Abstract
Advances in omics technologies — such as genomics, transcriptomics, proteomics and metabolomics — have begun to enable personalized medicine at an extraordinarily detailed molecular level. Individually, these technologies have contributed medical advances that have begun to enter clinical practice. However, each technology individually cannot capture the entire biological complexity of most human diseases. Integration of multiple technologies has emerged as an approach to provide a more comprehensive view of biology and disease. In this Review, we discuss the potential for combining diverse types of data and the utility of this approach in human health and disease. We provide examples of data integration to understand, diagnose and inform treatment of diseases, including rare and common diseases as well as cancer and transplant biology. Finally, we discuss technical and other challenges to clinical implementation of integrative omics.
The rapidly decreasing costs of high-throughput sequencing and other massively parallel technologies, such as mass spectrometry, are enabling their use in clinical research and clinical practice. Exome and genome sequencing are already being used to aid diagnoses, particularly of rare diseases1–3, to inform cancer treatment and progression and, in early efforts, to create predictive models of disease in healthy individuals4–6. Numerous research efforts and companies are focusing on genome-wide profiles of genetic, gene expression and other omics data, such as the microbiome (BOX 1), as biomarkers for disease (see TABLE 1 for details). For instance, genome-wide association studies (GWAS) have been successful in identifying risk loci for disease. However, in many cases, the causal variant or gene is not identified7. Here, other omics technologies can provide a useful glimpse into the precise pathophysiology of the disease. Experiments generating data that are more proximal to an organismal phenotype, such as proteomics, can be expensive and are often not comprehensive, and a challenge remains to distinguish the causal origin of a disease. Thus, except in rare cases, no single technology can capture the complexity of the molecular events that lead to human disease.
Box 1. Incorporating the microbiome into integrative omics.
The microbiome has been associated with many common human diseases; however, an additional complication exists, as the direction of causality is not clear. Whereas causality is simple in genomic data, where (with the exception of cancer processes that cause mutations) DNA influences phenotypes, it is more difficult to disentangle whether microbiome composition is a cause or effect of disease, as these studies require expensive longitudinal or interventional experiments or mouse models that may not provide comprehensive insight into human biology. Nonetheless, it is very clear that patients with diseases, such as inflammatory bowel disease110,111, type 2 diabetes112 and obesity113,114, have different microbiome profiles from those of healthy controls. In addition, the microbiome has a strong influence on immune function, which in some cases has been putatively causally linked to disease in animal models (reviewed in REF. 115).
As our understanding of the microbiome progresses, integrative analysis of this and other omics technologies is certain to advance our understanding of human disease. Recently, human genetic profiles have been shown to influence overall gut microbiota composition116,117, which could suggest putative causal explanations for some disease-associated genetic loci118 (for recent reviews, see REFS 119,120). Additionally, interactions between human genetics and microbiomes have been shown to influence disease, highlighting the potential for simultaneous interrogation of the two profiles121. Likewise, metabolic signalling between hosts and their microbiomes has become an area of active research, and there is increasing evidence that metabolite influences from gut bacteria may play a role in human disease122. Thus, it is likely that integrated analysis across genome, metabolome, microbiome and other omics profiles will prove beneficial for managing health and disease.
Table 1.
Data types for integrative omics
| Data type | Large-scale research efforts | Utility and advantages | Major caveats |
|---|---|---|---|
| Genetic variation | Many GWAS consortia, 1000 Genomes, gnomAD and UK Biobank | Unbiased source of genetic basis of disease and direct inference of causality | At least one step removed from the phenotype |
| Epigenetics | ENCODE and Roadmap Epigenomics Project | Functional impact and typically easy to infer causality | Not applicable for all phenotypes |
| Gene expression | GTEx and GEUVADIS | Inexpensive assay for an intermediate step towards the phenotype | Not applicable for all phenotypes |
| Proteomics and metabolomics | CPTAC, EDRN and Common Fund | Likely to be very close to the phenotype | Expensive and difficult to scale (proteomics) |
| Microbiome | Human Microbiome Project | Likely to be very close to the phenotype and measures a combination of genetic and environmental influences | Combination of genetic and environmental influences makes it difficult to infer the direction of causality |
In this table, ‘phenotype’ refers to an organismal phenotype. CPTAC, Clinical Proteomic Tumour Analysis Consortium; EDRN, Early Detection Research Network; ENCODE, Encyclopedia of DNA Elements; GEUVADIS, Genetic European Variation in Health and Disease; gnomAD, Genome Aggregation Database; GTEx, Genotype–Tissue Expression; GWAS, genome-wide association study.
Ideally, different technologies would be combined both to help diagnose disease and to create a holistic picture of human phenotypes and disease. However, implementation of multi-omics data introduces new informatics and interpretation challenges. Specifically, novel analytical and statistical methods are needed for combining disparate data sets, as well as standardized quality control metrics. Additionally, the field must address challenges in the interpretation of molecular events and, accordingly, their actionability and whether they can guide therapeutics and clinical care.
Below, we describe ways in which integrative omics can impact medicine by helping to manage health, as well as diagnose and treat disease. We discuss preclinical and clinical applications for rare Mendelian diseases, such as muscular dystrophy, and more common diseases, such as autism and Alzheimer disease. Furthermore, we investigate the use of multiple levels of omics technologies in cancer diagnosis and treatment. Throughout, we discuss the advantages of integrating multiple data sets, for instance, where one technology may address shortcomings of another to help provide insight into a mechanism of disease. Additionally, we discuss current methods as well as challenges in optimally combining and interpreting data from multiple sources, with some promising examples of their successful applications to elucidating mechanisms of human disease.
Dissecting Mendelian disease
In North America, approximately 10% of paediatric hospital admissions and 20% of infant deaths are attributable to Mendelian diseases8–10. In many cases, clinicians and families affected by Mendelian diseases are turning to exome and genome sequencing to find the causative mutations of their disease, which, depending on the disease and study design, has proved successful in 25–50% of cases previously not solved by targeted gene panels3,11–13. For diseases that typically act via a recessive mechanism, these investigations are most effective when the causal variant is either already in a variant–disease database, such as Clinvar, or a protein-truncating (for example, stop-gain, frameshift or essential splice site) variant in a known disease gene. However, in some cases, the effect of the variant may be more subtle (for example, an intronic variant creating a cryptic splice site), the variant may be difficult to detect owing to somatic mosaicism or several candidates are equally likely to be deemed causal. Furthermore, such diagnoses are additionally complicated when the genetic aetiology is not well known or when the candidate variants fall in genes that are less well described. Integrating additional information, such as RNA sequencing (RNA-seq) or network analyses, can be useful for detecting molecular events that prioritize among likely causal variants or provide additional evidence that a candidate mutation is causative. For instance, in a multi-omics analysis of patients with uncharacterized Fanconi anaemia, DNA sequencing and array comparative genomic hybridization (aCGH) were effective in identifying the mutations that were eventually deemed causal, whereas RNA-seq provided evidence of pathogenicity for some unsuspecting variants, including intronic and synonymous variants that affect splicing patterns, as well as a deletion of a non-coding exon and upstream region that resulted in ablated expression of a transcript14.
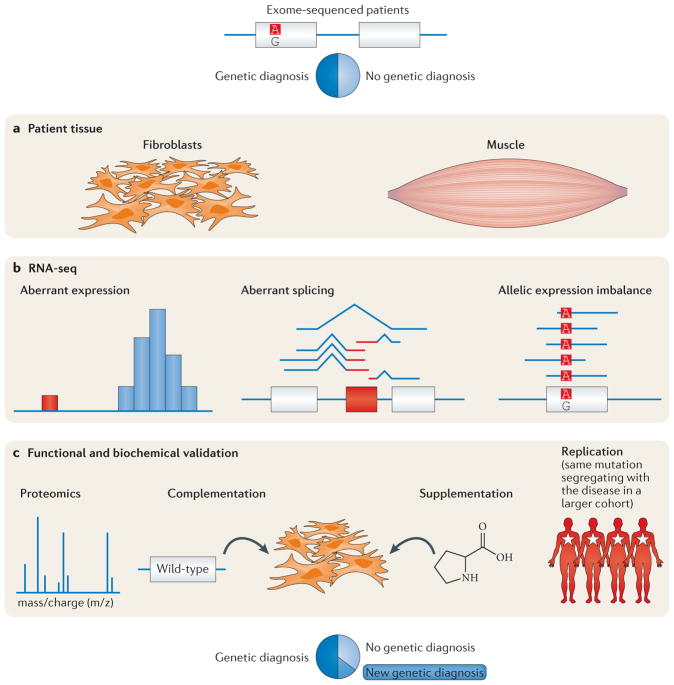
More recently, two systematic studies of approximately 50 patients each have provided estimates of the additional gain in diagnosis rate using RNA-seq and other technologies (FIG. 1), ranging from 10% to 35%15,16. In one of these studies, a diagnostic investigation of patients with muscular dystrophy (MD), no causal variants were identified through whole-exome sequencing (WES), but RNA-seq data identified splice anomalies that revealed variants with cryptic splicing effects. Notably, even if whole-genome sequencing (WGS) were performed on these patients, these variants would have been identified but likely not flagged as causal, as many of them were intronic or otherwise not predicted to affect splicing. Given its rapidly decreasing costs and substantial information gain, RNA-seq is likely to become a powerful tool in characterizing disease pathophysiology in clinical practice. Similarly, as proteomics technologies become cheaper and more accessible, they may be used to identify protein level changes brought about, for instance, by missense variants that affect protein stability or post-translational modifications.
