Abstract
Schlafen (SLFN) proteins have been suggested to play important functions in cell proliferation and immune cell development. In this study, we determined the antiviral activities of putative RNA-helicase domain-containing SLFN14. Murine SLFN14 expression was specifically induced by TLR3-mediated pathways and type I interferon (IFN) in RAW264.7 mouse macrophages. To examine the role of SLFN during viral infection, cells were infected with either wild-type PR8 or delNS1/PR8 virus. SLFN14 expression was specifically induced following influenza virus infection. Overexpression of SLFN14 in A549 cells reduced viral replication, whereas knockdown of SLFN14 in RAW264.7 cells enhanced viral titers. Furthermore, SLFN14 promoted the delay in viral NP translocation from cytoplasm to nucleus and enhanced RIG-I-mediated IFN-β signaling. In addition, SLFN14 over-expression promoted antiviral activity against varicella zoster virus (VZV), a DNA virus. In conclusion, our data suggest that SLFN14 is a novel antiviral factor for both DNA and RNA viruses.
Keywords: SLFN14, Influenza, VZV, Interferon, Anti-viral
1. Introduction
Schlafen (SLFN) family genes were first described as having an important regulatory function affecting thymocyte maturation in mice (Schwarz et al., 1998). SLFN family genes are differentially regulated and expressed in a cell type-specific pattern (Mavrommatis et al., 2013a,b; Neumann et al., 2008). In mice, ten SLFNs have been identified, whereas there are six human SLFN isoforms (SLFN5, SLFN11, SLFN12, SLFN12L, SLFN13, and SLFN14). All of these human SLFNs, except SLFN12 and 12L, possess a putative ATPases-Associated with various cellular activities (AAA) domain and a putative RNA helicase motif, which is similar to the DNA/RNA helicase domains of nucleic acid sensors such as retinoic acid inducible gene-I (RIG-I) and melanoma differentiation associate gene 5 (MDA5) (de la Casa-Esperon, 2011). Recent reports suggested that SLFNs are involved in important cellular functions, such as immune cell development (Berger et al., 2010; Ahmadi and Veinotte, 2011) and regulation of tumorigenesis (Companioni Napoles et al., 2017; Mavrommatis et al., 2013a,b). Additionally, SLFN11 has been suggested to be an essential restriction factor for the replication of retroviruses such as human immunodeficiency virus and equine infectious anemia virus (Li et al., 2012; Lin et al., 2016). However, the antiviral activities of other SLFN members are currently unknown.
Influenza virus is an important human respiratory pathogen that can cause severe morbidity and high mortality rates, resulting in 250,000–500,000 deaths annually worldwide. The induction of type I and III interferons (IFNs) and IFN-stimulated genes (ISGs) by activation of antiviral immune responses represent an early and immediate defense in the battle against influenza infection (Hale et al., 2010; Pulendran and Maddur, 2015). RIG-I is a well-characterized influenza virus sensor that is responsible for the induction of IFN antiviral signaling in response to the recognition of a 5′ triphosphorylated RNA virus structure (Pichlmair et al., 2006; Yoneyama et al., 2004). The RIGI helicase domain binds viral dsRNA, and the c-terminal domain (CTD) binds the 5′-triphosphate end. RNA binding through the helicase and CTD domains releases the caspase activation and recruitment domains (CARD), which then recruit and activate the signaling adaptor mitochondrial antivirus signaling protein (MAVS). Upon activation, RIG-I-mediated signaling leads to the phosphorylation and activation of transcription factors such as IFN regulatory factor 3 (IRF3), IRF7, and NF-κB. In addition to RIG-I, MDA5 has been identified as an antiviral effector that suppresses influenza A virus replication (Kato et al., 2006). Additionally, several other helicases have been identified as sensors of influenza A virus (Fullam and Schroder, 2013). These sensors/receptors then trigger the expression of IFN and ISGs, which are important for the control of viral replication (Schneider et al., 2014). The potential role of SLFNs as viral sensors during influenza virus infection has not been examined.
Given that some members of SLFNs have a putative RNA helicase motif that could serve as a virus sensor, we examined the expression of SLFNs in response to Toll-like receptor (TLR)/Rig I-like receptor (RLR) agonists and RNA or DNA viruses. We show that SLFN14 is specifically induced by TLR3 or IFN-β stimulation and influenza virus infection in mouse macrophages. Overexpression of SLFN14 led to the suppression of viral replication of both influenza A virus and varicella zoster virus (VZV), suggesting that it has antiviral activities against a broad array of viruses. These data provide new insights into a novel function of SLFNs in viral infections and suggest that SLFNs could be targets for antiviral therapies.
2. Materials and methods
2.1. Cell culture and reagents
Human lung adenocarcinoma cells (A549), human immortalized keratinocytes (HaCaTs), mouse macrophage RAW 264.7 cells and Madin-Darby canine kidney (MDCK) cells were obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA). A549 cells were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin. MDCK and mouse macrophage RAW 264.7 cells were grown in DMEM supplemented with 10% FBS and 1% penicillin/streptomycin. Normal human dermal fibroblasts (HDFs) were purchased from Lonza, Basel, Switzerland, and grown in fibroblast basal medium (FBM) supplemented with FGM SingleQuots (Lonza). THP-1 human leukemia monocytes were cultured in RPMI 1640 medium supplemented with 10% FBS and 1% penicillin/streptomycin and differentiated into macrophages in the presence of 100 ng/mL PMA (Sigma-Aldrich, St. Louis, MS, USA).
Recombinant human IFN-α, λ1, and λ2 were purchased from R & D Systems (Minneapolis, MI, USA), and IFN-β protein was obtained from PBL Assay Science (Piscataway, NJ, USA). Human IFN neutralizing monoclonal antibody and murine recombinant IFN-β protein were obtained from R & D Systems.
2.2. Virus infection and plaque assay
Human influenza virus A/Puerto-Rico/8/34 (H1N1) PR8 and influenza virus lacking the NS1 open reading frame (delNS1), generated by reverse genetics from PR8 as previously described, was provided by Dr. Adolfo Garcia-Sastre (Icahn School of Medicine at Mount Sinai, NY, USA) (Garcia-Sastre et al., 1998). Seasonal A/H3N2, B/Yamagata, B/Victoria clinical strains were obtained from Korea Bank for Pathogenic Viruses (KBPV) and influenza B/Malaysia/2506/04 (B/Victoria) strains were used. Virus titers were determined by standard plaque assay in MDCK cells with few modifications (Kim et al., 2016; Seong et al., 2016). Briefly, viral supernatants diluted in DMEM were added to MDCK cells in 6-well plates. After 2 h of attachment, viral supernatants were removed, and cells were overlaid with Eagle’s minimum essential medium (EMEM) (without phenol red and with l-glutamine) (Lonza), 1.5% LE agarose (Lonza), and 2 µg/mL (TPCK)-trypsin (Sigma) and then incubated for another 3 days. After incubation, the infected cells were fixed with 4% formaldehyde in phosphate-buffered saline (PBS) and stained with 0.5% crystal violet (JUNSEI, Japan) solution. Plaque forming units (PFUs) were counted.
VZV strain YC01 (GenBank Accession No. KJ808816) has been described previously, and plaque assays were performed with modifications as described (Choi et al., 2015; Kim et al., 2015). Cells were infected with cell-associated VZV at an MOI of 0.01. After 1 h of absorption, the medium was removed, washed, and replaced with new medium. At the indicated times, cells were harvested, diluted 2 fold, and inoculated into confluent monolayer of human fetal fibroblasts (HFFs). After 7 days, cells were fixed with 4% formaldehyde and stained with 0.03% crystal violet. Plaques were counted under a phase-contrast microscope.
2.3. Immunofluorescence assay for influenza NP staining
Recombinant adenovirus expressing pAd-PL (empty vector) or hSLFN14 (pAd-hSLFN14-mStrawberry) was generated by Sirion Biotech (Germany). Cells were infected with Ad-hSLFN14-mStrawberry at an MOI of 1000 and incubated for a few days. A549 cells were infected with PR8 virus at an MOI of 3. At the indicated times, cells were fixed with 4% paraformaldehyde for 20 min and permeablized with 0.1% Triton X-100. Cells were then stained with rabbit polyclonal anti-NP antibodies (1:500 dilution), followed by anti-rabbit FITC conjugated antibody. Coverslips were mounted on glass slides using mounting media containing 4,6-diamidino-2-phenylindole (DAPI) and were examined by confocal microscopy (LSM700; Carl Zeiss).
