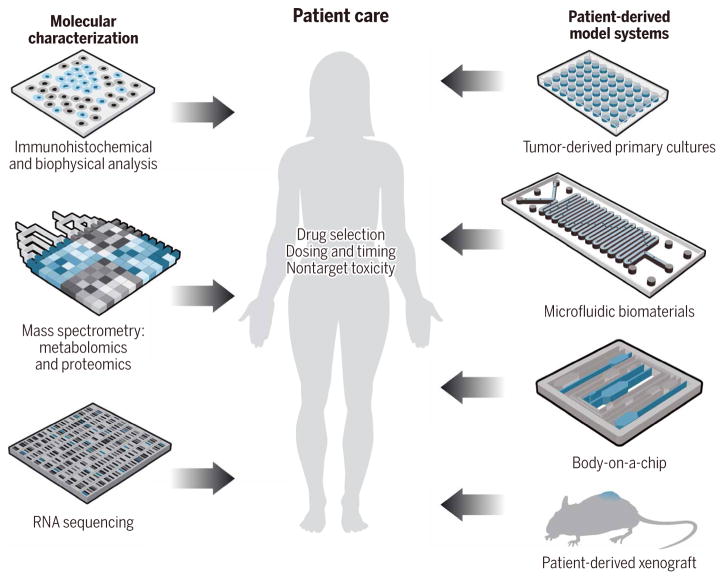
The precision medicine paradigm has been championed as a strategy to account for the dynamic, patient-specific variations in tumor behavior. Originally, precision oncology used molecular information to prioritize therapies for patients who expressed specific biomarkers. Now, comprehensive approaches integrate pathology, -omics analyses, and functional diagnostics (such as rapid drug screening and animal modeling) to predetermine the safety and efficacy of personalized treatments for individual patients (Fig. 2) (47, 48). Information about the genetic aberrations, the gene expression profiles, and the patient-specific drug responses helps guide new off-label uses of U.S. Food and Drug Administration–approved therapies, which could not have been predicted from sequencing alone. By integrating data from multiple independent methodologies, these approaches can cross-validate personalized therapies for cancer patients. These approaches have particular relevance for patients with advanced disease or multidrug-resistant cancers, where standard therapies have failed.
Fig. 2. A comprehensive precision medicine toolkit.
A comprehensive approach to precision medicine oncology integrates pathology and sequencing analyses (left) with functional diagnostic platforms (right). More complete information about individual patient susceptibility will help guide personalized treatment strategies (middle).
To formulate and test patient-specific combination therapies, clinicians require model systems that accurately recapitulate the salient characteristics of the native tumor. Microenvironmental and systemic metabolic feedback mechanisms (such as increases in insulin secretion upon treatment with PI3K inhibitors) have the capacity to markedly alter tumor responses to therapy but are frequently not accounted for in culture platforms being applied in the precision medicine setting (49). Such shortcomings may account for divergence between in vitro and in vivo responses observed in the precision medicine context (47). Therefore, model systems must be attentive toward the constellation of interdependent variables that influence drug sensitivity, including the genetic composition, proximal microenvironment, and systemic factors.
Patient-derived xenograft (PDX) models seem to be the most suitable platform to preserve the clonal architecture of the original patient sample, as well as its gene expression, histology, and epigenetic profile. PDXs, for which pieces of a patient’s primary tumor are propagated in immunodeficient mice, enable highly representative co-clinical testing of drug combinations (48). However, this approach is extremely time-consuming, is expensive, and lacks the capacity for high-throughput screening or precise experimental manipulation. Furthermore, PDXs select for specific cell types over time and lack a functional immune system. Notable differences between humans and animals can result in discrepancies with regard to drug metabolism and signaling mechanisms (50). These challenges might be overcome by complementing PDX models with physiologically relevant in vitro assays that can be used to quickly and accurately perform high-throughput screening with potential therapeutic agents and provide mechanistic information to guide treatment. As researchers develop new methods for maintaining the full complement of immune cells in cultured systems, advanced in vitro models can be used to examine the impact of immune cells and immune targeted therapies on tumor metabolism and the microenvironment as well as their role in tumor clearance, questions that cannot be readily addressed in immunodeficient murine systems (9, 51, 52).