Figure 1. Identifying a causal variant to diagnose a patient with a rare disease.
In Kremer et al.15 and Cummings et al.16, multi-omics approaches were used to aid in the diagnosis of patients with undiagnosed disease. Although exome and genome sequencing can be effective in identifying causal genetic variation between 20% and 50% of the time, depending on the mode of inheritance and phenotype, the majority of cases cannot be solved by these technologies alone. a,b | Using RNA sequencing (RNA-seq) data from patient tissue, these approaches were able to make a molecular diagnosis for many patients, identifying genes with aberrant expression, splicing or allele-specific expression, which would suggest a molecular mechanism for the disease progression. c | In some cases, functional validation, such as proteomics, can lend additional support to these diagnoses. Figure is adapted from REF. 15, Macmillan Publishers Limited.
Genetic architecture of common disease
Most common diseases such as diabetes17, obesity18, schizophrenia19,20 and autism21 are complex and a result of a combination of multiple genetic and environmental factors. Thus far, thousands of genomic loci have been significantly associated with human diseases (for a recent review, see REF. 22); however, once established as bona fide associations, the difficult task remains of characterizing the genes in the context of the molecular pathophysiology of the disease and its interacting genes and pathways. To this end, a number of methods have arisen to analyse multiple omics data sets, including network and enrichment analysis.
Network analyses
Integration of multiple orthogonal data types can be used to narrow the search space for disease genes and identify causal mechanisms of disease. Specifically, network models, including protein–protein interaction, regulatory and co-expression networks, have proved to be a valuable resource for prioritizing and identifying disease genes and pathways (for recent reviews, see REFS 23–26). These networks can be used with any genome-scale data set, including single-nucleotide polymorphism (SNP) or gene expression data, to investigate the topological properties of the most significantly disease-associated genes in a study, particularly when no or few hits reach genome-wide significance. In the case of genetic variation data, a challenge exists in mapping SNPs to the affected gene: in some cases, the effect of the variant is clear — such as a frameshift variant in an immune-response-related gene, NOD2, in Crohn’s disease27 — but more often, the affected gene for a variant may be ambiguous28. Additionally, SNPs may be grouped into genes to increase power, but patterns of linkage disequilibrium must be addressed29.
Despite these challenges, network methods have yielded successful insights into human disease. For instance, in patients with autism spectrum disorder (ASD), genes harbouring de novo missense or nonsense mutations are enriched for genes with high degrees of connectivity in protein–protein interaction networks to all other genes and particularly previously ASD-implicated genes30. In this way, such approaches provide a mechanism to prioritize among putative disease genes, either by suggesting a greater functional impact due to their presence as a hub gene in a network or through guilt-by-association with previously associated genes.
Additionally, two recent studies from our laboratory integrating genomic, RNA-seq and proteomic data have identified new genes and complexes involved in autism and characterized their function31,32. Specifically, analysis of protein–protein interaction networks revealed a module (or coherent community of interacting genes) that was enriched for known genes involved in autism, as well as genes harbouring copy number mutations and rare mutations in autism cases. This module was enriched for genes involved in synaptic transmission, and RNA-seq revealed that many of the genes in a submodule were differentially expressed in the corpus callosum in patients with ASD, providing a putative molecular explanation for the observation that many individuals with ASD have a smaller corpus callosum than controls32. Similarly, mapping of rare variants in patients with autism onto protein complexes revealed both novel proteins and novel molecular machinery involved in autism, including the histone deacetylase (HDAC) chromatin remodelling complexes and other protein complexes31. Thus, integrating protein interaction data with WGS and WES data can provide new insights into important diseases, including autism, type 2 diabetes33 and heart disease34 (additionally reviewed in REF. 35).
Enrichment analysis
Recently, numerous large-scale enrichment analyses have been performed in order to understand the global mechanisms of information flow from DNA to physiology. Protein-coding variation is fundamental to many traits and, as such, associated loci from GWAS for many traits are enriched for protein- sequence-disrupting (non-synonymous) variation36. However, only a small fraction of associations fall into this category and, therefore, integration of non-coding regulatory annotations with disease association data can be valuable for identifying disease genes and disease aetiology (reviewed in REF. 37). In particular, assays for measuring gene expression (RNA-seq) as well as regulatory activity in regions that control gene expression (such as chromatin immunoprecipitation followed by sequencing (ChIP–seq) for transcription factor binding sites or DNase-seq for detecting regions of open chromatin) have been valuable in identifying tissue-specific signatures of genomic regulation. Accordingly, disease-associated variants are enriched among expression quantitative trait loci (eQTLs) as well as in transcription factor binding sites38–41 and, thus, it is likely that many disease aetiologies may act through regulatory mechanisms. Indeed, a recent study of 108 loci associated with schizophrenia provided evidence for 20 of these loci having changes in gene expression that could at least partially explain their associations20.
Recently, partitioning heritability methods using GWAS summary statistics and functional annotation data elucidated the relative contribution of coding and regulatory variants, suggesting that the bulk of heritability of many common traits stems from variants in regulatory regions (regions of open chromatin as measured by DNase hypersensitivity)42, as well as many cell type-specific enhancers43. Additionally, such enrichment information can be used to discern causal variation as well as to identify novel genes for diseases and traits by increasing the weight of annotations that are specific to each trait36. As of this writing, such methods are not yet in clinical practice but have been invaluable in revealing the aetiology of many common diseases.
Narrowing causal mechanisms in common disease
As previously mentioned, GWAS have been successful in identifying loci that are statistically associated with disease, but they rarely identify causal variation. Integration of multiple data types, such as functional annotation data, can also provide insight into the potential function of specific disease-associated variants.
Indirect integration across individuals
Currently, a cost-effective method to ascertain the causality of variants associated with a trait is using multiple independent data sets to pinpoint causal mechanisms from a set of candidate loci with biological evidence44. Such a process may begin with a GWAS, after which, a set of genomewide significant loci are assayed for functional follow-up; the specific experiment may depend on the types of loci identified or the genetic architecture of the disease. For coding variants, follow-up experiments that ascertain the effect of the variant on protein structure or function are ideally performed to demonstrate causality45. For non-coding variants, the effects are often more difficult to interpret, but recent large-scale epigenetic studies, such as the Encyclopedia of DNA elements (ENCODE)46 and the Roadmap Epigenomics47 projects, can suggest possible mechanisms for regulatory control, as well as transcription factors to target for follow-up experiments. For instance, a detailed study on a variant associated with systemic lupus erythematosus (SLE) showed that the variant also affects nuclear factor-κB (NF-κB) binding and is associated with expression of tumour necrosis factor-α induced protein 3 (TNFAIP3) at both the mRNA and protein level48.
