Abstract
Global concerns have been paid to the potential hazard of traditional herbal medicinal products (THMPs). Substandard and counterfeit THMPs, including traditional Chinese patent medicine, health foods, dietary supplements, etc. are potential threats to public health. Recent marketplace studies using DNA barcoding have determined that the current quality control methods are not sufficient for ensuring the presence of authentic herbal ingredients and detection of contaminants/adulterants. An efficient biomonitoring method for THMPs is of great needed. Herein, metabarcoding and single-molecule, real-time (SMRT) sequencing were used to detect the multiple ingredients in Jiuwei Qianghuo Wan (JWQHW), a classical herbal prescription widely used in China for the last 800 years. Reference experimental mixtures and commercial JWQHW products from the marketplace were used to confirm the method. Successful SMRT sequencing results recovered 5416 and 4342 circular-consensus sequencing (CCS) reads belonging to the ITS2 and psbA-trnH regions. The results suggest that with the combination of metabarcoding and SMRT sequencing, it is repeatable, reliable, and sensitive enough to detect species in the THMPs, and the error in SMRT sequencing did not affect the ability to identify multiple prescribed species and several adulterants/contaminants. It has the potential for becoming a valuable tool for the biomonitoring of multi-ingredient THMPs.
KEY WORDS: Traditional herbal medicinal products (THMP), Species mixture, Authentication, DNA metabarcoding, Single molecule real-time (SMRT) sequencing, Circular-consensus sequencing (CCS)
Graphical abstract
In this paper, metabarcoding and single-molecule, real-time (SMRT) sequencing were used to detect the multiple ingredients in Jiuwei Qianghuo Wan (JWQHW), a classical herbal prescription widely used in China for the last 800 years. Successful SMRT sequencing results recovered 5416 and 4342 circular-consensus sequencing (CCS) reads belonging to the ITS2 and psbA-trnH regions, suggesting the combination of metabarcoding and SMRT sequencing is a valuable tool for the biomonitoring of multi-ingredient THMPs.
1. Introduction
The recent increased profile and commercial use of herbal medicines requires pharmacovigilance in the traditional herbal medicinal products (THMP) industry. There are more than 1300 traditional herbal medicinal products registered by the health regulator authorities of the European Union1. The demand for herbal medicine increased with compound annual growth rate (CAGR) of 6%—10% and this market is forecasted to reach $107 billion by the year 2017; according to a report from Global Industry Analysts2. Drug safety should not be considered secondary to efficacy or be compromised by the increased demand for THMP1, 3. The regulation of medical products must be strengthened, especially in low- and middle-income countries4. In 2009, the World Health Organization (WHO) clearly defined “substandard drugs” and “counterfeit drugs”5, as they can cause serious health issues. A review reported by Chan6 showed that the contamination and substitution of herbal medicines have induced seriously anticholinergic poisoning in Hong Kong during 1989–2012. Chan said “pharmacovigilance of herbal medicines should help determine the incidence and causes of adverse reactions and monitor the effectiveness of preventive measures”6. Due to adulterations of the supply chain, and/or inadequate quality control during manufacturing, poor-quality herbal preparations may enter the market and threaten public health7. Consumer reports indicate there are growing demands for safe THMP, and the need for regulators to monitor and recall the problematic herbal products in a timely manner.
There are considerable gaps in the commercial technology available to support pharmacovigilance in the THMP industry. The quality control methods recorded in the pharmacopoeias from different countries consist of the methods for testing preparations of crude drugs. For instance, analytical chemical methods focus on the target compounds, while physical methods mainly test the physical properties of different dosage forms. Species identity is limited to either DNA-based molecular diagnostics or morphological techniques that are not useful for processed herbal products. Molecular diagnostic tools that incorporate DNA identification techniques are useful for single ingredient authentication8, 9, 10. Multiplex PCR11, Roche 454 sequencing12, 13, and nucleotide signature14 have been involved in multiple ingredient authentication. The combination of analytical chemistry with molecular diagnostic DNA based tools is very useful for some herbal products that have challenging adulteration issues. For instance, researchers assessed the extent of adulteration in the raw herbal trade of Saraca asoca using DNA barcoding validated by NMR spectroscopic techniques11, and a DNA barcoding polymerase chain reaction (PCR) and a UHPLC-HR-MS integrated system were used to detect the existence of aristolochic acids-containing species in traditional medicines15. However, these methods are not sufficient for the oversight of THMPs with multiple herbal ingredients and there is an immediate need for the development of novel molecular diagnostic tools.
The use of a single herb for the treatment of a specific disease is uncommon; the vast majority of cases need to use THMPs to cure disease, which presents a considerable challenge for molecular diagnostic tools. DNA barcoding technology is an efficient method for the identification of THMP with one herbal species, which is now supported by considerable research of which it has been dubbed “a renaissance in herbal medicine identification”16. Several studies have documented the use of DNA barcoding in differentiation of traditional medicines with toxic adulterants15, the fraudulent market substitution of commercial Rhodiola products17, and overcoming authentication challenges in the herbal market18. Nowadays, DNA barcoding technology has been recorded in the Chinese Pharmacopoeia (2015 edition) and the British Pharmacopoeia (2017 edition) for the identification of herbal drugs19, 20. To date, most studies have been based on Sanger sequencing, which has high accuracy but low throughput and is limited to single species detection. This is a considerable problem, given that most Chinese, Ayurvedic, Japanese, Korean, and North American THMPs contain more than one species. The shortcomings of Sanger sequencing is due to the difficulty of obtaining results from a complex DNA template; multiple sequence cause overlapping trace files with poor resolution for base pair determination. There is a distinct need to develop a new sequencing technique to solve the issue of testing THMP with mixtures of different herbal species.
Third-generation sequencing may provide a solution for testing multiple species ingredients in THMP. This novel technology is called single molecule, real-time (SMRT) DNA sequencing and has been developed on platforms such as the PACBIO RS II (Pacific Biosciences of California, Inc., http://www.pacb.com/). The SMRT sequencing biotechnology has been used in sequencing various plants, including the whole-genome of Oropetium thomaeum21, the chloroplast genomes of three Fritillaria species (Beimu)22, the full-length transcriptome of Zea mays23 and Salvia miltiorrhiza (Danshen)24, and the potato late blight-resistance gene25, due to its extraordinarily long reads and high consensus accuracy for DNA sequencing. Moreover, through correction with next-generation sequencing (NGS) reads and/or self-correction via circular-consensus sequencing (CCS) reads, researchers have addressed the problem of the high error rate observed with SMRT sequencing26.