2.4. Western blot analysis
Protein lysates were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on 10–15% acrylamide gels and transferred to polyvinyldifluoride (PVDF) membranes. The membranes were then incubated in a blocking buffer comprised of 5% (w/v) bovine serum albumin (BSA), 0.2 M Tris base, 1.36 M NaCl, and 0.1% Tween 20 (TBS/T) for 1 h at 25 °C and washed three times (5 min each) with 5 mL of TBS/T. Membranes were incubated overnight with primary antibodies against SLFN14 (SC-248648, Santa Cruz Biotechnology for murine SLFN14; ab106406, Abcam for human SLFN14), SLFN13 (SC-137776, Santa Cruz Biotechnology), RIG-I, MDA5, MAVS (8348, Cell Signaling), phospho-MAPK/total MAPK (9910, 9926, Cell Signaling), phospho-STAT1/total STAT1 (9914, 9939, Cell Signaling), β-actin (AM1021B, Abgent), VZV gE (ab52549, Abcam), VZV IE62 (SC-17525, Santa Cruz Biotechnology), SOCS (8343, Cell Signaling), and anti-myc (2278, Cell Signaling) at 4 °C. For influenza A virus protein expression, anti-NS1 (SC-130568, Santa Cruz Biotechnology), and anti- NP (11675, Sino Biological Inc., Beijing, China) antibodies were used, and for influenza B NP expression, monoclonal antibody (M148, Takara) was used. VZV gE (ab52549) and VZV IE62 (SC-17525) were used to measure VZV protein levels. After washing three times with TBS/T, membranes were incubated with HRP-conjugated anti-rabbit or mouse IgG secondary antibody (Cell Signaling Technology) for 1 h at 25 °C. After washing three times with TBS/T, membranes were incubated with Western Lumi Pico solution (ECL solution kit) (DoGen, Korea). Signals were determined using a Fusion Solo Imaging System (Vilber Lourmat, France). Band intensities were quantified by Fusion- Capt analysis software. A representative image of two to three independent experiments is shown.
2.5. Plasmids, SLFN siRNA, and transfection
The SLFN14-myc plasmid was previously described (Fletcher et al., 2015). For overexpression experiments, plasmids were transfected using Lipofectamine 2000 (Invitrogen) or, for HDF cells, HDF Avalanche transfection reagent (EZ Biosystems, College Park, MD, USA) according to the manufacturers’ instructions.
Cells were seeded in 6-well plates and allowed to grow to over 70% confluency over 24 h. Transient transfections with either scrambled control, human SLFN13, RIG-I, or MDA5 siRNA (Bioneer, Daejeon, Korea), or murine SLFN14 siRNA (Thermo Scientific) were performed with Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol.
2.6. Luciferase reporter assays
HEK293T cells were obtained from ATCC and were seeded into 96-well plates. Cells were transiently transfected with the IFN-β luciferase reporter plasmid (Promega, Madison, WI, USA), together with various expression plasmids. pCMV-NS1-Flag plasmid was purchased from Sino Biological, and pEF-RIG-I-Flag plasmid was a kind gift from Dr. Takashi Fujita (Kyoto University, Japan). As an internal control, 10 ng of pRLTK plasmid was transfected simultaneously with the other plasmids. A dual-Glo luciferase reporter assay system (Promega) was used to measure IFN-β luciferase activity according to the manufacturer’s instructions.
2.7. qRT-PCR
Total cellular RNA was prepared using Trizol reagent (Invitrogen). First-strand synthesis of cDNA from 1 µg of total RNA was performed using M-MLV Reverse Transcription system (Promega) according to the manufacturer’s instructions. Changes in mRNA expression levels were calculated using the comparative Ct method as described previously. Data were normalized to glyceraldehyde 3-phosphate dehydrogenase (GAPDH) expression. Primer sequences are listed in Supplementary Table I. Quantification of cDNA was performed by qRT-PCR using SYBR Green PCR mix (Applied Biosystems, Foster City, CA, USA). Cycling parameters were 95 °C for 10 min, followed by 40 cycles of 95 °C for 30 s and 60 °C for 1 min. The specificity of each reaction was validated by melt curve analysis and agarose gel electrophoresis of PCR products. Expression was normalized using the ΔCt method, in which the amount of target, normalized to an endogenous reference and relative to a calibrator, is calculated as 2−ΔΔCt, where Ct is the cycle number of the detection threshold.
2.8. Statistical analysis
All experiments were repeated independently at least three times. Paired comparisons were performed with Student's t-test. Differences were considered statistically significant at p < 0.05. All analyses were carried out using Prism software (GraphPad Software, Inc., La Jolla, CA, USA).
3. Results
3.1. SLFN expression induced by TLR agonists and IFN in mouse macrophages
In the SLFN family, a C-terminal extension, characterized by a motif that is homologous to the RNA helicase superfamily, exists only in group III SLFNs. For mice, group III includes SLFN5, SLFN8, SLFN9, SLFN10, and SLFN14, whereas for humans, SLFN5, SLFN11, SLFN12, SLFN13, and SLFN14 belong to group III (Mavrommatis et al., 2013a,b).
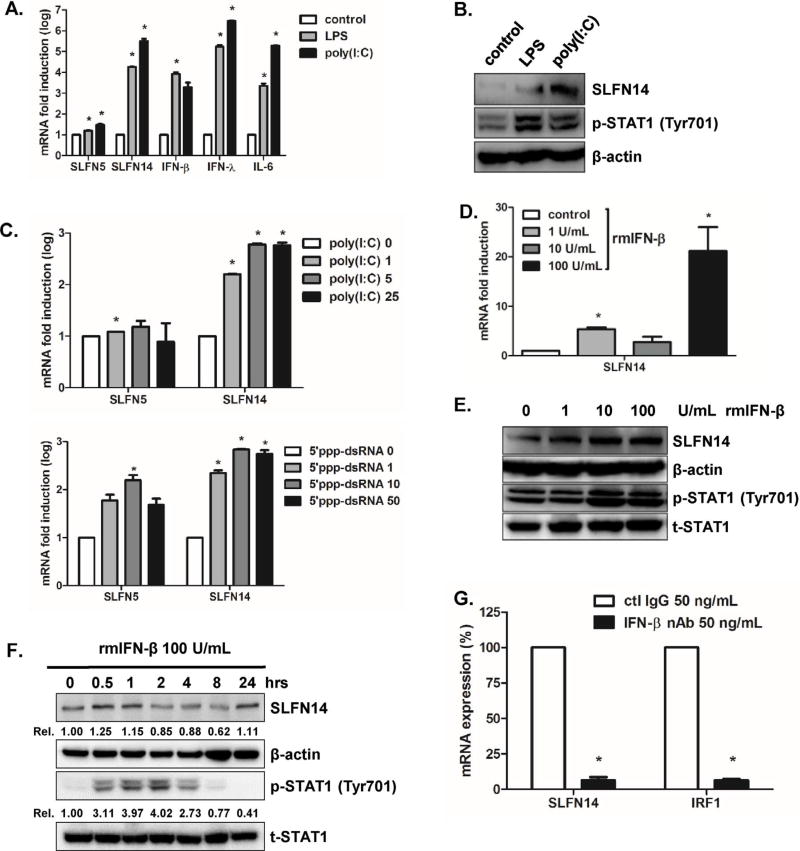
First, we wanted to measure the expression of SLFNs in mouse macrophage RAW 264.7 cells in response to TLR agonists such as lipopolysaccharides (LPS) and polyinosinic-polycytidylic acid (poly [I:C]). Activation of the LPS-mediated TLR4 and poly I:C-stimulated TLR3 pathways resulted in increased transcript levels of both type I and III IFNs (IFN-β and IFN-λ) and proinflammatory cytokine IL-6. In addition, LPS-mediated SLFN14 expression was upregulated more than 4 fold, whereas poly I:C-mediated SLFN14 expression was upregulated more than 5.5 fold (Fig. 1A). Western blot results also confirmed TLR-induced upregulation of SLFN14 protein expression induced by LPS and poly I:C stimulation (Fig. 1B).
Fig. 1.