Patient-derived microphysiological systems combine the biological complexity of primary tissue samples with the simplicity of in vitro analysis. The fast timing, low cost, and small tissue volumes permit testing large numbers of drug permutations and dose regimens, thereby ranking optimal combinations to be tested in vivo. Furthermore, engineered, clinically derived culture models might help improve the use of antimetabolic agents in precision oncology. Efforts to study cancer metabolism in vitro rarely consider the ubiquitous effects of vascular transport phenomena and matrix mechanical properties. This poses a challenge, because both parameters affect not only the local concentrations of biochemical factors but also the bioavailability of drugs, which is viewed as a major determinant of resistance to therapy but is often overlooked in molecular analyses of clinical specimens and in chemosensitivity testing. Functional screening platforms that incorporate a full range of biophysical cues might help predict whether a specific patient is likely to benefit from treatments with drugs that target the specific vulnerabilities afforded by tumor metabolism. Furthermore, incorporating patient samples into sophisticated micro-physiological systems may provide spatial information about how the activity of cells in different regions of a controlled microenvironment might simulate heterogeneous responses to targeted therapies. As micro-physiological culture platforms move toward clinical settings, they offer a promising strategy for guiding patient-specific drug selection and treatment modalities.
CONCLUSIONS
Physical scientists and engineers can contribute new tools and insights into the mechanisms by which cells, tissues, and organ systems regulate metabolic processes. Such outcomes require the expertise of biologists and clinical oncologists, who are most directly familiar with the physiology of patient tissues. The cross-disciplinary exchange of knowledge ensures the advancement of technologies and treatments to preserve or restore human health. This dialogue is especially critical for developing patient-derived model systems that faithfully recapitulate matrix mechanics and transport properties to evaluate antimetabolic therapies in precision medicine settings.
Acknowledgments
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the NIH.
Funding: This work was supported by the Center on the Physics of Cancer Metabolism through award number 1U54CA210184-01 from the National Cancer Institute.
REFERENCES AND NOTES
- 1.Bass J. Circadian topology of metabolism. Nature. 2012;491:348–356. doi: 10.1038/nature11704. [DOI] [PubMed] [Google Scholar]
- 2.DeBerardinis RJ, Chandel NS. Fundamentals of cancer metabolism. Sci Adv. 2016;2:e1600200. doi: 10.1126/sciadv.1600200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pavlova NN, Thompson CB. The emerging hallmarks of cancer metabolism. Cell Metab. 2016;23:27–47. doi: 10.1016/j.cmet.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Jr, Kinzler KW. Cancer genome landscapes. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anastasiou D. Tumour microenvironment factors shaping the cancer metabolism landscape. Br J Cancer. 2017;116:277–286. doi: 10.1038/bjc.2016.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hensley CT, Faubert B, Yuan Q, Lev-Cohain N, Jin E, Kim J, Jiang L, Ko B, Skelton R, Loudat L, Wodzak M, Klimko C, McMillan E, Butt Y, Ni M, Oliver D, Torrealba J, Malloy CR, Kernstine K, Lenkinski RE, DeBerardinis RJ. Metabolic heterogeneity in human lung tumors. Cell. 2016;164:681–694. doi: 10.1016/j.cell.2015.