Abstract
Acquired genomic structural variants (SVs) are major hallmarks of the cancer genome, but they are challenging to reconstruct from short-read sequencing data. Here, we exploit the long-reads of the nanopore platform using our customized pipeline, Picky (https://github.com/TheJacksonLaboratory/Picky), to reveal SVs of diverse architecture in a breast cancer model. We identified the full spectrum of SVs with superior specificity and sensitivity relative to short-read analyses and uncovered repetitive DNA as the major source of variation. Examination of the genome-wide breakpoints at nucleotide-resolution uncovered micro-insertions as the common structural features associated with SVs. Breakpoint density across the genome is associated with propensity for inter-chromosomal connectivity and enriched in promoters and transcribed regions of the genome. Furthermore, an over-representation of reciprocal translocations from chromosomal double-crossovers was observed through phased SVs. We demonstrated that Picky analysis is an effective tool to uncover comprehensive SVs in cancer genomes from long-read data.
Introduction
Genomic structural variation is prevalent in the human genome1 and includes deletions, insertions, duplications, inversions, and translocations. Collectively, these structural variants (SVs) account for a significant portion of genome heterogeneity between individuals2 and human populations3. Many cancer genomes have been found to harbor significant structural variation, and specific SVs are considered to be instrumental in promoting tumor progression by disrupting gene structures, dysregulating gene expression, creating fusing transcription units or increasing gene copy number4–6. The detection of specific SVs can be used as the basis for tumor classification and potentially of prognostic value for tumor severity and therapeutic response4–7. However, the molecular organization of various SV classes and the mechanisms that generate them are not well understood.
Advances in sequencing technology coupled with improvements in computational algorithms have greatly enhanced our understanding of the abundance, diversity, and molecular features of SVs across human populations3 and disease8,9. However, short-read sequencing approaches, although perform well on subset of SVs types10,11, are limited to fully disclose the complexity and spectrum of SVs1,12,13. Specifically, paired-end short reads are not sufficiently sensitive to detect small SVs, and lack the nucleotide-level of detail for analysis of the breakpoints that flank SVs. They are also unable to decipher complex SV patterns. Therefore, long-read sequencing approaches and analytic methods are essential to facilitate comprehensive and unbiased SV profiling, particularly useful for resolving complex structural rearrangements in cancer genomes14–17.
Recent progress in nanopore single-molecule sequencing offers to extend sequencing read length and throughput18–21. Here we introduce a computational analysis pipeline, named Picky, to detect the full spectrum of SVs and characterize their genomic breakpoints with high specificity and sensitivity. Applying Picky to a moderate coverage of nanopore sequences in a well-studied breast cancer cell line HCC118722, we classified a wide range of SVs and characterized the breakpoints in detail.
Results
Applying nanopore long read sequencing and Picky analysis pipeline to detect SVs
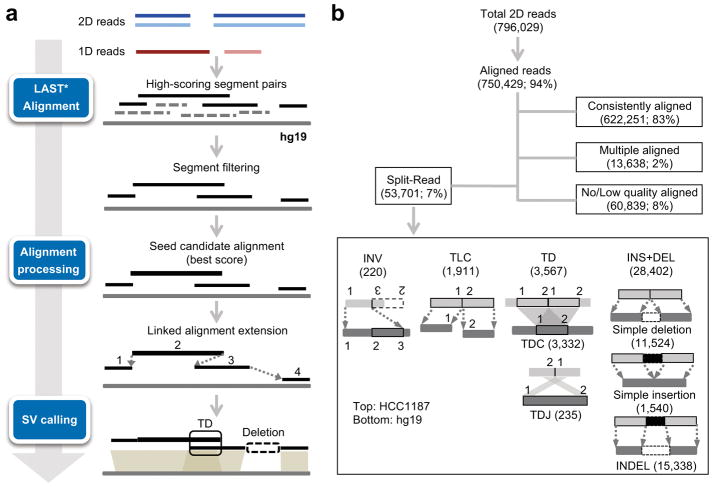
We performed total of 15 MinION runs and generated 7.9 Gb of the aligned 2D reads of different sizes (3–4 Kb and 12 Kb) for the HCC1187 genome (Supplementary Table 1, see Online Methods). Details of the read length distribution, yield and accuracy of nanopore long read sequencing were provided in Supplementary Note. Picky probes long reads in three consecutive steps: read alignment to a reference genome, optimal alignment merge/selection, and SV classification (Fig. 1a). Picky was designed to enable SV calls from alignments from different aligners including NGMLR17 and minimap223 (https://github.com/TheJacksonLaboratory/Picky/wiki/Using-an-Alternative-Aligner). Here, we adopts LAST24,25 to perform genome alignment. Alignments for each read were then evaluated for their quality and spurious alignments were filtered out based on poor alignment score or low percentage identity. Next, alignments for different segments of a long read were picked and merged. We applied a greedy seed-and-extension algorithm to stitch segments together and then combined segments that maximized coverage for each long read. Only reads with ≥ 70% genome alignments across their total length were used for further analysis. Based on the order and the distance between the non-contiguous alignments, Picky assigned split reads into seven classes of SVs: inversion (INV), translocation (TLC), tandem duplication (TD), complete tandem duplication resided within a read segment (TDC) or a duplication junction spanned across a read segment (TDJ), simple insertion (INS) or deletion (DEL), and INDEL (both INS and DEL resided within a read segment) (see Online Methods).
Figure 1. A customized SV pipeline designed for long-read SV analysis.
(a) Overview of the Picky pipeline. Blue and light blue lines indicate the template and the complementary reads from nanopore 2D reads, while brown and light brown lines indicate different strands of nanopore 1D reads. * indicates non-default parameterization. (b) Read alignment summary and different classes of SVs detected by algorithmic design of Picky.