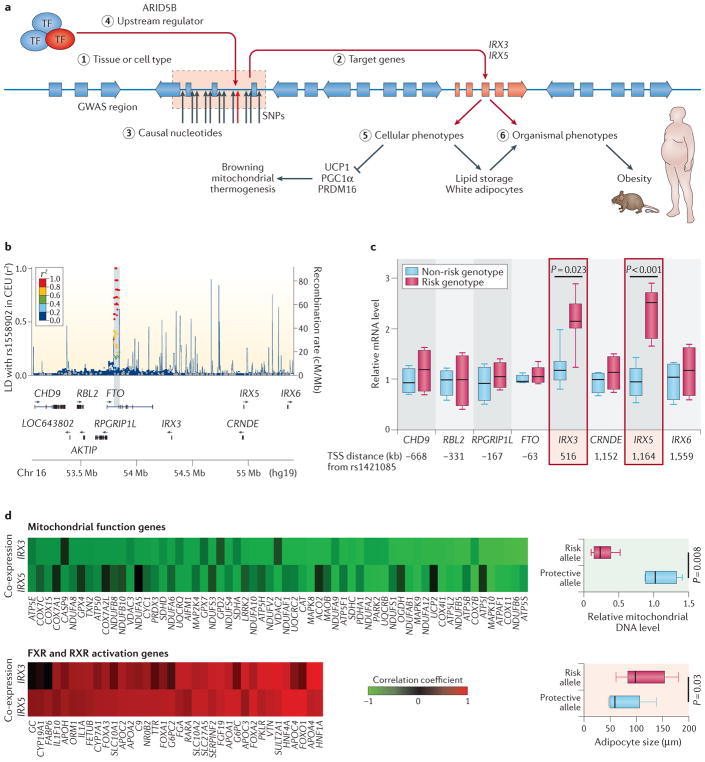
Recently, two investigations from Manolis Kellis and colleagues integrating multiple data types have yielded fruitful insights into the molecular pathology of Alzheimer disease and obesity. First, combining gene expression and epigenomic data, the group showed that genes that are upregulated in an Alzheimer disease mouse model show immune cell enhancer signatures49. Crucially, whereas a link between immune system genes and Alzheimer disease had long been previously established, multiple omics data types proved useful in this scenario to establish a direction of effect, showing that there is a concerted increase in expression and regulatory activity at immune system genes in Alzheimer disease. Similarly, integrating epigenome and chromosomal conformation data, as well as expression information from patients with an FTO obesity allele and a number of other data types, provided a mechanistic explanation for the risk allele50 (FIG. 2). Genome editing of the risk allele using CRISPR–Cas9 restored aberrant expression and thermogenesis, suggesting a potential therapeutic avenue for obesity phenotypes.
Figure 2. From genome-wide association studies to mechanism.
In a recent study, Claussnitzer and colleagues present a comprehensive approach50 to identifying a causal mechanism for an obesity-associated variant in the FTO gene. Part a shows an overview of the deciphered biological mechanisms and the numbered steps of the strategy referred to below. From the initial genome-wide association study (GWAS), the significant association of the FTO region with obesity is shown in the Manhattan plot (part b). First, the researchers established the relevant tissue or cell type (step 1) as well as the downstream target genes using regulatory genomics, including chromatin state information and chromosomal conformation (Hi-C) data. Here, they established the variant as an expression quantitative trait locus (eQTL) for the developmental genes iroquois homeobox 3 (IRX3) and IRX5 (step 2), where the risk allele shows increased expression of these genes but not others in the vicinity (part c). They demonstrate that expression of IRX3 and IRX5 is anti-correlated and correlated with genes involved in mitochondrial function and adipocyte size, respectively (part d). Next, they established the causal nucleotide variant (step 3) in an AT-rich interactive domain-containing protein 5B (ARID5B) motif (step 4) using CRISPR–Cas9 to show its molecular effects, including altered signatures of expression and phenotypic effects on the regulation of energy balance (step 5). Finally, they establish causality of the variant on an organismal level using mouse models (step 6). AKTIP, AKT interacting protein; CEU, Utah residents (CEPH) with northern and western European ancestry; CHD9, chromodomain helicase DNA binding protein 9; CRNDE, colorectal neoplasia differentially expressed; FXR, farnesoid X-activated receptor; LD, linkage disequilibrium; PGC1α, peroxisome proliferator-activated receptor-γ co-activator 1-α; PRDM16, PR domain zinc-finger protein 16; RBL2, RB transcriptional co-repressor like 2; RXR, retinoid X receptor; SNPs, single-nucleotide polymorphisms; TF, transcription factor; TSS, transcription start site; UCP1, mitochondrial brown fat uncoupling protein 1. Figure is adapted from The New England Journal of Medicine, Claussnitzer, M. et al., FTO obesity variant circuitry and adipocyte browning in humans, 373, 895–907, Copyright© (2015) Massachusetts Medical Society, REF. 50. Reprinted with permission from Massachusetts Medical Society.
Direct integration within an individual
Whereas synthesizing data from multiple disparate technologies can create a link between layers of biological mechanism, characterizing multiple omics profiles in a single individual will be a powerful tool for creating a holistic view of the molecular effects that lead to physiological phenotypes. However, these approaches can be expensive, as they require multiple interventions and technologies on the same individual and, as such, thus far have had limited sample sizes. The first such study was performed in our laboratory and followed a single individual for over 7 years6 (and M.P.S., unpublished observations), whereas a similar study followed another individual for 1 year51. In Chen et al.6, genomic analyses predicted an elevated risk of type 2 diabetes, which was subsequently revealed through detailed omics analyses, including transcriptomics, proteomics, metabolomics and other measurements. In particular, genes involved in insulin signalling and response were found to be downregulated by RNA-seq and by liquid chromatography–tandem mass spectrometry (LC–MS/MS) proteomics during a respiratory syncytial virus infection, which coincided with increased blood glucose concentration to diabetic levels. These approaches are advantageous in their ability to track a mechanistic link across a shared genetic and individual background, as one can follow a progression of molecular events, such as the differential expression of a GWAS-identified disease-associated gene leading to differences in RNA and protein levels and their corresponding metabolites.
However, as omics profiling experiments have a high multiple hypothesis testing burden (for example, across all genes in the genome or thousands of metabolites), larger sample sizes will be useful to determine the generality of such correlations. A recent study monitoring various omics profiles across 23 individuals identified inflammatory signatures during weight gain, and found that certain metabolic pathways did not return to baseline after subsequent weight loss52. This analysis highlights the extent of similarities in longitudinal omics profiles across individuals, as well as individual-specific signatures at steady state and under experimental perturbations. To further qunatify these differences, projects have been initiated to extend such analyses to thousands of individuals, characterizing preterm births, inflammatory bowel disease and type 2 diabetes53. In a similar vein, two separate groups recently profiled genetic and metabolomics data: one of these calculated polygenic risk scores for over 100 individuals and correlated these with measurements of metabolites54, whereas the other identified rare deleterious variants in healthy volunteers that correlated with outliers of individual metabolites and metabolic pathways55. Additionally, as reference databases of omics data for healthy individuals become available (as are already available for exome56, genome (for example, the Genome Aggregation Database (gnomAD)) and RNA-seq57 data), it will become easier to interpret individual-level data in the context of these control cohorts.
Other efforts include the Framingham Heart Study and genome characterization studies, such as the Genotype–Tissue Expression (GTEx)57 project, with its proposed extension to analyses beyond gene expression in the enhanced GTEx (eGTEx) project58. These projects have adopted a breadth-first strategy for omics profiling, in which a high number of individuals are characterized with a limited panel of technologies that assay a single set of molecular markers (for example, whole-genome DNA methylation assays).
Cancer
One area where multiple omics analyses have had and will continue to have enormous impact is in cancer profiling, diagnosis and treatment. Indeed, many of the previously discussed strategies (for example, network methods) will be effective in identifying genetic mechanisms of cancers. However, there are conceptual differences in cancers that complicate their analyses and require special handling. In addition to the technical challenges of calling somatic variants (see the ‘Accuracy and validation’ subsection in the ‘Challenges’ section below), the majority of genetic changes evident in cancer cases are benign and do not drive cancerous cell growth; therefore, determining which mutations are drivers or which pathways are involved remains a considerable challenge. Additionally, although some cancers share genetic signatures across individuals, there is still a high level of diversity among driver mutations, which can lead to differences in prognosis and therapeutics.
Identifying driver mutations
A typical process to identify driver mutations involves WGS of multiple tumours to identify recurrently mutated genes59. Overlaying functional data can help to prioritize this information, as driver mutations are more likely to be in genes that are expressed in a given cancer. For instance, in an analysis of driver mutations identified using WES coupled with copy number variation (CNV) microarray data, RNA-seq data were used to identify an expressed gene fusion of EGFR–SEPT14, which was functionally validated to affect glioma growth60. In a different analysis using similar technologies, the driver mutations and processes underlying multiple metastases within an individual were shown to be largely similar across metastases, suggesting that a single metastasis is sufficient for downstream analysis61. In this way, using additional omics data complements genetic data, providing a mechanism to filter the deluge of genetic variation to functionally relevant causal variants.