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davidson SM, Papagiannakopoulos T, Olenchock BA, Heyman JE, Keibler MA, Luengo A, Bauer MR, Jha AK, O’Brien JP, Pierce KA, Gui DY, Sullivan LB, Wasylenko TM, Subbaraj L, Chin CR, Stephanopolous G, Mott BT, Jacks T, Clish CB, Vander Heiden MG. Environment impacts the metabolic dependencies of Ras-driven non-small cell lung cancer. Cell Metab. 2016;23:517–528. doi: 10.1016/j.cmet.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liotta LA, Kohn EC. The microenvironment of the tumour-host interface. Nature. 2001;411:375–379. doi: 10.1038/35077241. [DOI] [PubMed] [Google Scholar]
- 9.Hotamisligil GS. Foundations of immunometabolism and implications for metabolic health and disease. Immunity. 2017;47:406–420. doi: 10.1016/j.immuni.2017.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Munn LL. Aberrant vascular architecture in tumors and its importance in drug-based therapies. Drug Discov Today. 2003;8:396–403. doi: 10.1016/s1359-6446(03)02686-2. [DOI] [PubMed] [Google Scholar]
- 11.Nakazawa MS, Keith B, Simon MC. Oxygen availability and metabolic adaptations. Nat Rev Cancer. 2016;16:663–673. doi: 10.1038/nrc.2016.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xie J, Wu H, Dai C, Pan Q, Ding Z, Hu D, Ji B, Luo Y, Hu X. Beyond Warburg effect—Dual metabolic nature of cancer cells. Sci Rep. 2014;4:4927. doi: 10.1038/srep04927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hardie DG. AMPK—Sensing energy while talking to other signaling pathways. Cell Metab. 2014;20:939–952. doi: 10.1016/j.cmet.2014.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saga I, Shibao S, Okubo J, Osuka S, Kobayashi Y, Yamada S, Fujita S, Urakami K, Kusuhara M, Yoshida K, Saya H, Sampetrean O. Integrated analysis identifies different metabolic signatures for tumor-initiating cells in a murine glioblastoma model. Neuro Oncol. 2014;16:1048–1056. doi: 10.1093/neuonc/nou096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feron O. Pyruvate into lactate and back: From the Warburg effect to symbiotic energy fuel exchange in cancer cells. Radiother Oncol. 2009;92:329–333. doi: 10.1016/j.radonc.2009.06.025. [DOI] [PubMed] [Google Scholar]
- 16.Pisarsky L, Bill R, Fagiani E, Dimeloe S, Goosen RW, Hagmann J, Hess C, Christofori G. Targeting metabolic symbiosis to overcome resistance to anti-angiogenic therapy. Cell Rep. 2016;15:1161–1174. doi: 10.1016/j.celrep.2016.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hopkins BD, Goncalves MD, Cantley LC. Obesity and cancer mechanisms: Cancer metabolism. J Clin Oncol. 2016;34:4277–4283. doi: 10.1200/JCO.2016.67.9712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wiig H, Tenstad O, Iversen PO, Kalluri R, Bjerkvig R. Interstitial fluid: The overlooked component of the tumor microenvironment? Fibrogenesis Tissue Repair. 2010;3:12. doi: 10.1186/1755-1536-3-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stylianopoulos T, Martin JD, Snuderl M, Mpekris F, Jain SR, Jain RK. Coevolution of solid stress and interstitial fluid pressure in tumors during progression: Implications for vascular collapse. Cancer Res. 2013;73:3833–3841. doi: 10.1158/0008-5472.CAN-12-4521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen CS. Mechanotransduction—A field pulling together? J Cell Sci. 2008;121:3285–3292. doi: 10.1242/jcs.023507. [DOI] [PubMed] [Google Scholar]