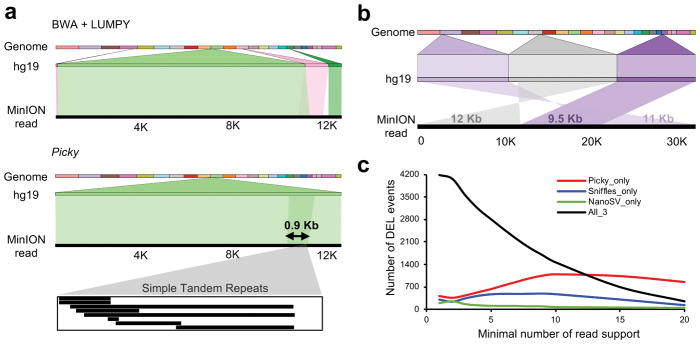
We applied Picky to 796,029 2D reads to detect and classify SVs (Fig. 1b). From 53,701 split reads, Picky detected a total of 34,100 unique SVs and their corresponding 66,660 breakpoints, classified them into 220 inversions, 1,911 translocations, 3,567 tandem duplications and 28,402 insertions, deletions and INDEL (Supplementary Tables 2 and 3). The percentage of reads containing breakpoints (i.e. split reads) positively correlated with DNA read lengths (Supplementary Fig. 1). Furthermore, 4% (2,177 of 53,701) of split reads contained more than two breakpoints, which indicates the presence of multiple SVs on the same chromosomes. This phasing information is uniquely provided by long read sequencing. In contrast to short read SV callers such as LUMPY26, which assumes no more than two uniquely mapped segments and one breakpoint per split read, Picky incorporates an algorithmic rationale to interpret long reads spanning multiple breakpoints that may even encompass the entire SV. Examples of a 0.9-Kb dinucleotide (TA)n microsatellite TD (Fig. 2a) and a complex TLC identified by Picky but were either misclassified or undetected by short-read based analysis (Fig. 2b). Through examination of adjacent SVs via multiple breakpoints within individual long reads, we found 67 co-occurring translocations (TLC-TLC), representing an enrichment of 3.09 (obs/exp) over the background (Supplementary Fig. 2a,b). Interestingly, 25 of these dual-translocation events were reciprocal (i.e. chromosome A-B-A), suggesting that double-crossovers between two non-homologous chromosomes could be a common mode to generate translocation.
Figure 2. The sensitivity of the Picky pipeline in SV detection.
(a) Picky accurately defines a short TD. A 12.9 Kb nanopore read aligns to reference genome as two overlapping segments of 11.4 Kb and 2.4 Kb with 86% and 83% identities (e-value = 0). This TD was misclassified as a translocation by the short-read aligner BWA and SV caller LUMPY. (b) A complex SV of two translocations detected by Picky from a 32.5 Kb nanopore read. The alignments were visualized by Ribbon (https://github.com/MariaNattestad/ribbon). (c) Numbers of reference deletions supported by each versus all pipelines among different thresholds.
The accuracy of Picky in SV detection
To confirm the accuracy of Picky, we validated over 200 SV events via PCR across either identified breakpoint junctions (TLCs, INVs and TDJs) or full-length rearranged regions (INSs, DELs and TDCs) (Supplementary Fig. 3a–f and Supplementary Table 4, see Online Methods). 100% and 79% of the predicted SVs supported by multiple and single reads were validated, respectively (Supplementary Fig. 4a). We also observed multiple PCR fragments amplified from both normal and rearranged haplotypes, suggesting the presence of heterozygosity in SV containing loci (Supplementary Fig. 5a). To quantify the extent of heterozygosity, we examined 50 randomly selected rearranged loci covered by multiple reads from each SV class and determined the number of loci spanned by reads from both the normal and variant haplotypes found in the same locus (Supplementary Fig. 5b). We observed extensive heterozygosity of SVs among different classes, ranging from 44% in TDC to 100% in INV, INDEL and TLC (Supplementary Fig. 5c). Using the 176 validated SVs as the reference data set, we further compared the specificity between nanopore long read and short read analyses. As shown, short-read sequencing analysis by LUMPY could only accurately detect a subset of SV classes (TLC, TDJ and DEL) given sufficient coverage (> 30X), but drastically underperformed in classifying short-span TDs (TDC) and inversions, and was unable to unambiguously define insertion events (Supplementary Fig. 4b). To evaluate the sensitivity of Picky, we compared SV detected between the nanopore long-read (LR) and illumina short-read (SR) data in HCC1187 at either lower depth (10X)27 (Supplementary Fig. 4c) or higher depth (102X) (SRA accession SRX969058). Comparing with the SVs found by high depth SR data, we observed that vast majority (93%–95%) of SVs found in the long-read data were unique and LR specific SVs exhibit distinct features like short span size (Supplementary Fig. 6a, b).
Comparison of Picky with other long-read based SV analysis tools
Next, we compared Picky to two long-read SV analysis tools NanoSV16 and Sniffles17 to evaluate their relative sensitivity as well as accuracy. Picky comprises unique features in both the alignment scheme and SV detection (Supplementary Fig. 7). We analyzed the nanopore data from the well-studied NA12878 human genome21 and used the insertions and deletions determined by the PacBio long-read data from the same genome28 as the reference call sets. We focused our assessment based on: 1) the sensitivity and precision of Picky to recall known SVs; 2) the ability of Picky to uncover new SVs. Under the identical comparison parameters (see Online Methods), Picky recalls 5,246 (66%) of the 7,903 reference deletion calls, comparable to 5,251 (66%) and 5,102 (65%) deletions recalled by Sniffles and NanoSV, respectively. While the deletions recalled by each of the three programs were largely overlapped, Picky recaptured highest numbers of deletions not found by other two methods across different thresholds (Fig. 2c), suggesting its highly sensitive nature in deletion detection. This was also evident by the total numbers of deletions called by each of the pipelines. Picky detected 338,701 deletions, 3- and 13-fold higher than the numbers of deletions uncovered by Sniffles and NanoSV. Of which, 77% of them were Picky-specific, significantly higher than the 36% and 11% of the Sniffles- and NanoSV-specific deletions. In insertions, Picky recalled 3,222 (28%) of the reference INS, while the Sniffles and NanoSV called 4,429 (39%) and 3,815 (33%), respectively. The slightly lower number of INS supported by Picky was resulted from the priority of SV classification used by different programs when multiple types of SVs were detected within the same locus (Supplementary Fig. 7). In Picky, TD was assigned over INS while in NanoSV and Sniffles, INS was chosen over duplication. When all the INS and TD were combined from each program, Picky again showed better sensitivity with 5–8-fold more calls than Sniffles and NanoSV. Majority of the TD and DUP (72–78%) detected by NanoSV and Sniffles overlapped with those found in Picky, while 87% of the Picky calls were new.
