Abstract
The development of safe and effective vaccines for the prevention of elusive infectious diseases remains a public health priority. Immunization, characterized by adaptive immune responses to specific antigens, can be raised by an array of delivery vectors. However, current commercial vaccination strategies are predicated on the retooling of archaic technology. This review will discuss current and emerging strategies designed to elicit immune responses in the context of genetic vaccination. Selected strategies at the biomaterial-biological interface will be emphasized to illustrate the potential of coupling both fields towards a common goal.
1. Introduction
Vaccines are undeniably one of the most significant medical advancements of mankind, as the prevention of infectious diseases results in the sparing of more than 3 million lives per year and economic savings on the order of tens of billions of US dollars.1–3 Recent shortages of influenza vaccines and the inability to overcome elusive diseases such as HIV-1 have provided an essential reminder of the tenuous nature of the world’s vaccine supply and the strategies for its delivery. The development and assessment of new strategies is required to overcome such barriers, as opposed to the refinement of current approaches, to create safer and more effective vaccines.
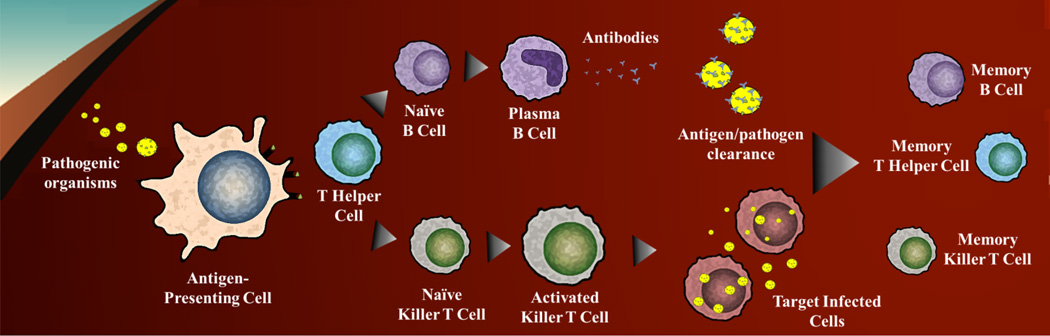
The principle of controlled immunization (vaccination) was pioneered by Edward Jenner and others in the early 19th century and has remained relatively unchanged in clinical development and practice. These early endeavors led to the vaccine methodologies available today. Despite success, the underlying processes required to produce immunity were largely unknown. Thus, research in the past quarter century has focused on the assembly of working models of adaptive immunity. Briefly, adaptive immunity consists of a cellular response of T and B cells upon exposure to antigens. The cascade is initiated by the interaction of these cells with antigen-presenting cells (APCs; macrophages, dendritic cells, and neutrophils) displaying processed antigens (epitopes) on major histocompatibility complexes (MHCs). T cells are stimulated by epitope presentation in MHC-I or MHC-II and accompanying co-stimulatory molecules (e.g., CD40, CD80, and CD86) through T cell receptors (TCRs), leading to activation and maturation of various T cell subsets. MHC-II presentation is recognized by TH2 cells which subsequently activate B cells through the cross-linkage of B cell receptors (BCRs), prompting maturation into long-lived antibody-producing plasma cells or memory B cells.4 Similarly, MHC-I presentation is recognized by TH1 cells which then activate macrophages, CD8 T cells, and other cells responsible for cell-mediated immunity and phagocyte-dependent protective mechanisms.
Generally, adaptive responses can be segmented into two distinct protection mechanisms. First, antibody-mediated (humoral) responses are instigated by exposure to externally localized antigens such as bacterial and viral surface proteins or secreted products such as tetanus toxoid (Figure 1), resulting in MHC-II presentation and plasma cell antibody production.4 Antibody production facilitates the neutralization and clearance of complimentary antigens. Conversely, cell-mediated immunity is characterized by responses that do not result in antibody production but rather involve the activation of macrophages, cytotoxic T-lymphocytes, and antigen-specific cytotoxic T cells in conjunction with the release of proflammatory cytokines (Figure 1).4 Unlike humoral responses, cell-mediated immunity is directed towards the eradication of intracellular infections that affect both phagocytic and nonphagocytic cells by promoting destruction of infected host cells. Examples of cell-mediated responses include any antigen of intracellular origin, produced or residing within the APC itself, such as viral infections (HIV, influenza, and smallpox) and intracellular bacteria. Recently, it has been recognized that TH17 responses can elicit protective immunity against various bacterial and fungal pathogens.5 In this case, protection is mediated through the recruitment of neutrophils, subsequent release of anti-microbial peptides, and IL-17-driven TH1 responses. Additionally, TH17 cells have been documented in regulating/augmenting B cell antibody generation and germinal center and ectopic inducible bronchus-associated lymphoid tissue (iBALT) formation.6, 7
Figure 1.
General antigen processing. Upon entry into the body, a pathogenic organism encounters an antigen presenting cell (APC; dendritic cells or macrophages) and is internalized for intracellular processing. The APCs will present the antigen to T Helper Cells via major histocompatibility complexes (MHCs), which will then present either to naïve B cells or killer T cells. Subsequent activation steps result in either antibody production (humoral response) or cell mediated responses. After an ‘active’ phase of immunity, long-term (adaptive immunity) is established through the formation of memory B and T cells.
Despite centuries of development, only 27 human diseases are recognized by the CDC as preventable by vaccination.8 However, these examples do not account for rapidly emerging and immunologically evasive entities (malaria, HIV) or where basal regulatory mechanisms have gone awry (cancer). To this end, some sub-optimal vaccines have been replaced with improved alternatives and new strategies. Notably, more than half of all new vaccines have been developed in the past 35 years.2 However, infectious diseases still cause significant morbidity and mortality, especially in countries that do not have access to modern medicine.9 Thus, new and broadly accessible vaccines are needed to improve current rates of vaccination against diseases amongst individuals who may be at risk because of their age, medical condition, occupation, or geographical location.
This review will provide an overview of the main types of vaccines as they pertain to current and emerging applications and production schemes. An emphasis will be placed on recent genetic vaccination strategies. In particular, genetic vaccine design and vector-mediated delivery routes will be described in the context of improving immune responses and therapeutic outcomes. Finally, the types of genetic antigen delivery vectors discussed and compared will include biological and biomaterial carriers and the prospect of combining respective advantages of each.
2. Vaccine strategies and production schemes
Vaccines can broadly be classified into four groups, each representing a different approach to eliciting adaptive immunity. These include: 1) live attenuated or 2) inactivated (killed) organisms; 3) purified components (toxoid, subunit, conjugate); and 4) DNA-based vaccines.
2.1 Live attenuated organisms
Since Edward Jenner’s use of bovine poxvirus (a viral entity closely resembling smallpox but non-pathogenic in humans) to elicit protection against smallpox, the concept of live organisms promoting protective immunity has become the paradigm of vaccine design (Figure 1). Fundamentally, this approach is predicated on the identification and neutralization of virulence factors (attenuation) while maintaining beneficial immunogenicity. The strategy has been successfully utilized to combat such pathogens and diseases as measles, mumps, rubella, varicella zoster (chickenpox), influenza, rotavirus, polio, yellow fever, and rabies.
Attenuated vaccines are successful due to their array of antigens for immune induction, innate adjuvant properties, and evolutionarily optimized invasive properties. Thus, the use of live vaccines triggers immune responses that are similar to what occurs during the natural progression of infection, prompting strong cell-mediated and humoral responses as a result. In addition, outcomes typically include long-term immunity with a minimal regimen of vaccine dosages (only one to two).
Once an infectious organism has been identified, there are numerous methods of attenuation in preparation for dosage scale up. Some examples include the use of mutagenic agents (e.g., formaldehyde) or serial passages in cell culture or animal embryos (e.g., egg-based attenuation of viruses). Using egg-based attenuation as an example, a target virus is grown in a series of eggs and becomes increasingly adapted with each passage. In this case, by using evolutionary adaptation as a tool, the virus becomes less etiologically fit while still being recognizable by the human immune system. More generally, the manufacturing processes for live organism-based vaccines are straight-forward as production processes are already in place or existing unit operation-based infrastructure allows the accumulation of high concentrations of cultured microbial/viral organisms.
Important limitations of live organism-based vaccines include the potential for reversion to a form capable of causing disease, delays associated with egg-based production schemes in the face of pandemic needs, and refrigeration requirements. Some organisms have the potential to revert back to a pathogenic state due to the failure to properly identify and remove virulence factors or when mutations occur during the production and immunization processes resulting in more pathogenic organisms, exemplified by the oral polio vaccine (OPV), an ingested live vaccine that has a propensity to mutate. These mutations have resulted in rare cases of paralytic polio, and for these reasons, the process is no longer used in the US and has been replaced by a fully inactivated polio vaccine (IPV).10 Today’s egg-based vaccine manufacturing has the capacity to annually produce up to 400 million dosages of a trivalent flu vaccine.9 However, time delays, egg allergy, and scalability issues are causes for concern for many experts.9 To address these items, alternatives such as cell-culture and vaccine-strategy redesign have the potential to provide broader immune coverage and improved public distribution as compared to current practices. Another limitation is the standard requirement of refrigeration to maintain live vaccine effectiveness.9 Thus, live vaccines may not be viable options in developing countries, remote locations, or climates lacking widespread refrigeration capabilities.
2.2 Inactivated organisms
An alternative to live vaccines are fully inactive (dead) organisms incapacitated using chemicals (e.g., formaldehyde, formalin, or β-propiolactone), heat, or radiation. These attenuation processes remove the pathogen’s ability to replicate but keep it physically intact, permitting immune modulation. Inactive vaccines are more stable and do not pose virulence reversion risks. The approach is most suitable when the immune response is provoked by the organism itself (as opposed to a product that must be biosynthesized and secreted) and when inactivation does not reduce antigen immunogenicity.
Due to the inability to replicate in vivo, inactivated vaccines are often administered concomitantly with an adjuvant (immune-potentiator; e.g., aluminium salts or an oil-in-water emulsion of squalene [MF59]) to increase potency. In addition, these vaccines typically are not effective in eliciting cell-mediated immunity (due to the lack of endogenously produced antigens required for MHC-I presentation) and result in shorter lengths of protection, necessitating the use of boosters to create long-term immunity. Manufacturing of inactivated vaccines is similar to those of live, attenuated vaccines prior to applying the inactivation method of choice. Since the inactivated organism does not (usually) require delicate transport, these vaccines typically do not require refrigeration and can be lyophilized, which increases accessibility.
2.3 Purified components or subunit and conjugate vaccines
When an immunogenic antigen is known to confer protective immunity, it is safer and more efficient to focus on the synthesis and/or isolation of this particular target for vaccine production as compared to using either entire live (attenuated) or killed organisms.
Sometimes a bacterial-based disease is not directly linked to the physical makeup of a bacterium, but rather to an internally produced toxin that leads to disease in the infected host. Immunization with an inactivated variant of the toxin, called a toxoid, leads to a neutralizing antibody-mediated (humoral) response. For example, Clostridium tetani produces a neurotoxin (tetanospasmin) which causes tetanus. Purified toxin is inactivated using formaldehyde prior to immunization. This strategy results in nearly 100% clinical efficacy against tetanus. Production of toxoid vaccines are simple, as in the example of C. tetani, where bacteria are cultured, the toxin purified, and inactivation completed prior to additional purification and sterilization. To boost immune responses, toxoids are typically formulated with adjuvants, such as aluminium or calcium salts.
Similarly, use of only part of a target antigen is designated as a subunit vaccine. These acellular forms of vaccines include those for diphtheria and pertussis, which are commonly packaged together with C. tetani toxoid to create the ‘DTP’ vaccine. As with toxoid-based vaccines, subunit immunization results primarily in a humoral response.
When poorly-immunogenic virulence factors are used as targets for immunization, conjugate vaccines are utilized to create more powerful combined immune responses. This is conducted by the conjugation of a molecule of interest to an immunogenic carrier protein. Conjugate vaccines such as the Streptococcus pneumoniae polysaccharide vaccine are composed of an assortment of pneumococcal polysaccharides attached to carrier proteins such as CRM197 (diphtheria toxin) or the outer membrane protein complex from Neisseria meningitides.