We selected Jiuwei Qianghuo Wan (JWQHW), a classical herbal prescription widely used in China for 800 years (Yuan Dynasty, 13th century), as the model for the application of the herbal species composition monitoring method through SMRT sequencing. As recorded in the Chinese Pharmacopoeia27, the methods applied to the evaluation of JWQHW are mainly microscopic identification, thin-layer chromatography (TLC) identification, and high performance liquid chromatography (HPLC) determination. However, molecular diagnostic related method is still relatively rare for detecting the herbal species of this traditional Chinese patent medicine (TCPM). To enhance the biomonitor of THMP, the metabarcoding and SMRT sequencing were conducted in this study.
2. Materials and methods
2.1. Reference sample collection and identification of the herbal ingredients of JWQHW
Nine herbal materials composing the JWQHW herbal preparation were collected from a drug store, including Notopterygii Rhizoma et Radix (Qianghuo), Saposhnikoviae Radix (Fangfeng), Atractylodis Rhizoma (Cangzhu), Asari Radix et Rhizoma (Xixin), Chuanxiong Rhizoma (Chuanxiong), Angelicae dahuricae Radix (Baizhi), Scutellariae Radix (Huangqin), Rehmanniae Radix (Dihuang), and Glycyrrhizae Radix et Rhizoma (Gancao) (Table 1). Besides, Panax ginseng C. A. Meyer was chosen as a positive control28, 29. All species were authenticated using morphological identification and DNA barcoding according to the record of the Chinese Pharmacopoeia27 and with reference to a vouchered Kew-IMPLAD (Institute of Medicinal Plant Development) set of authentic Chinese Materia Medica.
Table 1.
The botanical species source of each of the herbal materials in JWQHW (Chinese Pharmacopoeia, 2015 edition).
| Herbal material name (CP 2015) | Plant part | Botanical source | Family | Robustness of authentication methods used for the reference ingredients |
|
|---|---|---|---|---|---|
| DNA Barcoding | Morphological | ||||
| Notopterygii Rhizoma et Radix (Qianghuo) | Rhizome and root | Notopterygium franchetii H. Boissieu | Apiaceae | To species level (Notopterygium incisum) | To species level (Notopterygium incisum) |
| Notopterygium incisum K. C. Ting ex H. T. Chang | |||||
| Saposhnikoviae Radix (Fangfeng) | Root | Saposhnikovia divaricata (Turcz..) Schischk. | Apiaceae | To species level (Saposhnikovia divaricata) | To species level (Saposhnikovia divaricata) |
| Atractylodis Rhizoma (Cangzhu) | Rhizome | Atractylodes lancea (Thunb.) DC. | Asteraceae | – | To species level (Atractylodes lancea) |
| Atractylodes chinensis (DC.) Koidz. | |||||
| Asari Radix et Rhizoma (Xixin) | Root and rhizome | Asarum heterotropoides Fr. Schmidt var. mandshuricum (Maxim.) Kitag. | Aristolochiaceae | To genus level (Asarum heterotropoides var. mandshuricum, Asarum sieboldii var. seoulense, or Asarum sieboldii) | To genus level (Asarum) |
| Asarum sieboldii Miq. | |||||
| Asarum sieboldii Miq. var. seoulense Nakai | |||||
| Chuanxiong Rhizoma (Chuanxiong) | Rhizome | Ligusticum chuanxiong Hort. | Apiaceae | To species level (Ligusticum chuanxiong) | To species level (Ligusticum chuanxiong) |
| Angelicae dahuricae Radix (Baizhi) | Root | Angelica dahurica (Fisch. ex Hoffm.) Benth. et Hook. f. | Apiaceae | – | To species level (Angelica dahurica) |
| Angelica dahurica (Fisch. ex Hoffm.) Benth. et Hook. f. var. formosana (Boiss.) Shan et Yuan | |||||
| Rehmanniae Radix (Dihuang) | Root | Rehmannia glutinosa Libosch. | Scrophulariaceae | To species level (Rehmannia glutinosa) | To species level (Rehmannia glutinosa) |
| Scutellariae Radix (Huangqin) | Root | Scutellaria baicalensis Georgi | Lamiaceae | To species level (Scutellaria baicalensis) | To species level (Scutellaria baicalensis) |
| Glycyrrhizae Radix et Rhizoma (Gancao) | Root and rhizome | Glycyrrhiza uralensis Fisch. | Fabaceae | To species level (Glycyrrhiza uralensis) | To species level (Glycyrrhiza uralensis) |
| Glycyrrhiza inflata Bat. | |||||
| Glycyrrhiza glabra L. | |||||
2.2. Biomonitoring pipeline for THMPs using DNA metabarcoding and SMRT sequencing
2.2.1. Reference experimental mixtures production
The reference experimental mixture of JWQHW was formulated in the laboratory according to the methods described in the Chinese Pharmacopoeia27. It was processed as follows: first, the nine herbal materials were crushed into powder; second, the powder was sieved and mixed evenly; finally, the powder was mixed with water and molded into pills, which was marked RF01. Specifically, 10 g mixed powder was marked RF02; Panax ginseng powder was added to it with an equal amount of Asari Radix et Rhizoma (Xixin), which is present at the lowest percentage in the mixture (Supplementary Information Table S1), and the sample was mixed to homogeneity and molded into pills as above mentioned.
2.2.2. DNA extraction
DNA extraction was performed according to the DNA barcoding protocol recorded in the Chinese Pharmacopoeia with some changes in the beginning steps19. First, the sample quantity of RF02 was 120 mg per 2.0 mL EP tube. Second, the samples were washed in pre-wash buffer (composition: 100 mmol/L Tris–HCl, pH 8.0; 20 mmol/L EDTA, pH 8.0; 700 mmol/L NaCl; 2% PVP-40; 0.4% β-mercaptoethanol) as follows: 1000 μL pre-wash buffer was added to the tube containing the samples, which was then vibrated for 5 min and centrifuged for 3 min at 7500 rpm (Sigma I-14, Sigma Laborzentrifugen GmbH, Germany(Sigma I-14, Sigma Laborzentrifugen GmbH, Germany). Removed the supernatant and then repeated these steps until the supernatant was almost colorless (about 5—7 times). Genomic DNA was extracted using the Plant Genomic DNA Extraction Kit (Tiangen Biotech (Beijing) Co., Ltd., China). The water-bath time of the DNA extraction step was extended to 12 h at 56 °C. The genomic DNA was dissolved in 120 μL sterile ultrapure water. The other steps were the same as those recorded in the Chinese Pharmacopoeia19. DNA quality and concentration were quantified on a NanoDrop 2000 (Thermo Fisher Scientific Inc., USA).