Nanopore long reads uncovered short-span SVs enriched in repetitive DNA sequences
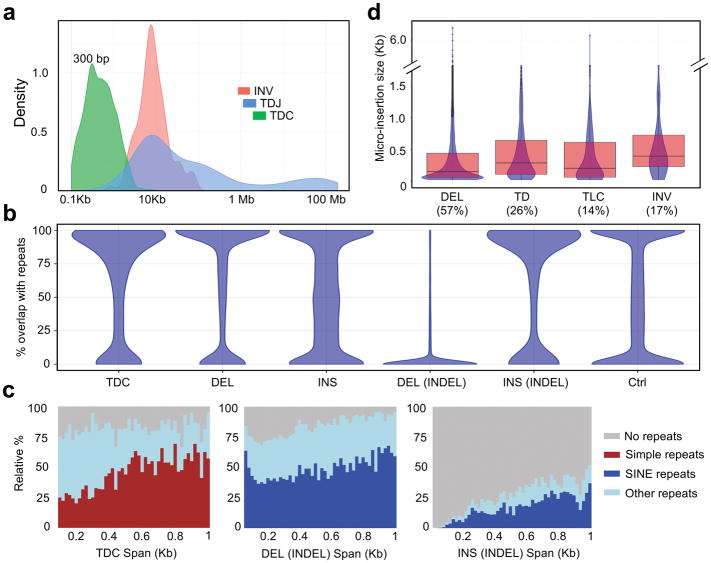
SVs identified in HCC1187 genome exhibit a broad span distribution, ranging from 20 bp up to 100 Mb (Fig. 3a and Supplementary Table 3). Comparing to the short-read analysis, Picky uniquely detects short SVs through long reads spanning across the entire variable regions. Vast majority of the INSs, DELs and TDCs span less than 1Kb with notable peaks around 300 bp (Fig. 3a and Supplementary Fig. 8a), suggesting that they are enriched with repetitive sequences. When we examined their repeat content, simple INSs, DELs and TDCs exhibited a bimodal distribution based on the fraction of the SVs overlapping with repetitive sequence regions (Fig. 3b) and the SINE and simple repeats are the predominant enriched repeat classes (Fig. 3c and Supplementary Fig. 8b,c). Therefore, small-size INSs, DELs and TDs account for the majority of the SVs detected in HCC1187 genome and they are predominantly copy number variation in repetitive sequence regions.
Figure 3. Long reads uncover repeat-rich SVs and the presence of micro-insertions within SV junctions.
(a) Span distribution of INV, TDJ and TDC. (b) Fraction of regions overlapped with repeats among different SV classes. TDC, n = 3,332. DEL, n = 11,524. INS, n = 1,540. DEL(INDEL), n = 15,338. INS(INDEL), n = 15,338. Ctrl, n = 47,072. (c) Relative percentages of the major repeat types across different span sizes. (d) The percentages of SVs with micro-insertions and the size distribution of the inserted sequences. DEL, n = 26,862. TD, n = 3,567. TLC, n = 1,911. INV, n = 220. Center line, median; boxes, first and third quartiles; whiskers, 1.5 interquartile range (IQR) from the box.
Micro-insertion is a prevalent structural feature found within breakpoint junctions
Nanopore split reads aligned across SV junctions enable us to characterize breakpoint junctions in their entirety at nucleotide-resolution. We observed additional non-aligned inserted sequences within the 66,660 breakpoints from different SV types, ranged from 57% (DELs) to 14% (TLCs) (Fig. 3d). The majority of these inserted sequences was less than 500 bp, although some of them can be as long as 6 Kb (Fig. 3d), and their validity was confirmed by PacBio SMRT sequencing method (Supplementary Fig. 9a,b, see Online Methods). BLASTing these insertions against the NCBI non-redundant nucleotide database reveals that 90% of the inserted DNA pieces are completely novel with no homology to any known sequences. The small size of these sequences and their lack of significant homology with known sequences, are consistent with the “genomic shards” resulted from non-templated DNA synthesis at the rearrangement junctions observed in a few selective rearrangement events29,30. Besides the micro-insertions, short stretches (usually 2–6 nucleotides) of identical sequence, known as overlapping microhomology31, were frequently spotted at SV breakpoint junctions (highlighted in Supplementary Fig. 3). The microhomology, together with the micro-insertions, resided within breakpoint junctions suggest that de novo DNA synthesis is a potential mechanism used in the non-homologous end-joining (NHEJ) during genomic rearrangement repair process.
Breakpoint landscapes is associated with chromatin organization and transcriptional regulation
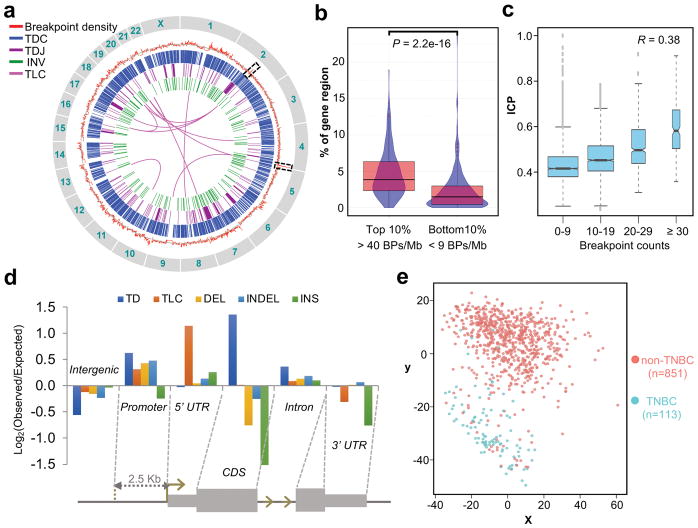
The distribution of the large numbers of SVs reflects the highly jumbled nature of HCC1187 genome (Fig. 4a). High frequency translocations between t(2;19), t(2;17), t(1;8) and t(10:13) were consistent with the translocations previously described by SKY (spectral karyotyping)32. The density of breakpoint was found to associate with the fraction of genome annotated with gene coding regions. Using 1 Mb genomic span as the bin size, the top 10% breakpoint dense regions have a significantly higher percent of nucleotides coding for genes than the bottom 10% breakpoint poor regions (P = 2.2e-16) (Fig. 4b); suggesting high transcription activity could be one of the mechanisms impacting genome fragility. Interestingly, two of the hyper-density breakpoint loci (2q21.3–2q22.1; 65 breakpoints/Mb and 4q35; 139 breakpoints/Mb; highlighted in Fig. 4a) had very high (within top 10 percentile) inter-chromosomal contact probability (ICP) values, the propensity of a region to form inter-chromosomal contacts within interphase nucleus33, found in the GM12878 cell line. Further analysis on the whole genome level indicated a positive correlation between ICP and breakpoint count (Fig. 4c). This correlation raises the possibility that chromosome regions with frequent exposure to other chromosome and/or residing at the exterior of a chromosome territory, could be more prone to chromosomal breaks compared to the protected regions deeper inside the chromosome territory. These observations further support that intermingling of chromatin organization directly influences the structural properties associated with elevated frequency of DNA double strand breaks34,35.