Since many of the purified antigens presented above are bacterially derived, production can be adjusted to utilize an industrial microbial host such as Escherichia coli via recombinant DNA strategies. Genetic transfer into well-characterized hosts will facilitate the replacement of current pathogenically-derived vaccine targets with safer production routes and the potential for more potent antigens. The new antigen route also offers the advantage of established process engineering schemes associated with the new host that can be easily adapted to vaccine production.
2.4 DNA vaccines
DNA-based vaccination is an emerging immunization strategy utilizing the administration of one or more genes encoding functionally active antigens from a targeted pathogen.11–13 Upon uptake by immune effector cells, host-mediated expression of antigenic targets results in the induction of both cellular and humoral immunity. The ability to combine the simplicity and specificity of the response from purified component vaccines (humoral) with cell-mediated immunity induction, the hallmark of live vaccines, has established genetic vaccination as an attractive alternative to traditional vaccine strategies.11
DNA-based technology, unlike other antigenic immunization strategies, is highly malleable, as it combines the tools and power of molecular biology with the increasing information available through genome sequencing projects. This synergistic combination allows for the rapid design and assembly of potential antigenic targets and eliminates the requirement of isolating pathogen-derived antigens for vaccine production. In addition, manufacturing of plasmid DNA vaccines is conducted using E. coli as a production host, thus, taking advantage of standard fermentation methodologies and production infrastructure. Since plasmids encoding different antigens are produced and processed in the same manner, rapid alterations in the production line can be implemented to accommodate emerging diseases or any unexpected shortages.9, 14 In a demonstration of the potential scale and flexibility of DNA-based vaccine production, previous studies have suggested that using established infrastructure will allow DNA vaccines to meet global demands.15 DNA vaccines are also relatively stable, reducing and possibly removing the need for cold chain transport and storage.16
Despite production advantages, genetic antigens are themselves not immunogenic, which necessitates the use of adjuvants and carriers, but allows for repeated homologous vaccination (i.e., successive treatment with the same antigen) without fear of declining efficacy.17 The remainder of this article will cover current strategies and vectors designed to improve and direct DNA vaccine outcomes.
3. Enhancing immune potency of DNA vaccines
DNA vaccine technology is a highly flexible platform that is routinely applied to influence in vivo responses. Approaches include optimizations of the antigen expression cassette, inclusion of adjuvants (traditional and molecular), utilization of alternative immunization strategies, and the development and formulation of next-generation delivery vectors (Table 1).
Table 1.
DNA vaccination strategies to improve efficacy.
| Strategies | Examples | Purpose | References |
|---|---|---|---|
| Plasmid modifications | |||
| • Backbone size reduction | Minicircle and MIP plasmids |
Reduce in vivo silencing, enhance gene delivery efficiency | 18, 19 |
| • Antibiotic resistance marker removal/replacement |
RNA-out | Reduce amount of bacterial backbone DNA | 19 |
| • Origin of replication | pUC | Provide high copy levels without being able to replicate in other hosts |
34 |
| • Promoter choice | CMV, CAGG, RSV | Facilitate desired constitutive expression levels | 27, 29, 30 |
| • Enhancer elements | SV40 enhancer | Activate transcription of genes | 29 |
| • Leader sequences | Introns | Positively augment promoter activity (increased transcription) and enhance polyadenylation |
19 |
| Sequence modifications | |||
| • Codon optimization | Modification of GC/AT content |
Enhance translation | 180 |
| • Strong translation start sequence | Kozak consensus sequence | Enhance translation | 44 |
| • Multiple termination sequences | Prevent read-through translation | 34 | |
| • Splicing signal | Splicing enhancers | Enhance transcription | 41–43 |
| • Polyadenylation | Increase stability of mRNA product | 34 | |
| • Localization sequences | LAMP1, TPA | Bias antigenic processing towards desired response (Th1 vs Th2) |
46, 47 |
| Molecular Adjuvants | |||
| • Cytokines | IL-1, IL-12, IL-15, GM- CSF |
Secreted signaling molecules that control the duration and strength of an immune response |
58–63 |
| • Chemokines (specialized type of cytokine) |
XCLs, CCLs, CXCLs, and CX3CLs |
Chemo-attractants for the induction of ‘immunocompetent’ local environment |
55–57 |
| • Signaling ligands | TRIF | Secreted ligands bind desired receptors to instigate desired immune pathways |
67, 68 |
| • Transcription factors | IRF1, IRF3, IRF7 | Regulate immune responses at the genetic level | 49, 69–72 |
| • Co-stimulatory molecules | CD40, CD80, CD86 | Provide additional control over the activation of T cells | 64–66 |
| Immunization | |||
| • Naked | With or without general adjuvants |
Direct injection of pDNA | 181 |
| • Delivery assisted | Electroporation, gene gun, jet injector, topical patch |
Enhanced delivery of pDNA delivery utilizing methods designed to increase localized concentrations (pooled regions and intracellular) |
181 |
| • Carrier assisted | Cationic polymers and lipids, biological carriers, inorganic material |
Enhanced delivery and expression of pDNA | 181 |
| • Prime boost | Homologous/Heterologous | Alternative administration route designed to boost responses by first priming the immune system. |
182, 183 |
3.1 Expression cassette engineering
An ideal plasmid for DNA vaccination should be easily produced at the commercial scale, unable to integrate into the human genome, and mediate high levels of transgene expression. Various steps have been taken to address these criteria, including optimization of the plasmid backbone (origin of replication, backbone size and content, and antibiotic resistance markers),18–24 engineering of the promoter and enhancer regions,25–29 development of improved DNA and mRNA sequences,13, 30–32 and the incorporation of leader and polyadenylation (poly A) sequences.33
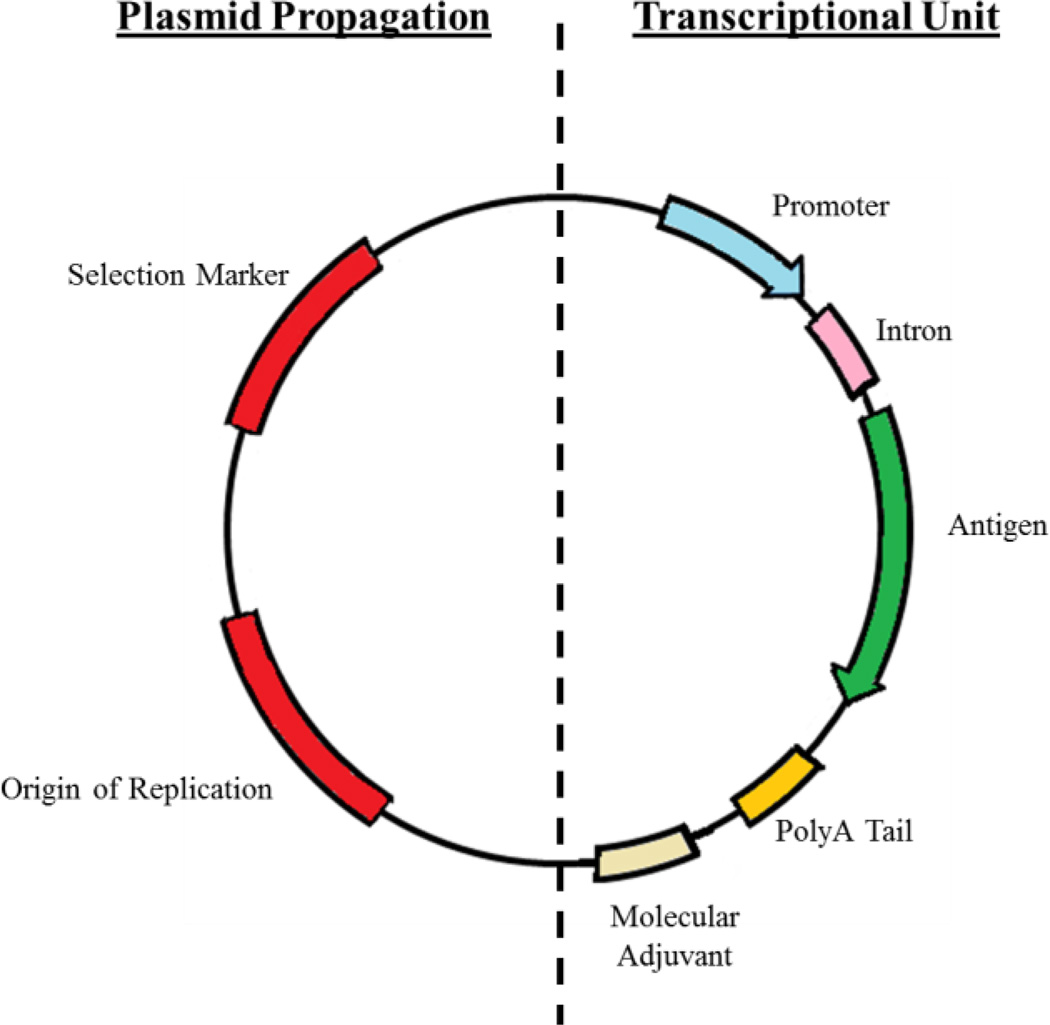
Commonly overlooked, design of the plasmid backbone is the first and arguably most critical step in the development of a potent genetic vaccine. Since large quantities of plasmid DNA (pDNA) are required per dose, production is generally carried out using high-copy bacterial plasmids, such as pUC-based vectors. This family of vectors contains a modified pBR322 origin of replication (mutated to remove regulatory constraints on plasmid number) allowing bacterial maintenance of over 500 copies per bacterium, effectively increasing plasmid yields and lowering production cost.34 Equally important, plasmid size and bacterial components (bacterial plasmid elements such as origins of replication and antibiotic resistance markers) significantly influence the magnitude and duration of transgene expression.35 Reduction in gene expression (silencing) is believed to occur at the nuclear stage by trimethylated-mediated chromatin-linked transcriptional blockage.23 Thus, expression vectors have been designed to drastically improve transgene expression by the removal of any bacterial pDNA sequence.18–23 For example, minicircle (MC) pDNA vectors are small expression vectors devoid of all bacterial DNA (including origin of replication and resistance markers). This is made possible by the inclusion of several engineered sites on the plasmid backbone that result in recombination-based removal of bacterial backbone DNA but not the transcriptional unit. However, current MC vectors utilize temperature-sensitive lower copy origins of replication, reducing the potential for large scale production when compared to pUC-based vectors. Additionally, since recombination occurs within the bacterial production host, additional purification steps are also required to recover the final MC plasmid. Similarly, transgene silencing is also initiated when greater than 1 kb of non-coding DNA is placed outside the transcription unit (regardless of whether it is bacterial DNA).36 To simultaneously overcome both bacterial backbone and non-coding DNA silencing, a new expression system, termed mini-intronsic plasmid (MIP), was developed.19 This system shifts the position of the bacterial replication origin and selection marker from the backbone to an intron within the transgene cassette. The same study demonstrated that transgene expression was stronger and extended when compared to either plasmid or minicircle systems. Finally, to enable the ability to rapidly customize vaccine antigens and retain optimized transgene expression, generalized cloning sites must be inserted to locations that will permit the ‘drop-in’ of all required expression sequences.
Secondary to physical plasmid construction is the choice and design of the transcriptional unit. A typical (monovalent) transcriptional unit is arranged with a promoter upstream and a polyA signal downstream of a genetic antigen (Figure 2). The selection of a strong constitutive promoter, usually in the form of a viral promoter, to mediate high levels of transgene expression is required to achieve an effective immune response. Traditionally, transcriptional units have been driven by the human cytomegalovirus immediate-early (CMV) or the chicken-β actin with CMV early enhancer (CAGG) promoters.34, 37, 38 If reduced levels of transgene expression are desired, alternative viral promoters such as the simian vacuolating virus 40 (SV40) or cellular-specific promoters such as the human elongation factor 1-α (EF-1α) may be considered. Additional engineering of the CMV promoter (incorporation of a downstream HTLV-1 R-U5 region) has been demonstrated to drive higher levels of transgene expression and ultimately improve cellular immune responses in mice and nonhuman primates.27 After the promoter, intron sequences are typically included between the promoter and gene construct as their presence has demonstrated elevated expression.33 It is thought that the inclusion of introns positively augments promoter activity39 and the rate of polyadenylation and/or nuclear transport associated with RNA splicing.40 Transgene expression may be further elevated by the addition of splicing enhancers internally and flanking the intron.41–43 The spliced untranslated leader region (UTR) is designed to be less than 150 base-pairs long, contain no open reading frames (ORFs), and reduce occurrences of secondary structures. Consequently, the translation initiation sequence (Kozak consensus sequence) is generally inserted at the start of the secondary exon and immediately prior to the ATG start codon.44
Figure 2.