SLFN expression patterns in response to TLR ligand and IFN stimulation. (A) Mouse macrophage RAW 264.7 cells were stimulated with TLR3 ligand poly I:C (5 µg/mL) or TLR4 ligand lipopolysaccharide (LPS) (100 ng/mL) for 4 h. Total cellular RNA was isolated and murine SLFN5, SLFN14, IFN-β, IFN-λ, and IL-6 mRNA expression was measured using realtime qRT-PCR. The results are shown as the fold induction compared to expression levels in the mock control and are representative of three independent experiments. (B) SLFN14 and phospho-STAT1 protein levels were measured by western blot. Results are representative of three independent experiments. (C) Various concentrations of poly I:C and 5′ triphosphate double-stranded RNA (5′ ppp-dsRNA) were incubated with RAW 264.7 cells for 4 h, and realtime qRT-PCR was performed to determine the fold induction mRNA levels of SLFN5 and SLFN14. (D, E) recombinant murine IFN-β protein were added to cells at different doses, and SLFN14 mRNA and protein expression was measured. (F) Recombinant murine IFN-β protein was added to cells for 0.5, 1, 2, 4, 8, and 24 h and analyzed by western blotting with antibodies specific for SLFN14, phospho-STAT1, and total STAT1 using total cell lysates. Levels of cellular actin are shown as loading controls. Results are representative of three independent experiments. (G) Expression of the SLFN14 gene was quantified by real-time qRT-PCR in A549 cells following treatment with an IFN- β neutralizing monoclonal antibody (nAb). The expression levels of SLFN14 and interferon regulatory factor 1 (IRF1) were normalized to that of GAPDH. The expression level in the control IgG-treated group was arbitrarily set to 100%, and relative expression is shown in the graph. Statistical analysis: *p<0.05 vs. the control IgG-treated group.
5′ triphosphate double-stranded RNA (5′ ppp-dsRNA) is a synthetic ligand for retinoic acid-inducible protein I (RIG-I), whereas poly I:C is a synthetic analog of double stranded RNA (dsRNA). We next examined whether poly I:C and 5′ ppp-dsRNA stimulation could modulate the expression of SLFN14. Increasing concentrations of poly I:C and 5′ ppp-dsRNA positively correlated with the fold induction levels of SLFN14 mRNA by real-time qRT-PCR, whereas the induction level of SLFN5 expression was only slightly, and not significantly, increased (Fig. 1C).
In addition to TLR activation, we wanted to determine whether type I IFN pretreatment could modulate SLFN14 expression. Various doses of recombinant mouse IFN-β protein were added to RAW 264.7 cells. Realtime qRT-PCR and western blot results indicated dose-dependent elevation of SLFN14 mRNA and protein expression levels, respectively (Fig. 1D and E). Furthermore, SLFN14 expression was increased at early time points, but decreased slightly at 2 h post IFN-β treatment, indicating the early induction of SLFN14 expression following IFN signaling (Fig. 1F). To determine whether SLFN14 expression was dependent on the IFN-β pathway, we treated A549 cells with IFN-β blocking antibody and performed realtime qRT-PCR to measure the relative expression levels of SLFN14 and IRF1. As shown in Fig. 1G, there was significant suppression of SLFN14 and IRF1 mRNA expression following IFN-β-blocking antibody treatment.
3.2. Cell type-specific expression patterns of SLFN family members in human cells
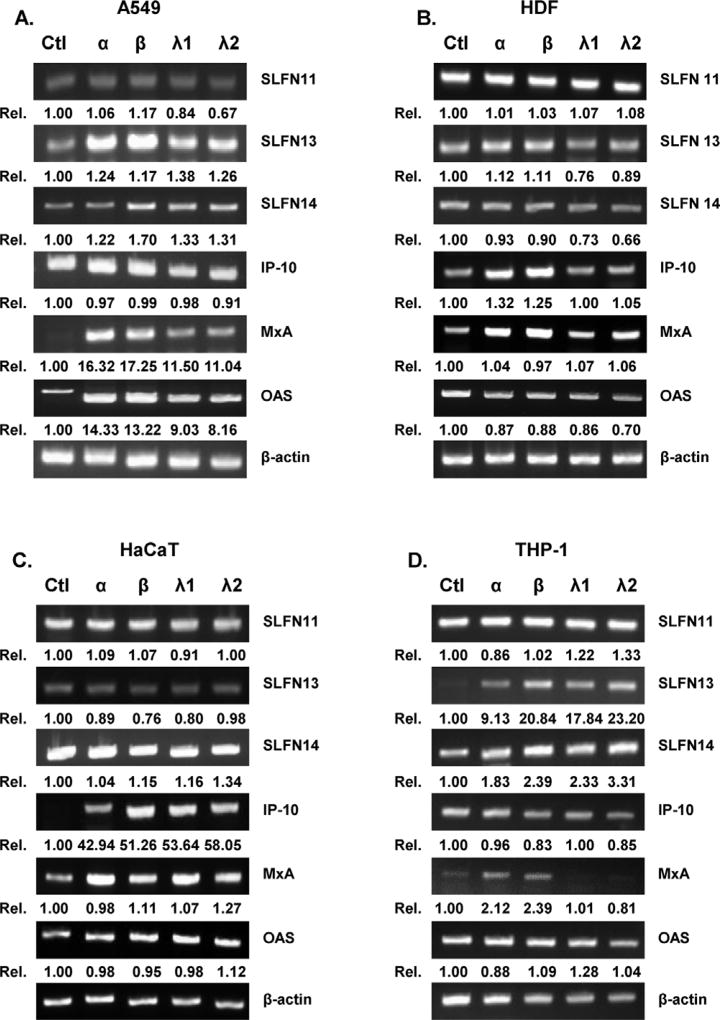
To determine whether SLFN14 expression is induced by type I and III IFN stimulation, various types of cells were exposed to recombinant IFN proteins, and qRT-PCR was performed. Recombinant human IFNs α and β were used as representative type I IFNs, and IFNs λ1 (IL-29) and λ2 (IL-28A) were used as representative type III IFNs. Interestingly, SLFN11, 13, and 14 expression levels in human dermal fibroblasts (HDFs) and immortalized HaCaT keratinocytes were similar, even in the presence of the IFNs. However, in human lung adenocarcinoma A549 and differentiated macrophage THP-1 cells, all four IFN treatments resulted in increased expression of SLFN13 and SLFN14 (Fig. 2A–D).
Fig. 2.
IFN-induced SLFN expression patterns in human cells. (A) Recombinant human IFN-α (10 ng/mL), IFN-β (10 ng/mL), IFN-λ1 (10 ng/mL), and IFN-λ2 (10 ng/mL) were added to the following cells: A549 human lung adenocarcinoma (A549) (A), HDFs (human dermal fibroblasts) (B), HaCaTs (human immortalized keratinocytes) (C), and THP-1 (human monocytes differentiated by PMA treatment) (D). RT-PCR results indicated the expression levels of SLFN11, SLFN13, SLFN14, IFN-γ inducible protein 10 (IP-10), myxovirus resistance gene (MxA), 2′5′ oligoadenylate synthetase 1 (OAS1), and β-actin. Quantitative densitometric analysis of RT-PCR is presented, with normalized densitometric units plotted against treatment (shown as numbers).
3.3. Influenza infection results in the upregulation of Schlafen family
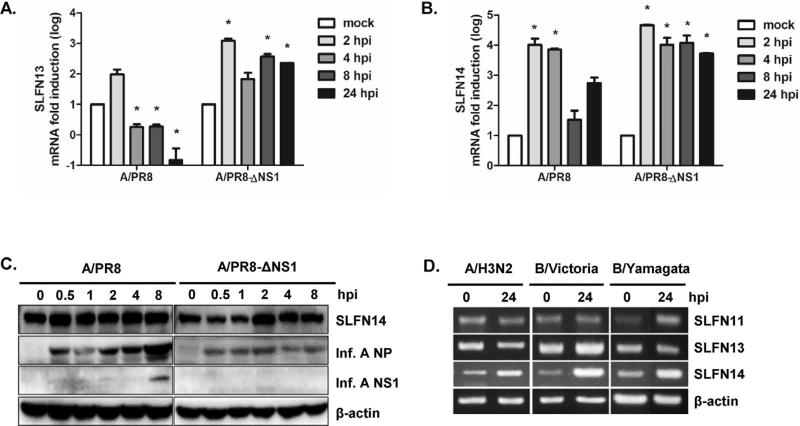
Next, we determined the effect of influenza infection on SLFN gene expression. A549 cells were infected with wildtype PR8 or delNS1/PR8 influenza A (H1N1) virus, and total RNA was isolated at 0, 2, 4, 8, and 24 h post infection (hpi). PR8 infection of A549 cells resulted in significant induction of SLFN13 and SLFN14 gene expression at 2 hpi, and then a gradual reduction in expression levels in the late phases of infection (4–24 hpi) (Fig. 3A and B). On the other hand, persistently elevated mRNA levels of both SLFN13 and SLFN14 were observed in A549 cells infected with delNS1/PR8 virus.