Figure 4. The analysis of genomic distribution of breakpoints and their affected genes.
(a) Genome-wide distribution of the SVs and their associated breakpoints density. From outer circle to inner circle: Red: measured breakpoint density; Grey: reference average density of 22 breakpoints/Mb; Blue: TDC; Purple: TDJ (with span size < 20 Mb); Green: INV; Magenta: TLC (with pair counts ≥ 4) at 1Mb bin size. (b) Fraction of regions with transcribed genes in regions of the top 10% (n = 250) and the bottom 10% (n = 234) breakpoint densities. BP, breakpoint. P value was calculated by Mann-Whitney test. Center line, median; boxes, first and third quartiles; whiskers, 1.5 interquartile range (IQR) from the box. (c) The trend of increasing ICP with increasing number of breakpoints. The breakpoints and ICP were calculated for every chromosome segments of 138 HindIII sites (see Online Methods). The ICP values were then grouped based on the breakpoint counts as indicated on the x-axis. 0–9, n = 2,981; 10–19, n = 2,202; 20–29, n = 622; ≥ 30, n = 197. The R value is the Pearson’s correlation coefficient on the underlying, ungrouped data. Center line, median; boxes, first and third quartiles; whiskers, 1.5 interquartile range (IQR) from the box; notch, 95% confidence interval of the median. (d) Enrichment of breakpoint from each SV class distributed along different genomic features. (e) The multidimensional scaling (MDS) plot of SV-genes in the BRCA dataset within the TCGA. Non-TNBC, n = 851; TNBC, n = 113.
Enrichment of SVs in the regulatory repertoire of the genome with impacts on gene expression
We investigated the distribution of different SV associated-breakpoints among intergenic, coding sequence (CDS), promoter (2.5 Kb upstream of TSS), 5′ and 3′ un-translated regions (UTRs) and introns (Fig. 4d). Enrichment of breakpoints was found in promoters and 5′ UTR for most of the SV types, particularly the TLCs and TDs (Supplementary Fig. 10a). Interestingly, repeat-rich versus repeat-poor TDs exhibited contrasting distribution patterns (Supplementary Fig. 10b). SVs occurring in promoters/regulatory elements can selectively lead to oncogene activation or tumor suppressor gene inactivation, which are likely to be cancer specific. To test this, we determined the expressions of genes affected by the SVs from 113 TNBC and 851 non-TNBC tissues of the breast carcinoma (BRCA) dataset within the cancer genome atlas (TCGA)36. In contrast to the data from the permutation and the control genes, the expression of 1,260 coding genes disrupted by the major SV classes (DEL, INDEL and TD) effectively distinguished TNBC from non-TNBC types (Fig. 4e and Supplementary Fig. 11), highlighting the functional impacts of SV analysis and its link to tumor molecular classification.
Discussion
Existing short read sequencing and SV analysis tools are limited in resolving complex structural variation and delineating molecular structures of breakpoints, particularly within repetitive regions.
Our breakpoint analysis suggests the linkage between the propensity of inter-chromosomal connectivity and frequency of genomic lesions. Nuclear regions with high transcriptional activity are shown to have extensive inter-chromatin contacts33. Therefore, the accessibility and conformation in the active chromatin domains may provide the structural basis of genome fragility. The enrichment of breakpoints in the regulatory repertoire of the genome further implicates that genome rearrangement can reconfigure the transcriptional program in the cancer transformation process. Expanding the high-resolution of SV analysis should further our understanding of the homeostasis between genome architecture, variation and carcinogenesis.
Long-read sequencing possesses many unique features that improve from the current state of SV detection. Yield and accuracy from nanopore has been dramatically improved for the past year, including a new basecaller Scrappie21. Given the superior alignment specificity, higher resolution and broader utility of single molecule long-read data, we anticipate that there will be soon a paradigm shift on the sequencing approaches for genome-wide, haplotype-specific structural analysis; which will reveal new insights in the diversity and complexity of human genome variation and the mechanisms of their generation during tumorigenesis.
Online Methods
Nanopore long read sequencing
High molecular weight DNA was extracted from HCC1187 cells by MagAttract HMW DNA Kit (Qiagen, 67563) followed the manufacturer’s instructions. Briefly, 1 × 106 frozen cells were lysed with 220 μL of Buffer ATL, 20 μL Proteinase K and incubated overnight at 56°C with 900 rpm. 4 μL RNase A was added to cleave RNA. 150 μL Buffer AL, 280 μL Buffer MB and 40 μL MagAttract Suspension G beads were then added to capture the HMW DNA. Next, the beads were cleaned up by 700 μL Buffer MW1, Buffer PE and NFW followed by elution with 150 μL Buffer AE. Nanopore sequencing libraries were prepared according to the target size and the sequencing kits supplied by Oxford Nanopore Technologies (ONT) (Supplementary Table 5). HMW genomic DNA was fragmented by either miniTUBE Blue (Covaris, 520065, for 3 Kb), miniTUBE Red (Covaris, 520066, for 5 Kb) or g-TUBE (Covaris, 520079, for 8 Kb and 12 Kb). For libraries targeted at 12 Kb, size selection was performed for sheared fragments larger than 10 Kb using 0.75% agarose cassette (Sage Science, BLF7510) by Blue Pippin™ DNA Size Selection System. For libraries of less than 10 Kb, AMPure XP beads (Beckman Coulter, A63881) were used for clean-up. Next, libraries were prepared according to recommendation by ONT. Briefly, NEBNext FFPE RepairMix (NEB, M6630) was added to repair nicks in the DNA. Then end-repair and dA-tailing were performed using NEBNext Ultra II End-Repair/dA-tailing Module (NEB, E7546). For 2D libraries (WTD01-WTD13), we prepared the ligation reaction as below: 38 μL water (DNA), 10 μL Adapter Mix (AMX), 2 μL Hairpin Adapter (HPA) and 50 μL Blunt/TA Ligase Master Mix (New England Biolabs, M0367). Ligation was performed at room temperature for 15 minutes. 1 μL Hairpin Tether (HPT) was added to the reaction and incubated at room temperature for 15 minutes. Then 50 μL MyOne C1 beads (Thermo Fisher, 65001) beads were prewashed twice with 100 μL Bead Binding Buffer (BBB). The MyOne C1 beads resuspended in 100 μL BBB were added to the ligated DNA reaction and incubated on a rotator at room temperature for 15 minutes. The beads were washed twice with 150 μL BBB and eluted in 25 μL Elution Buffer (ELB). For 1D libraries (WTD14-WTD15), we prepared the ligation reaction as below: 30 μL water (DNA), 20 μL Adapter Mix (AMX) and 50 μL Blunt/TA Ligase Master Mix (New England Biolabs, M0367). Ligation was also performed at room temperature for 15 minutes. The AMPure XP beads were resuspended at room temperature by vortex. 40 μL of beads were added into the ligation product. The beads were washed twice with 140 μL Adapter Bead Binding (ABB) buffer and eluted in 25 μL Elution Buffer (ELB). The eluted product was the adaptor-ligated library as the Pre-sequencing Mix (PSM) used in nanopore sequencing.