Generic design of a plasmid DNA vaccine.
A genetic vaccine’s immune potency can be significantly elevated by codon optimization of the transgene sequence to match the target organism. This increase is related to the GC enrichment of nucleotides present in mammalian DNA as compared to AT-rich sequences in bacterial DNA. Therefore, the availability of tRNAs for translation and the variation in codon usage leads to distinct translation differences between the antigenic source and the transgene host cells. Additionally, dual stop codons are often utilized to prevent read through translation. Following an additional 3’ UTR, a polyA signal is inserted to mediate mRNA processing and to limit enzymatic degradation.33 Alternatively, targeting and signalling sequences can be incorporated to direct and enhance antigen processing towards either MHC-I or MHC-II presentation.45 For example, endosomal/lysosomal targeting promotes an MHC-II (predominately humoral) response; thus, the addition of lysosomal-associated membrane protein 1 (LAMP1) or tissue plasminogen activator (TPA; to promote antigen secretion) signal sequences are commonly used strategies.46, 47 Conversely, N-terminal ubiquitin tags are utilized to direct antigen entry into the proteasomal degradation pathway which results in predominately MHC-I responses.48
When designing each of the previously described genetic sequences, optimization to promote and prolong transcribed mRNA is essential to further improve immune responses. Specifically, modifications of the mRNA sequence that do not interfere with the antigen’s final protein structure and resulting presentation are required. Alterations include removal of unstable secondary structures (usually caused by GC-rich regions), cis-acting motifs (e.g., TATA boxes), repeat sequences, cryptic splice sites, undesired ribosomal binding sites, and cruciforms (multiple hairpin loops which confer sensitivity to nucleases).33
A major advantage of the DNA platform is the flexibility and ease in customization of antigens to address disease-specific barriers. Furthermore, although the text above describes the creation of monovalent DNA platforms, the systems can easily be adapted to deliver and express a broad coverage of antigens, which do not necessarily have to target the same disease. With this in mind, the development of an all-encompassing genetic vaccine platform that has the potential to provide coverage of numerous antigens in a single dose is an emerging trend in the field of DNA vaccines.
3.2 Adjuvants
Although first-generation DNA vaccines were noted for their poor immunogenicity, recent molecular adjuvants (plasmid expressed immune-potentiator molecules) have dramatically increased the ability to induce a more robust and directed response.49 Historically, DNA vaccination was conducted using administration of naked DNA either with or without traditional adjuvant addition. When adjuvants were included, aluminium salts (alum) or oil-based (MF59) adjuvants were common options. These choices operate through the creation of ‘immunocompetent’ local environments which leads to the recruitment of respective immune-effector cells (alum attracts macrophages and monocytes; whereas, MF59 also attracts granulocytes), accelerates monocyte differentiation, and augments antigen uptake.50, 51 However, these adjuvants serve as general immunostimulatory elements that have little influence upon the direction and specificity of an immune response.
In general, DNA vaccines have a ‘built-in’ adjuvant in the form of the CpG motif (i.e., a cytosine triphosphate deoxynucleotide (C) linked to a guanine triphosphate deoxynucleotide (G) by a phosphodiester linkage (p)). Immunostimulatory properties arise due to aberrant lack of methylation, leading to recognition as a pathogen-associated molecular pattern (PAMP) by the pattern recognition receptor Toll-Like Receptor 9 (TLR9).52 Addition of several CpG motifs in DNA vaccines has resulted in improved immunogenicity, presumably by increased innate immune activation.53, 54
Interestingly, due to the modularity of DNA vaccines, genetic adjuvants are often included (either on the antigen-encoding plasmid or on a separate expression vector) to further increase the resulting immune response. Upon vaccination, cells transfected with plasmids encoding molecular adjuvants will express and secrete the desired product (generally a signalling molecule) into the surrounding region creating a focused ‘immunocompetent’ local environment. Unlike general adjuvants, the result is long-lasting and can be easily tailored to the specific vaccination strategy. These adjuvants operate by biasing the cellular immune response towards TH1 induction (effective against viruses, intracellular bacteria, and cancer), external antigens (neutralizing antibody response), or by the recruitment of immune effector cells. Since these adjuvants are derived from known immune-modulating agents, a higher level of control can be exercised in the direction of the desired response. Examples have included peptides (flagellin and influenza A virus nucleoprotein), chemokines (XCLs, CCLs, CXCLs, and CX3CLs),55–57 cytokines (GM-CSF, interleukins),58–63 surface expressed co-stimulatory molecules,64–66 receptor ligands,67, 68 and transcription factors.49, 69–72 Molecular adjuvants that bias towards intracellular antigenic trafficking (i.e., stimulating cell-mediated immunity) include the use of IL-2, IL-10, IL-12, IL-15, IL-18, TGFβ, and IFNγ.62, 63, 73 Alternatively, use of IL-4, IL-5, and IL-15 can bias a humoral-based response.58, 63
As opposed to the direct expression of a dedicated molecular adjuvant, an alternative route towards a desired immunological response involves the use of transcription factors to shift gene expression patterns. These adjuvants operate by controlling (or inducing) the production of immunogenic cytokines and chemokines. There has been significant commercial development of transcription factor-based vectors including TBK1 (TANK-binding kinase 1; general immunopotentiator71; InvivoGen), IRF1 (interferon regulatory factor 1; increases humoral responses72, limited cell-mediated response69; InvivoGen), IRF3 (increases cell-mediated responses72; InvivoGen), and chimeric IRF7/3 (increases both cell-mediated and humoral responses70; InvivoGen).
Of the previously described adjuvants, GM-CSF (granulocyte-macrophage colony-stimulating factor) was one of the first co-delivery cytokine plasmids supporting DNA-induced immunity. Accordingly, GM-CSF has become a common and widely studied molecular adjuvant that transitioned to human use.74 Unfortunately, human application was unable to replicate the same level of efficacy that was present during animal trials. This cytokine molecular adjuvant operates by recruiting, activating, and/or enhancing the response of professional phagocytic cells. Nonetheless, other recruitment-oriented molecular adjuvants are being developed and can potentially coordinate the triggered movement and functionality of adaptive immunity-mediated effector cells.75
Alternatively, immunogenicity can be enhanced by the delivery of surface-expressed co-stimulatory molecules, such as CD80 (B7-1) and CD86 (B7-2)76, that are required as a secondary signal for the induction of T cells. These adjuvants have successfully been used to potentiate cytotoxic lymphocyte (CTL) activity in cancer,77, 78 HIV,79 and tuberculosis studies.80 Similarly, expression of CD40, a surface receptor on B cells and dendritic cells, has demonstrated improved humoral immunity with a switch to a TH1 response.81–83 Emphasizing co-stimulatory molecules, blockage of the receptors through expression of inhibitors or siRNA can assist in the regulation of immune responses that are present during chronic viral infection and cancer.84, 85 In summary, the diversity and specificity of molecular adjuvants holds enormous potential and will likely continue to receive significant attention on the basis of encouraging preliminary results reported in the clinic.
3.3 Immunization strategies
Significant effort has been devoted to patient administration of pDNA for increased localized concentrations (i.e., a ‘depot’ effect) and intracellular flux (using methods such as electroporation).35 These strategies have resulted in elevated responses; however, even when administered with adjuvants and carriers, the immune response is typically weaker than that produced by comparable viral vectors or proteinaceous/toxoid antigens.35 These limitations have driven the development of a consecutive immunization strategy, known as ‘prime-boost,’ which combines the small but focused response of the DNA platform with the expansive immune activation of live recombinant vaccines.86 The process involves immunization with a genetic antigen with adjuvant or carrier, followed by a secondary administration of the same antigen using either the original vector or a new biological/synthetic vector. This combination was heralded as an innovative administration strategy that promotes both humoral and cell-mediated vaccine-specific immunity. The underpinning of this successful strategy is based upon the powerful secondary response that is elicited following primary memory formation mediated by DNA immunization.
Initial studies were predicated on the utilization of the same vaccination vector during re-administration (i.e., homologous prime-boost) which, though successful in generating and boosting the humoral response to an antigen, failed to significantly strengthen cell-mediated immunity.87 Presumably this occurs due to the rapid clearance of homologous vaccination agents.86 In contrast, heterologous prime-boost strategies utilizing a different vector for the secondary administration of the antigen resulted in significantly higher cell-mediated responses.88 Furthermore, heterologous prime-boost studies have demonstrated that responses are fundamentally different than those induced by consecutive administration of either vaccine modality89 – eliciting responses 10 times higher than either platform in isolation.90
4. Vector delivery options
The goal of DNA vaccination is the successful transfection of immune effector cells leading to the production of preventative immunity. Initial efforts prioritized injection of naked pDNA boosted with adjuvants (alum, complete Freund’s adjuvant [CFA], and MF59), physical methods (gene gun, electroporation, ultrasound, magnetofection), and viral-encapsulation as unassisted DNA vaccination resulted in limited success.91 However recently, emerging delivery vectors have demonstrated potential to elicit robust and better directed immune responses. An ideal vector-based strategy is one that is easily modified and manufactured (at a large scale) and sturdy during long-term storage.
4.1 Viral vectors
Viral vectors are often heralded for their high transfection efficiency both in vitro and in vivo.35 Efficient delivery is attributed to the natural evolution of viral vectors towards improving gene transfer into the host for self-replication purposes. An array of viral vectors for which both replicating and non-replicating forms exist including lenti, adeno, pox, herpes, alpha, and measles viruses.92 However, vectors are primarily designed to be non-integrative (eliminating lentivirus) and non-replicative. Other vectors based on vaccine candidates have been underutilized (poliovirus and yellow fever) despite the formation of life-long protective immunity.92 To date, viral vectors are not commonly utilized outside of providing vaccine targets. This limited interest in vaccine-associated use arises from small genetic cargo capacity, genetic instability, difficulty in manufacturing and production, and prior vector-associated immunity.35 Although viral vectors are often associated with detrimental levels of immunogenic activations leading to significant cytotoxicity, well-constructed vaccine strategies can reduce off-target effects by modifying tropism and tissue/cell-specific promoter activity.93
4.2 Bacterial vectors
Unlike viral vectors, innate biological properties of bacteria reduce ubiquitous delivery and bias delivery towards immunological presentation mechanisms. The concept of utilizing bacteria to deliver genetic material was first reported in the early 1980s94 and has continually improved to produce vectors possessing native or heterologous properties aimed to overcome specific barriers associated with in vivo administration and subsequent immune responses. This classification of vector falls within the ‘nonviral’ category of delivery systems but still retains beneficial biological traits of enhanced delivery.95 Strains of bacteria currently used include Listeria monocytogenes,96–99 Escherichia coli,94, 100–109 Bifidobacterium longum,110–112 Salmonella spp.,113–117 Vibrio cholera,118–121 Shigella spp.,122–124 Mycobacteria bovis,125 Yersinia enterocolitica,126 and Lactococcus lacti.127 Although only a small fraction of the highlighted vectors have been used in the context of DNA vaccines, most have engineering tools to facilitate the development towards a gene delivery context. However, the reason for the usage of particular bacterial species and strains is predicated on safety and economical concerns. For example, strains such as L. monocytogenes possess all the innate properties (ideal size for APC uptake, adjuvant-like physical composition, and an endosomal escape mechanism) to become a powerful gene delivery vector. However, the organism is severely hindered by slow growth rates, under-developed microbiology techniques (compared to model organisms like E. coli), and the potential reversion of pathogenicity.
