Summary
Objective
The severe epilepsies of infancy (SEI) are a devastating group of disorders that pose a major care and economic burden on society; early diagnosis is critical for optimal management. This study sought to determine the incidence and aetiologies of SEI, and model the yield and cost-effectiveness of early genetic testing.
Methods
Population-based study of the incidence, etiologies and cost-effectiveness of a whole exome sequencing-based gene panel (targeted-WES) in infants with SEI born 2011–2013, identified through EEG and neonatal databases. SEI was defined as seizure onset before age 18 months, frequent seizures, epileptiform EEG and failure of ≥2 anti-epileptic drugs. Medical records, investigations, MRIs and EEGs were analysed, and genetic testing performed if no etiology was identified. Economic modelling was performed to determine yield and cost-effectiveness of investigation of infants with unknown etiology at epilepsy onset, incorporating targeted-WES at different stages of the diagnostic pathway.
Results
Of 114 infants with SEI (incidence 54/100,000 live births/year), the etiology was determined in 76 (67%): acquired brain injuries (14), focal cortical dysplasias (14), other brain malformations (17), channelopathies (11), chromosomal (9), metabolic (6) and other genetic (5) disorders. Modelling showed that incorporating targeted-WES increased diagnostic yield compared to investigation without targeted-WES (48/86 vs 39/86). Early targeted-WES had lower total cost ($677,081USD vs $738,136USD) than late targeted-WES. A pathway with early targeted-WES and limited metabolic testing yielded seven additional diagnoses compared to investigation without targeted-WES (46/86 vs 39/86), with lower total cost ($455,597USD vs $661,103USD), lower cost per diagnosis ($9,904USD vs $16,951USD) and a dominant cost-effectiveness ratio.
Significance
Severe epilepsies occur in 1/2,000 infants, with the etiology identified in two-thirds, most commonly malformative. Early use of targeted-WES yields more diagnoses for lower cost. Early genetic diagnosis will enable timely administration of precision medicines, once developed, with the potential to improve long-term outcome.
Keywords: epilepsy, infancy, incidence, etiology, genomic, health economic
Introduction
Epilepsy affects more than 50 million people worldwide.1 Severe epilepsies of infancy (SEI) are characterized by frequent seizures, epileptiform EEG abnormalities and anti-epileptic drug (AED) resistance. They include epileptic syndromes such as early infantile epileptic encephalopathy, epilepsy of infancy with migrating focal seizures and infantile spasms (West syndrome).2–4 Developmental outcome is often poor, comorbidities frequent, and mortality high due to the effects of seizures and consequences of severe neurological impairment. SEI consume significant diagnostic and therapeutic healthcare resources.
SEI are caused by numerous genetic and acquired disorders, though the etiology often remains unknown.3,4 Current diagnostic testing includes imaging, chromosomal and metabolic testing, typically performed in a tiered or staged fashion (Table 1), with variable use of genetic testing.5,6 The yield of whole exome sequencing (WES), a next generation sequencing technology that enables interrogation of large numbers of genes in parallel, in research cohorts of severe epilepsies of childhood varies from 10 to 72%. However, use of WES and other genetic testing in clinical practice is limited by availability, cost and lack of evidence of cost-effectiveness in population-based settings.7,8 Early diagnosis of etiology is critical, as prompt, optimal treatment may improve outcomes.3,9,10 This is already established for surgically-remediable epilepsies and will become increasingly important as novel therapies are developed for genetic epilepsies.
Table 1.
Tiers of imaging, chromosomal and metabolic tests currently performed in Victorian pediatric centres (Pathway 1), and their associated costs including admission and procedural costs where relevant in US dollars*.
| Tests | Cost* | |
|---|---|---|
| Tier 1 | MRI-brain under general anaesthetic Blood tests: chromosomal microarray, full blood count, electrolytes, urea and creatinine, glucose, calcium, magnesium, phosphate, liver function tests, lactate, ammonia, amino acids, acylcarnitines, biotinidase, uric acid Urine tests: organic acids, amino acids, piperideine-6-carboxylate, S-sulphocysteine, guanidinoacetic acid, purines and pyrimidines |
$3,202 |
| Tier 2 | Blood tests: common mitochondrial mutations, POLG common mutations, transferrin isoforms, copper and caeruloplasmin, very long chain fatty acids, white cell enzymes, glucose^, lactate^, pyruvate^, amino acids^ Cerebrospinal fluid tests: cell count, protein, glucose, lactate, pyruvate, amino acids, neurotransmitters |
$2,184 |
| Repeat MRI-brain | Performed at 3 Tesla using epilepsy protocol sequences under general anaesthetic | $1,565 |
| Tier 3 | Skin biopsy: electron microscopy for changes of neuronal ceroid lipofuscinosis, lysosomal and mitochondrial disorders, fibroblast culture (for DNA source) Liver and muscle biopsies: histopathology, histochemistry, electron microscopy, respiratory chain enzyme analysis |
$5,309 |
See Tables S3 and S4 for detailed breakdown of costs and their sources
Paired with cerebrospinal fluid
This population-based study aimed to establish the incidence and etiologies of SEI, and model the diagnostic yield, cost and cost-effectiveness of WES-based gene panel (targeted-WES) performed at various points along the diagnostic pathway.