Fig. 3.
Induction of SLFN13 and SLFN14 following influenza virus infection. A549 cells were infected with the human influenza virus strain PR8 or PR8/delNS1 (multiplicity of infection = 1) for the indicated lengths of time. Host mRNA expression was measured by real-time qRT-PCR for SLFN13 (A) and SLFN14 (B). The expression of target genes was normalized to that of GAPDH. Expression in the mock-infected control was set to 1, and other samples were normalized to this value. Data are shown as the mean ± SEM of three independent experiments. Statistical analysis: *p < 0.05 compared with mock-infected cells at each timepoint. (C) SLFN14 protein levels were measured in influenza A virus-infected mouse macrophages RAW 264.7 cells. The images shown are representative of three independent experiments. (D) A549 cells were infected with clinical influenza virus strains, including seasonal A/H3N2, B/Victoria, and B/Yamagata, at a multiplicity of infection of 0.1 for 24 h, and RT-PCR was performed.
In RAW 264.7 cells, SLFN14 protein expression was upregulated at 2 h after delNS1/PR8 virus infection, whereas the level of SLFN14 protein expression remained similar after PR8 infection (Fig. 3C). We also examined whether various clinical strains of influenza could modulate SLFN expression. Infection with each clinical virus strain (A/H3N2, B/Victoria, and B/Yamagata) resulted in increased SLFN14 mRNA expression (Fig. 3D). These results suggest that expression kinetics of SLFN14 may differ depending on cell type, and that NS1 may suppress the induction of SLFN genes at later time points in infection.
3.4. SLFN14 overexpression moderates viral replication
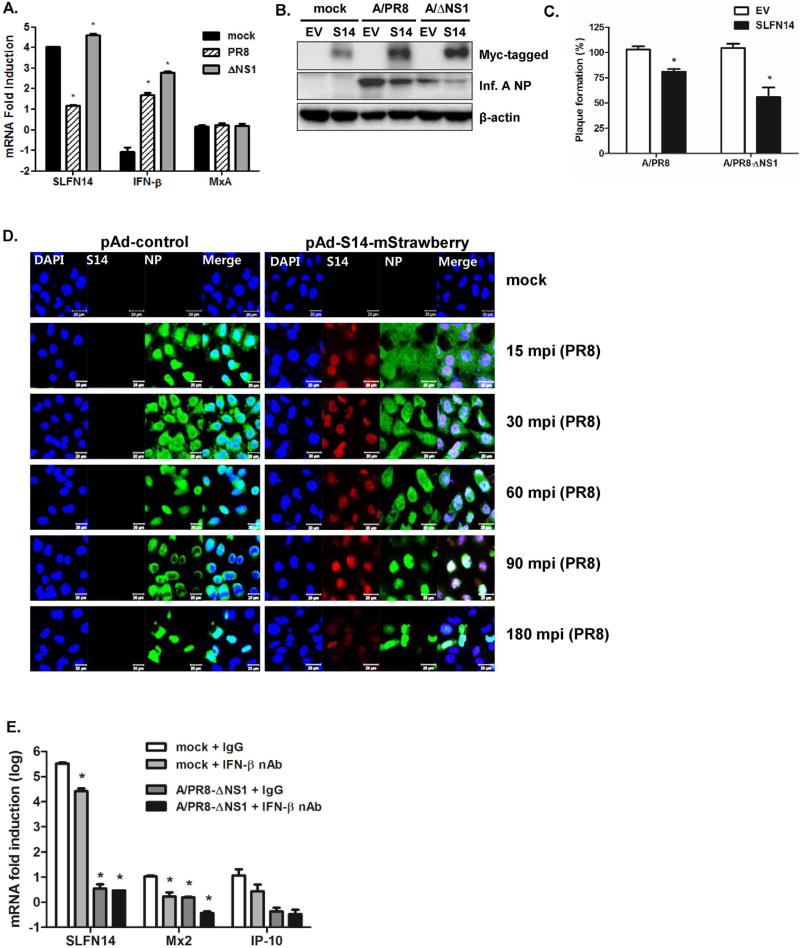
Next, we investigated whether SLFN14 overexpression affects viral replication efficiency and the anti-viral immune response. Realtime qRT-PCR was performed to measure the mRNA fold induction levels of SLFN14, IFN-β, and myxovirus resistance protein A (MxA) in response to mock control, PR8, and delNS1/PR8 infection. The SLFN14 expression level was more significantly induced in response to delNS1/PR8 than PR8 virus (Fig. 4A). SLFN14 overexpression did not increase MxA expression; however, PR8 or delNS1/PR8-induced IFN-β expression was increased by more than 4–6 folds in SLFN14-overexpressing cells, compared with that in control vector-transfected cells. To test whether SLFN14 overexpression modulates viral protein expression, a western blot was performed. Influenza A NP expression was significantly reduced in SLFN14-overexpressing cells (Fig. 4B). Consistent with this result, plaque assays confirmed that SLFN14 overexpression significantly suppressed the PR8/delNS1 virus titer by approximately 43.6% (Fig. 4C).
Fig. 4.
Effect of SLFN14 overexpression on influenza virus replication. (A) A549 cells were transiently transfected with either empty vector (EV) or the SLFN14-myc expression plasmid for 24 h. The next day, cells were infected with mock control, PR8, or delNS1/PR8 virus at a multiplicity of infection (MOI) of 1.0. Changes in the transcriptional expression of MxA, IFN-β, and SLFN14 were measured using real-time qRT-PCR. Transcript expression levels were calculated in relation to the expression level of GAPDH and expressed as a fold-change in comparison with the expression level in EV-transfected control cells. *p < 0.05 vs. mock-infected control cells. (B) Western blotting was performed with antibodies specific for influenza NP. The transfection efficiency of SLFN14 was confirmed by measuring myc tag expression levels. Levels of cellular actin are shown as loading controls. Results are representative of three independent experiments. (C) Plaque assays were performed. Data are presented as the percentage (%) decrease in the number of plaque-forming units with respect to that of the control-treated cells, which was normalized to 100%. The average of all three experiments is shown. *p < 0.05 vs. EV-transfected cells. (D) A549 cells were pAd control (empty vector)-infected or infected with pAd-hSLFN14-mStrawberry (red) at a multiplicity of infection (MOI) of 1000 and subsequently infected with PR8 viruses at an MOI of 3 for the indicated times. Anti-influenza NP antibodies were used to detect influenza A virus NP protein (green) by confocal microscopy. Data shown are representative of results from three independent experiments; minutes post influenza virus infection (mpi); scale bar = 20 µM (E) Expression of the SLFN14, Mx2 and IP-10 gene was quantified by real-time qRT-PCR in A549 cells following treatment with an IFN- β neutralizing monoclonal antibody (nAb). *p < 0.05 vs. EV-transfected cells. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
To further confirm the results of transient SLFN14 overexpression, a recombinant plasmid adenovirus (pAd) expressing human SLFN14 and mStrawberry (red) tag (pAd-SLFN14-mStrawberry) was generated. A549 cells were infected with control adenovirus containing empty vector (pAd-control) or pAd-SLFN14-mStrawberry virus at a multiplicity of infection (MOI) of 1000 plaque-forming units (pfu)/cell. After 72 h, cells were infected with PR8 at an MOI of 3, and viral NP protein was stained to visualize virus entry into nucleus. Starting at 15 min post infection (mpi), we detected green fluorescence inside the nucleus of control adenovirus-treated cells. However, pAd-SLFN14-mStrawberry-expressing cells showed delayed accumulation (beginning at 90 mpi) of NP staining in the nucleus, suggesting that SLFN14 overexpression delayed translocation of viral NP into the nucleus (Fig. 4D). Additionally, we examined the effect of IFN blocking on SLFN14-mediated signaling and found that IFN blocking reduces the expression levels of Mx2 and IP-10 in SLFN14-overexpressing cells (Fig. 4E).