2.2.3. DNA amplification and PCR product purification
To mark the sequences obtained from different regions, two 5-bp tags were designed and added to the 5′ end of the universal ITS2 and psbA-trnH primers (Supplementary Information Table S2). The PCRs of the ITS2 and psbA-trnH regions were carried out according to the Chinese Pharmacopoeia19 using 2 × Taq MasterMix (AidLab Biotechnologies Co., Ltd., China) with the annealing temperature increasing to 58 °C and 40 cycles. To improve the amplification efficiency, 1 μL Mg2+ (10 mmol/L, SBS Genetech Co., Ltd., China) was added per sample. The PCR products were electrophoresed on 2% agarose gel and purified with QIAquick Gel Extraction Kit (QIAGEN N. V., Germany). The purified PCR products were quantified and assessed using the Agilent 2100 Bioanalyzer (Agilent Technologies, Inc., USA) and Qubit (Thermo Fisher Scientific, Inc., USA).
2.2.4. Library construction and SMRT sequencing
The PACBIO RSII SMRT sequencing platform (Pacific Biosciences of California, Inc., USA, http://www.pacificbiosciences.com/) was used to sequence the ITS2 and psbA-trnH amplicons. In order to construct an SMRT sequencing library of the purified PCR amplicons, a SMRTbellTM Template Prep Kit 1.0 (part #100–259-100) was used. The templates were bound to P6-DNA polymerase and V2 primers using the DNA/polymerase Binding Kit P6 v2 (part #100-372-700). The MagBeads (part #100-133-600) were used to bind the complexes and transfer them to a 96-well PCR plate for one SMRT cell sequencing using C4 reagents (part # 100–356-200). The sequencing procedures were carried out according to the previous report22.
2.2.5. Sequencing data analysis and herbal species detection
Subreads were filtered using the SMRT Analysis Server 2.3.0 (Pacific Biosciences of California, Inc., USA). The filtered reads were run through the RS_ReadsOfInsert.1 protocol to produce CCS reads according to the following parameters: minimum full passes at 10, minimum prediction accuracy at 99%, minimum reads length of insert at 250 bp and maximum read length of insert at 600 bp. According to the tags, the CCS reads from RF02 were separated and data libraries were constructed using Perl scripts. The CCS reads from each library were clustered and redundant sequences were removed by CD-HIT software (http://weizhongli-lab.org/cd-hit/). The clustered reads were then identified in the DNA Barcoding System for Identifying Herbal Medicine (http://www.tcmbarcode.cn/en/) using Basic Local Alignment Search Tool (BLAST) to evaluate the species composition. MEGAN6, a powerful interactive species analysis tool, was used to cluster BLAST files and construct networks to identify the species composition according to the taxonomy30.
2.3. Reliability and repeatability of the pipeline
To test the reliability and repeatability of the pipeline, a parallel experiment was conducted with another reference experimental mixture, RF01, the composition of which was almost exactly the same as that of RF02, with Panax ginseng omitted. To distinguish between PCR amplicons derived from different samples, another two 5-bp tags were designed and added to the 5′ end of the universal primers of the ITS2 and psbA-trnH sequences.
2.4. Practical applications on the commercial JWQHW samples
Randomly purchased from different drug stores, three commercial JWQHW samples, which marked as JWQHW01 (Lot No.: 20141101), JWQHW02 and JWQHW03 (Lot No.: 20140501), were produced by the same manufacturer. Similarly, they were sampled and tested using the same method applied to RF02. Honey was used as the adhesive in the commercial JWQHW herbal preparation and was removed at the pre-wash step. As a result, the commercial samples were much larger than the RF02, at 300 mg per 2.0 mL EP tube. Additionally, six more 5-bp tags were designed and added to the primers. The remaining steps were exactly consistent with above mentioned method.
The definitions of terms used when describing different kind of samples are as outlined below. Herbal materials: the decoction pieces of the herbal ingredients of JWQHW which bought from drug store. The herbal materials were used to produce the reference JWQHW sample; Reference JWQHW sample: formulated by the herbal materials in the laboratory according to the methods described in the Chinese Pharmacopoeia27; Commercial JWQHW sample: different batches of samples produced by the same manufacturer, randomly purchased in different drug stores.
3. Results
3.1. Authentication of the nine herbal materials in Jiuwei Qianghuo Wan (JWQHW)
The nine herbal materials of JWQHW bought from the drug store were authenticated by DNA barcoding and morphological identification to ensure the accurate identification of the reference samples. The ITS2 and psbA-trnH regions were amplified and bi-directionally sequenced using Sanger sequencing (Supplementary Information Table S3). The results showed that seven of the herbal materials—Notopterygii Rhizoma et Radix (Qianghuo), Saposhnikoviae Radix (Fangfeng), Scutellariae Radix (Huangqin), Rehmanniae Radix (Dihuang), Asari Radix et Rhizoma (Xixin), Chuanxiong Rhizoma (Chuanxiong) and Glycyrrhizae Radix et Rhizoma (Gancao)—yielded perfect ITS2 sequences. All sequences obtained by Sanger sequencing were identified through the DNA Barcoding System for Identifying Herbal Medicine using BLAST. Among these herbal materials, Asari Radix et Rhizoma (Xixin) was identified as Asarum heterotropoides var. mandshuricum, As. sieboldii var. seoulense, or As. sieboldii. That was because the ITS2 sequences of the three species were the same. The results confirmed that the samples were of the species recorded in the Chinese Pharmacopoeia27 (Table 1). Atractylodis Rhizoma (Cangzhu) and Angelicae dahuricae Radix (Baizhi) failed due to the lack of PCR amplification. A morphologist authenticated these two herbal materials. Thus, the accurate identification of the reference samples was guaranteed.
3.2. SMRT pipeline for monitoring the herbal species composition of THMP
In this study, a method of the monitoring of the herbal species composition of natural herbal products was established. The pipeline of this method is shown in Fig. 1. The two reference samples (RF01–02) and three commercial samples (JWQHW01–03) were treated according to this method to obtain the ITS2 and psbA-trnH sequences PCR products. All PCR products from the five samples were purified and quantified exactly, and then mixed equivalently for single molecule real-time sequencing.
Figure 1.