- 21.Desgrosellier JS, Cheresh DA. Integrins in cancer: Biological implications and therapeutic opportunities. Nat Rev Cancer. 2010;10:9–22. doi: 10.1038/nrc2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang JB, Erickson JW, Fuji R, Ramachandran S, Gao P, Dinavahi R, Wilson KF, Ambrosio AL, Dias SM, Dang CV, Cerione RA. Targeting mitochondrial glutaminase activity inhibits oncogenic transformation. Cancer Cell. 2010;18:207–219. doi: 10.1016/j.ccr.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mitra SK, Schlaepfer DD. Integrin-regulated FAK–Src signaling in normal and cancer cells. Curr Opin Cell Biol. 2006;18:516–523. doi: 10.1016/j.ceb.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 24.Rubashkin MG, Cassereau L, Bainer R, DuFort CC, Yui Y, Ou G, Paszek MJ, Davidson MW, Chen YY, Weaver VM. Force engages vinculin and promotes tumor progression by enhancing PI3K activation of phosphatidylinositol (3,4,5)-triphosphate. Cancer Res. 2014;74:4597–4611. doi: 10.1158/0008-5472.CAN-13-3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paszek MJ, Zahir N, Johnson KR, Lakins JN, Rozenberg GI, Gefen A, Reinhart-King CA, Margulies SS, Dembo M, Boettiger D, Hammer DA, Weaver VM. Tensional homeostasis and the malignant phenotype. Cancer Cell. 2005;8:241–254. doi: 10.1016/j.ccr.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 26.Kenny PA, Bissell MJ. Tumor reversion: Correction of malignant behavior by microenvironmental cues. Int J Cancer. 2003;107:688–695. doi: 10.1002/ijc.11491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fraley SI, Feng Y, Krishnamurthy R, Kim DH, Celedon A, Longmore GD, Wirtz D. A distinctive role for focal adhesion proteins in three-dimensional cell motility. Nat Cell Biol. 2010;12:598–604. doi: 10.1038/ncb2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arnold H, Pette D. Binding of glycolytic enzymes to structure proteins of the muscle. Eur J Biochem. 1968;6:163–171. doi: 10.1111/j.1432-1033.1968.tb00434.x. [DOI] [PubMed] [Google Scholar]
- 29.Hu H, Juvekar A, Lyssiotis CA, Lien EC, Albeck JG, Oh D, Varma G, Hung YP, Ullas S, Lauring J, Seth P, Lundquist MR, Tolan DR, Grant AK, Needleman DJ, Asara JM, Cantley LC, Wulf GM. Phosphoinositide 3-kinase regulates glycolysis through mobilization of aldolase from the actin cytoskeleton. Cell. 2016;164:433–446. doi: 10.1016/j.cell.2015.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haikala HM, Marques E, Turunen M, Klefström J. Myc requires RhoA/SRF to reprogram glutamine metabolism. Small GTPases. 2016;9:274–282. doi: 10.1080/21541248.2016.1224287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bertero T, Oldham WM, Cottrill KA, Pisano S, Vanderpool RR, Yu Q, Zhao J, Tai Y, Tang Y, Zhang YY, Rehman S, Sugahara M, Qi Z, Gorcsan J, III, Vargas SO, Saggar R, Saggar R, Wallace WD, Ross DJ, Haley KJ, Waxman AB, Parikh VN, De Marco T, Hsue PY, Morris A, Simon MA, Norris KA, Gaggioli C, Loscalzo J, Fessel J, Chan SY. Vascular stiffness mechanoactivates YAP/TAZ-dependent glutaminolysis to drive pulmonary hypertension. J Clin Invest. 2016;126:3313–3335. doi: 10.1172/JCI86387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Erler JT, Bennewith KL, Nicolau M, Dornhöfer N, Kong C, Le QT, Chi JT, Jeffrey SS, Giaccia AJ. Lysyl oxidase is essential for hypoxia-induced metastasis. Nature. 2006;440:1222–1226. doi: 10.1038/nature04695. [DOI] [PubMed] [Google Scholar]