3.5. SLFN14 enhances RIG-I-mediated signaling pathways
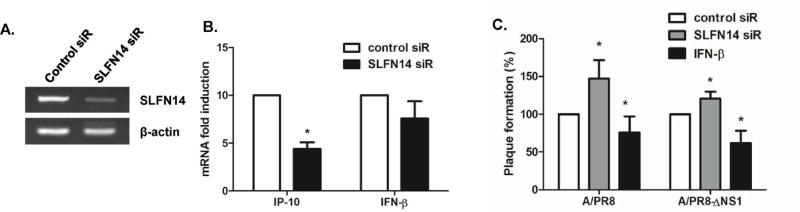
To further evaluate the effects of SLFN on viral replication efficiency, we used mouse SLFN14 siRNA to transfect RAW 264.7 cells. SLFN knockdown efficiency was determined by measuring SLFN14 mRNA levels, and qRT-PCR results revealed downregulation of SLFN14 expression following siRNA treatment (Fig. 5A). To characterize the effects of SLFN14 knockdown on the expression of IFN and ISGs, we measured IFN-β and IFN-g inducible protein 10 (IP-10) by qRT-PCR. SLFN14 knockdown significantly decreased SLFN14 mRNA levels and reduced expression levels of both IFN-β and IP-10, although differences in IFN-β mRNA expression levels were not significant (Fig. 5B). In addition, there was a significant increase in the numbers of viral plaques detected in supernatants from SLFN14 siRNA-transfected PR8-infected cells, suggesting that viral yield was affected by SLFN14 expression (Fig. 5C). As a positive control, recombinant murine IFN-β-treated samples were used; IFN-β pre-treatment in RAW264.7 cells led to suppression of plaque formation. In addition, SLFN14 knockdown in delNS1/PR8-infected cells also enhanced virus titers. These results revealed that SLFN14 is involved in the control of viral replication in mouse macrophages.
Fig. 5.
Effect of SLFN14 knockdown on antiviral responses (A) RAW 264.7 cells were treated with either control siRNA or SLFN14 siRNA. After 24 h transfection, cells were mock infected or infected with A/PR8 or PR8/delNS1 at a multiplicity of infection of 1. The knockdown efficiency of SLFN14 siRNA was determined by measuring the expression levels of SLFN14 mRNA. (B) The effect of SLFN14 knockdown on IP-10 and IFN-β gene expression was also measured by realtime qRT-PCR. (C) Cells were pre-treated with recombinant IFN-β or transfected with either control siRNA or SLFN14 siRNA. Progeny viral titers were calculated and expressed as plaque-forming units (PFU)/mL. Data are shown as means ± SEM of three different experiments and are presented as the percentage relative to the control siRNA sample. Statistical analysis: *p < 0.05 compared to control siRNA-transfected cells.
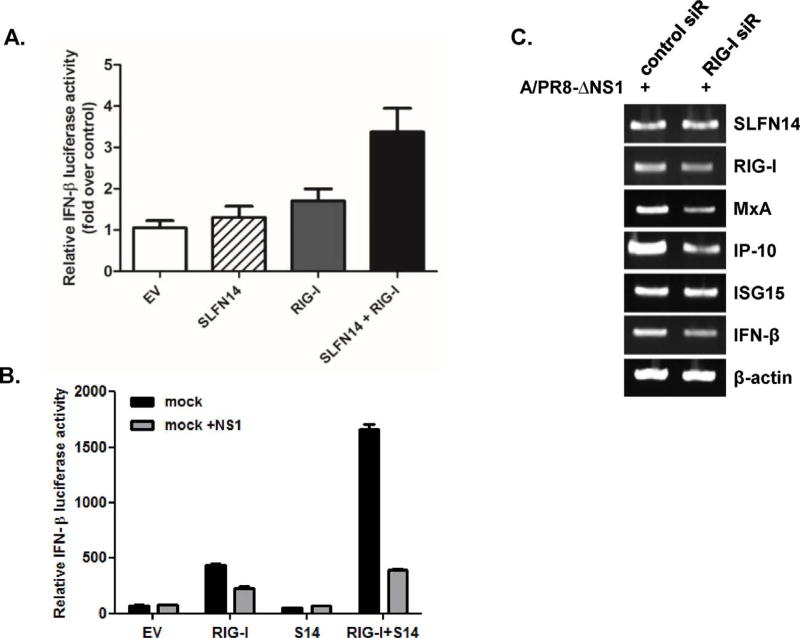
Next, we wanted to test whether SLFN14 enhances RIG-I-mediated signaling. HEK293T cells were transiently transfected with an IFN-β promoter reporter gene along with RIG-I or SLFN14. RIG-I-mediated IFN-β promoter activation was enhanced by SLFN14 expression (Fig. 6A). The interaction of influenza NS1 gene with RIG-I has been well characterized, and NS1 is known to inhibit RIG-I-mediated activation of the IFN-β promoter (Ruckle et al., 2012). We also tested whether SLFN14-promoted RIG-I signaling is dependent on NS1. Cotransfection of the influenza NS1 gene in RIG-I- and SLFN14-transfected cells led to a significant decrease in IFN-β luciferase activity (Fig. 6B). Next, to determine whether SLFN expression is dependent on viral RNA sensor RIG-I, A549 cells were transfected with siRNAs targeting RIG-I prior to infection with PR8/delNS1 virus. Knockdown of RIG-I led to suppression of the expression of RIG-I and downstream molecules such as MxA, IP-10, ISG15, and IFN-β, but there was only a minimal change in SLFN14 expression, suggesting that SLFN14 expression is not dependent on RIG-I expression (Fig. 6C).
Fig. 6.
SLFN14 promotes RIG-I-mediated signaling (A) HEK293T cells were transiently transfected with empty vector (EV), SLFN14, RIG-I, or SLFN14/RIG-I, along with reporter plasmids and a Renilla luciferase plasmid (internal control). Cells were stimulated with either DMSO control or poly I:C (5 µg/mL) Relative IFN-β luciferase activity is shown as fold induction over DMSO control. A representative of three independent experiments is presented. (B) Plasmids expressing RIG-I, SLFN14 (SL14) and influenza NS1 were transfected into HEK293T cells, together with the IFN-β reporter plasmids and Renilla luciferase plasmid (internal control). After 24 h, luciferase activity was measured. (C) Knockdown of RIG-I was performed with siRNA transfection. mRNA expression of SLFN14, RIG-I, MxA, IP-10, ISG15, and IFN-β was measured by RT-PCR.