Methods
Inclusion/exclusion criteria
We studied infants with SEI born in Victoria, Australia during 2011–2013. SEI was defined as <18 months old of 1) frequent seizures (≥ daily for one week or ≥ weekly for one month), 2) ongoing seizures despite trials of two appropriate AEDs, and 3) epileptiform EEG abnormality. Infantile spasms were automatically included.
Ascertainment
Victoria had a population of 5,582,670 people and 71,444 live births in 2011 (www.abs.gov.au, www.bdm.vic.gov.au). Government funded health care is available to all residents. Newborn screening for metabolic disorders is performed routinely unless declined by parents. There are two pediatric tertiary hospitals with neurology departments and four neonatal intensive care units (NICUs) in Victoria, all in Melbourne. Nine EEG laboratories perform EEGs on children <2 years old. Seventeen pediatric neurologists provide care to children with seizures, with general pediatricians. The advanced, centralised nature of the Victorian health system provides an ideal environment to conduct an epidemiological study, as illustrated by other population-based studies (e.g. www.neuroscience.org.au/australian-epilepsy-pregnancy-register, www.auscr.com.au, NEMESIS stroke study).11
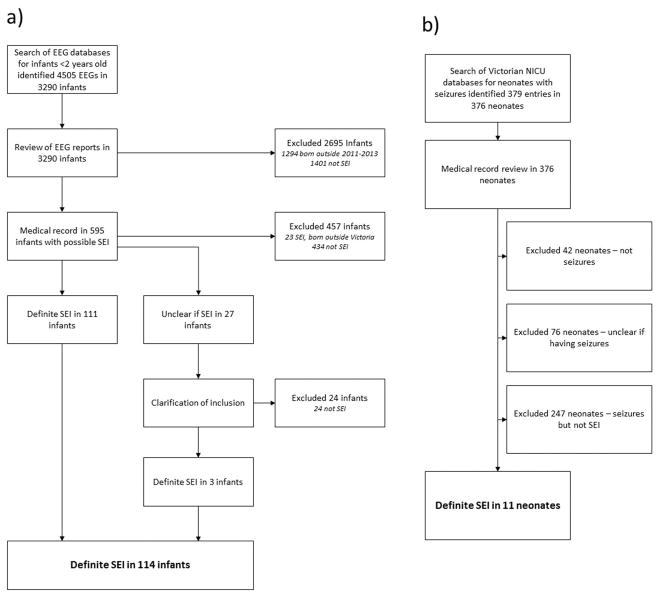
The study began in 2013; ascertainment was retrospective for infants presenting in 2011–2012 and prospective for 2013–2015. Infants with potential SEI were identified by review of all EEG reports in children <2 years in Victoria during 2011–2015 (n=4505), search of NICU databases for neonates with seizures born 2011–2013 (n=379), and regular questioning of pediatric neurologists (Figure 1). Medical records of infants with potential SEI were reviewed to determine if inclusion criteria were met.
Figure 1.
Flow diagram showing the process of identifying infants with SEI from a) EEG reports and b) neonatal intensive care unit database entries
Assessment
All infants meeting SEI inclusion criteria (n=114) were studied to determine their electroclinical phenotype and underlying etiology.2 History, examination and investigation findings were obtained from clinical assessment and medical records (available on all 114 infants). All EEG recordings and seizure videos were reviewed by two pediatric epileptologists (KBH, ASH). All brain MRIs were reviewed by KBH and pediatric neuroradiologist SM.
Research genetic testing in infants whose etiology was unknown included: targeted-WES (40 infants), molecular inversion probes (MIPS) with panels of 39–65 epilepsy genes (32) (Tables S1 and S2),12,13 single gene sequencing (1), and whole genome sequencing (WGS) (1). Singleton WES was performed during 2015–2017 as detailed in the supplementary material. Analysis was confined to a panel of 341 infantile-onset epilepsy genes. Research genetic testing to pursue the genetic basis of known etiologies was not performed (eg tuberous sclerosis, lissencephaly).