- 33.Baker AM, Bird D, Lang G, Cox TR, Erler JT. Lysyl oxidase enzymatic function increases stiffness to drive colorectal cancer progression through FAK. Oncogene. 2013;32:1863–1868. doi: 10.1038/onc.2012.202. [DOI] [PubMed] [Google Scholar]
- 34.DelNero P, Lane M, Verbridge SS, Kwee B, Kermani P, Hempstead B, Stroock A, Fischbach C. 3D culture broadly regulates tumor cell hypoxia response and angiogenesis via pro-inflammatory pathways. Biomaterials. 2015;55:110–118. doi: 10.1016/j.biomaterials.2015.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cantor JR, Abu-Remaileh M, Kanarek N, Freinkman E, Gao X, Louissaint A, Jr, Lewis CA, Sabatini DM. Physiologic medium rewires cellular metabolism and reveals uric acid as an endogenous inhibitor of UMP synthase. Cell. 2017;169:258–272.e17. doi: 10.1016/j.cell.2017.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vickerman V, Blundo J, Chung S, Kamm R. Design, fabrication and implementation of a novel multi-parameter control microfluidic platform for three-dimensional cell culture and real-time imaging. Lab Chip. 2008;8:1468–1477. doi: 10.1039/b802395f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morgan JP, Delnero PF, Zheng Y, Verbridge SS, Chen J, Craven M, Choi NW, Diaz-Santana A, Kermani P, Hempstead B, López JA, Corso TN, Fischbach C, Stroock AD. Formation of microvascular networks in vitro. Nat Protoc. 2013;8:1820–1836. doi: 10.1038/nprot.2013.110. [DOI] [PubMed] [Google Scholar]
- 38.Miller JS, Stevens KR, Yang MT, Baker BM, Nguyen DHT, Cohen DM, Toro E, Chen AA, Galie PA, Yu X, Chaturvedi R, Bhatia SN, Chen CS. Rapid casting of patterned vascular networks for perfusable engineered three-dimensional tissues. Nat Mater. 2012;11:768–774. doi: 10.1038/nmat3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nashimoto Y, Hayashi T, Kunita I, Nakamasu A, Torisawa Y-s, Nakayama M, Takigawa-Imamura H, Kotera H, Nishiyama K, Miura T, Yokokawa R. Integrating perfusable vascular networks with a three-dimensional tissue in a microfluidic device. Integr Biol. 2017;9:506–518. doi: 10.1039/c7ib00024c. [DOI] [PubMed] [Google Scholar]
- 40.Sobrino A, Phan DTT, Datta R, Wang X, Hachey SJ, Romero-López M, Gratton E, Lee AP, George SC, Hughes CCW. 3D microtumors in vitro supported by perfused vascular networks. Sci Rep. 2016;6:31589. doi: 10.1038/srep31589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ingber DE. Reverse engineering human pathophysiology with organs-on-chips. Cell. 2016;164:1105–1109. doi: 10.1016/j.cell.2016.02.049. [DOI] [PubMed] [Google Scholar]
- 42.Esch MB, King TL, Shuler ML. The role of body-on-a-chip devices in drug and toxicity studies. Annu Rev Biomed Eng. 2011;13:55–72. doi: 10.1146/annurev-bioeng-071910-124629. [DOI] [PubMed] [Google Scholar]
- 43.Oleaga C, Bernabini C, Smith AST, Srinivasan B, Jackson M, McLamb W, Platt V, Bridges R, Cai Y, Santhanam N, Berry B, Najjar S, Akanda N, Guo X, Martin C, Ekman G, Esch MB, Langer J, Ouedraogo G, Cotovio J, Breton L, Shuler ML, Hickman JJ. Multi-organ toxicity demonstration in a functional human in vitro system composed of four organs. Sci Rep. 2016;6:20030. doi: 10.1038/srep20030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sung JH, Kam C, Shuler ML. A microfluidic device for a pharmacokinetic–pharmacodynamic (PK–PD) model on a chip. Lab Chip. 2010;10:446–455. doi: 10.1039/b917763a. [DOI] [PubMed] [Google Scholar]