Molecular signatures of cancer
In addition to identifying driver mutations, multiple types of omics data can reveal general biochemical pathways that are active in individual cancers and classify them into subtypes. As such, this can be a valuable tool for ascertaining which pathways to target within a patient, even if strong candidate mutations are not detected in those pathways — for example, owing to difficult to characterize non-coding mutations or indirect effects. For instance, clusters of transcriptomics and DNA methylation patterns have been used to identify subtypes of cancers, which have varying survival prognoses59,62. More recently, three studies of the Clinical Proteomic Tumour Analysis Consortium (CPTAC) have used proteomic approaches to identify cancer subtypes for colorectal, ovarian and breast cancer based on protein expression signatures63–65. Importantly, the proteomics data revealed overlapping but not identical correlation with the transcriptome and genetic data, indicating that the different data types expose different types of information. These studies demonstrated the distinct genetic and transcriptional processes that translate into proteomic alterations. Finally, integration of imaging information with omics information is expected to be valuable in cancer diagnosis and prognosis66,67.
Recent developments in characterizing the non-coding regions that regulate gene expression have become increasingly valuable for understanding the regulatory landscape of cancer. Studies integrating reference data sets of regulatory information46,47 with WGS data from The Cancer Genome Atlas (TCGA) revealed a number of regulatory regions that are enriched for mutations in patients with cancer68–71. In these cases, causal genetic variation in these non-coding regions is still difficult to pinpoint, highlighting the continuing need for research into prioritizing such variation; nevertheless, shared network topology across individuals with the same cancer can inform cancer subtypes that may have different prognoses and therapeutic strategies. Finally, given the strong dependence of cancerous growth on metabolic changes, it is likely that metabolomics will also play an important role in cancer diagnostics or prognosis in the future.
Challenges
Until now, most integrative models have been reported and published in research settings. However, the adoption of clinical genomics has expanded rapidly over the past few years from the first successful diagnosis1 to multi-institutional and international adoption72. In the same vein, longitudinal multi-omics profiling, with its first recent research examples6,54, may similarly emerge as a clinical tool.
However, for clinical adoption of any technology to occur, high specificity and sensitivity are required, both in detection and interpretation. At present, aside from the use of WES or WGS in exceptional cases, such technologies are not regularly used across clinical practices because for many diseases, they have not been proved superior to current tests. Going forward, clinical guidelines must be established to ensure accuracy and efficacy, and tests to show non-inferiority and cost-effectiveness must be performed.
Nonetheless, omics profiling can be an effective way of detecting large-scale or pathway-level alterations — cheaper and often more comprehensive than performing thousands of individual tests — and longitudinal profiling can show patient-specific trends and add statistical support through repeated measurements6. Although challenges remain in establishing clinical guidelines, many of the concepts surrounding the interpretation of genetic variants (particularly rare or novel variants) may apply to a general molecular event (such as a differentially expressed gene, novel protein phosphorylation or unique metabolome signature) as our understanding of the biology and reference databases mature.
Analytical challenges
There are various analytical challenges that must be addressed to enable the widespread adoption of integrative omics in clinical practice, particularly those of statistical methods for data aggregation, scalability and integration into electronic health records (EHRs). Most importantly, a robust and reproducible statistical framework is needed to properly analyse multiple disparate data sets, each with their own variances and biases. Multi-omics data can be analysed in a multistage or meta-dimensional fashion (reviewed in REF. 73). Briefly, one option for drawing inferences from these data involves pairwise analyses of data sets, mounting evidence to support a signal. However, analysing three or more data sets simultaneously requires more sophisticated multi-dimensional methods, such as Bayesian models74, neural networks75 or dimensionality reduction76. This is further complicated by the fact that various omics data types are fundamentally different: for instance, genetic variation data are discrete and static, whereas RNA-seq measurements are continuous and can provide longitudinal information.
Although the data analysis methods described above are effective for learning about biology and disease, they are not specifically designed to apply this information to individual-level data for clinical purposes. In the genomics space, with an individual’s genotype and a database of results from GWAS, one can compute a polygenic risk score to assess an individual’s risk of disease4,77 (for recent reviews on methodology, see REFS 22,78). A major obstacle remains in building such frameworks for multiple omics profiles, which is likely to face some of the same challenges, such as the difficulty in applying results discovered in one population to individuals in another79,80.
In addition to challenges with analytical approaches, these analyses and the storage of all associated data will require tremendous computational resources: although the amount of data for multiple omics technologies on a single individual may be manageable (for example, terabyte-scale (1012 bytes)), these data must be put into a larger context to understand deviations from the background distribution, which requires data from thousands of samples (exabyte-scale (1018 bytes)). Fortunately, cloud-computing-based options have begun to alleviate these concerns81, providing elastic computation and storage facilities based on specific requirements from each hospital or healthcare provider system while simultaneously promoting reproducibility in computational processes82.
At present, such integrative data sets often do not have a standard format for research use, let alone for use in a structured clinical system; therefore, the infrastructure to house and manage these data will be required, which introduces financial and administrative burdens. In particular, health informaticians will be tasked with building a robust infrastructure for storing genetic and transcriptomic data in the EHR. Moreover, determining which information will be reported back to a patient and incorporated into an EHR will require concerted efforts from clinicians and researchers.
Accuracy and validation
Individually, genome-wide data sets carry inherent error rates83, and structural variants are still difficult to detect and, as such, are rarely called. The accuracy of more continuous and longitudinal data, such as mRNA expression and proteomic data, may be more difficult to assess depending on the specific tissue assayed, but these technologies are highly reproducible for technical and biological replicates84,85. In some situations, these technologies independently identify different aspects of the same biological process and thus can validate each other: for instance, RNA-seq can internally replicate exonic variants identified through WES or WGS, whereas proteomic expression can validate expression from RNA-seq. However, in a clinical setting, where high confidence is required, these tests are currently validated by independent technologies, potentially including established clinical tests, such as enzymatic or single-assay tests.
For cancer genomics, disentangling heterogeneous data is a substantial challenge. As each tumour is a mosaic of cells with varying degrees of somatic mutation, variant detection is difficult, even before attempting to discern driver mutations from passenger mutations. In particular, cancers display signatures of somatic mutations that are clonal or found in only a subset of cells in a tissue, complicating their discovery, and high-coverage and high-quality data are necessary to distinguish these from sequencing errors (for a recent review on the computational methods to do so, see REF. 86). Ultra-deep sequencing of cell-free DNA to follow the presence of cancer mutations and single-cell sequencing to detect cancer heterogeneity are emerging as powerful methods. However, cell-free DNA for early cancer detection requires robust methods to distinguish genuine low-frequency events from sequencing errors, and single-cell sequencing is still expensive. Nonetheless, such methods have already been used to disentangle tumour heterogeneity87 and identify a secondary finding of cancer in a prenatal test88. As additional omics data sets are integrated with ultra-deep sequencing, we expect the advantages of each of these methods to complement each other and provide a uniquely powerful method for molecular interrogation in the clinic.
Interpretation
Even with highly accurate data, another difficulty lies in the interpretation of genome-scale results, particularly rare and novel molecular events, which often vastly outnumber the number of events that can be reasonably functionally validated. Many variants in an individual genome, especially if they have not been seen before, do not have a clear functional effect and are known as ‘variants of uncertain significance’ (VUS)89. This problem is compounded for other data types, such as transcriptomic or proteomic data, and decisions for what constitutes a clinically significant molecular event, for example, an RNA expression threshold, are difficult to determine across disparate data types. Fortunately, large reference population data sets, which are already available for exome56 and genome sequencing (gnomAD) and gene expression57,90, will aid in the interpretation of rare events by providing a quantitative context as to their actual frequency in a population. In particular, a causal variant would be expected to have a significantly higher frequency in affected individuals than in a wider asymptomatic population, which can lend support to or negate previous suggestions of pathogenicity91,92. Additionally, physicians may discover additional pathogenic molecular events for unrelated conditions, which are known as secondary or incidental findings93, over which there is still considerable debate as to the extent to which results should be returned to patients (for a recent review, see REF. 94).