Interestingly, an advantage of bacterial vectors is the potential to protect against pathogens that enter through the mucosa. This is accomplished through the use of strains capable of replicating at mucosal membranes.103, 117, 128 In this case, the generation of a humoral response does not require inclusion of a genetic vaccine but rather can be elicited by bacterially-expressed antigens presented with the vector.117 Considering the duality of bacterial vectors (i.e., the ability to deliver genetic cargo while simultaneously simulating humoral immunity through the presence or production of immunogenic molecules), both humoral and cell-mediated protection can be obtained through administration of one vector.
Despite efforts to attenuate bacterial vectors and documented successful cytotoxicity panels,129 lingering safety concerns have prevented widespread use. Biosafety issues have been addressed through various means including antibiotic pre-treatment109 and auxotroph-conferring genetic manipulations.100, 130–132 As a result, it is likely that additional engineering will allow the generation of safe and effective vectors. Furthermore, if a safe alternative is obtained, bacteria offer unequivocal potential in the individual or simultaneous delivery of all antigenic types (DNA, RNA, protein, toxoid, and cellular components) without the need for additional adjuvants.
4.3 Biomaterial vectors
A myriad of biocompatible materials have been developed that are capable of electrostatically binding to or encapsulating genetic cargo. Upon packaging genetic cargo, these synthetic carriers enter host cells through general or specialized endocytosis mechanisms before facilitating cytoplasmic release. Vectors include cationic lipids and polymers, polysaccharides, polypeptides, and inorganic particles. Historically, researchers have utilized poly(lactide-co-glycolic acid) (PLGA) and variants thereof for antigen delivery.11 Use of PLGA was driven by a safe biocompatibility profile and a wealth of information available describing delivery and degradation properties. However, as more information was gained with respect to structure-function relationships, a number of other polymer systems have been proposed and tested. Of these, liposomes have been widely studied, but stability of these preparations limits clinical application.133 Alternatively, polymers such as pH-sensitive poly(beta-amino ester),134, 135 acid-labile polyketals,136 and cationic polylactides (CPLAs)137, 138 have been developed to facilitate cargo release in the acidic endosomal compartment. Other naturally derived biomaterials have been adapted for vaccination purposes including polysaccharides139, in addition to polypeptides in the context of synthetic peptide epitope-based polymers140 and self-adjuvating polymer-peptide conjugates.141 Alternatively, inorganic molecules used as vaccination agents include aluminium phosphate and aluminium hydroxide, which operate by facilitating a depot effect for antigen release and activation of APCs. Analogously, emulsions (either oil-in-water or water-in-oil) utilizing Freund’s adjuvant and MF59 are also capable of providing depot-type antigen release and triggering APC induction.
Biomaterial particle-based vaccine carriers can be readily tuned to fully engage desired immune effector cells. Furthermore, there are three important compositional characteristics of biomaterials that influence the formation of adaptive immune responses and can be applied liberally to various circumstances (cancer, vaccines, wound healing, and tissue engineering). These include particle size, antigen selection and valency, and the inclusion/coating of targeting ligands and/or immune-potentiators.
For particulate delivery systems, size determines biodistribution and residence time,142, 143 cellular uptake mechanisms,35, 142 and passive targeting of cell types.144, 145 In the context of vaccines, it is important for vectors to reach immunologically-important anatomical regions such as the lymph nodes in order to interact with APCs.143 This region can be accessed through direct intranodal injections146 and trafficking through the lympathic vessels.143 Furthermore, 45 nm is the particle cut-off size for effective drainage into the lymph nodes,145 which is greater than the 5.5 nm and less than the 1 µm size limits for rapid renal elimination and reticuloendothelial system (RES) clearance, respectively.147, 148 However, particles >100 nm are less effective at passive transport through the lymphatic system by interstitial flow.143 In other studies, while investigating size-dependent lymph node targeting, fluorescently labelled particles less than 30 nm were detected within 2 hours; whereas, larger particles (100, 500, 1,000 nm) were sparingly detected in lymph tissue and were observed to be pooled near the injection site.144, 149, 150
Aside from biodistribution, size largely determines the manner a particle will enter the cell. General endocytosis occurs on the order of 200 nm or less (smaller particles are taken up more frequently), macropinocytosis on the order of 500 nm (up to 10 µm), and phagocytosis accommodating sizes up to 10 µm.35 The last mechanism provides an alternative means of directing a biomaterial carrier to the immune system. Namely, particles designed to only be engulfed by professional phagocytes offer a targeted means of carriage and subsequent presentation by tissue APCs present at the administration site.144, 145 Thus, in the design of biomaterial vectors for vaccine purposes, small particles appear to be more effective at targeting and pooling at desired sites but larger particles offer the prospect of targeted APC delivery.
Considerations for antigen selection and valency are perhaps the most critical parameters for biomaterials-driven vaccine strategies. As before, the selection of antigen will largely determine the response elicited, but given the basis of genetic vaccines, most immune responses will be mixed (TH1, TH2, and TH17). However, using polymer chemistry tools, it is possible to structurally graft and/or encapsulate different forms (pDNA, protein, subunit, toxoid) of the antigen of choice to synergistically harness the duality of different vaccine classes.151 Analogously, high-density surface grafting of antigenic material (not DNA) can result in increased crosslinking of antigen-specific TCRs and BCRs, which leads to lower thresholds for activations.152, 153 For example, Irvine and co-workers synthesized layered lipid particles composed of entrapped malarial antigens and multivalent surface coatings.154 This strategy resulted in increased antibody production (over an order of magnitude) as compared to non-surface-coated particles.
Similarly, the inclusion of targeting moieties or immune-potentiator molecules can improve and/or direct an immune response. For example, delivery specificity to APCs can be improved by inclusion of mannose, an antagonist of CD206.155, 156 Similarly, inclusion of different surface ligands, such as MHC-II ligand and CCR1/3/5 molecules, have demonstrated the ability to polarize immunological responses towards the development of tailor-made vaccines.157 In addition to the summary provided above, comprehensive discussions of biomaterials in the context of gene delivery and vaccination have been covered in previously reviews.11, 35
4.4 Biomaterial-interfaced vectors
4.4.1 Virus-mimicking strategies
Virus-like particles (VLPs) are self-assembled particles of viral-derived components (capsid and envelop proteins) that are promising alternatives to traditional vaccine strategies. Although closely mimicking the natural structure of viruses, VLPs are unable to replicate or revert to a prior virulent form (due to the lack of genomic DNA).158 In addition, VLPs are easily modified, produced, and scaled at a low cost. VLPs have the ability to deliver various cargo including genetic material, peptides, proteins, small molecules, adjuvants, and other antigenic material.159 However, VLPs do not usually contain all the needed molecules to fully mediate an effective gene delivery response, and thus, require the inclusion of polymeric components to elevate gene delivery. VLPs often contain the capsid that recognizes and binds viral-associated cellular receptors and a biological and/or synthetic endolysosomal escape mediator. Escape from the lysosome has been accomplished through the addition of biomaterials to VLPs. Specifically, viral particles have been complexed with cationic polymers and lipids to improve endolysosomal release (via proton sponge and lipid mixing escape mechanisms, respectively).160–162 Analogously, virosomes (reconstituted virion-like phospholipid bilayer spherical vesicles) contain all the glycoproteins derived from viruses but are devoid of both capsid proteins and genetic material. They have gained attention as potential gene delivery carriers due to their ease of production and modification, coupled with their low cytotoxicity profile.163, 164 However, their in vivo applications are limited by the lingering risk of unwanted innate immunogenicity. Thus, numerous biomaterial-mediated modifications, including additions of polyethylene glycol (PEG) and targeting molecules,165 have been conducted to overcome this obstacle and reduce off-target effects
Alternatively, rather than simply modifying viral-like particles with biomaterials, entirely biomaterial-based devices have been used to mimic biophysical properties (size, shape, and surface antigens) of viruses. Of interest, self-assembled liposomes that mimic viral structures have been synthesized through the assembly of lipids, transferrin, and DNA.166 This strategy is marked by multicentre lamellar nanostructures with a transferrin coating which closely resembles the influenza and herpes viruses.167 As of yet, this strategy has not been applied in the context of genetic vaccination. Similarly, pH-sensitive nanogel systems that structurally and functionally resemble traits of viruses have also been developed.168 These gels mimic a viral structure through the use of a hydrophobic core and two layers of hydrophilic shells. Additionally, serum albumin-linked PEG was grafted onto the surface to create a capsid-like structure.168 Unlike traditional viral particles, these biomimetics demonstrated pH-sensitive swelling from physiological (pH 7.4) to the endosomal (pH 6.4) level, which in turn facilitated endosomal escape and release of cargo. Nanogels have been synthesized utilizing several natural and synthetic US Food and Drug Administration (FDA)-approved biopolymers including, poly(methyl methacrylate), poly(l-lactic acid), poly(glycolic acid), PLGA, poly(ε-caprolactone), chitosan, poly-L-lysine, poly(γ-glutamic acid), dextran, dextrin, mannan, pullulan, heparin, hyaluronic acid, and alginate.169
To date, most studies of biomaterial-interfaced virus-mimicking strategies have been performed in vitro, and their in vivo efficacy has not been well-established. Combining the innate properties of viral particles with the tool-set available for biomaterial synthesis allows for limitless opportunities in the design and modification of vaccine strategies.
4.4.3 Microbial-like particles
Although recombinant bacteria have been actively investigated for gene delivery and vaccination, a novel approach has been demonstrated with the use of bacteria-mediated delivery of nanoparticle systems, termed ‘microbots’.170 This study utilized an attenuated strain of L. monocytogenes and surface conjugated nanoparticles that were loaded with pDNA. Surface conjugation was mediated through biotin-streptavidin interactions.170 Transfection with these hybrid vectors resulted in high levels of gene delivery and production of target proteins. Separately, our group has developed a hybrid gene delivery vector containing bacterial and biomaterial components. A bacterial core, E. coli, was surface-modified using a cationic polymer (poly(beta-amino ester)) in an effort to combine normally disparate vector-associated properties and tools sets to systematically overcome gene delivery barriers.171 This approach facilitated tailored APC cellular responses to levels significantly exceeding individual vector components or commercially-available transfection agents in vitro. In addition, in vivo ovalbumin gene delivery mouse models demonstrated that the hybrid device can mediate significant antibody responses without the need of adjuvants. Furthermore, the system is primed for an expansive combinatorial effort to vary both the biomaterial and biological components of the hybrid design. Here, the variety of cationic biomaterials commonly used in gene delivery offer new capabilities in the context of hybrid vector construction. As one example, we have recently tested the use of CPLA as an alternative biomaterial component (data submitted). Analogously, E. coli has been used in conjunction with commercial reagents (lipofectamine) to improve gene delivery.172
Outer membrane vesicles (OMVs) are naturally occurring proteoliposomes that bud from Gram-negative bacteria.173 These single lipid bilayer particles range in size from 50–250 nm. After discovery of OMVs, it was quickly realized that in vivo administration could prompt protective humoral and mucosal immune responses without the need of adjuvants.173 Presumably this arises from the presence of lingering immunoactive virulence factors on the vesicle surface.174 Using molecular biology tools, targeting ligands and additional immune-potentiator molecules have been attached to the OMV surface to improve resulting responses.174 Similar to synthetic vectors, OMVs are acellular, making them promising alternatives to live or attenuated pathogen-based vaccines. Most notably, Neisseria meningitides-derived OMVs have been commercialized by Novartis for the synthesis of European-approved vaccine Bexero.175 Similarly, non-denatured cell envelopes from Gram-negative bacteria (bacterial ghosts [BGs]) have demonstrated the ability to mediate gene delivery.167 BGs are produced through the heterologous expression of the bacteriophage-derived lysis gene E. These vectors contain all intrinsic surface components (polysaccharides, flagella, and fimbriae) but are substantially larger than OMVs (retaining size characteristics of the host bacterium). Additionally, as with OMVs, BGs retain intrinsic adjuvant properties derived from residual lipopolysaccharides (LPSs). Although this set of vectors has documented success in vivo and possesses a robust tool-set for vector engineering, biomaterial-functionalization has yet to occur but is a promising potential future avenue.
5. Current and emerging trends
Genetic vaccination has the potential for combating and eventually overcoming a variety of diseases that have overwhelmed current standard-of-cares. Yet challenges remain, and despite decades of research, the elicitation of a significant DNA-mediated immune response in a safe and scalable manner is still elusive. However, these barriers are beginning to fall. As the use of biological and biomaterial vectors, in the wake of advancements in adaptive immunity understanding, evolves past archaic design strategies towards tailoring and functionalization that more effectively instigates and directs immune responses, DNA vaccination will continue to grow in terms of impact and potential.