The libraries were sequenced on MinION Mk1b device (ONT) using R9 and R9.4 flow cells following the standard 48 h run scripts (Supplementary Table 5). Real-time basecalling was performed on EPI2ME cloud platform (ONT). Read sequences were extracted from base-called fastf5 files by Poretools (version 0.5.1) to generate fastq file. All 2D reads from WTD01-13 (2D ligation libraries) and all 1D reads from WTD14 and WTD15 (1D ligation libraries) were used for subsequent analysis.
PacBio sequencing
High molecular weight DNA was mechanically sheared and size selected followed by dividing into two aliquots; one was used to prepare template for PacBio sequencing on RSII and one was used to prepare template for nanopore sequencing on MinION. The PacBio genomic DNA library prep was performed according to the manufacturer’s instruction (http://www.pacb.com/support/documentation/). In brief, 2 μg of purified DNA was taken into library construction by the SMRTbellTM Template Prep Kit 1.0 (Pacific Biosciences). DNA fragments were repaired using the DNA damage repair solution at 37°C for 60 minutes and at 4°C for 1 minute. DNA ends were end-repaired by adding 2.0 μL End Repair Mix to the reaction, which was incubated at 25°C for 5 minutes and at 4°C for 1 minute, followed by a 0.55X AMPure XP purification step. 32 μL End-repaired DNA was added 1.0 μL 20 μM Annealed Blunt Adapter, 4.0 μL Template Prep Buffer, 2.0 μL 1 mM ATP low and 1.0 μL 30 U/μL Ligase. The reaction was incubated at 25°C for 16 hours (overnight) and then at 65°C for 10 minutes to inactive the ligase. To remove failed ligation fragments, 1.0 μL 100 U/μL Exo III and 1.0 μL 10 U/μL Exo VII were directly added to the ligation product incubated at 37°C for 60 minutes and at 4°C for 1 minute. A 0.55X AMPure XP purification step was performed to remove all adapter dimers and contaminants, then followed additional two 0.4X AMPure XP purification steps to remove the fragments less than 2 Kb. The profile of the library was checked by Agilent High Sensitivity DNA chip (Agilent Technologies). The library was sequenced on PacBio RS II instrument using MagBead OneCellPerWell protocol (movie length of 300 minutes, on-plate loading concentration of 0.15 nM). The PacBio sequencing data was processed by the PacBio SMRT Portal pipeline of Read of Insert with the parameters “Minimum Number of Passes = 0” and “Minimum Predicted Accuracy = 0.75”.
Picky pipeline for SV detection
The reads (in fastq format) were processed with an in-house assembled analysis pipeline Picky (Fig. 1a). Briefly, Picky is composed of three steps: aligning nanopore reads to the reference genome, picking the best alignments and calling structural variants (SVs).
Picky uses the LAST aligner (last755)24,25 to produce all high-scoring segment pairs (HSPs) of each nanopore read against the human genome (hg19). For high sensitivity, we adopted the scoring scheme (reward = 1, penalty = −1, gap open = 0, gap extension = 2) used in NCBI megaBLAST (https://www.ncbi.nlm.nih.gov/books/NBK279678/).
Next, Picky (command: selectRep) produces the read alignment by stitching the segments from LAST with a greedy seed-and-extend strategy to maximize the coverage of the read by the selected co-linear segments. Spurious aligned segments (%Identity < 55 or EG2 > 1.7e-12) were discarded. EG2 is defined by LAST as the expected number of alignments with greater or equal score, between two randomly-shuffled sequences of length 1 billion each. The remaining segments were ranked according to probability of random hit and alignment score. Picky selectRep then performed the seed-and-extend process by selecting as seed candidate alignment a highest ranked segment among the remaining segments, and linking this seed candidate alignment with the remaining segments whose read coordinates were in the vicinity of the candidate seed alignment read coordinates. The linking of segment(s) produced a linked alignment extension, equivalent to a read alignment, when its total coverage spanned ≥ 70% of the read. The seed-and-extend process repeated until all segments had been selected as seed candidate alignment. Linked alignment extensions with a combined score within 90% of the best combined score were all consider putative read alignments. The implementation specifics can be found in public site https://github.com/TheJacksonLaboratory/Picky/wiki.
Finally, SV calling was performed with Picky’s callSV command. Read with a single putative read alignment with linked segment(s), known as split-read, contains putative SV. Picky computed the distances between adjacent pair of segments in a split-read in both the reference genome-coordinate (sDiff) and the read-coordinates (qDiff). The distances sDiff and qDiff along with chromosome and alignment strand was used to detect the SV present as in Supplementary Figure 12. Picky assigned split reads into seven classes of SVs: inversion (INV): segments aligned to the same chromosome but in different orientations; translocation (TLC): segments aligned to different chromosomes; tandem duplication (TD): segments contain a complete duplicated region (TDC) or only span across a duplication junction (TDJ); simple insertion (INS) or deletion (DEL), segments correspond to the same chromosomal region in the same orientation but either flank a sequence that does not match the reference genome or lack an intervening sequence observed in the reference genome, respectively; and (INDEL), segments indicate both INS and DEL for the same split read. Full list of called SVs can be found in Supplementary Table 3.