When integrating multiple omics technologies, these problems are occasionally ameliorated, particularly for rare and novel molecular events for which statistical analysis is not feasible. In particular, direct integration of omics technologies that expose orthogonal information may provide additional evidence for a molecular event: for instance, if a VUS is shown by RNA-seq to affect splicing of a key disease gene, this can corroborate a potential pathogenic mechanism16. This way, multiple technologies can establish a chain of causality that a single technology cannot.
Finding the relevant tissue
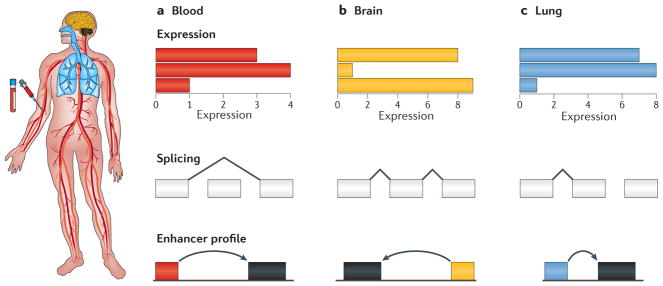
In order to maintain consistency across samples, many large-scale research studies have been performed on readily available samples, such as blood or cell lines, including transformed lymphoblastoid cell lines90,95. However, for clinical applications, it is ideal to study tissues that are relevant for a particular disease as gene expression varies considerably across tissues96,97 (FIG. 3). The GTEx, Roadmap Epigenomics and Functional Annotation of Mammalian Genome 5 (FANTOM5) projects provide reference data sets for multi-tissue gene expression and epigenomics data47,57,98. In many cases, the disease-relevant tissue may be well described, such as muscle tissue for MD; however, if the disease is less well defined or the tissue is not available, a tissue may be identified from a network analysis of the disease99. Indeed, using the disease-relevant tissue has proved beneficial in diagnosis of patients with MD, where transcriptome analysis of the muscle tissue resulted in diagnoses that would not have been made via easily accessible proxy tissue, such as blood or fibroblasts, owing to the relatively low expression of disease-relevant genes16. In using such data for clinical utility, care should be taken to ensure that data from patient samples are comparable to reference data sets, which will be crucial going forward for additional omics data, such as metabolomics and proteomics. Of course, such analyses are further complicated where there is substantial cellular heterogeneity in the tissue, such as the brain: in these cases, technologies with single-cell resolution will provide valuable insights into resolving each individual cell type. In cases where primary tissue is difficult to obtain or maintain in culture, introduction of a mutation into induced pluripotent stem (iPS) cells using CRISPR systems can provide a powerful framework for molecular validation100.
Figure 3. Finding the relevant tissue.
Although blood (part a) is often the most convenient tissue to assay owing to its availability and ease of procurement, it is often not the ideal tissue for observing a molecular phenotype for a given disease, which may primarily affect other tissues such as brain (part b) or lung (part c). In particular, its transcriptional landscape, including expression levels, splicing patterns and enhancer usage, may not be amenable to detecting differential uses of these patterns compared with a tissue that is more proximally affected by a disease, such as muscle tissue in muscular dystrophy.
Actionability and therapeutics
Perhaps most important to the discussion of the use of any technology in the clinic is that of actionability. Indeed, a piece of information does not need to inform a course of action to be useful: having the knowledge of a diagnosis and ending a diagnostic odyssey can be invaluable to patients and families101 (for a thorough perspective on the purpose of genetic testing for diagnoses, see REF. 102). However, data that can inform an intervention are additionally beneficial, in a framework that has been termed ‘precision medicine’ or ‘personalized medicine’. In particular, classifying a patient’s subtype of a disease to recommend a specific drug, determining whether a potential transplant is a good match on the basis of omics profiling (BOX 2) or identifying a causal mechanism for a novel disease (and developing a therapeutic that can target the direct molecular outcome) can improve outcomes and prolong the lives of patients. However, even non-causal molecular events that are statistically associated with an outcome can be actionable, particularly in the form of lifestyle change recommendations, including diet, monitoring and preventive treatments; indeed, individuals with high genetic risk of coronary heart disease experience greater benefits from statin treatment103,104.
Box 2. Multiple omics profiling of transplant donors and recipients.
Every year, thousands of patients are given organ and haematopoietic stem cell transplants. However, mortality among transplant patients remains very high. A standard practice for matching donors with recipients involves human leukocyte antigen (HLA) typing, for which methods have been recently developed using high-throughput sequencing technologies123,124. However, it is becoming increasingly clear that non-HLA factors can considerably affect prognosis and development of graft-versus-host disease (GVHD), as HLA-matched sibling donor transplants convey a lower risk of GVHD than HLA-matched but unrelated donor transplants125, and common non-HLA polymorphisms have been associated with GVHD126.
Accordingly, many omics applications may be used to determine optimal donor–recipient matches, as well as to monitor markers of rejection127. For instance, sequencing cell-free DNA can detect circulating donor DNA128, the levels of which are correlated with the severity of organ rejection129. Additionally, sequencing this cell-free DNA can simultaneously detect viral DNA to indicate a marker of infection130. Additional omics data, such as RNA or protein expression, may also be used to assess compatibility of donor–recipient pairs, as well as monitor for markers of rejection (for a recent review, see REF. 131). Integration across multiple omics technologies may well emerge as a useful tool for transplant biology.
Conclusions and future perspectives
At present, only in very few cases have omics technologies (particularly genome sequencing and, to a lesser extent, RNA-seq) been shown to outperform traditional clinical tests and, therefore, substantial technical and regulatory hurdles exist to incorporating these technologies into clinical practice. However, as the use of multiple technologies enables a clearer picture of health and disease, it is likely that integration of these technologies will become commonplace in future clinical practice. Additionally, as recent large biobank initiatives, such as the UK Biobank, Million Veterans Project and All of Us, collect biological data and perform multiple layers of omics assays on millions of individuals, they will yield profound insights into human disease and serve as valuable reference databases for additional studies and clinical applications.
Predictive models of disease risk for healthy individuals and early detection of disease
As with traditional clinical tests, molecular measurements from large-scale omics data can be integrated into models of disease risk. In particular, recently, a set of methods has been developed for calculating the genetic risk of a particular disease, known as a polygenic risk score (recently reviewed in REF. 105). These methods have been successful in stratifying patients into high-risk and low-risk categories for diseases such as cardiovascular disease77, as well as for predicting traits such as educational attainment106. Single-assay tests are often performed to follow up the results of predictions of disease risk, whether derived from genetics or family history. For example, if a patient is predicted to be at risk of type 2 diabetes, then assays for glucose and glycosylated haemoglobin (HbA1c) levels and other tests, such as a glucose tolerance test, are performed. However, in the future, if a metabolomics panel could be performed simultaneously at high quality and low cost, this would obviate the need for the single follow-up assay. In addition, data from wearable devices that continuously collect data are likely to be very powerful in combination with omics data for early detection of disease before symptom onset107.
Disease management
In addition to prediction and early diagnosis, integrative omics is expected to become increasingly powerful for disease treatment and prognosis. Information from the transcriptome, epigenome, microbiome, proteome and metabolome as well as imaging and wearable data will all be used to help decipher disease to facilitate prognosis and thereby guide treatment. In cancer, DNA and RNA sequencing of tumour–normal pairs has identified translocation and gene expression signatures, which has suggested targeted therapies that resulted in disease regression108,109. In the future, as multiple omics measurements are associated with prognosis in other diseases, it is likely that such data-driven paradigms will be powerful tools for medical research and also facilitate clinical diagnosis and treatment.
Acknowledgments
K.J.K. is supported by the US National Institute of General Medical Sciences (NIGMS) Fellowship F32GM115208. M.P.S. is supported by grants from the National Institutes of Health (NIH).
Abbreviations
- Actionability
The property of a molecular finding that would result in a specific medical recommendation that is expected to improve a disease outcome.
- Mendelian diseases
Diseases caused by a single locus or gene and that follow Mendelian patterns of inheritance (for example, dominant or recessive).
- Genetic aetiology
The genetic factors that cause a particular disease.
- Expression quantitative trait loci
(eQTLs). Genetic variants that are statistically associated with gene expression.
- Heritability
The fraction of phenotypic variability of a trait that can be attributed to additive genetic variation.