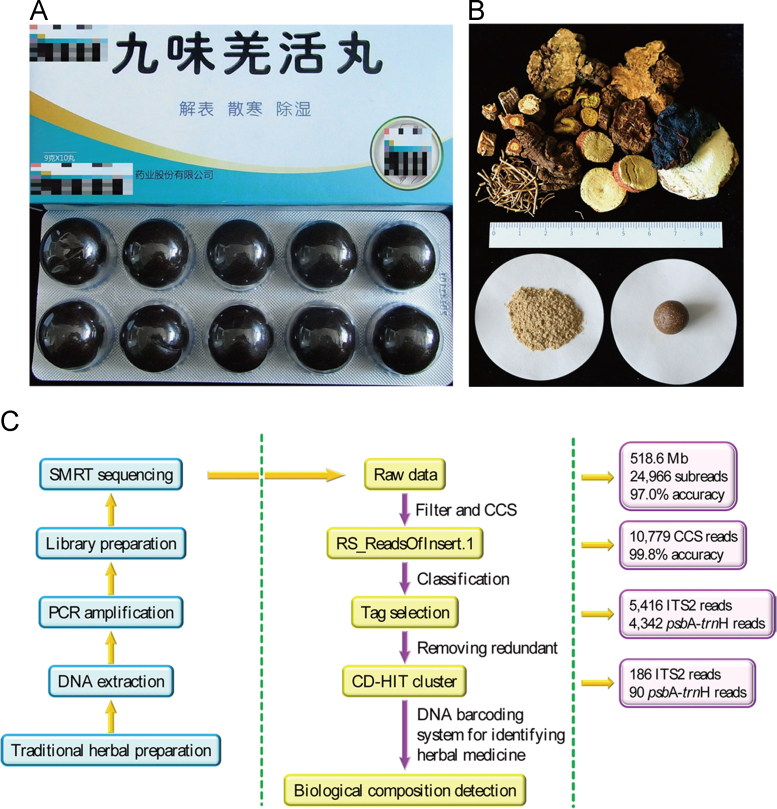
Biomonitoring method for traditional herbal medical products. (A) Commercial samples of JWQHW used in the experiment. (B) Reference samples of JWQHW used in the experiment. (C) Pipeline of the method.
A total of 518.6 MB bases in 24,996 reads were obtained using SMRT sequencing. After quality control, the SMRT sequencer generated 10,779 trimmed and filtered CCS reads for all samples. Details of the CCS reads are given in Supplementary Information Table S4. Among all reads, 1,021 were unmatched and deleted after tag selection. Ultimately, 5,416 and 4,342 CCS reads were identified as ITS2 and psbA-trnH regions, respectively (Table 2). After redundant sequences were removed, 276 clustered sequences were obtained, included 186 and 90 sequences of the ITS2 and psbA-trnH regions, respectively.
Table 2.
Summary of sequence data generated on the PacBio RS platform.
| Sample ID | CCS reads number |
Cluster number |
||||
|---|---|---|---|---|---|---|
| Total | ITS2 | psbA-trnH | Total | ITS2 | psbA-trnH | |
| JWQHW01 | 1330 | 893 | 437 | 70 | 43 | 27 |
| JWQHW02 | 1194 | 1187 | 7 | 57 | 53 | 4 |
| JWQHW03 | 1570 | 1208 | 362 | 79 | 54 | 25 |
| RF01 | 2895 | 1015 | 1880 | 33 | 18 | 15 |
| RF02 | 2769 | 1113 | 1656 | 37 | 18 | 19 |
| Total | 9758 | 5416 | 4342 | 276 | 186 | 90 |
3.3. Analysis of the herbal species composition of the reference JWQHW samples
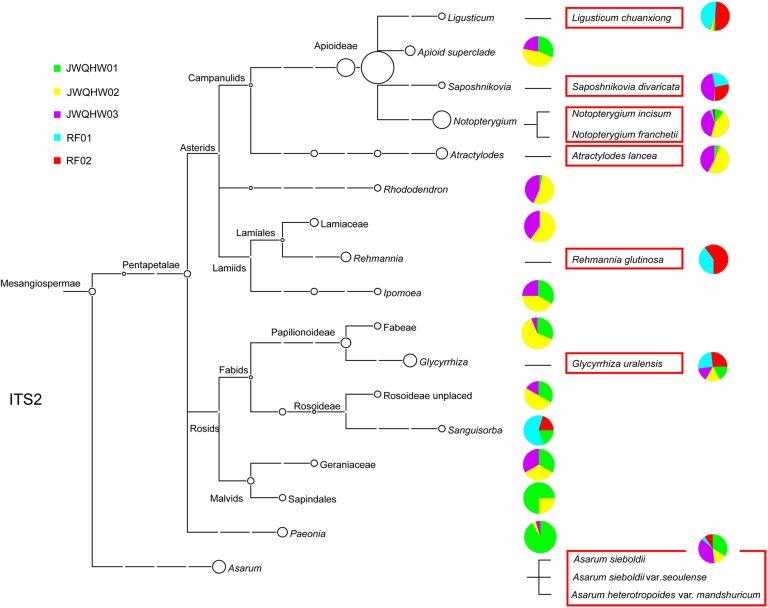
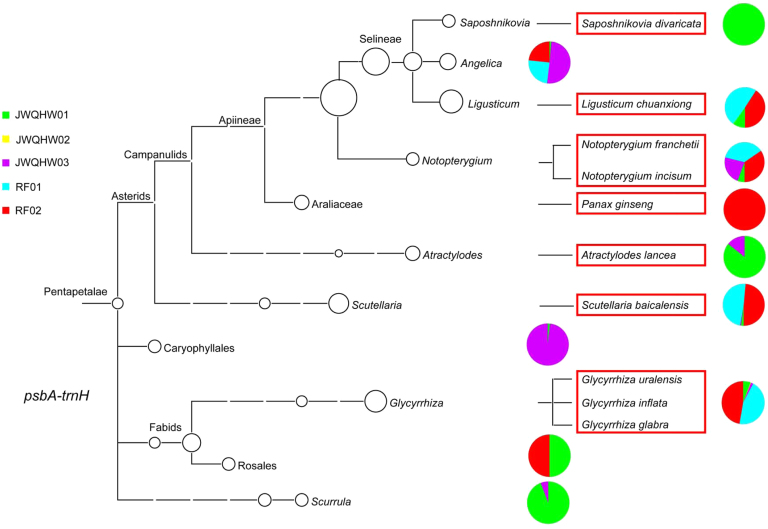
We obtained 5,664 CCS reads from the two reference JWQHW samples (RF01 and RF02) for the ITS2 and psbA-trnH loci. Using the ITS2 sequence, six prescribed herbal materials were detected. Notopterygium incisum (Qianghuo), Saposhnikovia divaricata (Fangfeng), Ligusticum chuanxiong (Chuanxiong), Rehmannia glutinosa (Dihuang), Asarum heterotropoides var. mandshuricum, As. sieboldii var. seoulense, or As. sieboldii (Xixin), and Glycyrrhiza uralensis (Gancao), were detected in both of the reference samples. Scutellariae Radix (Huangqin), Atractylodis Rhizoma (Cangzhu), Angelicae dahuricae Radix (Baizhi), and Panax ginseng were not found (Table 3, Fig. 2 and Supplementary Information Table S5). The auxiliary DNA barcode was used for the detection of the psbA-trnH sequence of Scutellaria baicalensis (Huangqin) and thus augmented the testing results. The positive control, Panax ginseng, was also detected via the psbA-trnH sequence in RF02 (Table 3, Fig. 3 and Supplementary Information Table S5). Neither the ITS2 nor the psbA-trnH sequence of Atractylodis Rhizoma (Cangzhu) and Angelicae dahuricae Radix (Baizhi) was detected using SMRT sequencing, which was consistent with the findings obtained from the herbal materials using Sanger sequencing. The phylogenetic trees constructed by the ITS2 and psbA-trnH regions show the genetic relationship and species coverage of the detected species (Figure 2, Figure 3).