Picky is also conscious of the homopolymer undercalling issue observed in nanopore sequences decoded by the Metrichor RNN basecalling algorithm. Specifically, homopolymers beyond five identical bases in all four nucleotide contexts (An, Tn, Cn and Gn) were significantly underrepresented relative to expectation (Supplementary Fig. 13a), an observation commonly reported in nanopore sequences base-called by RNN basecaller21. The underrepresentation of homopolymer in these reads was a major source of false positives in deletion detection (Supplementary Fig. 13b). To remove the false positive deletions defined within the compressed homopolymer regions, we implemented an optional filtering step to annotate and remove the homopolymer-associated deletions. As a result of this adjustment, the numbers of SVs classified as insertions, deletions or INDEL decreased from 29,977 to 28,402. This filter/flag is an optional step and the standard Picky should be used directly with the homopolymer-aware basecaller like Scrappie without additional post-flag/filtering.
Comparison between long read and short read data
Nanopore reads aligned uniquely (possibly multiple fragments) to the human genome (hg19) were extracted for genome coverage computation with BedTools (v2.25.0). Similarly, HCC1187 Illumina paired-end sequencing data (SRA accession SRX969058) was mapped to human genome with BWA-MEM (v0.7.12). The mapped reads were sampled at 2.5X, 10X, 30X and 60X. Genome coverage was computed on reads with a mapping quality of 60. To compare our long-read (LR) SV calls with the short-read (SR) SV calls, SV from LR and SR are overlapping if 1) their genomic spans overlap and 2) the ratio of the larger SV length over the smaller SV length does not exceed 3. Density plots were generated from the specific SV spans.
NA12878 nanopore data SVs comparison
We downloaded NA12878 nanopore reads21. NanoSV16 was used to call SVs (Set N) with the parameter “-c 2” on LAST alignment. LAST alignments were generated based on the last-train scoring parameters established16. Sniffles17 was used to call SVs (Set S) with the parameters “-n-1 -s 2” on NGM-LR alignments. Picky was used to call SVs (Set P) on LAST alignment generated with the parameters “-C2 -K2 -r1 -q3 -a2 -b1”. All alignments were performed against the human reference genome hg19. The insertions and deletions determined by the PacBio long-read data from the same genome28 were used as the reference call set (Set R). Only SVs with length > 30 bases were used for comparison. To determine the overlap between SV Set X and SV Set Y, we count the number of SV call x that overlap SV call y, and the number of SV call y that overlap SV call x. SV call x and y are overlapping if their genomic spans overlap and that the ratio of the larger SV length over the smaller SV length does not exceed 3. To determine the sensitivity in deletion calling, the overlap calculations are repeated with the required minimum read support enumerated from 2 to 20.
Phased adjacent SVs from multi-breakpoint long reads
We counted the pair of adjacent SVs called in all the multi-breakpoint long reads. We assumed that the expected count would follow the distribution of independently drawing two SVs from the population of the SVs from all the multi-breakpoint long reads. The log-likelihood was then computed.
SVs span size distribution
To explore the genomic feature of SVs, we applied different methods to determine the distributions of their span size. For DEL, INS and INDEL, we calculated the total numbers of SVs from each genomic bin (bin size = 20 bp). For INDEL, we used sDiff and qDiff as deletion and insertion spans, respectively. For TDC, TDJ and INV, density plots were generated from their span distributions to show their large size variations.
Genomic distribution of the SVs and the breakpoints
Breakpoint density was computed from the numbers of breakpoints per Mb across the genome. The density of TLC pair was calculated using the TLC breakpoint distribution in pair per Mb across the genome. Circos plot was performed using the breakpoint densities, spans of TDC, TDJ (< 20 Mb), INV and TLC pairs (counts > 3).
Association between gene coding and breakpoint density
Gene density was computed as the fraction of bases overlapping with annotated gene regions (exons and introns; GenCode V24) in each Mb bin across the genome. Violin plots of the gene density from the top 10% breakpoint dense regions (breakpoint density > 40) and the bottom 10% least dense regions (breakpoint density < 9) were generated. Mann-Whitney test was performed.
Breakpoint landscape analysis
Each breakpoint was stepwise assigned to different class of genomic features based on the GENCODE v24 gene model (Supplementary Fig. 14). The promoter was defined as the upstream 2.5 Kb region of the transcription start site. The fraction of reference genome in each class was taken as the background distribution to compute the expected number of breakpoints for each SV type. For SVs with two breakpoints, the pair was considered independent. The ratio of number of breakpoints between observed and expected was log2 transformed.
Repeat analysis
To determine whether the inserted DNA fragments from INS and INDEL as well as TD regions contained repetitive sequences, if so, which class of repeats, we extracted the inserted or duplicated DNA sequences from their corresponding nanopore reads and annotated them to different repeat classes by aligning them to public annotated repeat sequences using RepeatMasker-open-4-0-6 (http://www.repeatmasker.org/). Violin plot was generated with the percentages of the SV fragments annotated to repeats (hg19). The relative ratios of the most predominant repeat class, all other repeats and no repeat from each of the five SV types (TDC, deletion regions in INDEL, insertion regions in INDEL, DEL and INS) were produced in 20 bp span size intervals.
Distribution of genomic micro-insertions
Un-aligned DNA sequences found between each breakpoint junction (nanopore reads alignment with qDiff > 20) were extracted from their corresponded nanopore reads from INDEL, TD, TLC and INV. Their size distribution was plotted for these 4 classes.
ICP analysis
ICP was defined as the sum of a region’s inter-chromosomal contact frequencies divided by its total contact frequencies. We downloaded the TCC (Tethered Conformation Capture) interaction matrix from NCBI SRA accession SRX03011035 and computed the ICP on the whole genome33. The counts of breakpoints were partitioned into four groups from low to high. The correlation was plotted from ICPs found in different partitions.