- DNase hypersensitivity
A measure of openness of chromatin, as measured by its sensitivity to cleavage by DNase I.
- Structural variants
A class of genetic variation that is typically 1 kb or larger, which includes copy number duplications, insertions or deletions, as well as translocations and inversions.
Footnotes
Author contributions
Both authors contributed to all aspects of the manuscript, including researching data, discussing content, writing and editing.
Competing interests
The authors declare competing interests. See Web version for details.
FURTHER INFORMATION
Genome Aggregation Database: http://gnomad.broadinstitute.org
ALL LINKS ARE ACTIVE IN THE ONLINE PDF
References
- 1.Worthey EA, et al. Making a definitive diagnosis: successful clinical application of whole exome sequencing in a child with intractable inflammatory bowel disease. Genet Med. 2011;13:255–262. doi: 10.1097/GIM.0b013e3182088158. This is the first paper describing the treatment-changing diagnosis in an individual patient using exome sequencing, paving the way for clinical applications of genomics. [DOI] [PubMed] [Google Scholar]
- 2.Ng SB, et al. Exome sequencing identifies the cause of a mendelian disorder. Nat Genet. 2010;42:30–35. doi: 10.1038/ng.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Taylor JC, et al. Factors influencing success of clinical genome sequencing across a broad spectrum of disorders. Nat Genet. 2015;47:717–726. doi: 10.1038/ng.3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ashley EA, et al. Clinical assessment incorporating a personal genome. Lancet. 2010;375:1525–1535. doi: 10.1016/S0140-6736(10)60452-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dewey FE, et al. Phased whole-genome genetic risk in a family quartet using a major allele reference sequence. PLoS Genet. 2011;7:e1002280. doi: 10.1371/journal.pgen.1002280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen R, et al. Personal omics profiling reveals dynamic molecular and medical phenotypes. Cell. 2012;148:1293–1307. doi: 10.1016/j.cell.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Visscher PM, Brown MA, McCarthy MI, Yang J. Five years of GWAS discovery. Am J Hum Genet. 2012;90:7–24. doi: 10.1016/j.ajhg.2011.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scriver CR, Neal JL, Saginur R, Clow A. The frequency of genetic disease and congenital malformation among patients in a pediatric hospital. Can Med Assoc J. 1973;108:1111–1115. [PMC free article] [PubMed] [Google Scholar]
- 9.Buehler JW, Strauss LT, Hogue CJ, Smith JC. Birth weight-specific causes of infant mortality, United States, 1980. Public Health Rep. 1987;102:162–171. [PMC free article] [PubMed] [Google Scholar]
- 10.Kochanek KD, Kirmeyer SE, Martin JA, Strobino DM, Guyer B. Annual summary of vital statistics: 2009. Pediatrics. 2012;129:338–348. doi: 10.1542/peds.2011-3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang Y, et al. Clinical whole-exome sequencing for the diagnosis of Mendelian disorders. N Engl J Med. 2013;369:1502–1511. doi: 10.1056/NEJMoa1306555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jacob HJ, et al. Genomics in clinical practice: lessons from the front lines. Sci Transl Med. 2013;5:194cm5. doi: 10.1126/scitranslmed.3006468. [DOI] [PubMed] [Google Scholar]
- 13.Lee H, et al. Clinical exome sequencing for genetic identification of rare Mendelian disorders. JAMA. 2014;312:1880–1887. doi: 10.1001/jama.2014.14604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chandrasekharappa SC, et al. Massively parallel sequencing, aCGH, and RNA-Seq technologies provide a comprehensive molecular diagnosis of Fanconi anemia. Blood. 2013;121:e138–e148. doi: 10.1182/blood-2012-12-474585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kremer LS, et al. Genetic diagnosis of Mendelian disorders via RNA sequencing. Nat Commun. 2017;8:15824. doi: 10.1038/ncomms15824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cummings BB, et al. Improving genetic diagnosis in Mendelian disease with transcriptome sequencing. Sci Transl Med. 2017;9:eaal5209. doi: 10.1126/scitranslmed.aal5209. References 15 and 16 use transcriptome sequencing to provide molecular diagnoses that were missed by exome sequencing for patients with rare disease. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flannick J, Florez JC. Type 2 diabetes: genetic data sharing to advance complex disease research. Nat Rev Genet. 2016;17:535–549. doi: 10.1038/nrg.2016.56. [DOI] [PubMed] [Google Scholar]
- 18.Locke AE, et al. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518:197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ripke S, et al. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fromer M, et al. Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nat Neurosci. 2016;19:1442–1453. doi: 10.1038/nn.4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grove J, et al. Common risk variants identified in autism spectrum disorder. 2017 bioRxiv https://doi.org/10.1101/224774.
- 22.Manolio TA. Bringing genome-wide association findings into clinical use. Nat Rev Genet. 2013;14:549–558. doi: 10.1038/nrg3523. [DOI] [PubMed] [Google Scholar]
- 23.Moreau Y, Tranchevent LC. Computational tools for prioritizing candidate genes: boosting disease gene discovery. Nat Rev Genet. 2012;13:523–536. doi: 10.1038/nrg3253. [DOI] [PubMed] [Google Scholar]
- 24.Ryan CJ, et al. High-resolution network biology: connecting sequence with function. Nat Rev Genet. 2013;14:865–879. doi: 10.1038/nrg3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mitra K, Carvunis AR, Ramesh SK, Ideker T. Integrative approaches for finding modular structure in biological networks. Nat Rev Genet. 2013;14:719–732. doi: 10.1038/nrg3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cowen L, Ideker T, Raphael BJ, Sharan R. Network propagation: a universal amplifier of genetic associations. Nat Rev Genet. 2017;18:551–562. doi: 10.1038/nrg.2017.38. [DOI] [PubMed] [Google Scholar]
- 27.Ogura Y, et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn’s disease. Nature. 2001;411:603–606. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- 28.Huang H, et al. Fine-mapping inflammatory bowel disease loci to single-variant resolution. Nature. 2017;547:173–178. doi: 10.1038/nature22969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu JZ, et al. A versatile gene-based test for genome-wide association studies. Am J Hum Genet. 2010;87:139–145. doi: 10.1016/j.ajhg.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Neale BM, et al. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature. 2012;485:242–245. doi: 10.1038/nature11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li J, et al. Identification of human neuronal protein complexes reveals biochemical activities and convergent mechanisms of action in autism spectrum disorders. Cell Systems. 2015;1:361–374. doi: 10.1016/j.cels.2015.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li J, et al. Integrated systems analysis reveals a molecular network underlying autism spectrum disorders. Mol Syst Biol. 2014;10:774–774. doi: 10.15252/msb.20145487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Replication TDG, et al. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat Genet. 2012;44:981–990. doi: 10.1038/ng.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lage K, et al. Genetic and environmental risk factors in congenital heart disease functionally converge in protein networks driving heart development. Proc Natl Acad Sci USA. 2012;109:14035–14040. doi: 10.1073/pnas.1210730109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lage K. Protein-protein interactions and genetic diseases: the interactome. Biochim Biophys Acta. 2014;1842:1971–1980. doi: 10.1016/j.bbadis.2014.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pickrell JK. Joint analysis of functional genomic data and genome-wide association studies of 18 human traits. Am J Hum Genet. 2014;94:559–573. doi: 10.1016/j.ajhg.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Albert FW, Kruglyak L. The role of regulatory variation in complex traits and disease. Nat Rev Genet. 2015;16:197–212. doi: 10.1038/nrg3891. [DOI] [PubMed] [Google Scholar]