Table 3.
Detection of the prescribed herbal materials in five JWQHW samples by SMRT sequencing based on the ITS2 and psbA-trnH regions.
| Herbal material name | JWQHW01 |
JWQHW02 |
JWQHW03 |
RF01 |
RF02 |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| ITS2 | psbA-trnH | ITS2 | psbA-trnH | ITS2 | psbA-trnH | ITS2 | psbA-trnH | ITS2 | psbA-trnH | |
| Notopterygii Rhizoma et Radix (Qianghuo) | √ | √ | √ | √ | √ | √ | √ | √ | √ | |
| Saposhnikoviae Radix (Fangfeng) | √ | √ | √ | √ | √ | |||||
| Atractylodis Rhizoma (Cangzhu) | √ | √ | √ | √ | √ | |||||
| Asari Radix et Rhizoma (Xixin) | √ | √ | √ | √ | √ | |||||
| Chuanxiong Rhizoma (Chuanxiong) | √ | √ | √ | √ | √ | √ | √ | √ | √ | √ |
| Scutellariae Radix (Huangqin) | √ | √ | √ | √ | ||||||
| Rehmanniae Radix (Dihuang) | √ | √ | ||||||||
| Glycyrrhizae Radix et Rhizoma (Gancao) | √ | √ | √ | √ | √ | √ | √ | √ | √ | √ |
| Angelicae Dahuricae Radix (Baizhi) | ||||||||||
| Panax ginseng | √ | |||||||||
Figure 2.
Phylogeny and relative abundances of species detected in five JWQHW samples based on ITS2 sequences. Pie chart shows the fraction of CCS reads of each species in five samples.
Figure 3.
Phylogeny and relative abundances of species detected in five JWQHW samples based on psbA-trnH sequences. Pie chart shows the fraction of CCS reads of each species in five samples.
3.4. Biomonitoring for the commercial JWQHW samples
A total of 4,094 CCS reads, including the ITS2 and psbA-trnH regions, from the three commercial samples (JWQHW01-03) were obtained in this study. Of all CCS reads, 2791 were identified as authentically derived from the prescribed herbal ingredients; the others were considered contaminants.
Using the ITS2 sequence, Notopterygium incisum and N. franchetii (Qianghuo), Ligusticum chuanxiong (Chuanxiong), Asarum heterotropoides var. mandshuricum, As. sieboldii var. seoulense, or As. sieboldii (Xixin), Atractylodes lancea (Cangzhu), and Glycyrrhiza uralensis (Gancao) were detected in all three samples. Scutellaria baicalensis (Huangqin), Rehmannia glutinosa (Dihuang), and Angelica dahurica or An. dahurica var. formosana (Baizhi) were not found (Table 3, Fig. 2 and Supplementary Information Table S5). The ITS2 sequence of Saposhnikovia divaricata (Fangfeng) was detected in JWQHW02 and JWQHW03 while the psbA-trnH sequence was detected in JWQHW01. Similarly, Scutellaria baicalensis (Huangqin) was detected in JWQHW01 and 03 based on the psbA-trnH sequence (Table 3, Fig. 3 and Supplementary Information Table S5). With the combination of the two DNA barcodes, all prescribed herbal ingredients were detected in the commercial JWQHW samples except Rehmanniae Radix (Dihuang) and Angelicae dahuricae Radix (Baizhi). This result suggests that these species might be highly processed such that the genomic DNA was too degraded to be amplified.
In addition to these prescribed herbal ingredients, several other potential contaminant species were detected. The CCS reads mapping to the ITS2 and psbA-trnH regions were classified into 22 families with 47 genera and 12 families with 18 genera, respectively (Supplementary Information Table S6). For instance, Atractylodes koreana (Nakai) Kitamura, an adulterant of Atractylodis Rhizoma (Cangzhu), was found in JWQHW01. Atractylodes macrocephala Koidzumi was found in JWQHW02. Species belonging to Angelica, Peucedanum, Paeonia, Ipomoea, Rhododendron and Vicia were found in all three commercial samples. Some Scurrula and Taxillus species were found in JWQHW01 and 03. Among these potential contaminant species, Angelica amurensis Schischkin, Peucedanum ledebourielloides K. F. Fu, and Angelica polymorpha Maximowicz were three most common species detected.