Specifically, tradition vector studies utilized simple antigen encapsulation (through innate, physical, or electrostatic capture) and systemic administration methodologies. Use of these systems has resulted in some success, but due to systemic effects and/or low efficacy, they do not transition well to non-model-based applications. Thus, current research is investigating the following topics: 1) targeted delivery; 2) cellular specificity; 3) vector combination and interfacial design; 4) response-modulation; 5) designed antigen release and activation kinetics. Each of these examples are classical research areas in gene and drug delivery but have not been readily applied in the field of immunology. Ultimately, investigating the underlying biological mechanisms of DNA vaccines arising from chemical- and structure-properties of individual delivery agents must be the focus of future studies. Doing so will facilitate the transition from empirical experimentation to an era of rational design.
6. Clinical trials and commercial products
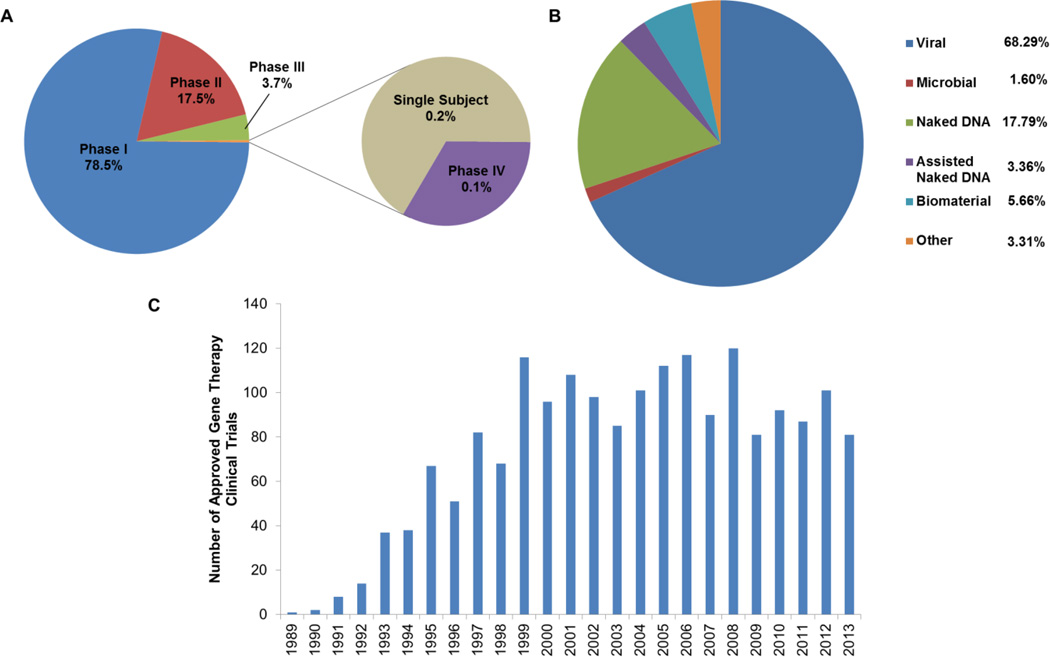
Although substantial progress has been made in the preclinical development of gene-based vaccines, the ultimate goal is to translate observed animal model efficacy to human studies. However, given the early stage of the field of gene therapy, no naked DNA vaccines have been used in a randomised clinical trial.176 As such, since genetic vaccine clinical data is limited, the remainder of the review will emphasize the development of general gene-based therapies that have advanced to clinical trials and corresponding commercial entities. Numerous trials of various phases are being conducted using a myriad of strategies (Fig. 3). Despite their limitations, viral vectors still remain the primary vector of choice. In addition, the overall number of trials has grown over the years (Fig. 3C).
Figure 3.
Global gene therapy clinical trial information. Breakdown of trials by phase (A), vector (B), and year (C). Information gathered from pooled clinical data (http://www.wiley.com/legacy/wileychi/genmed/clinical/).
It is probable that the approval and subsequent commercialization of these gene-based trials remains several years away. This is evident by the small number of approved gene-based products. These include Gendicine™ (SiBiono Gene Tech Co.; first globally approved clinical gene therapy), Oncorine™ (Sunway Biotech Co. Ltd), Cerepro® (Ark Therapeutics Group; first GMP certification in the EU; first and only adenoviral vector that has completed a phase III clinical trial), and Glybera (UniQure). However, no gene-based therapies are approved by the FDA, though three gene-based products are available for veterinary use.177–179 Interestingly, all of these products are vaccines – demonstrating the potential of genetic vaccination. We surmise that the insights provided by on-going and approved gene-based therapies will help shape improved studies that will result in a ‘burst’ of successful trials (including those in vaccination) in the future.
7. Conclusions
Despite initial challenges, emerging technological advancements have generated renewed interest in the development of rationally-designed genetic vaccination strategies. Specifically, the efficacy of genetic vaccination strategies can be improved by systematically engineering and optimizing the expression cassette (plasmid), delivery vector, and immunization/administration method. These new strategies coupled with the various inherent advantages of DNA vaccines – their ease of design and manufacturing, strong safety record, and stability – provide the framework for immunologically-effective and economically-palatable vaccine production schemes. However, delivery vector selection will require a thorough understanding of the biology of the vaccine target and the resultant disease pathology. Through the continual refinement of DNA vaccine technology, the prospects for prophylactic treatment of human and animal disease will help shape the future of vaccinology.
Acknowledgments
The authors recognize support from NIH awards AI088485 (BAP) and DC013554 (APH) and a SUNY-Buffalo Schomburg fellowship (CHJ).
Biographies

Charles H. Jones received B.S. degrees in Chemical & Biomolecular Engineering and Biochemistry in 2011 from North Carolina State University in Raleigh, NC. He is currently pursuing his Ph.D. in Chemical Engineering at the State University of New York at Buffalo. His thesis work focuses on the development of novel gene delivery vectors and their resulting impact upon adaptive immunity. His academic interests lie at the intersection of biomaterials and immunology, with a special focus on developing innovative vaccine strategies and elucidating underlying immunological pathways.

Anders P. Hakansson is an Assistant Professor of Microbiology and Immunology at the State University of New York at Buffalo. He received degrees from Lund University in Medicine (B.S.) and Medical Microbiology (Ph.D.). He was a part of numerous research fellowships including positions at David E. Briles’ laboratory at the University of Alabama, Birmingham; the Channing Laboratory at Brigham and Women’s Hospital (Harvard Medical School); and the Division of Infectious Disease at Boston Children’s Hospital (Harvard Medical School). His research seeks to understand how the respiratory pathogen Streptococcus pneumoniae (pneumococcus) causes disease in humans and how human defense mechanisms, including breast milk, can provide disease-protective properties.

Blaine Pfeifer is an Associate Professor of Chemical Engineering at the State University of New York at Buffalo. He received Chemical Engineering degrees from Colorado State University (B.S.) and Stanford University (M.S. and Ph.D.) and completed his postdoctoral work at the Massachusetts Institute of Technology. His research seeks to apply cellular, metabolic, and process engineering in the context of natural product biosynthesis and the development of therapies for infectious disease and cancer.
Notes and references
- 1.Ehreth J. Vaccine. 2003;21:596–600. doi: 10.1016/s0264-410x(02)00623-0. [DOI] [PubMed] [Google Scholar]
- 2.Ulmer JB, Valley U, Rappuoli R. Nature Biotechnology. 2006;24:1377–1383. doi: 10.1038/nbt1261. [DOI] [PubMed] [Google Scholar]
- 3.Andre FE, Booy R, Bock HL, Clemens J, Datta SK, John TJ, Lee BW, Lolekha S, Peltola H, Ruff TA, Santosham M, Schmitt HJ. Bulletin of the World Health Organization. 2008;86:140–146. doi: 10.2471/BLT.07.040089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pulendran B, Ahmed R. Nature Immunology. 2011;12:509–517. doi: 10.1038/ni.2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kumar P, Chen K, Kolls JK. Current Opinion in Immunology. 2013;25:373–380. doi: 10.1016/j.coi.2013.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moyron-Quiroz JE, Rangel-Moreno J, Kusser K, Hartson L, Sprague F, Goodrich S, Woodland DL, Lund FE, Randall TD. Nat Med. 2004;10:927–934. doi: 10.1038/nm1091. [DOI] [PubMed] [Google Scholar]
- 7.Mitsdoerffer M, Lee Y, Jager A, Kim HJ, Korn T, Kolls JK, Cantor H, Bettelli E, Kuchroo VK. Proc Natl Acad Sci. 2010;107:14292–14297. doi: 10.1073/pnas.1009234107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. http://www.cdc.gov/vaccines/vpd-vac/vpd-list.htm.