- 38.Nicolae DL, et al. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet. 2010;6:e1000888. doi: 10.1371/journal.pgen.1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schaub MA, Boyle AP, Kundaje A, Batzoglou S, Snyder M. Linking disease associations with regulatory information in the human genome. Genome Res. 2012;22:1748–1759. doi: 10.1101/gr.136127.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Karczewski KJ, et al. Systematic functional regulatory assessment of disease-associated variants. Proc Natl Acad Sci USA. 2013;110:9607–9612. doi: 10.1073/pnas.1219099110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maurano MT, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gusev A, et al. Partitioning heritability of regulatory and cell-type-specific variants across 11 common diseases. Am J Hum Genet. 2014;95:535–552. doi: 10.1016/j.ajhg.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Finucane HK, et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat Genet. 2015;47:1228–1235. doi: 10.1038/ng.3404. References 42 and 43 elucidate the relative contribution of regulatory (non-protein-coding) variation to human diseases and traits. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Spain SL, Barrett JC. Strategies for fine-mapping complex traits. Hum Mol Genet. 2015;24:R111–R119. doi: 10.1093/hmg/ddv260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Majithia AR, et al. Rare variants in PPARG with decreased activity in adipocyte differentiation are associated with increased risk of type 2 diabetes. Proc Natl Acad Sci USA. 2014;111:13127–13132. doi: 10.1073/pnas.1410428111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.ENCODE Project Consortium et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kundaje A, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Adrianto I, et al. Association of a functional variant downstream of TNFAIP3 with systemic lupus erythematosus. Nat Genet. 2011;43:253–258. doi: 10.1038/ng.766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gjoneska E, et al. Conserved epigenomic signals in mice and humans reveal immune basis of Alzheimer’s disease. Nature. 2015;518:365–369. doi: 10.1038/nature14252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Claussnitzer M, et al. FTO obesity variant circuitry and adipocyte browning in humans. N Engl J Med. 2015;373:895–907. doi: 10.1056/NEJMoa1502214. This paper provides a mechanistic explanation for the influence of the FTO locus on human obesity, using multiple omics technologies to bridge GWAS results to physiology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Poldrack RA, et al. Long-term neural and physiological phenotyping of a single human. Nat Commun. 2015;6:8885. doi: 10.1038/ncomms9885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Piening BD, et al. Integrative personal omics profiles during periods of weight gain and loss. Cell Syst. 2018 doi: 10.1016/j.cels.2017.12.013. https://doi.org/10.1016/j.cels.2017.12.013. [DOI] [PMC free article] [PubMed]
- 53.Integrative HMP (iHMP) Research Network Consortium. The Integrative Human Microbiome Project: dynamic analysis of microbiome–host omics profiles during periods of human health and disease. Cell Host Microbe. 2014;16:276–289. doi: 10.1016/j.chom.2014.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Price ND, et al. A wellness study of 108 individuals using personal, dense, dynamic data clouds. Nat Biotechnol. 2017;57:289–756. doi: 10.1038/nbt.3870. References 6 and 54 report on the use of multiple omics assays longitudinally within the same individual or individuals to influence health outcomes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guo L, et al. Plasma metabolomic profiles enhance precision medicine for volunteers of normal health. Proc Natl Acad Sci USA. 2015;112:E4901–E4910. doi: 10.1073/pnas.1508425112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lek M, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.The GTEx Consortium. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.eGTEx Project. Enhancing GTEx by bridging the gaps between genotype, gene expression, and disease. Nat Genet. 2017;49:1664–1670. doi: 10.1038/ng.3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Frattini V, et al. The integrated landscape of driver genomic alterations in glioblastoma. Nat Genet. 2013;45:1141–1149. doi: 10.1038/ng.2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kumar A, et al. Substantial interindividual and limited intraindividual genomic diversity among tumors from men with metastatic prostate cancer. Nat Med. 2016;22:369–378. doi: 10.1038/nm.4053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sato Y, et al. Integrated molecular analysis of clearcell renal cell carcinoma. Nat Genet. 2013;45:860–867. doi: 10.1038/ng.2699. References 59 and 62 characterize molecular signatures of cancers using genome, transcriptome and methylome sequencing to identify driver genes and subtypes of cancers. [DOI] [PubMed] [Google Scholar]
- 63.Wang J, et al. Proteogenomic characterization of human colon and rectal cancer. Nature. 2014;513:382–387. doi: 10.1038/nature13438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mertins P, et al. Proteogenomics connects somatic mutations to signalling in breast cancer. Nature. 2016;534:55–62. doi: 10.1038/nature18003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu T, et al. Integrated proteogenomic characterization of human high-grade serous ovarian cancer. Cell. 2016;166:755–765. doi: 10.1016/j.cell.2016.05.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yu KH, et al. Predicting non-small cell lung cancer prognosis by fully automated microscopic pathology image features. Nat Commun. 2016;7:12474. doi: 10.1038/ncomms12474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yu KH, Snyder M. Omics profiling in precision oncology. Mol Cell Proteom. 2016;15:2525–2536. doi: 10.1074/mcp.O116.059253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Polak P, et al. Cell-of-origin chromatin organization shapes the mutational landscape of cancer. Nature. 2015;518:360–364. doi: 10.1038/nature14221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yao L, Tak YG, Berman BP, Farnham PJ. Functional annotation of colon cancer risk SNPs. Nat Commun. 2014;5:5114. doi: 10.1038/ncomms6114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Melton C, Reuter JA, Spacek DV, Snyder M. Recurrent somatic mutations in regulatory regions of human cancer genomes. Nat Genet. 2015;47:710–716. doi: 10.1038/ng.3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Araya CL, et al. Identification of significantly mutated regions across cancer types highlights a rich landscape of functional molecular alterations. Nat Genet. 2015;48:117–125. doi: 10.1038/ng.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chong JX, et al. The genetic basis of Mendelian phenotypes: discoveries, challenges, and opportunities. Am J Hum Genet. 2015;97:199–215. doi: 10.1016/j.ajhg.2015.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ritchie MD, Holzinger ER, Li R, Pendergrass SA, Kim D. Methods of integrating data to uncover genotype-phenotype interactions. Nat Rev Genet. 2015;16:85–97. doi: 10.1038/nrg3868. [DOI] [PubMed] [Google Scholar]
- 74.Fridley BL, Lund S, Jenkins GD, Wang LA. Bayesian integrative genomic model for pathway analysis of complex traits. Genet Epidemiol. 2012;36:352–359. doi: 10.1002/gepi.21628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Holzinger ER, Dudek SM, Frase AT, Pendergrass SA, Ritchie MD. ATHENA: the analysis tool for heritable and environmental network associations. Bioinformatics. 2014;30:698–705. doi: 10.1093/bioinformatics/btt572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Argelaguet R, et al. Multi-omics factor analysis disentangles heterogeneity in blood cancer. 2017 bioRxiv https://doi.org/10.1101/217554.