3.6. SLFN13 knockdown results in enhanced viral replication of influenza B virus
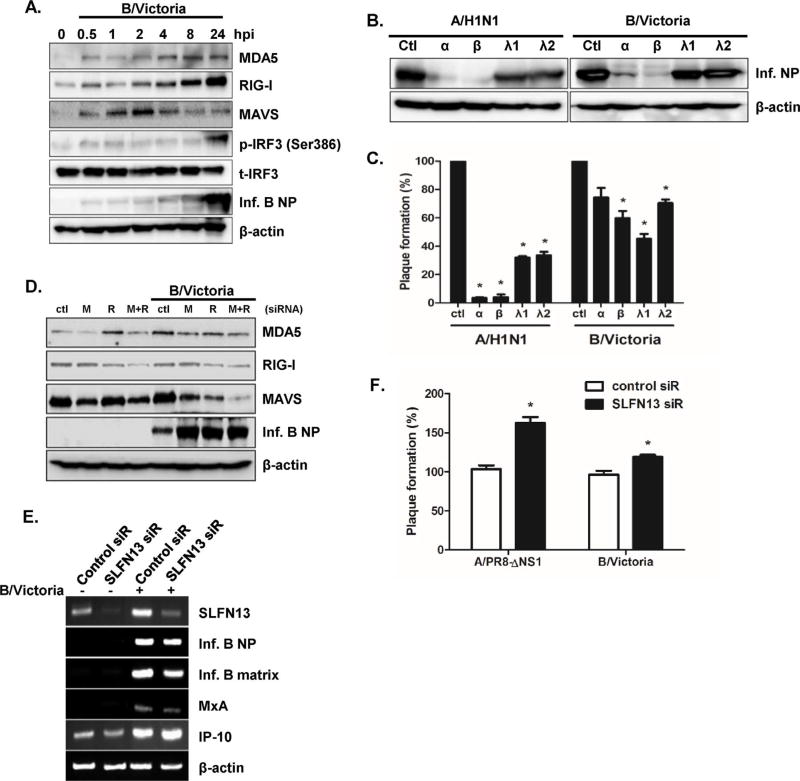
Although both influenza A and B viruses cause flu epidemics and show strong structural similarities, recent studies suggest that host immune responses to these viruses are different (Osterlund et al., 2012). Thus, we investigated the role of SLFN in influenza B virus infections, using B/Victoria lineage virus. A549 cells were infected with B/Victoria virus, and expression levels of MDA5, RIG-I, MAVS, and phospho-IRF3 were measured by western blotting. Increased expression of MDA5, RIG-I, MAVS, and phospho-IRF3 was observed following B/Victoria infection at the indicated time points (Fig. 7A). We also examined whether IFN treatment of B/Victoria-infected cells inhibited viral replication. Similar to A/H1N1-infected cells, B/Victoria-infected cells showed lower levels of viral NP expression after IFN treatment (Fig. 7B). Both type I and III IFN pre-treatments led to a significant reduction in influenza A and B virus plaques, compared to the number in control-treated cells (Fig. 7C). Interestingly, IFN-α and −β treatments caused greater reductions in plaque formation than that of IFN-λ1 or λ2 treatment in influenza-infected cells. Moreover, we examined the effect of MDA5 (M) or RIG-I (R) knockdown on influenza B virus NP expression. Attenuation of RIG-I or MDA5 in response to siRNA treatment led to a significant increase in influenza B NP protein expression (Fig. 7D). We also tested the effect of SLFN13 knockdown on B/Victoria infection. Although there was no significant change in influenza B NP and matrix gene expression, SLFN13 knockdown led to higher numbers of plaque-forming units (Fig. 7E and F). These data suggest that SLFN13 mediates antiviral responses to both influenza A and B virus infections.
Fig. 7.
SLFN13 knockdown results in increased viral replication in response to influenza B virus infection (A) A549 cells were infected with human influenza B/Victoria virus (MOI = 0.1) for 24 h. Protein levels of RIG-I, MDA5, MAVS, phospho-IRF3, and viral NP were analyzed by western blotting. The images shown are representative of three independent experiments. (B, C) Recombinant human IFN-α (10 ng/mL), IFN-β (10 ng/mL), IFN-λ1 (10 ng/mL), and IFN-λ2 (10 ng/mL) were added to cells. The next day, cells were infected with A/H1N1 or B/Victoria virus, and viral NP expression and viral titers were measured. Progeny viral titers were calculated and expressed as plaque-forming units (PFU)/mL. Control-treated cells were normalized to 100%, and data are presented as the percentage relative to the control treated samples. Data are shown as means ± SEM of two independent experiments. (D) A549 cells were transfected with control or RIG-I (R) or MDA5 (M)-specific siRNA, and the knockdown efficiency was measured by western blotting. Levels of viral NP proteins were measured, and anti-actin monoclonal antibody was used as a loading control. The images shown are representative of three independent experiments. (E) Knockdown of SLFN13 was performed with SLFN13-specific siRNA transfection. mRNA expression of SLFN13, MxA, IP-10, influenza B NP, and influenza B matrix genes was measured by RT-PCR. (F) Progeny viral titers were calculated and expressed as plaque-forming units (PFU)/mL. Data are shown as means ± SEM of three different experiments and are presented as the percentage relative to the control siRNA sample. Statistical analysis: *p < 0.05 compared with control siRNA-transfected cells.
3.7. SLFN14 overexpression affects DNA virus antigen expression
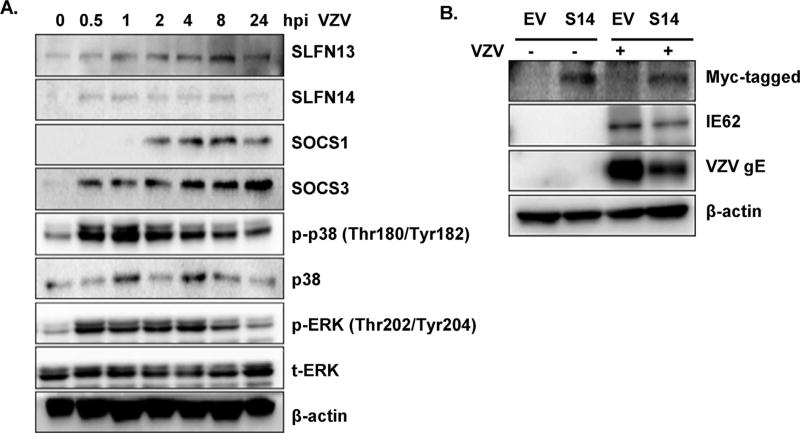
To evaluate whether SLFN also plays an important role in DNA virus-mediated antiviral signaling pathways, the effects of SLFN on varicella zoster virus (VZV), an alphaherpesvirus that causes skin rashes in the form of shingles, were examined. SLFN expression was measured following VZV infection in primary human dermal fibroblasts (HDFs). Interestingly, VZV infection in HDFs resulted in the increased expression of both SLFN13 and SLFN14, although SLFN14 expression levels appeared low compared with SLFN13 (Fig. 8A). Similar to the results with influenza viruses, overexpression of SLFN14 in VZV-infected HDFs resulted in decreased expression of two major VZV proteins, glycoprotein E (gE) and immediate early protein 62 (IE62), required for viral replication (Fig. 8B). These data suggest that SLFN family members can also play an important role in DNA virus sensing and pathogenesis.
Fig. 8.
Antiviral effect of SLFN14 against varicella zoster virus (VZV). (A) Human dermal fibroblasts (HDFs) were mock-infected or infected with VZV (multiplicity of infection = 0.01) for the indicated times. Protein levels of SLFN13, SLFN14, SOCS1, SOCS3, phospho-p38 MAPK, and p-ERK were analyzed by western blotting. Anti-actin monoclonal antibody was used as a loading control. The blot shown is representative of three independent experiments. (B) Empty Vector (EV) and SLFN14-myc plasmids were overexpressed in HDFs, followed by VZV infection (multiplicity of infection = 0.01). Overexpression of SLFN14 was confirmed by myc tag expression levels. VZV immediate early 62 (IE62) and glycoprotein E (gE) expression were measured by western blotting. The blot shown is representative of three independent experiments.
4. Discussion
Although emerging evidence suggests that SLFN family members have important functions in the control of cell proliferation and immune cell development, the effects of SLFNs on viruses are just beginning to be examined. Here, we report that SLFN13 and SLFN14 are novel antiviral factors in influenza virus infections. Furthermore, we show that knockdown of SLFN14 reduces the expression of IP-10, one of the major ISGs, after influenza infection, and that co-expression of RIG-I with SLFN14 enhances RIG-I-mediated IFN signaling.
Given that SLFN family members have motifs homologous to those in the superfamily of RNA helicases, we postulated that these SLFN helicase domains may bind to viral RNA and DNA. Recent studies have highlighted an important function of other viral RNA helicases such as DDX3, DDX21, and DDX60 in virus sensing (Fullam and Schroder, 2013; Miyashita et al., 2011; Thulasi Raman et al., 2016). DDX3 was shown to interact with influenza NS1 and NP prote ins and to act as an antiviral protein by regulating stress granule formation, whereas DDX21 was shown to inhibit viral RNA and protein synthesis during infection through sequential interactions with PB1 and NS1 (Chen et al., 2014). In addition, the DDX60 helicase domain binds to viral RNA and DNA, and knockdown of DDX60 was shown to reduce type I IFN and ISGs after viral infection. Similar to these studies, which showed DDX60 regulation of IFN-β and ISGs in response to TLR ligands or various virus infections, we also observed that SLFN14 overexpression led to significantly increased IFN-β expression in response to infection with viruses.
SLFNs are divided into three groups based on size and structure. SLFNs with small molecular masses, ranging from 37 to 42 kDa, belong to Group I, 58–68 kDa proteins belong to group II, and 100–104 kDa belong to group III. C-terminal extensions are present only in group III SLFNs and are characterized by a motif that is homologous to those in the superfamily of RNA helicases (Geserick et al., 2004). Furthermore, these extensions were shown to each have a nuclear localization signal, which suggests that group III SLFNs may affect nuclear mechanisms (Neumann et al., 2008). In support of this, we observed SLFN14 expression mainly in the nucleus during influenza virus infection and restricts influenza NP expression, suggesting the possible interaction of SLFN14 with nuclear proteins.