- 9.Schmidt C. Nature Biotechnology. 2013;31:957–960. doi: 10.1038/nbt.2733. [DOI] [PubMed] [Google Scholar]
- 10.Blume S, Geesink I. Science. 2000;288:1593–1594. doi: 10.1126/science.288.5471.1593. [DOI] [PubMed] [Google Scholar]
- 11.Nguyen DN, Green JJ, Chan JM, Langer R, Anderson DG. Adv Mater. 2009;21:847–867. doi: 10.1002/adma.200801478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ulmer JB, Wahren B, Liu MA. Trends in Molecular Medicine. 2006;12:216–222. doi: 10.1016/j.molmed.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 13.Kutzler MA, Weiner DB. Nature Reviews. Genetics. 2008;9:776–788. doi: 10.1038/nrg2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saade F, Petrovsky N. Expert Review of Vaccines. 2012;11:189–209. doi: 10.1586/erv.11.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoare M, Levy MS, Bracewell DG, Doig SD, Kong S, Titchener-Hooker N, Ward JM, Dunnill P. Biotechnol Progr. 2005;21:1577–1592. doi: 10.1021/bp050190n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gurunathan S, Klinman DM, Seder RA. Annual Review of Immunology. 2000;18:927–974. doi: 10.1146/annurev.immunol.18.1.927. [DOI] [PubMed] [Google Scholar]
- 17.MacGregor RR, Boyer JD, Ciccarelli RB, Ginsberg RS, Weiner DB. The Journal of Infectious Diseases. 2000;181:406. doi: 10.1086/315199. [DOI] [PubMed] [Google Scholar]
- 18.Kay MA, He CY, Chen ZY. Nature Biotechnology. 2010;28:1287–1289. doi: 10.1038/nbt.1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu J, Zhang F, Kay MA. Mol Ther. 2013;21:954–963. doi: 10.1038/mt.2013.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen ZY, He CY, Ehrhardt A, Kay MA. Mol Ther. 2003;8:495–500. doi: 10.1016/s1525-0016(03)00168-0. [DOI] [PubMed] [Google Scholar]
- 21.Chen ZY, He CY, Kay MA. Human Gene Therapy. 2005;16:126–131. doi: 10.1089/hum.2005.16.126. [DOI] [PubMed] [Google Scholar]
- 22.Osborn MJ, McElmurry RT, Lees CJ, DeFeo AP, Chen ZY, Kay MA, Naldini L, Freeman G, Tolar J, Blazar BR. Mol Ther. 2011;19:450–460. doi: 10.1038/mt.2010.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gracey Maniar LE, Maniar JM, Chen ZY, Lu J, Fire AZ, Kay MA. Mol Ther. 2013;21:131–138. doi: 10.1038/mt.2012.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luke J, Carnes AE, Hodgson CP, Williams JA. Vaccine. 2009;27:6454–6459. doi: 10.1016/j.vaccine.2009.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luke JM, Vincent JM, Du SX, Gerdemann U, Leen AM, Whalen RG, Hodgson CP, Williams JA. Gene Therapy. 2011;18:334–343. doi: 10.1038/gt.2010.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qin L, Ding Y, Pahud DR, Chang E, Imperiale MJ, Bromberg JS. Human Gene Therapy. 1997;8:2019–2029. doi: 10.1089/hum.1997.8.17-2019. [DOI] [PubMed] [Google Scholar]
- 27.Barouch DH, Yang ZY, Kong WP, Korioth-Schmitz B, Sumida SM, Truitt DM, Kishko MG, Arthur JC, Miura A, Mascola JR, Letvin NL, Nabel GJ. Journal of Virology. 2005;79:8828–8834. doi: 10.1128/JVI.79.14.8828-8834.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li HS, Liu Y, Li DF, Zhang RR, Tang HL, Zhang YW, Huang W, Peng H, Xu JQ, Hong KX, Shao YM. Chinese Medical Journal. 2007;120:496–502. [PubMed] [Google Scholar]
- 29.Kong W, Brovold M, Koeneman BA, Clark-Curtiss J, Curtiss R. Proceedings of the National Academy of Sciences. 2012;109:19414–19419. doi: 10.1073/pnas.1217554109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gustafsson C, Govindarajan S, Minshull J. Trends Biotechnol. 2004;22:346–353. doi: 10.1016/j.tibtech.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 31.Muthumani K, Kudchodkar S, Zhang D, Bagarazzi ML, Kim JJ, Boyer JD, Ayyavoo V, Pavlakis GN, Weiner DB. Vaccine. 2002;20:1999–2003. doi: 10.1016/s0264-410x(02)00086-5. [DOI] [PubMed] [Google Scholar]
- 32.Holmstrom F, Pasetto A, Nahr V, Brass A, Kriegs M, Hildt E, Broderick KE, Chen M, Ahlen G, Frelin L. J Immunol. 2013;190:1113–1124. doi: 10.4049/jimmunol.1201497. [DOI] [PubMed] [Google Scholar]
- 33.Garmory HS, Brown KA, Titball RW. Genetic Vaccines and Therapy. 2003;1:2. doi: 10.1186/1479-0556-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Williams JA, Carnes AE, Hodgson CP. Biotechnology Advances. 2009;27:353–370. doi: 10.1016/j.biotechadv.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jones CH, Chen CK, Ravikrishnan A, Rane S, Pfeifer BA. Mol Pharm. 2013;10:4082–4098. doi: 10.1021/mp400467x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lu J, Zhang F, Xu S, Fire AZ, Kay MA. Mol Ther. 2012;20:2111–2119. doi: 10.1038/mt.2012.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chatellard P, Pankiewicz R, Meier E, Durrer L, Sauvage C, Imhof MO. Biotechnol Bioeng. 2007;96:106–117. doi: 10.1002/bit.21172. [DOI] [PubMed] [Google Scholar]
- 38.Qin JY, Zhang L, Clift KL, Hulur I, Xiang AP, Ren BZ, Lahn BT. Plos One. 2010;5:e10611. doi: 10.1371/journal.pone.0010611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brinster RL, Allen JM, Behringer RR, Gelinas RE, Palmiter RD. Proc Natl Acad Sci. 1988;85:836–840. doi: 10.1073/pnas.85.3.836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang MT, Gorman CM. Nucleic Acids Research. 1990;18:937–947. doi: 10.1093/nar/18.4.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu HX, Zhang M, Krainer AR. Genes & Development. 1998;12:1998–2012. doi: 10.1101/gad.12.13.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fairbrother WG, Yeh RF, Sharp PA, Burge CB. Science. 2002;297:1007–1013. doi: 10.1126/science.1073774. [DOI] [PubMed] [Google Scholar]
- 43.Wang Y, Ma M, Xiao X, Wang Z. Nature Structural & Molecular Biology. 2012;19:1044–1052. doi: 10.1038/nsmb.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Su Y, Wang S, Shao J, Zhang B, Wei H. Chinese Journal of Biotechnology. 2013;29:458–465. [PubMed] [Google Scholar]
- 45.Leifert JA, Rodriguez-Carreno MP, Rodriguez F, Whitton JL. Immunological Reviews. 2004;199:40–53. doi: 10.1111/j.0105-2896.2004.0135.x. [DOI] [PubMed] [Google Scholar]
- 46.Li Z, Howard A, Kelley C, Delogu G, Collins F, Morris S. Infect Immun. 1999;67:4780–4786. doi: 10.1128/iai.67.9.4780-4786.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu TC, Guarnieri FG, Staveley-O’Carroll KF, Viscidi RP, Levitsky HI, Hedrick L, Cho KR, August JT, Pardoll DM. Proc Natl Acad Sci. 1995;92:11671–11675. doi: 10.1073/pnas.92.25.11671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Delogu G, Howard A, Collins FM, Morris SL. Infect Immun. 2000;68:3097–3102. doi: 10.1128/iai.68.6.3097-3102.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shedlock D, Tingey C, Mahadevan L, Hutnick N, Reuschel E, Kudchodkar S, Flingai S, Yan J, Kim J, Ugen K, Weiner D, Muthumani K. Vaccines. 2014;2:196–215. doi: 10.3390/vaccines2020196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.O’Hagan DT, Ott GS, De Gregorio E, Seubert A. Vaccine. 2012;30:4341–4348. doi: 10.1016/j.vaccine.2011.09.061. [DOI] [PubMed] [Google Scholar]
- 51.Seubert A, Monaci E, Pizza M, O’Hagan DT, Wack A. J Immunol. 2008;180:5402–5412. doi: 10.4049/jimmunol.180.8.5402. [DOI] [PubMed] [Google Scholar]
- 52.Spies B, Hochrein H, Vabulas M, Huster K, Busch DH, Schmitz F, Heit A, Wagner H. J Immunol. 2003;171:5908–5912. doi: 10.4049/jimmunol.171.11.5908. [DOI] [PubMed] [Google Scholar]
- 53.Kojima Y, Xin KQ, Ooki T, Hamajima K, Oikawa T, Shinoda K, Ozaki T, Hoshino Y, Jounai N, Nakazawa M, Klinman D, Okuda K. Vaccine. 2002;20:2857–2865. doi: 10.1016/s0264-410x(02)00238-4. [DOI] [PubMed] [Google Scholar]
- 54.Coban C, Ishii KJ, Gursel M, Klinman DM, Kumar N. Journal of Leukocyte Biology. 2005;78:647–655. doi: 10.1189/jlb.1104627. [DOI] [PubMed] [Google Scholar]
- 55.Song R, Liu S, Leong KW. Mol Ther. 2007;15:1007–1015. doi: 10.1038/mt.sj.6300129. [DOI] [PubMed] [Google Scholar]
- 56.Kim SJ, Suh D, Park SE, Park JS, Byun HM, Lee C, Lee SY, Kim I, Oh YK. Virology. 2003;314:84–91. doi: 10.1016/s0042-6822(03)00417-3. [DOI] [PubMed] [Google Scholar]
- 57.Williman J, Young S, Buchan G, Slobbe L, Wilson M, Pang P, Austyn J, Preston S, Baird M. Vaccine. 2008;26:5153–5158. doi: 10.1016/j.vaccine.2008.03.084. [DOI] [PubMed] [Google Scholar]
- 58.Ramanathan MP, Kutzler MA, Kuo YC, Yan J, Liu H, Shah V, Bawa A, Selling B, Sardesai NY, Kim JJ, Weiner DB. Vaccine. 2009;27:4370–4380. doi: 10.1016/j.vaccine.2009.01.137. [DOI] [PubMed] [Google Scholar]
- 59.Kutzler MA, Robinson TM, Chattergoon MA, Choo DK, Choo AY, Choe PY, Ramanathan MP, Parkinson R, Kudchodkar S, Tamura Y, Sidhu M, Roopchand V, Kim JJ, Pavlakis GN, Felber BK, Waldmann TA, Boyer JD, Weiner DB. J Immunol. 2005;175:112–123. doi: 10.4049/jimmunol.175.1.112. [DOI] [PubMed] [Google Scholar]
- 60.Geissler M, Gesien A, Tokushige K, Wands JR. J Immunol. 1997;158:1231–1237. [PubMed] [Google Scholar]
- 61.Nobiron I, Thompson I, Brownlie J, Collins ME. Veterinary Microbiology. 2000;76:129–142. doi: 10.1016/s0378-1135(00)00238-8. [DOI] [PubMed] [Google Scholar]
- 62.O’Hagan D, Singh M, Ugozzoli M, Wild C, Barnett S, Chen M, Schaefer M, Doe B, Otten GR, Ulmer JB. Journal of Virology. 2001;75:9037–9043. doi: 10.1128/JVI.75.19.9037-9043.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li W, Li S, Hu Y, Tang B, Cui L, He W. Vaccine. 2008;26:3282–3290. doi: 10.1016/j.vaccine.2008.03.081. [DOI] [PubMed] [Google Scholar]
- 64.Kim JJ, Bagarazzi ML, Trivedi N, Hu Y, Kazahaya K, Wilson DM, Ciccarelli R, Chattergoon MA, Dang K, Mahalingam S, Chalian AA, Agadjanyan MG, Boyer JD, Wang B, Weiner DB. Nature Biotechnology. 1997;15:641–646. doi: 10.1038/nbt0797-641. [DOI] [PubMed] [Google Scholar]
- 65.Flo J, Tisminetzky S, Baralle F. Immunology. 2000;100:259–267. doi: 10.1046/j.1365-2567.2000.00041.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Boyle JS, Brady JL, Lew AM. Nature. 1998;392:408–411. doi: 10.1038/32932. [DOI] [PubMed] [Google Scholar]
- 67.Wan C, Yi L, Yang Z, Yang J, Shao H, Zhang C, Pan Z. Veterinary Immunology and Immunopathology. 2010;137:47–53. doi: 10.1016/j.vetimm.2010.04.008. [DOI] [PubMed] [Google Scholar]