- 77.Khera AV, et al. Genetic risk, adherence to a healthy lifestyle, and coronary disease. N Engl J Med. 2016;375:2349–2358. doi: 10.1056/NEJMoa1605086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wray NR, et al. Research review: polygenic methods and their application to psychiatric traits. J Child Psychol Psychiatry. 2014;55:1068–1087. doi: 10.1111/jcpp.12295. [DOI] [PubMed] [Google Scholar]
- 79.Martin AR, et al. Human demographic history impacts genetic risk prediction across diverse populations. Am J Hum Genet. 2017;100:635–649. doi: 10.1016/j.ajhg.2017.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Manrai AK, et al. Genetic misdiagnoses and the potential for health disparities. N Engl J Med. 2016;375:655–665. doi: 10.1056/NEJMsa1507092. References 79 and 80 highlight the challenges of using polygenic risk scores on under-studied populations. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Stein LD. The case for cloud computing in genome informatics. Genome Biol. 2010;11:207. doi: 10.1186/gb-2010-11-5-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dudley JT, Butte AJ. In silico research in the era of cloud computing. Nat Biotechnol. 2010;28:1181–1185. doi: 10.1038/nbt1110-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wall JD, et al. Estimating genotype error rates from high-coverage next-generation sequence data. Genome Res. 2014;24:1734–1739. doi: 10.1101/gr.168393.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 86.Ding L, Wendl MC, McMichael JF, Raphael BJ. Expanding the computational toolbox for mining cancer genomes. Nat Rev Genet. 2014;15:556–570. doi: 10.1038/nrg3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gerlinger M, et al. Ultra-deep T cell receptor sequencing reveals the complexity and intratumour heterogeneity of T cell clones in renal cell carcinomas. J Pathol. 2013;231:424–432. doi: 10.1002/path.4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bianchi DW, et al. Noninvasive prenatal testing and incidental detection of occult maternal malignancies. JAMA. 2015;314:162–169. doi: 10.1001/jama.2015.7120. [DOI] [PubMed] [Google Scholar]
- 89.Cheon JY, Mozersky J, Cook-Deegan R. Variants of uncertain significance in BRCA: a harbinger of ethical and policy issues to come? Genome Med. 2014;6:121. doi: 10.1186/s13073-014-0121-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lappalainen T, et al. Transcriptome and genome sequencing uncovers functional variation in humans. Nature. 2013;501:506–551. doi: 10.1038/nature12531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Minikel EV, et al. Quantifying prion disease penetrance using large population control cohorts. Sci Transl Med. 2016;8:322ra9. doi: 10.1126/scitranslmed.aad5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Whiffin N, et al. Using high-resolution variant frequencies to empower clinical genome interpretation. Genet Med. 2017;19:1151–1158. doi: 10.1038/gim.2017.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Green RC, et al. ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet Med. 2013;15:565–574. doi: 10.1038/gim.2013.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Knoppers BM, Zawati MH, Sénécal K. Return of genetic testing results in the era of whole-genome sequencing. Nat Rev Genet. 2015;16:553–559. doi: 10.1038/nrg3960. [DOI] [PubMed] [Google Scholar]
- 95.Kasowski M, et al. Variation in transcription factor binding among humans. Science. 2010;328:232–235. doi: 10.1126/science.1183621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mele M, et al. The human transcriptome across tissues and individuals. Science. 2015;348:660–665. doi: 10.1126/science.aaa0355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wang ET, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Andersson R, et al. An atlas of active enhancers across human cell types and tissues. Nature. 2014;507:455–461. doi: 10.1038/nature12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Marbach D, et al. Tissue-specific regulatory circuits reveal variable modular perturbations across complex diseases. Nat Methods. 2016;13:366–370. doi: 10.1038/nmeth.3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Osborn MJ, et al. CRISPR/Cas9 targeted gene editing and cellular engineering in Fanconi anemia. Stem Cells Dev. 2016;25:1591–1603. doi: 10.1089/scd.2016.0149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Carmichael N, Tsipis J, Windmueller G, Mandel L, Estrella E. ‘Is it going to hurt?’: the impact of the diagnostic Odyssey on children and their families. J Genet Counsel. 2014;24:325–335. doi: 10.1007/s10897-014-9773-9. [DOI] [PubMed] [Google Scholar]
- 102.Burke W, Zimmern RL, Kroese M. Defining purpose: a key step in genetic test evaluation. Genet Med. 2007;9:675–681. doi: 10.1097/gim.0b013e318156e45b. [DOI] [PubMed] [Google Scholar]
- 103.Mega JL, et al. Genetic risk, coronary heart disease events, and the clinical benefit of statin therapy: an analysis of primary and secondary prevention trials. Lancet. 2015;385:2264–2271. doi: 10.1016/S0140-6736(14)61730-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Natarajan P, et al. Polygenic risk score identifies subgroup with higher burden of atherosclerosis and greater relative benefit from statin therapy in the primary prevention setting. Circulation. 2017 doi: 10.1161/CIRCULATIONAHA.116.024436. https://doi.org/10.1161/CIRCULATIONAHA.116.024436. References 77, 103 and 104 discuss using polygenic risk scores to stratify patients with heart disease into risk groups that respond differently to statin treatment. [DOI] [PMC free article] [PubMed]
- 105.Maier RM, Visscher PM, Robinson MR, Wray NR. Embracing polygenicity: a review of methods and tools for psychiatric genetics research. Psychol Med. 2017 doi: 10.1017/S0033291717002318. https://doi.org/10.1017/S0033291717002318. [DOI] [PMC free article] [PubMed]
- 106.Rietveld CA, et al. GWAS of 126,559 individuals identifies genetic variants associated with educational attainment. Science. 2013;340:1467–1471. doi: 10.1126/science.1235488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Li X, et al. Digital health: tracking physiomes and activity using wearable biosensors reveals useful health-related information. PLoS Biol. 2017;15:e2001402. doi: 10.1371/journal.pbio.2001402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Craig DW, et al. Genome and transcriptome sequencing in prospective metastatic triple-negative breast cancer uncovers therapeutic vulnerabilities. Mol Cancer Ther. 2013;12:104–116. doi: 10.1158/1535-7163.MCT-12-0781. [DOI] [PubMed] [Google Scholar]
- 109.Borad MJ, et al. Integrated genomic characterization reveals novel, therapeutically relevant drug targets in FGFR and EGFR pathways in sporadic intrahepatic cholangiocarcinoma. PLoS Genet. 2014;10:e1004135. doi: 10.1371/journal.pgen.1004135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kostic AD, Xavier RJ, Gevers D. The microbiome in inflammatory bowel disease: current status and the future ahead. Gastroenterology. 2014;146:1489–1499. doi: 10.1053/j.gastro.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Morgan XC, et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012;13:R79. doi: 10.1186/gb-2012-13-9-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Larsen N, et al. Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PLoS ONE. 2010;5:e9085. doi: 10.1371/journal.pone.0009085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Turnbaugh PJ, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Turnbaugh PJ, et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 115.Levy M, Kolodziejczyk AA, Thaiss CA, Elinav E. Dysbiosis and the immune system. Nat Rev Immunol. 2017;17:219–232. doi: 10.1038/nri.2017.7. [DOI] [PubMed] [Google Scholar]
- 116.Benson AK, et al. Individuality in gut microbiota composition is a complex polygenic trait shaped by multiple environmental and host genetic factors. Proc Natl Acad Sci USA. 2010;107:18933–18938. doi: 10.1073/pnas.1007028107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Goodrich JK, et al. Human genetics shape the gut microbiome. Cell. 2014;159:789–799. doi: 10.1016/j.cell.2014.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Knights D, et al. Complex host genetics influence the microbiome in inflammatory bowel disease. Genome Med. 2014;6:119. doi: 10.1186/s13073-014-0107-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Goodrich JK, Davenport ER, Clark AG, Ley RE. The relationship between the human genome and microbiome comes into view. Annu Rev Genet. 2017;51:413–433. doi: 10.1146/annurev-genet-110711-155532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hall AB, Tolonen AC, Xavier RJ. Human genetic variation and the gut microbiome in disease. Nat Rev Genet. 2017;18:690–699. doi: 10.1038/nrg.2017.63. [DOI] [PubMed] [Google Scholar]
- 121.Chu H, et al. Gene-microbiota interactions contribute to the pathogenesis of inflammatory bowel disease. Science. 2016;352:1116–1120. doi: 10.1126/science.aad9948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Holmes E, Li JV, Athanasiou T, Ashrafian H, Nicholson JK. Understanding the role of gut microbiome–host metabolic signal disruption in health and disease. Trends Microbiol. 2011;19:349–359. doi: 10.1016/j.tim.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 123.Wang C, et al. High-throughput, high-fidelity HLA genotyping with deep sequencing. Proc Natl Acad Sci USA. 2012;109:8676–8681. doi: 10.1073/pnas.1206614109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wittig M, et al. Development of a high-resolution NGS-based HLA-typing and analysis pipeline. Nucleic Acids Res. 2015;43:e70. doi: 10.1093/nar/gkv184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zhang MJ, et al. Comparison of outcomes after HLA-matched sibling and unrelated donor transplantation for children with high-risk acute lymphoblastic leukemia. Biol Blood Marrow Transplant. 2012;18:1204–1210. doi: 10.1016/j.bbmt.2012.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.McCarroll SA, et al. Donor-recipient mismatch for common gene deletion polymorphisms in graft-versus-host disease. Nat Genet. 2009;41:1341–1344. doi: 10.1038/ng.490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Li YR, Levine JE, Hakonarson H, Keating BJ. Making the genomic leap in HCT: application of second-generation sequencing to clinical advances in hematopoietic cell transplantation. Eur J Hum Genet. 2014;22:715–723. doi: 10.1038/ejhg.2013.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Snyder TM, Khush KK, Valantine HA, Quake SR. Universal noninvasive detection of solid organ transplant rejection. Proc Natl Acad Sci USA. 2011;108:6229–6234. doi: 10.1073/pnas.1013924108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.De Vlaminck I, et al. Circulating cell-free DNA enables noninvasive diagnosis of heart transplant rejection. Sci Transl Med. 2014;6:241ra77. doi: 10.1126/scitranslmed.3007803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.De Vlaminck I, et al. Noninvasive monitoring of infection and rejection after lung transplantation. Proc Natl Acad Sci USA. 2015;112:13336–13341. doi: 10.1073/pnas.1517494112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Yang JYC, Sarwal MM. Transplant genetics and genomics. Nat Rev Genet. 2017;2003:449. doi: 10.1038/nrg.2017.12. [DOI] [PubMed] [Google Scholar]