4. Discussion
4.1. A reliable DNA authentication test for the biomonitor of THMP
The Herbal industry has been lacking a biotechnological tool for detecting multiple species ingredients in natural herbal products. Although there are reports of the application of SMRT sequencing to whole-genome21, chloroplast genome22, 31, transcriptome sequencing23, and other uses, this method has not been widely used in sequencing THMP. Regulators and consumers are becoming familiar with the application of Next Generation Sequencing (NGS) or high-throughput sequencing (HTS) detecting adulteration in THMP12, 13, 32. Published literature on NGS indicates there are considerable problems with HTS that present an immediate impediment to generating scientifically valid test results, as are necessary for commercial use of this tool to verify herbal ingredient identity; HTS may indicate presence of species in a sample due only to detection of incidental DNA, and may also over-estimate the amounts of incidental DNA due to polymerase chain reaction (PCR) amplification bias, which may skew estimates of species abundance33, 34, 35, 36. However, use of HTS can be useful for making a list of herbal species that needs further verification. A study of traditional Chinese medicines (TCMs) seized by Australia border protection officials showed that HTS is an efficient and cost-effective way to oversee the legality and safety of highly processed TCM products12. Similar use of HTS has been used to authenticate the species ingredient of TCM Liuwei Dihuang Wan13. However, while HTS has sufficient sequencing depth, the read length for the most common platform (approximately 2 × 100 − 250 bp), it is too short to cover a whole DNA barcode, making it necessary to assemble short reads. Comparatively, the read length obtained from SMRT sequencing can reach 15 kb, which is adequate to cover any DNA barcode. This study used SMRT sequencing to sequence the ITS2 and psbA-trnH amplicons, and the raw data were filtered directly without subread assembly to obtain CCS reads to ensure high data quality.
Combining the ITS2 and psbA-trnH sequences revealed seven of the prescribed herbal ingredients in reference sample RF02 (Table 3). The positive control, Panax ginseng, was also detected successfully. These results indicated that SMRT sequencing was sensitive enough to detect all the prescribed herbal ingredients theoretically. In addition, the psbA-trnH sequence was a perfect supplementary to ITS2 sequence for species identification16, 37. The repeat experiment with RF01 obtained consistent results with those of RF02, which confirmed the reliability, repeatability and stability of the SMRT sequencing to monitor the herbal species composition of THMPs. This pipeline needs commercial validation in a ring study before it can be used commercially.
The attempt of commercial sample validation using this pipeline was successful. All experimental and data analysis steps were in strict accordance with the pipeline. The results showed that with the combination of ITS2 and psbA-trnH, six, five, and seven prescribed herbal materials were detected in JWQHW01, JWQHW02, and JWQHW03, respectively (Table 3). These results indicate that the biomonitoring method can be applied to commercial samples and consistent with the latest report of our team. In previous study, Jia et al. 38 performed a primary study of Yimu Wan using SMRT sequencing, which only contained four ingredients. While in resent study, we tried to apply this method on THMPs which contained nearly ten ingredients or more to test the identification ability. The results showed that it has the potential as a powerful tool for THMP quality control. There are 1493 TCPMs recorded in Chinese Pharmacopoeia, nearly 900 of which contain herb powders. This assortment indicates that the practical application of this biomonitoring method might be potentially implemented on approximately 60% of TCPMs, making it a golden standard for quality control. Further testing with the labs of our collaborators in India, USA and Canada will soon follow. In addition, the Roche 454 and Ion Torrent sequencing platforms could also be used in the identification of THMPs12, 13, 32. Nucleotide signature is a good option in the case of boiled or steamed ingredients; the genomic DNA of these ingredients may be highly degraded, resulting in ITS2 and psbA-trnH amplicons that are difficult to obtain14. The biomonitoring method is also applicable for products of unknown compositions. The species composition determination is based on the ITS2 and psbA-trnH sequences that result from the CCS reads. A list of all potential species that are present in all of the samples could be provided after data analysis. The identities of actual ingredients and contaminants could be logically achieved using the pharmacopoeias, which is based on the standard DNA barcoding reference database that was constructed in a previous study16.
4.2. Satisfactory identification ability of the biomonitoring method
In this study, nine herbal materials of JWQHW were authenticated to ensure the validity of the reference samples. Considering that all the materials were decoction pieces (i.e., sliced or cut dried herbs), the respective genomic DNA may have been damaged to different degrees, which would influence the PCR and sequencing steps. To ensure the reliability of the reference samples, identification was also supported by morphological examination.
As shown above, this method could be used to detect the prescribed herbal materials and the positive control in the reference samples, a finding consistent with the Sanger sequencing results. Seven of nine prescribed herbal materials were detected in both of the reference samples; Chinese Pharmacopoeia27 indicates that the herbal materials in JWQHW may be identified by several techniques: microscopy is used to test for five herbal materials, Angelicae dahuricae Radix (Baizhi), Saposhnikoviae Radix (Fangfeng), Scutellariae Radix (Huangqin), Rehmanniae Radix (Dihuang), and Glycyrrhizae Radix et Rhizoma (Gancao); TLC is used to test for four herbal materials, Notopterygii Rhizoma et Radix (Qianghuo), Atractylodis Rhizoma (Cangzhu), Chuanxiong Rhizoma (Chuanxiong), and Glycyrrhizae Radix et Rhizoma (Gancao); and HPLC is used to test for the content of baicalin (Scutellariae Radix (Huangqin)). There is no specific method for the detection of Asari Radix et Rhizoma (Xixin), while the biomonitoring method detected the existence of Asari Radix et Rhizoma (Xixin). This result indicated the powerful detection ability of the biomonitoring method. Moreover, Atractylodis Rhizoma (Cangzhu), which could not be detected by Sanger sequencing and was not found in the reference samples, was detected in all three commercial samples using SMRT sequencing. Although few CCS reads were found in the commercial samples, their presence suggests that the genomic DNA quality of Atractylodis Rhizoma (Cangzhu) from the commercial samples was better than the reference samples, enabling it to be amplified and sequenced in the experiment. Additionally, Rehmanniae Radix (Dihuang) was not found in any of the commercial samples, suggesting that the manufacturer may have added processed rather than crude materials to the herbal preparation. These results indicate that the quality of genomic DNA was the most important factor influencing the outcome of this method.
Furthermore, the SMRT sequencing results indicated that the adulterant species were present in the reference samples, although the herbal materials had been authenticated. Sanguisorbae Radix (Diyu, Sanguisorba officinalis L.), an adulterant of Notopterygii Rhizoma et Radix (Qianghuo), was detected in RF01 and RF02. One possible explanation is that when we authenticated the samples, we chose one small piece at random; thus, contaminants were not detected. Similar results were found in JWQHW01. Notopterygii Rhizoma et Radix (Qianghuo, 125.0–150.0 RMB/kg) is an endangered herbal drug, the price of which was 16 to 25 times that of Sanguisorbae Radix (Diyu, 6.0–8.0 RMB/kg). This suggests possible fraudulent product substitution.