- 68.Takeshita F, Tanaka T, Matsuda T, Tozuka M, Kobiyama K, Saha S, Matsui K, Ishii KJ, Coban C, Akira S, Ishii N, Suzuki K, Klinman DM, Okuda K, Sasaki S. Journal of Virology. 2006;80:6218–6224. doi: 10.1128/JVI.00121-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Castaldello A, Sgarbanti M, Marsili G, Brocca-Cofano E, Remoli AL, Caputo A, Battistini A. J Cell Physiol. 2010;224:702–709. doi: 10.1002/jcp.22169. [DOI] [PubMed] [Google Scholar]
- 70.Bramson JL, Dayball K, Hall JR, Millar JB, Miller M, Wan YH, Lin R, Hiscott J. Vaccine. 2003;21:1363–1370. doi: 10.1016/s0264-410x(02)00694-1. [DOI] [PubMed] [Google Scholar]
- 71.Ishii KJ, Kawagoe T, Koyama S, Matsui K, Kumar H, Kawai T, Uematsu S, Takeuchi O, Takeshita F, Coban C, Akira S. Nature. 2008;451:725–729. doi: 10.1038/nature06537. [DOI] [PubMed] [Google Scholar]
- 72.Sasaki S, Amara RR, Yeow WS, Pitha PM, Robinson HL. Journal of Virology. 2002;76:6652–6659. doi: 10.1128/JVI.76.13.6652-6659.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Flingai S, Czerwonko M, Goodman J, Kudchodkar SB, Muthumani K, Weiner DB. Frontiers in Immunology. 2013;4:354. doi: 10.3389/fimmu.2013.00354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Barouch DH, Letvin NL, Seder RA. Immunological Reviews. 2004;202:266–274. doi: 10.1111/j.0105-2896.2004.00200.x. [DOI] [PubMed] [Google Scholar]
- 75.Ferraro B, Morrow MP, Hutnick NA, Shin TH, Lucke CE, Weiner DB. Clinical Infectious Diseases: an Official Publication of the Infectious Diseases Society of America. 2011;53:296–302. doi: 10.1093/cid/cir334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lanier LL, O’Fallon S, Somoza C, Phillips JH, Linsley PS, Okumura K, Ito D, Azuma M. J Immunol. 1995;154:97–105. [PubMed] [Google Scholar]
- 77.Loukinov D, Ghochikyan A, Mkrtichyan M, Ichim TE, Lobanenkov VV, Cribbs DH, Agadjanyan MG. Journal of Cellular Biochemistry. 2006;98:1037–1043. doi: 10.1002/jcb.20953. [DOI] [PubMed] [Google Scholar]
- 78.Corr M, Tighe H, Lee D, Dudler J, Trieu M, Brinson DC, Carson DA. J Immunol. 1997;159:4999–5004. [PubMed] [Google Scholar]
- 79.Tsuji T, Hamajima K, Ishii N, Aoki I, Fukushima J, Xin KQ, Kawamoto S, Sasaki S, Matsunaga K, Ishigatsubo Y, Tani K, Okubo T, Okuda K. European Journal of Immunology. 1997;27:782–787. doi: 10.1002/eji.1830270329. [DOI] [PubMed] [Google Scholar]
- 80.Maue AC, Waters WR, Palmer MV, Whipple DL, Minion FC, Brown WC, Estes DM. Vaccine. 2004;23:769–779. doi: 10.1016/j.vaccine.2004.07.019. [DOI] [PubMed] [Google Scholar]
- 81.Gurunathan S, Irvine KR, Wu CY, Cohen JI, Thomas E, Prussin C, Restifo NP, Seder RA. J Immunol. 1998;161:4563–4571. [PMC free article] [PubMed] [Google Scholar]
- 82.Mendoza RB, Cantwell MJ, Kipps TJ. J Immunol. 1997;159:5777–5781. [PubMed] [Google Scholar]
- 83.Xu H, Zhao G, Huang X, Ding Z, Wang J, Wang X, Cheng Y, Kang Y, Wang B. The Journal of Gene Medicine. 2010;12:97–106. doi: 10.1002/jgm.1412. [DOI] [PubMed] [Google Scholar]
- 84.Sharpe AH, Wherry EJ, Ahmed R, Freeman GJ. Nature immunology. 2007;8:239–245. doi: 10.1038/ni1443. [DOI] [PubMed] [Google Scholar]
- 85.Song MY, Park SH, Nam HJ, Choi DH, Sung YC. J Immunother. 2011;34:297–306. doi: 10.1097/CJI.0b013e318210ed0e. [DOI] [PubMed] [Google Scholar]
- 86.Lu S. Current Opinion in Immunology. 2009;21:346–351. doi: 10.1016/j.coi.2009.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Woodland DL. Trends in Immunology. 2004;25:98–104. doi: 10.1016/j.it.2003.11.009. [DOI] [PubMed] [Google Scholar]
- 88.Estcourt MJ, Ramsay AJ, Brooks A, Thomson SA, Medveckzy CJ, Ramshaw IA. International Immunology. 2002;14:31–37. doi: 10.1093/intimm/14.1.31. [DOI] [PubMed] [Google Scholar]
- 89.Ratto-Kim S, Currier JR, Cox JH, Excler J-L, Valencia-Micolta A, Thelian D, Lo V, Sayeed E, Polonis VR, Earl PL, Moss B, Robb ML, Michael NL, Kim JH, Marovich MA. Plos One. 2012;7:e45840. doi: 10.1371/journal.pone.0045840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mulligan MJ, Russell ND, Celum C, Kahn J, Noonan E, Montefiori DC, Ferrari G, Weinhold KJ, Smith JM, Amara RR, Robinson HL. AIDS Research and Human Retroviruses. 2006;22:678–683. doi: 10.1089/aid.2006.22.678. [DOI] [PubMed] [Google Scholar]
- 91.Wang R, Doolan DL, Le TP, Hedstrom RC, Coonan KM, Charoenvit Y, Jones TR, Hobart P, Margalith M, Ng J, Weiss WR, Sedegah M, de Taisne C, Norman JA, Hoffman SL. Science. 1998;282:476–480. doi: 10.1126/science.282.5388.476. [DOI] [PubMed] [Google Scholar]
- 92.Robert-Guroff M. Current Opinion in Biotechnology. 2007;18:546–556. doi: 10.1016/j.copbio.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kumar P, Woon-Khiong C. Current Gene Therapy. 2011;11:144–153. doi: 10.2174/156652311794940782. [DOI] [PubMed] [Google Scholar]
- 94.Schaffner W. Proc Natl Acad Sci. 1980;77:2163–2167. doi: 10.1073/pnas.77.4.2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Baban CK, Cronin M, O’Hanlon D, O’Sullivan GC, Tangney M. Bioengineered. 2010;1:385–394. doi: 10.4161/bbug.1.6.13146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Stritzker J, Pilgrim S, Szalay AA, Goebel W. BMC Cancer. 2008;8:94. doi: 10.1186/1471-2407-8-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Grillot-Courvalin C, Goussard S, Huetz F, Ojcius DM, Courvalin P. Nature Biotechnology. 1998;16:862–866. doi: 10.1038/nbt0998-862. [DOI] [PubMed] [Google Scholar]
- 98.van Pijkeren JP, Morrissey D, Monk IR, Cronin M, Rajendran S, O’Sullivan GC, Gahan CG, Tangney M. Human Gene Therapy. 2010;21:405–416. doi: 10.1089/hum.2009.022. [DOI] [PubMed] [Google Scholar]
- 99.Shen H, Kanoh M, Liu F, Maruyama S, Asano Y. Microbiology and Immunology. 2004;48:329–337. doi: 10.1111/j.1348-0421.2004.tb03514.x. [DOI] [PubMed] [Google Scholar]
- 100.Courvalin P, Goussard S, Grillot-Courvalin C. Comptes Rendus de l’Academie des Sciences. Serie III, Sciences de la Vie. 1995;318:1207–1212. [PubMed] [Google Scholar]
- 101.Grillot-Courvalin C, Goussard S, Courvalin P. Cellular Microbiology. 2002;4:177–186. doi: 10.1046/j.1462-5822.2002.00184.x. [DOI] [PubMed] [Google Scholar]
- 102.Radford KJ, Higgins DE, Pasquini S, Cheadle EJ, Carta L, Jackson AM, Lemoine NR, Vassaux G. Gene Ther. 2002;9:1455–1463. doi: 10.1038/sj.gt.3301812. [DOI] [PubMed] [Google Scholar]
- 103.Castagliuolo I, Beggiao E, Brun P, Barzon L, Goussard S, Manganelli R, Grillot-Courvalin C, Palu G. Gene Ther. 2005;12:1070–1078. doi: 10.1038/sj.gt.3302493. [DOI] [PubMed] [Google Scholar]
- 104.Laner A, Goussard S, Ramalho AS, Schwarz T, Amaral MD, Courvalin P, Schindelhauer D, Grillot-Courvalin C. Gene Ther. 2005;12:1559–1572. doi: 10.1038/sj.gt.3302576. [DOI] [PubMed] [Google Scholar]
- 105.Parsa S, Wang Y, Rines K, Pfeifer BA. J. Biotechnol. 2008;137:59–64. doi: 10.1016/j.jbiotec.2008.07.1815. [DOI] [PubMed] [Google Scholar]
- 106.Higgins DE, Shastri N, Portnoy DA. Mol Microbiol. 1999;31:1631–1641. doi: 10.1046/j.1365-2958.1999.01272.x. [DOI] [PubMed] [Google Scholar]
- 107.Huang L, Jin J, Deighan P, Kiner E, McReynolds L, Lieberman J. Nature Biotechnology. 2013;31:350–356. doi: 10.1038/nbt.2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Xiang S, Fruehauf J, Li CJ. Nature Biotechnology. 2006;24:697–702. doi: 10.1038/nbt1211. [DOI] [PubMed] [Google Scholar]
- 109.Jones CH, Rane S, Patt E, Ravikrishnan A, Chen CK, Cheng C, Pfeifer BA. Mol Pharm. 2013;10:4301–4308. doi: 10.1021/mp4003927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hidaka A, Hamaji Y, Sasaki T, Taniguchi S, Fujimori M. Bioscience, Biotechnology, and Biochemistry. 2007;71:2921–2926. doi: 10.1271/bbb.70284. [DOI] [PubMed] [Google Scholar]
- 111.Hamaji Y, Fujimori M, Sasaki T, Matsuhashi H, Matsui-Seki K, Shimatani-Shibata Y, Kano Y, Amano J, Taniguchi S. Bioscience, Biotechnology, and Biochemistry. 2007;71:874–883. doi: 10.1271/bbb.60502. [DOI] [PubMed] [Google Scholar]
- 112.Sasaki T, Fujimori M, Hamaji Y, Hama Y, Ito K, Amano J, Taniguchi S. Cancer Science. 2006;97:649–657. doi: 10.1111/j.1349-7006.2006.00221.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Palffy R, Gardlik R, Behuliak M, Jani P, Balakova D, Kadasi L, Turna J, Celec P. Exp Biol Med (Maywood) 2011;236:177–183. doi: 10.1258/ebm.2010.010277. [DOI] [PubMed] [Google Scholar]
- 114.Bartolome A, Herrero-Gil A, Horcajo P, Orden JA, de Fuente R, Dominguez-Bernal G. Veterinary Microbiology. 2010;141:81–88. doi: 10.1016/j.vetmic.2009.08.006. [DOI] [PubMed] [Google Scholar]
- 115.Qiu L, Wang X, Hao H, Mu G, Dang R, Wang J, Zhang S, Du E, Yang Z. Journal of Virological Methods. 2013;188:108–113. doi: 10.1016/j.jviromet.2012.12.007. [DOI] [PubMed] [Google Scholar]
- 116.Ning JF, Zhu W, Xu JP, Zheng CY, Meng XL. Vaccine. 2009;27:1127–1135. doi: 10.1016/j.vaccine.2008.11.075. [DOI] [PubMed] [Google Scholar]
- 117.Abdul-Wahid A, Faubert G. Vaccine. 2007;25:8372–8383. doi: 10.1016/j.vaccine.2007.10.012. [DOI] [PubMed] [Google Scholar]
- 118.Cheng C, Zhou Y, Kan B, Wang Q, Rui Y. Mol Med Rep. 2014;9:2239–2244. doi: 10.3892/mmr.2014.2065. [DOI] [PubMed] [Google Scholar]
- 119.Murugaiah C, Nik Mohd Noor NZ, Mustafa S, Manickam R, Pattabhiraman L. Plos One. 2014;9:e81817. doi: 10.1371/journal.pone.0081817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ketley JM, Michalski J, Galen J, Levine MM, Kaper JB. FEMS Microbiology Letters. 1993;111:15–21. doi: 10.1111/j.1574-6968.1993.tb06355.x. [DOI] [PubMed] [Google Scholar]
- 121.Tobias J, Lebens M, Bolin I, Wiklund G, Svennerholm AM. Vaccine. 2008;26:743–752. doi: 10.1016/j.vaccine.2007.12.009. [DOI] [PubMed] [Google Scholar]
- 122.Pasetti MF, Barry EM, Losonsky G, Singh M, Medina-Moreno SM, Polo JM, Ulmer J, Robinson H, Sztein MB, Levine MM. Journal of Virology. 2003;77:5209–5217. doi: 10.1128/JVI.77.9.5209-5217.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Xu F, Hong M, Ulmer JB. Vaccine. 2003;21:644–648. doi: 10.1016/s0264-410x(02)00573-x. [DOI] [PubMed] [Google Scholar]