Multidimensional scaling of gene expression analysis
SVs with spans range from 1Kb to 1 Mb were selected to compare their impacts on gene expressions between TNBC and non-TNBC samples. There are 1,260 coding and 711 non-coding genes in the selected 537 DEL, 2,383 INDEL and 188 TDJ events (SV-genes). Their expression in 113 TNBC and 851 non-TNBC samples was retrieved from gene expression data of the breast carcinoma (BRCA) of the cancer genome atlas (TCGA) based on our previous study36. Two data sets, the control set and the permutation set, were used to evaluate the significance of grouping analysis. The control data was selected from the non-SV genes of the exact number that were expressed at similar level as the SV-genes. For the permutation data, the expressions of SV-genes were individual-wise permutated in TNBC and non-TNBC samples. Multi-dimensional Scaling (MDS) was used to analyze the expression of SV-genes in the BRCA dataset and visualize the sample relationship.
Validation of SV candidates
We selected 3–46 SV events from each SV classification (Supplementary Fig. 4a). In detail, for INV, TLC and TDJ, the validation was done by PCR across the breakpoint junctions, candidate SVs in these classes were selected mainly based on: 1) the presence of sequence specific PCR primers on each side of the breakpoints; 2) amplicon sizes ranged from 0.3–1 Kb. For INS, DEL, TDC, validation was performed by amplifying the entire SV regions. SV candidates that allowed PCR primers designed in the non-repeat regions surrounding the SV junctions and enabled the amplicon sizes ranged between 0.3–1Kb (DEL), 0.3–5 Kb (INS) and 0.2–2.1 Kb (TDC) were chosen. To minimize the confounding effect of micro-insertions in PCR primer design and amplicon size confirmation, only SV candidates contained < 50 bp micro-inserted sequences were selected for PCR validation. The presence of micro inserted sequences was validated separately by PCR followed by sequencing to obtain nucleotide resolution sequence information (see below). All primers used are provided in Supplementary Table 6.
Validation of micro-insertions
Twelve SVs with micro-insertion were randomly selected from the SV list for validation. They included 6 INDELs, 5 TDCs and 1 TLC. The inserted sizes of these candidates ranged from 36 bp to 580 bp. PCR primers were designed around the breakpoint sites in the non-repeat regions with predicted amplicon sizes ranged from 1.2–1.8 Kb, which covered both the SV junctions and the inserted sequences. Amplicons were pooled and sequenced by PacBio SMRT-sequencing and the circular consensus sequence (CCS) reads were used to confirm the micro-insertions within the breakpoint junctions. All primers used are provided in Supplementary Table 6.
Checking of short reads called SV against PCR validated SV
HCC1187 Illumina paired-end sequencing data (SRA accession SRX969058) was mapped to human genome (hg19) with BWA-MEM (v0.7.12). The mapped reads were also sampled at 2.5X, 10X, 30X and 60X. LUMPY (v0.2.13) was used to call SVs using both non-redundant split-read and discordant paired-end reads with the minimum weight for a call (-mw) as 2, 3, 5, 10, and 16 for the subsampled sets 2.5X, 10X, 30X, and 60X and the whole dataset (102X), respectively. We then loaded the LUMPY generated vcf files in IGV browser to visually check the PCR validated SVs locus for the same SV type called by LUMPY.
Life Sciences Reporting Summary
Further information on experimental design is available in the Life Sciences Reporting Summary.
Code availability
Picky pipeline and associated documentation are available at https://github.com/TheJacksonLaboratory/Picky.
Data availability
All nanopore whole-genome sequencing data for HCC1187 described in this study have been deposited in the Sequencing Read Archive under the accession number SRP115881. Illumina short-read data at 102X depth for HCC1187 was obtained from the Sequencing Read Archive under the accession number SRX969058. Nanopore data for NA12878 was obtained as raw fastq files from https://github.com/nanopore-wgs-consortium/NA12878. All other data that support the findings of this study are available from the corresponding author upon request.
Supplementary Material
Supplementary Table 1. Summary of the 15 nanopore runs in this study.
Supplementary Table 2. Summary of the mapping and the SV calling results.
Supplementary Table 3. List of seven SV types detected in nanopore data.
Supplementary Table 4. SVs selected for validation analysis.
Supplementary Table 5. Details of nanopore sequencing kits, devices and software.
Supplementary Table 6. List of all primers used in this study.
Read length, throughput and accuracy of nanopore sequencing.
Acknowledgments
The authors thank P. Shreckengast for collecting the HCC1187 cells; and C. Robinett and A. Lau for their comments on the manuscript; and B. Hanson and M. Bolisetty for their help in setting up our initial nanopore runs. Research reported in this publication was partially supported by the National Cancer Institute of the National Institutes of Health under Award Number P30CA034196. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Author Contributions
L.G., C.-H.W. and C.-L.W. designed the experiment, analysed the data and wrote the manuscript. L.G. performed the experiments. C.-H.W. developed the Picky pipeline. W.-C.C. performed the analysis of the TCGA data. H.T. performed the ICP analysis. F.M., C.Y.N., E.T.L. and C.-L.W. contributed to the manuscript preparation.
Competing financial interests
L.G., C.-H.W. and C.-L.W. have received a few batches of reagent from Oxford Nanopore. C.-L.W. has received travel and accommodation support from Oxford Nanopore as an invited speaker at the Oxford Nanopore user meeting.