Moreover, a total of 57 plant genera (combining ITS2 and psbA-trnH) were found in the commercial samples, reflecting the powerful identification and detection ability of the SMRT method. The possible explanations for this contamination can be classified into the following categories: (1) intentional addition to the herbal preparations during processing procedure, as for Atractylodes koreana and Sanguisorba officinalis, the adulterants of Atractylodis Rhizoma (Cangzhu) and Notopterygii Rhizoma et Radix (Qianghuo), respectively; (2) unintentional mixing into the herbal preparation, as for the species belonging to Paeonia, Ipomoea, Scurrula, and Taxillus; these species were commonly used in many other herbal preparations produced by the same manufacturer, and thus could easily cause cross-contamination during crushing. The results indicated that contamination was very difficult to avoid in herbal preparations. However, if the degree of adulteration was small, it could hardly be detected by TLC, HPLC or other conventional methods. Hence, an evaluation of the herbal species composition was essential parts of pharmacovigilance.
4.3. Errors in SMRT sequencing do not affect species identification
Previous research has indicated that the quality value (QV) of each base call in SMRT sequencing was significantly increased in the CCS sub-reads, allowing highly confident single nucleotide polymorphism (SNP) detection at a low variant frequency22. This advantage made the sequencing results suitable for use in DNA barcoding. As the ITS2 region is present in multiple copies in the genome, there are potentially dozens of different sequences in a single sample28. All of these copies may be amplified by the PCR amplification, but only one or a few dominant mutations of the ITS2 sequence from each sample could be sequenced using the Sanger method. By contrast, SMRT sequencing could sequence all the copies easily, while CCS could ensure QV stability. In other words, SMRT sequencing reflects the real diversity of each template. Fortunately, this diversity does not affect the species identification. Considering, for example, Notopterygii Rhizoma et Radix (Qianghuo), although different haplotypes of the ITS2 sequences of Notopterygium incisum and N. franchetii were found in the sequence pool, there was no misidentification of these two species. This result could be explained by the multicopy nature and compensatory base changes of the ITS2 sequences. The sequence identity ranges were 91.5–99.6% and 92.4–100.0%, respectively. Although we identified the sequences through BLAST and sequence alignment, demonstrating that the error in SMRT sequencing does not affect the species identification, further verification is needed in studies of other THMP.
Acknowledgments
We thank Dr. Meng Lei for providing help in the sequencing task. This study was supported by the National Natural Science Foundation of China (Grant No. 81373922) and Chinese Academy of Medical Sciences (CAMS) Innovation Fund for Medical Sciences (Grant No. CIFMS, 2016-I2M-3–016).
Footnotes
Peer review under responsibility of Institute of Materia Medica, Chinese Academy of Medical Sciences and Chinese Pharmaceutical Association.
Supplementary data associated with this article can be found in the online version at http://dx.doi.org/10.1016/j.apsb.2017.10.001.
Contributor Information
Jingyuan Song, Email: songjingyuan@hotmail.com.
Steven G. Newmaster, Email: snewmast@uoguelph.ca.
Appendix A. Supplementary material
Supplementary material
.
References
- 1.Grollman A.P., Marcus D.M. Global hazards of herbal remedies: lessons from Aristolochia. The lesson from the health hazards of Aristolochia should lead to more research into the safety and efficacy of medicinal plants. Embo Rep. 2016;17:619–625. doi: 10.15252/embr.201642375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Global Industry Analysts. The global herbal supplements and remedies market: trends, drivers and projections 2015. Available from: 〈http://www.strategyr.com/MarketResearch/Herbal_Supplements_and_Remedies_Market_Trends.asp〉.
- 3.Barbour V., Clark J., Jones S., Norton M., Simpson P., Veitch E. Why drug safety should not take a back seat to efficacy. PLoS Med. 2011;8:e1001097. doi: 10.1371/journal.pmed.1001097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Preston C., Valdez M.L., Bond K. Strengthening medical product regulation in low- and middle-income countries. PLoS Med. 2012;9:e1001327. doi: 10.1371/journal.pmed.1001327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.World Health Organization. Counterfeit medicines: frequently asked questions. 2009:2016. Available from: 〈http://www.who.int/medicines/services/counterfeit/faqs/QACounterfeit-October2009.pdf〉.
- 6.Chan T.Y. Herbal medicines induced anticholinergic poisoning in Hong Kong. Toxins. 2016;8:80. doi: 10.3390/toxins8030080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Boer H.J., Ichim M.C., Newmaster S.G. DNA barcoding and pharmacovigilance of herbal medicines. Drug Saf. 2015;38:611–620. doi: 10.1007/s40264-015-0306-8. [DOI] [PubMed] [Google Scholar]
- 8.Shanmughanandhan D., Ragupathy S., Newmaster S.G., Mohanasundaram S., Sathishkumar R. Estimating herbal product authentication and adulteration in India using a vouchered, DNA-based biological reference material library. Drug Saf. 2016;39:1211–1227. doi: 10.1007/s40264-016-0459-0. [DOI] [PubMed] [Google Scholar]
- 9.Han J., Pang X., Liao B., Yao H., Song J., Chen S. An authenticity survey of herbal medicines from markets in China using DNA barcoding. Sci Rep. 2016;6:18723. doi: 10.1038/srep18723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Newmaster S.G., Grguric M., Shanmughanandhan D., Ramalingam S., Ragupathy S. DNA barcoding detects contamination and substitution in North American herbal products. BMC Med. 2013;11:222. doi: 10.1186/1741-7015-11-222. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 11.Dalmasso A., Fontanella E., Piatti P., Civera T., Rosati S., Bottero M.T. A multiplex PCR assay for the identification of animal species in feedstuffs. Mol Cell Probes. 2004;18:81–87. doi: 10.1016/j.mcp.2003.09.006. [DOI] [PubMed] [Google Scholar]