- 45.Novik E, Maguire TJ, Chao P, Cheng KC, Yarmush ML. A microfluidic hepatic coculture platform for cell-based drug metabolism studies. Biochem Pharmacol. 2010;79:1036–1044. doi: 10.1016/j.bcp.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ballesta A, Innominato PF, Dallmann R, Rand DA, Lévi FA. Systems chronotherapeutics. Pharmacol Rev. 2017;69:161–199. doi: 10.1124/pr.116.013441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pauli C, Hopkins BD, Prandi D, Shaw R, Fedrizzi T, Sboner A, Sailer V, Augello M, Puca L, Rosati R, McNary TJ, Churakova Y, Cheung C, Triscott J, Pisapia D, Rao R, Mosquera JM, Robinson B, Faltas BM, Emerling BE, Gadi VK, Bernard B, Elemento O, Beltran H, Demichelis F, Kemp CJ, Grandori C, Cantley LC, Rubin MA. Personalized in vitro and in vivo cancer models to guide precision medicine. Cancer Discov. 2017;7:462–477. doi: 10.1158/2159-8290.CD-16-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Friedman AA, Letai A, Fisher DE, Flaherty KT. Precision medicine for cancer with next-generation functional diagnostics. Nat Rev Cancer. 2015;15:747–756. doi: 10.1038/nrc4015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fruman DA, O’Brien S. Cancer: A targeted treatment with off-target risks. Nature. 2017;542:424–425. doi: 10.1038/nature21504. [DOI] [PubMed] [Google Scholar]
- 50.Byrne AT, Alférez DG, Amant F, Annibali D, Arribas J, Biankin AV, Bruna A, Budinská E, Caldas C, Chang DK, Clarke RB, Clevers H, Coukos G, Dangles-Marie V, Eckhardt SG, Gonzalez-Suarez E, Hermans E, Hidalgo M, Jarzabek MA, de Jong S, Jonkers J, Kemper K, Lanfrancone L, Mælandsmo GM, Marangoni E, Marine JC, Medico E, Norum JH, Palmer HG, Peeper DS, Pelicci PG, Piris-Gimenez A, Roman-Roman S, Rueda OM, Seoane J, Serra V, Soucek L, Vanhecke D, Villanueva A, Vinolo E, Bertotti A, Trusolino L. Interrogating open issues in cancer medicine with patient-derived xenografts. Nat Rev Cancer. 2017;17:632. doi: 10.1038/nrc.2017.85. [DOI] [PubMed] [Google Scholar]
- 51.Penny HL, Sieow JL, Adriani G, Yeap WH, See Chi Ee P, San Luis B, Lee B, Lee T, Mak SY, Ho YS, Lam KP, Ong CK, Huang RY, Ginhoux F, Rotzschke O, Kamm RD, Wong SC. Warburg metabolism in tumor-conditioned macrophages promotes metastasis in human pancreatic ductal adenocarcinoma. Oncoimmunology. 2016;5:e1191731. doi: 10.1080/2162402X.2016.1191731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jenkins RW, Aref AR, Lizotte PH, Ivanova E, Stinson S, Zhou CW, Bowden M, Deng J, Liu H, Miao D, He MX, Walker W, Zhang G, Tian T, Cheng C, Wei Z, Palakurthi S, Bittinger M, Vitzthum H, Kim JW, Merlino A, Quinn M, Venkataramani C, Kaplan JA, Portell A, Gokhale PC, Phillips B, Smart A, Rotem A, Jones RE, Keogh L, Anguiano M, Stapleton L, Jia Z, Barzily-Rokni M, Cañadas I, Thai TC, Hammond MR, Vlahos R, Wang ES, Zhang H, Li S, Hanna GJ, Huang W, Hoang MP, Piris A, Eliane JP, Stemmer-Rachamimov AO, Cameron L, Su MJ, Shah P, Izar B, Thakuria M, LeBoeuf NR, Rabinowits G, Gunda V, Parangi S, Cleary JM, Miller BC, Kitajima S, Thummalapalli R, Miao B, Barbie TU, Sivathanu V, Wong J, Richards WG, Bueno R, Yoon CH, Miret J, Herlyn M, Garraway LA, Van Allen EM, Freeman GJ, Kirschmeier PT, Lorch JH, Ott PA, Hodi FS, Flaherty KT, Kamm RD, Boland GM, Wong KK, Dornan D, Paweletz CP, Barbie DA. Ex vivo profiling of PD-1 blockade using organotypic tumor spheroids. Cancer Discov. 2018;8:196–215. doi: 10.1158/2159-8290.CD-17-0833. [DOI] [PMC free article] [PubMed] [Google Scholar]