Previous reports suggest diverse and redundant functions of SLFN family members, including the regulation of cellular proliferation, immune response induction, and viral replication control. For example, knockdown of SLFN5 in human cells resulted in the increased invasion of malignant melanoma cells, suggesting its anti-melanoma effect (Sassano et al., 2015), whereas mouse SLFN1 and 2 were shown to negatively regulate cellular replication by suppressing cyclin D1 expression (Katsoulidis et al., 2009; Zhao et al., 2008). Furthermore, SLFN2, 3, and 4 were shown to play an essential role in regulating T cell activation and differentiation (Berger et al., 2010; Ahmadi and Veinotte, 2011; Geserick et al., 2004). SLFN11, specifically, was shown to suppress retroviral replication by inhibiting human immunodeficiency virus (HIV) protein synthesis in humans, and over-expression of equine SLFN11 was shown to inhibit equine infectious anemia virus (EIAV) replication (Li et al., 2012; Lin et al., 2016). These studies indicate that SLFNs most likely restrict the synthesis of viral proteins. We observed that knockdown of SLFN13 in A549 cells did not significantly affect influenza NP or matrix gene expression, although it resulted in enhanced viral titers. Moreover, our results showed that overexpression of SLFN14 reduced influenza virus protein NP expression and virus replication, whereas knockdown of SLFN13 or SLFN14 increased viral replication, raising the possibility that SLFN’s antiviral activity mainly depends on the inhibition of viral protein expression. Further studies are required to identify the mechanisms by which SLFN14 counteract viral proteins and functions as a viral DNA/RNA sensor to activate innate signaling.
As reported by Puck et al., the expression of SLFN family members differs depending on cell type. For example, SLFN5, SLFN12L, and SLFN13 expression is highest in T cells, whereas SLFN11 expression was more prominent in monocytes and human monocyte-derived dendritic cells (moDCs) (Puck et al., 2015). Basal levels of SLFN12L and SLFN13 expression were relatively low in monocytes, but were upregulated following differentiation into moDCs. We also investigated the expression of SLFN proteins in different cell types. SLFN14 mRNA levels were easily determined by RT-PCR. However, basal levels of SLFN14 protein were very low in A549, THP-1, HDF, and HaCaT cells, but in mouse macrophages, we were able to detect a sufficient basal level of mouse SLFN14 protein. In addition, SLFN14 was significantly upregulated, both at the mRNA and protein levels, in response to influenza virus infection in mouse macrophages.
Well-known viral sensors such as DDX60 and RIG-I have been shown to bind to dsDNA in vitro and are required for type I IFN expression during infection with DNA viruses such as herpesvirus (Miyashita et al., 2011; Gack, 2014). We investigated whether SLFN overexpression would response similarly to a DNA virus, characterizing VZV protein expression following SLFN14 overexpression in HDFs. Protein expression of VZV IE62 (ORF62) and gE (ORF68), two major antigens of VZV, was attenuated in response to SLFN14 overexpression in HDFs, as shown by western blotting. It is possible that SLFN14 overexpression would enhance IFN-induced signaling, thereby, restricting VZV replication. Our current studies demonstrated that STING-mediated antiviral signaling is important in the restriction of VZV replication (Manuscript currently in review). Further studies are required to reveal the molecular mechanisms by which SLFN14 affects signaling induced by VZV and whether SLFN14 interacts with STING pathway.
Once inside the cell, the influenza virus genome enters the nucleus for transcription and replication of viral genes. Primary transcription of the viral genome is triggered by virion-associated polymerase complexes, which leads to the translation of early viral proteins in the cell cytoplasm. Newly synthesized polymerase, NP, and NS1 proteins are transported to the nucleus, where they initiate and regulate the replication and synthesis of cRNA and viral RNA (vRNA) complexes (Hutchinson and Fodor, 2013). To determine whether SLFN14 regulates viral NP transportation, we used an immunofluorescence-based assay to monitor viral NP translocalization from the cytoplasm to nucleus in pAd-SLFN14-mStrawberry-expressing cells. NP was still localized in the cytoplasm at 90 mpi in pAd-SLFN14-mStrawberry-expressing cells, whereas NP localization in the nucleus was observed at 15 mpi in pAd control-expressing cells, suggesting that SLFN14 overexpression causes a delayed translocation of viral NP into the nucleus (Fig. 4D). Given that SLFN14 expression was mainly observed in the nucleus, it will be interesting to test the potential interaction of SLFN14 with IFN-γ inducible protein 16 (IFI16), which is an essential nuclear sensor for the induction of IRF3 signaling during herpesvirus infection (Orzalli et al., 2012; Johnson et al., 2014)
A recent report suggested that SLFN14 mutations could be the cause for an inherited thrombocytopenia with excessive bleeding, highlighting an important novel function of SLFN14 in platelet formation and maintenance (Fletcher et al., 2015). Considering that patients with SLFN14 mutations show a phenotype of thrombocytopenia with enlarged platelets and decreased ATP secretion, it will be interesting to investigate the susceptibility of these patients to infectious diseases associated with symptoms of thrombocytopenia, with the recognition of SLFN14’s antiviral role.
Considering that SLFN can be induced by viral infection and potentially affect the host immune response, further studies using SLFN knockout or transgenic mice to determine the role of SLFN in the regulation of susceptibility to viral infections may shed light on the function of SLFN in ameliorating viral infection-associated symptoms. In conclusion, our data indicate that SLFN family members can contribute to the control of viral replication. A better understanding of the role of SLFN family members in viral infection will greatly improve our knowledge of influenza and VZV pathogenesis and provide insight into potential therapies for influenza and VZV infections.
Supplementary Material
Acknowledgments
We would like to thank Dr. Adolfo Garcia-Sastre (Icahn School of Medicine at Mount Sinai, NY, USA) for providing viruses.
Funding information
This research was supported by the Basic Science Research Program of the National Research Foundation of Korea (NRF), funded by the Ministry of Science, ICT & Future Planning (NRF-2016R1C1B2006493) and a grant (1R21NS099838-01) from National Institute of Neurological Disorders and Stroke, grant (P30GM114737) from the Centers of Biomedical Research Excellence, National Institute of General Medical Sciences, National Institutes of Health.
Footnotes
Conflict of interest
None.
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.imbio.2017.07.002.