- 12.Coghlan M.L., Haile J., Houston J., Murray D.C., White N.E., Moolhuijzen P. Deep sequencing of plant and animal DNA contained within traditional Chinese medicines reveals legality issues and health safety concerns. PLoS Genet. 2012;8:e1002657. doi: 10.1371/journal.pgen.1002657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng X., Su X., Chen X., Zhao H., Bo C., Xu J. Biological ingredient analysis of traditional Chinese medicine preparation based on high-throughput sequencing: the story for Liuwei Dihuang Wan. Sci Rep. 2014;4:5147. doi: 10.1038/srep05147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao Z., Liu Y., Wang X., Song J., Chen S., Ragupathy S. Derivative technology of DNA barcoding (nucleotide signature and SNP double peak methods) detects adulterants and substitution in Chinese Patent Medicines. Sci Rep. 2017;7:5858. doi: 10.1038/s41598-017-05892-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu L., Sun W., Wang B., Zhao H., Li Y., Cai S. An integrated system for identifying the hidden assassins in traditional medicines containing aristolochic acids. Sci Rep. 2015;5:11318. doi: 10.1038/srep11318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen S., Pang X., Song J., Shi L., Yao H., Han J. A renaissance in herbal medicine identification: from morphology to DNA. Biotechnol Adv. 2014;32:1237–1244. doi: 10.1016/j.biotechadv.2014.07.004. [DOI] [PubMed] [Google Scholar]
- 17.Xin T., Li X., Yao H., Lin Y., Ma X., Cheng R. Survey of commercial Rhodiola products revealed species diversity and potential safety issues. Sci Rep. 2015;5:8337. doi: 10.1038/srep08337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mishra P., Kumar A., Nagireddy A., Mani D.N., Shukla A.K., Tiwari R. DNA barcoding: an efficient tool to overcome authentication challenges in the herbal market. Plant Biotechnol J. 2016;14:8–21. doi: 10.1111/pbi.12419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chinese Pharmacopoeia Commission . Vol I. China Medical Science Press; Beijing: 2015. (Pharmacopoeia of the People's Republic of China). [Google Scholar]
- 20.The British Pharmacopoeia Commission. SC VII D. DNA Barcoding as a tool for Botanical Identification of Herbal Drugs. London: TSO: British Pharmacopoeia; 2017 Available from: 〈https://www.pharmacopoeia.com〉
- 21.VanBuren R., Bryant D., Edger P.P., Tang H., Burgess D., Challabathula D. Single-molecule sequencing of the desiccation-tolerant grass Oropetium thomaeum. Nature. 2015;527:508–511. doi: 10.1038/nature15714. [DOI] [PubMed] [Google Scholar]
- 22.Li Q., Li Y., Song J., Xu H., Xu J., Zhu Y. High-accuracy de novo assembly and SNP detection of chloroplast genomes using a SMRT circular consensus sequencing strategy. New Phytol. 2014;204:1041–1049. doi: 10.1111/nph.12966. [DOI] [PubMed] [Google Scholar]
- 23.Wang B., Tseng E., Regulski M., Clark T.A., Hon T., Jiao Y. Unveiling the complexity of the maize transcriptome by single-molecule long-read sequencing. Nat Commun. 2016;7:11708. doi: 10.1038/ncomms11708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu Z., Peters R.J., Weirather J., Luo H., Liao B., Zhang X. Full-length transcriptome sequences and splice variants obtained by a combination of sequencing platforms applied to different root tissues of Salvia miltiorrhiza and tanshinone biosynthesis. Plant J. 2015;82:951–961. doi: 10.1111/tpj.12865. [DOI] [PubMed] [Google Scholar]
- 25.Witek K., Jupe F., Witek A.I., Baker D., Clark M.D., Jones J.D. Accelerated cloning of a potato late blight-resistance gene using RenSeq and SMRT sequencing. Nat Biotechnol. 2016;34:656–660. doi: 10.1038/nbt.3540. [DOI] [PubMed] [Google Scholar]
- 26.Au KF, Sebastiano V, Sebastiano PT, Durruthy JD, Lee L, Williams BA, et al. Characterization of the human ESC transcriptome by hybrid sequencing. Proc Natl Acad Sci U S A 2013;110:E4821--30 [DOI] [PMC free article] [PubMed]
- 27.Chinese Pharmacopoeia Commission . Vol I. China Medical Science Press; Beijing: 2010. pp. 478–479. (Pharmacopoeia of the People's Republic of China). [Google Scholar]
- 28.Song J., Shi L., Li D., Sun Y., Niu Y., Chen Z. Extensive pyrosequencing reveals frequent intra-genomic variations of internal transcribed spacer regions of nuclear ribosomal DNA. PLoS One. 2012;7:e43971. doi: 10.1371/journal.pone.0043971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen X., Liao B., Song J., Pang X., Han J., Chen S. A fast SNP identification and analysis of intraspecific variation in the medicinal Panax species based on DNA barcoding. Gene. 2013;530:39–43. doi: 10.1016/j.gene.2013.07.097. [DOI] [PubMed] [Google Scholar]
- 30.Huson D.H., Auch A.F., Qi J., Schuster S.C. MEGAN analysis of metagenomic data. Genome Res. 2007;17:377–386. doi: 10.1101/gr.5969107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen X., Li Q., Li Y., Qian J., Han J. Chloroplast genome of Aconitum barbatum var. puberulum (Ranunculaceae) derived from CCS reads using the PacBio RS platform. Front Plant Sci. 2015;6:42. doi: 10.3389/fpls.2015.00042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ivanova N.V., Kuzmina M.L., Braukmann T.W., Borisenko A.V., Zakharov E.V. Authentication of herbal supplements using next-generation sequencing. PLoS One. 2016;11:e156426. doi: 10.1371/journal.pone.0156426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Coissac E., Riaz T., Puillandre N. Bioinformatic challenges for DNA metabarcoding of plants and animals. Mol Ecol. 2012;21:1834–1847. doi: 10.1111/j.1365-294X.2012.05550.x. [DOI] [PubMed] [Google Scholar]
- 34.Burns M., Wiseman G., Knight A., Bramley P., Foster L., Rollinson S. Measurement issues associated with quantitative molecular biology analysis of complex food matrices for the detection of food fraud. Analyst. 2016;141:45–61. doi: 10.1039/c5an01392e. [DOI] [PubMed] [Google Scholar]
- 35.Schrijver I., Aziz N., Farkas D.H., Furtado M., Gonzalez A.F., Greiner T.C. Opportunities and challenges associated with clinical diagnostic genome sequencing: a report of the association for molecular pathology. J Mol Diagn. 2012;14:525–540. doi: 10.1016/j.jmoldx.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thudi M., Li Y., Jackson S.A., May G.D., Varshney R.K. Current state-of-art of sequencing technologies for plant genomics research. Brief Funct Genom. 2012;11:3–11. doi: 10.1093/bfgp/elr045. [DOI] [PubMed] [Google Scholar]
- 37.Chen S., Yao H., Han J., Liu C., Song J., Shi L. Validation of the ITS2 region as a novel DNA barcode for identifying medicinal plant species. PLoS One. 2010;5:e8613. doi: 10.1371/journal.pone.0008613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jia J., Xu Z., Xin T., Shi L., Song J. Quality control of the traditional patent medicine Yimu Wan based on SMRT sequencing and DNA barcoding. Front Plant Sci. 2017;8:926. doi: 10.3389/fpls.2017.00926. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material