References
- 1.Sudmant PH, et al. An integrated map of structural variation in 2,504 human genomes. Nature. 2015;526:75–81. doi: 10.1038/nature15394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Feuk L, Carson AR, Scherer SW. Structural variation in the human genome. Nat Rev Genet. 2006;7:85–97. doi: 10.1038/nrg1767. [DOI] [PubMed] [Google Scholar]
- 3.1000 Genomes Project Consortium. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bochukova EG, et al. Large, rare chromosomal deletions associated with severe early-onset obesity. Nature. 2010;463:666–670. doi: 10.1038/nature08689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Diskin SJ, et al. Copy number variation at 1q21.1 associated with neuroblastoma. Nature. 2009;459:987–991. doi: 10.1038/nature08035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Edwards PA. Fusion genes and chromosome translocations in the common epithelial cancers. J Pathol. 2010;220:244–254. doi: 10.1002/path.2632. [DOI] [PubMed] [Google Scholar]
- 7.Menghi F, et al. The tandem duplicator phenotype as a distinct genomic configuration in cancer. Proc Natl Acad Sci USA. 2016;113:E2373–2382. doi: 10.1073/pnas.1520010113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weischenfeldt J, Symmons O, Spitz F, Korbel JO. Phenotypic impact of genomic structural variation: insights from and for human disease. Nat Rev Genet. 2013;14:125–138. doi: 10.1038/nrg3373. [DOI] [PubMed] [Google Scholar]
- 9.Stankiewicz P, Lupski JR. Structural variation in the human genome and its role in disease. Annu Rev Med Annu Rev Med. 2010;61:437–455. doi: 10.1146/annurev-med-100708-204735. [DOI] [PubMed] [Google Scholar]
- 10.Chaisson MJ, et al. Resolving the complexity of the human genome using single-molecule sequencing. Nature. 2015;517:608–611. doi: 10.1038/nature13907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goodwin S, McPherson JD, McCombie WR. Coming of age: ten years of next-generation sequencing technologies. Nat Rev Genet. 2016;17:333–351. doi: 10.1038/nrg.2016.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alkan C, Coe BP, Eichler EE. Genome structural variation discovery and genotyping. Nat Rev Genet. 2011;12:363–376. doi: 10.1038/nrg2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mills RE, et al. Mapping copy number variation by population-scale genome sequencing. Nature. 2011;470:59–65. doi: 10.1038/nature09708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sovic I, et al. Fast and sensitive mapping of nanopore sequencing reads with GraphMap. Nat Commun. 2016;7:11307. doi: 10.1038/ncomms11307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Spies N, et al. Genome-wide reconstruction of complex structural variants using read clouds. Nat Methods. 2017;14:915–920. doi: 10.1038/nmeth.4366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cretu Stancu M, et al. Mapping and phasing of structural variation in patient genomes using nanopore sequencing. Nat Commun. 2017;8:1326. doi: 10.1038/s41467-017-01343-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sedlazeck FJ, et al. Accurate detection of complex structural variations using single molecule sequencing. 2017 doi: 10.1038/s41592-018-0001-7. Preprint at bioRxiv https://doi:10.1101/169557. [DOI] [PMC free article] [PubMed]
- 18.Jain M, et al. Improved data analysis for the MinION nanopore sequencer. Nat Methods. 2015;12:351–356. doi: 10.1038/nmeth.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deamer D, Akeson M, Branton D. Three decades of nanopore sequencing. Nat Biotechnol. 2016;34:518–524. doi: 10.1038/nbt.3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jain M, Olsen HE, Paten B, Akeson M. The Oxford Nanopore MinION: delivery of nanopore sequencing to the genomics community. Genome Biol. 2016;17:239. doi: 10.1186/s13059-016-1103-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jain M, et al. Nanopore sequencing and assembly of a human genome with ultra-long reads. Nat Biotechnol. 2018 doi: 10.1038/nbt.4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gazdar AF, et al. Characterization of paired tumor and non-tumor cell lines established from patients with breast cancer. Int J Cancer. 1998;78:766–774. doi: 10.1002/(sici)1097-0215(19981209)78:6<766::aid-ijc15>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 23.Li H. Minimap2: fast pairwise alignment for long nucleotide sequences. 2017 doi: 10.1093/bioinformatics/bty191. Preprint at ArXiv https://arxiv.org/abs/1708.01492. [DOI] [PMC free article] [PubMed]
- 24.Kielbasa SM, Wan R, Sato K, Horton P, Frith MC. Adaptive seeds tame genomic sequence comparison. Genome research. 2011;21:487–493. doi: 10.1101/gr.113985.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frith MC, Hamada M, Horton P. Parameters for accurate genome alignment. BMC bioinformatics. 2010;11:80. doi: 10.1186/1471-2105-11-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Layer RM, Chiang C, Quinlan AR, Hall IM. LUMPY: a probabilistic framework for structural variant discovery. Genome Biol. 2014;15:R84. doi: 10.1186/gb-2014-15-6-r84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stephens PJ, et al. Complex landscapes of somatic rearrangement in human breast cancer genomes. Nature. 2009;462:1005–1010. doi: 10.1038/nature08645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pendleton M, et al. Assembly and diploid architecture of an individual human genome via single-molecule technologies. Nat Methods. 2015;12:780–786. doi: 10.1038/nmeth.3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bignell GR, et al. Architectures of somatic genomic rearrangement in human cancer amplicons at sequence-level resolution. Genome Res. 2007;17:1296–1303. doi: 10.1101/gr.6522707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Campbell PJ, et al. Identification of somatically acquired rearrangements in cancer using genome-wide massively parallel paired-end sequencing. Nat Genet. 2008;40:722–729. doi: 10.1038/ng.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cahill D, Connor B, Carney JP. Mechanisms of eukaryotic DNA double strand break repair. Front Biosci. 2006;11:1958–1976. doi: 10.2741/1938. [DOI] [PubMed] [Google Scholar]
- 32.Howarth KD, et al. Array painting reveals a high frequency of balanced translocations in breast cancer cell lines that break in cancer-relevant genes. Oncogene. 2008;27:3345–3359. doi: 10.1038/sj.onc.1210993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kalhor R, Tjong H, Jayathilaka N, Alber F, Chen L. Genome architectures revealed by tethered chromosome conformation capture and population-based modeling. Nat Biotechnol. 2012;30:90–98. doi: 10.1038/nbt.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Branco MR, Pombo A. Intermingling of chromosome territories in interphase suggests role in translocations and transcription-dependent associations. PLoS Biol. 2006;4:e138. doi: 10.1371/journal.pbio.0040138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tjong H, et al. Population-based 3D genome structure analysis reveals driving forces in spatial genome organization. Proc Natl Acad Sci USA. 2016;113:E1663–1672. doi: 10.1073/pnas.1512577113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chung IF, et al. DriverDBv2: a database for human cancer driver gene research. Nucleic Acids Res. 2016;44:D975–979. doi: 10.1093/nar/gkv1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. Summary of the 15 nanopore runs in this study.
Supplementary Table 2. Summary of the mapping and the SV calling results.
Supplementary Table 3. List of seven SV types detected in nanopore data.
Supplementary Table 4. SVs selected for validation analysis.
Supplementary Table 5. Details of nanopore sequencing kits, devices and software.
Supplementary Table 6. List of all primers used in this study.
Read length, throughput and accuracy of nanopore sequencing.
Data Availability Statement
All nanopore whole-genome sequencing data for HCC1187 described in this study have been deposited in the Sequencing Read Archive under the accession number SRP115881. Illumina short-read data at 102X depth for HCC1187 was obtained from the Sequencing Read Archive under the accession number SRX969058. Nanopore data for NA12878 was obtained as raw fastq files from https://github.com/nanopore-wgs-consortium/NA12878. All other data that support the findings of this study are available from the corresponding author upon request.