- 124.Vecino WH, Morin PM, Agha R, Jacobs WR, Jr, Fennelly GJ. Immunology Letters. 2002;82:197–204. doi: 10.1016/s0165-2478(02)00043-3. [DOI] [PubMed] [Google Scholar]
- 125.Colditz GA, Brewer TF, Berkey CS, Wilson ME, Burdick E, Fineberg HV, Mosteller F. JAMA: the Journal of the American Medical Association. 1994;271:698–702. [PubMed] [Google Scholar]
- 126.Al-Mariri A, Tibor A, Lestrate P, Mertens P, De Bolle X, Letesson JJ. Infect Immun. 2002;70:1915–1923. doi: 10.1128/IAI.70.4.1915-1923.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Chatel JM, Pothelune L, Ah-Leung S, Corthier G, Wal JM, Langella P. Gene Therapy. 2008;15:1184–1190. doi: 10.1038/gt.2008.59. [DOI] [PubMed] [Google Scholar]
- 128.Shata MT, Stevceva L, Agwale S, Lewis GK, Hone DM. Molecular Medicine Today. 2000;6:66–71. doi: 10.1016/s1357-4310(99)01633-0. [DOI] [PubMed] [Google Scholar]
- 129.Chart H, Smith HR, La Ragione RM, Woodward MJ. J Appl Microbiol. 2000;89:1048–1058. doi: 10.1046/j.1365-2672.2000.01211.x. [DOI] [PubMed] [Google Scholar]
- 130.Sizemore DR, Branstrom AA, Sadoff JC. Science. 1995;270:299–302. doi: 10.1126/science.270.5234.299. [DOI] [PubMed] [Google Scholar]
- 131.Darji A, Guzman CA, Gerstel B, Wachholz P, Timmis KN, Wehland J, Chakraborty T, Weiss S. Cell. 1997;91:765–775. doi: 10.1016/s0092-8674(00)80465-1. [DOI] [PubMed] [Google Scholar]
- 132.Dietrich G, Bubert A, Gentschev I, Sokolovic Z, Simm A, Catic A, Kaufmann SH, Hess J, Szalay AA, Goebel W. Nature Biotechnology. 1998;16:181–185. doi: 10.1038/nbt0298-181. [DOI] [PubMed] [Google Scholar]
- 133.Jones KS. Biotechnol Progr. 2008;24:807–814. doi: 10.1002/btpr.10. [DOI] [PubMed] [Google Scholar]
- 134.Lynn DM, Langer R. J. Am. Chem. Soc. 2000;122:10761–10768. [Google Scholar]
- 135.Little SR, Lynn DM, Ge Q, Anderson DG, Puram SV, Chen J, Eisen HN, Langer R. Proc Natl Acad Sci. 2004;101:9534–9539. doi: 10.1073/pnas.0403549101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Yang SC, Bhide M, Crispe IN, Pierce RH, Murthy N. Bioconjug Chem. 2008;19:1164–1169. doi: 10.1021/bc700442g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Jones CH, Chen CK, Jiang M, Fang L, Cheng C, Pfeifer BA. Mol Pharm. 2013;10:1138–1145. doi: 10.1021/mp300666s. [DOI] [PubMed] [Google Scholar]
- 138.Chen CK, Jones CH, Mistriotis P, Yu Y, Ma X, Ravikrishnan A, Jiang M, Andreadis ST, Pfeifer BA, Cheng C. Biomaterials. 2013;34:9688–9699. doi: 10.1016/j.biomaterials.2013.08.063. [DOI] [PubMed] [Google Scholar]
- 139.Rivas-Aravena A, Sandino AM, Spencer E. Biological Research. 2013;46:407–419. doi: 10.4067/S0716-97602013000400012. [DOI] [PubMed] [Google Scholar]
- 140.Sadler K, Zeng W, Jackson DC. The Journal of Peptide Research: Official Journal of the American Peptide Society. 2002;60:150–158. doi: 10.1034/j.1399-3011.2002.21009.x. [DOI] [PubMed] [Google Scholar]
- 141.Liu TY, Hussein WM, Jia Z, Ziora ZM, McMillan NA, Monteiro MJ, Toth I, Skwarczynski M. Biomacromolecules. 2013;14:2798–2806. doi: 10.1021/bm400626w. [DOI] [PubMed] [Google Scholar]
- 142.El-Sayed A, Harashima H. Mol Ther. 2013;21:1118–1130. doi: 10.1038/mt.2013.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Mora-Solano C, Collier JH. Journal of Materials Chemistry. B, Materials for Biology and Medicine. 2014;2:2409–2421. doi: 10.1039/C3TB21549K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Manolova V, Flace A, Bauer M, Schwarz K, Saudan P, Bachmann MF. European Journal of Immunology. 2008;38:1404–1413. doi: 10.1002/eji.200737984. [DOI] [PubMed] [Google Scholar]
- 145.Reddy ST, Rehor A, Schmoekel HG, Hubbell JA, Swartz MA. J Control Release. 2006;112:26–34. doi: 10.1016/j.jconrel.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 146.Jewell CM, Lopez SC, Irvine DJ. Proc Natl Acad Sci. 2011;108:15745–15750. doi: 10.1073/pnas.1105200108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Mitragotri S, Lahann J. Nature Materials. 2009;8:15–23. doi: 10.1038/nmat2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Choi HS, Liu W, Liu F, Nasr K, Misra P, Bawendi MG, Frangioni JV. Nat Nanotechnol. 2010;5:42–47. doi: 10.1038/nnano.2009.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Reddy ST, van der Vlies AJ, Simeoni E, Angeli V, Randolph GJ, O’Neil CP, Lee LK, Swartz MA, Hubbell JA. Nature Biotechnology. 2007;25:1159–1164. doi: 10.1038/nbt1332. [DOI] [PubMed] [Google Scholar]
- 150.De Geest BG, Willart MA, Hammad H, Lambrecht BN, Pollard C, Bogaert P, De Filette M, Saelens X, Vervaet C, Remon JP, Grooten J, De Koker S. ACS Nano. 2012;6:2136–2149. doi: 10.1021/nn205099c. [DOI] [PubMed] [Google Scholar]
- 151.Cho HJ, Han SE, Im S, Lee Y, Kim YB, Chun T, Oh YK. Biomaterials. 2011;32:4621–4629. doi: 10.1016/j.biomaterials.2011.03.004. [DOI] [PubMed] [Google Scholar]
- 152.Puffer EB, Pontrello JK, Hollenbeck JJ, Kink JA, Kiessling LL. ACS Chemical Biology. 2007;2:252–262. doi: 10.1021/cb600489g. [DOI] [PubMed] [Google Scholar]
- 153.Hinton HJ, Jegerlehner A, Bachmann MF. Current Topics in Microbiology and Immunology. 2008;319:1–15. doi: 10.1007/978-3-540-73900-5_1. [DOI] [PubMed] [Google Scholar]
- 154.Moon JJ, Suh H, Li AV, Ockenhouse CF, Yadava A, Irvine DJ. Proc Natl Acad Sci. 2012;109:1080–1085. doi: 10.1073/pnas.1112648109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Vyas SP, Goyal AK, Khatri K. Methods in Molecular Biology. 2010;605:177–188. doi: 10.1007/978-1-60327-360-2_12. [DOI] [PubMed] [Google Scholar]
- 156.Yao W, Peng Y, Du M, Luo J, Zong L. Mol Pharm. 2013;10:2904–2914. doi: 10.1021/mp4000053. [DOI] [PubMed] [Google Scholar]
- 157.Grodeland G, Mjaaland S, Tunheim G, Fredriksen AB, Bogen B. Plos One. 2013;8:e80008. doi: 10.1371/journal.pone.0080008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Noad R, Roy P. Trends in Microbiology. 2003;11:438–444. doi: 10.1016/s0966-842x(03)00208-7. [DOI] [PubMed] [Google Scholar]
- 159.Rosenthal JA, Chen L, Baker JL, Putnam D, DeLisa MP. Current Opinion in Biotechnology. 2013;28:51–58. doi: 10.1016/j.copbio.2013.11.005. [DOI] [PubMed] [Google Scholar]
- 160.Ramsey JD, Vu HN, Pack DW. J Control Release. 2010;144:39–45. doi: 10.1016/j.jconrel.2010.01.031. [DOI] [PubMed] [Google Scholar]
- 161.Keswani RK, Pozdol IM, Pack DW. Mol Pharm. 2013;10:1725–1735. doi: 10.1021/mp300561y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Singarapu K, Pal I, Ramsey JD. J Biomed Mater Res A. 2013;101:1857–1864. doi: 10.1002/jbm.a.34483. [DOI] [PubMed] [Google Scholar]
- 163.Huckriede A, Bungener L, Daemen T, Wilschut J. Methods in Enzymology. 2003;373:74–91. doi: 10.1016/S0076-6879(03)73005-5. [DOI] [PubMed] [Google Scholar]
- 164.Moser C, Muller M, Kaeser MD, Weydemann U, Amacker M. Expert Review of Vaccines. 2013;12:779–791. doi: 10.1586/14760584.2013.811195. [DOI] [PubMed] [Google Scholar]
- 165.Chams V, Bonnafous P, Stegmann T. FEBS Lett. 1999;448:28–32. doi: 10.1016/s0014-5793(99)00333-6. [DOI] [PubMed] [Google Scholar]
- 166.Xu L, Frederik P, Pirollo KF, Tang WH, Rait A, Xiang LM, Huang W, Cruz I, Yin Y, Chang EH. Human Gene Therapy. 2002;13:469–481. doi: 10.1089/10430340252792594. [DOI] [PubMed] [Google Scholar]
- 167.Yoo JW, Irvine DJ, Discher DE, Mitragotri S. Nat Rev Drug Discov. 2011;10:521–535. doi: 10.1038/nrd3499. [DOI] [PubMed] [Google Scholar]
- 168.Lee ES, Kim D, Youn YS, Oh KT, Bae YH. Angewandte Chemie. 2008;47:2418–2421. doi: 10.1002/anie.200704121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Ferreira SA, Gama FM, Vilanova M. Nanomedicine-UK. 2013;9:159–173. doi: 10.1016/j.nano.2012.06.001. [DOI] [PubMed] [Google Scholar]
- 170.Akin D, Sturgis J, Ragheb K, Sherman D, Burkholder K, Robinson JP, Bhunia AK, Mohammed S, Bashir R. Nat Nanotechnol. 2007;2:441–449. doi: 10.1038/nnano.2007.149. [DOI] [PubMed] [Google Scholar]
- 171.Jones CH, Ravikrishnan A, Chen M, Reddinger R, Kamal Ahmadi M, Rane S, Hakansson AP, Pfeifer BA. Proc Natl Acad Sci U S A. 2014 doi: 10.1073/pnas.1411355111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Narayanan K, Lee CW, Radu A, Sim EU. Anal Biochem. 2013;439:142–144. doi: 10.1016/j.ab.2013.04.010. [DOI] [PubMed] [Google Scholar]
- 173.Unal CM, Schaar V, Riesbeck K. Seminars in Immunopathology. 2011;33:395–408. doi: 10.1007/s00281-010-0231-y. [DOI] [PubMed] [Google Scholar]
- 174.Gujrati V, Kim S, Kim SH, Min JJ, Choy HE, Kim SC, Jon S. ACS Nano. 2014;8:1525–1537. doi: 10.1021/nn405724x. [DOI] [PubMed] [Google Scholar]
- 175.Gorringe AR, Pajon R. Human Vaccines & Immunotherapeutics. 2012;8:174–183. doi: 10.4161/hv.18500. [DOI] [PubMed] [Google Scholar]
- 176.Alam S, McNeel DG. Expert Review of Vaccines. 2010;9:731–745. doi: 10.1586/erv.10.64. [DOI] [PubMed] [Google Scholar]
- 177.Davis BS, Chang GJ, Cropp B, Roehrig JT, Martin DA, Mitchell CJ, Bowen R, Bunning ML. Journal of Virology. 2001;75:4040–4047. doi: 10.1128/JVI.75.9.4040-4047.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Anderson ED, Mourich DV, Leong JA. Molecular Marine Biology and Biotechnology. 1996;5:105–113. [PubMed] [Google Scholar]
- 179.Bergman PJ, McKnight J, Novosad A, Charney S, Farrelly J, Craft D, Wulderk M, Jeffers Y, Sadelain M, Hohenhaus AE, Segal N, Gregor P, Engelhorn M, Riviere I, Houghton AN, Wolchok JD. Clin Cancer Res. 2003;9:1284–1290. [PubMed] [Google Scholar]
- 180.Ko HJ, Ko SY, Kim YJ, Lee EG, Cho SN, Kang CY. Infect Immun. 2005;73:5666–5674. doi: 10.1128/IAI.73.9.5666-5674.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Luo D, Saltzman WM. Nature Biotechnology. 2000;18:33–37. doi: 10.1038/71889. [DOI] [PubMed] [Google Scholar]
- 182.Calvo-Pinilla E, Rodriguez-Calvo T, Sevilla N, Ortego J. Vaccine. 2009;28:437–445. doi: 10.1016/j.vaccine.2009.10.027. [DOI] [PubMed] [Google Scholar]
- 183.Elvang T, Christensen JP, Billeskov R, Thi Kim Thanh Hoang T, Holst P, Thomsen AR, Andersen P, Dietrich J. Plos One. 2009;4:e5139. doi: 10.1371/journal.pone.0005139. [DOI] [PMC free article] [PubMed] [Google Scholar]