References
- Ahmadi S, Veinotte LL. Effect of Schlafen 2 on natural killer and T cell development from common T/natural killer progenitors. Pak. J. Biol. Sci. 2011;14:1002. doi: 10.3923/pjbs.2011.1002.1010. [DOI] [PubMed] [Google Scholar]
- Berger M, Krebs P, Crozat K, Li X, Croker BA, Siggs OM, Popkin D, Du X, Lawson BR, Theofilopoulos AN, Xia Y, Khovananth K, Moresco EM, Satoh T, Takeuchi O, Akira S, Beutler B. An Slfn2 mutation causes lymphoid and myeloid immunodeficiency due to loss of immune cell quiescence. Nat. Immunol. 2010;11:335. doi: 10.1038/ni.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Liu CH, Zhou L, Krug RM. Cellular DDX21 RNA helicase inhibits influenza A virus replication but is counteracted by the viral NS1 protein. Cell Host Microbe. 2014;15:484. doi: 10.1016/j.chom.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi EJ, Lee C, Kim YC, Shin OS. Wogonin inhibits Varicella-Zoster (shingles) virus replication via modulation of type I interferon signaling and adenosine monophosphate-activated protein kinase activity. J. Funct. Foods. 2015;17:399. [Google Scholar]
- Companioni Napoles O, Tsao AC, Sanz-Anquela JM, Sala N, Bonet C, Pardo ML, Ding L, Simo O, Saqui-Salces M, Blanco VP, Gonzalez CA, Merchant JL. SCHLAFEN 5 expression correlates with intestinal metaplasia that progresses to gastric cancer. J. Gastroenterol. 2017 Jan;52(1):39–49. doi: 10.1007/s00535-016-1202-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Casa-Esperon E. From mammals to viruses: the Schlafen genes in developmental, proliferative and immune processes. Biomol. Concepts. 2011;2:159. doi: 10.1515/bmc.2011.018. [DOI] [PubMed] [Google Scholar]
- Fletcher SJ, Johnson B, Lowe GC, Bem D, Drake S, Lordkipanidze M, Guiu IS, Dawood B, Rivera J, Simpson MA, Daly ME, Motwani J, Collins PW, Watson SP, Morgan NV. U.K. Genotyping Phenotyping of Platelets study, g SLFN14 mutations underlie thrombocytopenia with excessive bleeding and platelet secretion defects. J. Clin. Invest. 2015;125:3600. doi: 10.1172/JCI80347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullam A, Schroder M. DExD/H-box RNA helicases as mediators of anti-viral innate immunity and essential host factors for viral replication. Biochim. Biophys. Acta. 2013;1829:854. doi: 10.1016/j.bbagrm.2013.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gack MU. Mechanisms of RIG-I-like receptor activation and manipulation by viral pathogens. J. Virol. 2014;88:5213. doi: 10.1128/JVI.03370-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Sastre A, Egorov A, Matassov D, Brandt S, Levy DE, Durbin JE, Palese P, Muster T. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology. 1998;252:324. doi: 10.1006/viro.1998.9508. [DOI] [PubMed] [Google Scholar]
- Geserick P, Kaiser F, Klemm U, Kaufmann SH, Zerrahn J. Modulation of T cell development and activation by novel members of the Schlafen (slfn) gene family harbouring an RNA helicase-like motif. Int. Immunol. 2004;16:1535. doi: 10.1093/intimm/dxh155. [DOI] [PubMed] [Google Scholar]
- Hale BG, Albrecht RA, Garcia-Sastre A. Innate immune evasion strategies of influenza viruses. Future Microbiol. 2010;5:23. doi: 10.2217/fmb.09.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson EC, Fodor E. Transport of the influenza virus genome from nucleus to nucleus. Viruses. 2013;5:2424. doi: 10.3390/v5102424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KE, Bottero V, Flaherty S, Dutta S, Singh VV, Chandran B. IFI16 restricts HSV-1 replication by accumulating on the hsv-1 genome, repressing HSV-1 gene expression, and directly or indirectly modulating histone modifications. PLoS Pathog. 2014;10:e1004503. doi: 10.1371/journal.ppat.1004503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJ, Yamaguchi O, Otsu K, Tsujimura T, Koh CS, Reis e Sousa C, Matsuura Y, Fujita T, Akira S. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- Katsoulidis E, Carayol N, Woodard J, Konieczna I, Majchrzak-Kita B, Jordan A, Sassano A, Eklund EA, Fish EN, Platanias LC. Role of Schlafen 2 (SLFN2) in the generation of interferon alpha-induced growth inhibitory responses. J. Biol. Chem. 2009;284:25051. doi: 10.1074/jbc.M109.030445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JA, Park SK, Kumar M, Lee CH, Shin OS. Insights into the role of immunosenescence during varicella zoster virus infection (shingles) in the aging cell model. Oncotarget. 2015;6:35324. doi: 10.18632/oncotarget.6117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JA, Seong RK, Shin OS. Enhanced viral replication by cellular replicative senescence. Immune Netw. 2016;16:286. doi: 10.4110/in.2016.16.5.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Kao E, Gao X, Sandig H, Limmer K, Pavon-Eternod M, Jones TE, Landry S, Pan T, Weitzman MD, David M. Codon-usage-based inhibition of HIV protein synthesis by human schlafen 11. Nature. 2012;491:125. doi: 10.1038/nature11433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YZ, Sun LK, Zhu DT, Hu Z, Wang XF, Du C, Wang YH, Wang XJ, Zhou JH. Equine schlafen 11 restricts the production of equine infectious anemia virus via a codon usage-dependent mechanism. Virology. 2016;495:112. doi: 10.1016/j.virol.2016.04.024. [DOI] [PubMed] [Google Scholar]
- Mavrommatis E, Fish EN, Platanias LC. The schlafen family of proteins and their regulation by interferons. J. Interferon Cytokine Res. 2013a;33:206. doi: 10.1089/jir.2012.0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrommatis E, Arslan AD, Sassano A, Hua Y, Kroczynska B, Platanias LC. Expression and regulatory effects of murine Schlafen (Slfn) genes in malignant melanoma and renal cell carcinoma. J. Biol. Chem. 2013b;288:33006. doi: 10.1074/jbc.M113.460741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyashita M, Oshiumi H, Matsumoto M, Seya T. DDX60, a DEXD/H box helicase, is a novel antiviral factor promoting RIG-I-like receptor-mediated signaling. Mol. Cell. Biol. 2011;31:3802. doi: 10.1128/MCB.01368-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann B, Zhao L, Murphy K, Gonda TJ. Subcellular localization of the Schlafen protein family. Biochem. Biophys. Res. Commun. 2008;370:62. doi: 10.1016/j.bbrc.2008.03.032. [DOI] [PubMed] [Google Scholar]
- Orzalli MH, DeLuca NA, Knipe DM. Nuclear IFI16 induction of IRF-3 signaling during herpesviral infection and degradation of IFI16 by the viral ICP0 protein. Proc. Natl. Acad. Sci. U. S. A. 2012;109:E3008. doi: 10.1073/pnas.1211302109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterlund P, Strengell M, Sarin LP, Poranen MM, Fagerlund R, Melen K, Julkunen I. Incoming influenza A virus evades early host recognition, while influenza B virus induces interferon expression directly upon entry. J. Virol. 2012;86:11183. doi: 10.1128/JVI.01050-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichlmair A, Schulz O, Tan CP, Naslund TI, Liljestrom P, Weber F, Reis e Sousa C. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5'-phosphates. Science. 2006;314:997. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- Puck A, Aigner R, Modak M, Cejka P, Blaas D, Stockl J. Expression and regulation of Schlafen (SLFN) family members in primary human monocytes, monocyte-derived dendritic cells and T cells. Results Immunol. 2015;5:23. doi: 10.1016/j.rinim.2015.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulendran B, Maddur MS. Innate immune sensing and response to influenza. Curr. Top. Microbiol. Immunol. 2015;386:23. doi: 10.1007/82_2014_405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruckle A, Haasbach E, Julkunen I, Planz O, Ehrhardt C, Ludwig S. The NS1 protein of influenza A virus blocks RIG-I-mediated activation of the noncanonical NF-kappaB pathway and p52/RelB-dependent gene expression in lung epithelial cells. J. Virol. 2012;86:10211. doi: 10.1128/JVI.00323-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassano A, Mavrommatis E, Arslan AD, Kroczynska B, Beauchamp EM, Khuon S, Chew TL, Green KJ, Munshi HG, Verma AK, Platanias LC. Human schlafen 5 (SLFN5) is a regulator of motility and invasiveness of renal cell carcinoma cells. Mol. Cell. Biol. 2015;35:2684. doi: 10.1128/MCB.00019-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider WM, Chevillotte MD, Rice CM. Interferon-stimulated genes: a complex web of host defenses. Annu. Rev. Immunol. 2014;32:513. doi: 10.1146/annurev-immunol-032713-120231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz DA, Katayama CD, Hedrick SM. Schlafen, a new family of growth regulatory genes that affect thymocyte development. Immunity. 1998;9:657. doi: 10.1016/s1074-7613(00)80663-9. [DOI] [PubMed] [Google Scholar]
- Seong RK, Choi YK, Shin OS. MDA7/IL-24 is an anti-viral factor that inhibits influenza virus replication. J. Microbiol. 2016;54:695. doi: 10.1007/s12275-016-6383-2. [DOI] [PubMed] [Google Scholar]
- Thulasi Raman SN, Liu G, Pyo HM, Cui YC, Xu F, Ayalew LE, Tikoo SK, Zhou Y. DDX3 interacts with influenza a virus NS1 and NP proteins and exerts antiviral function through regulation of stress granule formation. J. Virol. 2016;90:3661. doi: 10.1128/JVI.03010-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, Fujita T. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 2004;5:730. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- Zhao L, Neumann B, Murphy K, Silke J, Gonda TJ. Lack of reproducible growth inhibition by Schlafen1 and Schlafen2 in vitro. Blood Cells. Mol. Dis. 2008;41:188. doi: 10.1016/j.bcmd.2008.03.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.