Statistical and economic analyses
Incidence was calculated using live birth rates from the Australian Bureau of Statistics and the Victorian Births, Deaths and Marriages Registry (www.abs.gov.au, www.bdm.vic.gov.au). Incidence was corrected for population migration to account for infants who may have moved out of the state prior to seizure presentation (supplementary material).
Economic modelling and a cost-effectiveness analysis was performed, using standard economic evaluation methodology.14 The analysis compared the relative diagnostic yield and total cost of seven diagnostic pathways, with groups of investigations (called tiers) performed in a tiered fashion (Table 1), incorporating targeted-WES at different points to generate estimates of incremental cost-effectiveness. For two pathways in which WES was performed early, later tier (Tier 3 +/− Tier 2) investigations were removed (Table 2a). The time horizon began at epilepsy onset. In the seven modelled pathways, each infant with unknown etiology prior to epilepsy onset progressed through the tiers until the first investigation that would have yielded the diagnosis, regardless of whether seizures were ongoing (Model A).
Table 2.
a) Diagnostic pathways modelled in the health economic analysis^ and b) Cost and yield of diagnostic pathways if all 86 infants with unknown etiology at the time of presentation are investigated until an etiology is identified or they reach the end of the diagnostic pathway (Model A). Costs are listed in US dollars
| a | b | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Pathway | Step 1 | Step 2 | Step 3 | Step 4 | Step 5 | Total cost | Diagnoses made | Average cost per diagnosis | Cost per patient | ICER relative to Pathway 1 |
| 1 | Tier 1 | Tier 2 | Repeat MRI | Tier 3 | - | $661,103 | 39 | $16,951 | $7,687 | - |
| 2 | Tier 1 | Tier 2 | Repeat MRI | Tier 3 | WES | $738,136 | 48 | $15,378 | $8,583 | $8,559 |
| 3 | Tier 1 | Tier 2 | Repeat MRI | WES | Tier 3 | $690,356 | 48 | $14,382 | $8,027 | $3,250 |
| 4 | Tier 1 | Tier 2 | WES | Repeat MRI | Tier 3 | $693,951 | 48 | $14,457 | $8,069 | $3,650 |
| 5 | Tier 1 | WES | Tier 2 | Repeat MRI | Tier 3 | $677,081 | 48 | $14,106 | $7,873 | $1,775 |
| 6 | Tier 1 | WES | Repeat MRI | Tier 2 | - | $553,431 | 48 | $11,530 | $6,435 | Pathway 6 dominates Pathway 1! |
| 7 | Tier 1 | WES | Repeat MRI | - | - | $455,597 | 46 | $9,904 | $5,298 | Pathway 7 dominates Pathway 1! |
Tier 3 not performed if a brain malformation was strongly suspected as these infants would not typically undergo tissue biopsies in clinical practice
ICER = incremental cost-effectiveness ratio, WES = targeted WES
’Dominates’ refers to being more effective and less costly than the comparison
Targeted-WES was the only next generation sequencing (NGS) technology used in the modelling. The model considered targeted-WES to be diagnostic in all infants with single gene disorders with exonic mutations identified using any genetic technology as long as the gene was well-covered and analysed in the targeted-WES. In the model, WES was considered negative in only one infant with a single gene disorder (whose diagnosis was made on WGS following negative WES).
Infants with brain malformations visible on their initial MRI were considered to have a Tier 1 testing diagnosis, even if their malformation was only recognised after repeat imaging or imaging review (n=4). The model considered that Tier 3 testing would only be performed in infants with ongoing seizures (in both Models A and B), significant developmental delay and no clinical suspicion of an occult brain malformation.
The models used the infants’ actual diagnoses and a standard cost for each tier of testing (Table 1). For example, the cost of testing for an infant with a diagnosis made on Tier 1 testing was $3,202, the cost of testing for an infant who had no diagnosis made was $13,899 for Pathway 2 (Table 2a) (Tier 1 $3,202→Tier 2 $2,184→Repeat MRI $1,565→Tier 3 $,5,309→ Targeted-WES $1,639).
Costs were calculated using data from Australian Medicare Benefits Schedule, Royal Children’s Hospital (RCH) Decision Support Unit, Victorian Clinical Genetics Service and State Neuropathology Service. Costs to the hospital in 2016–7 of performing and interpreting the tests were used, including cost of investigation-related admission, anaesthesia or surgery (Tables S3, S4). Treatment-related costs were excluded. Costs were converted to US dollars (USD, exchange rate 1 AUD = 0·745 USD).
For infants with a suspected genetic diagnosis at a particular point along the pathway, the costs of targeted testing were added to the total costs (Table S5), for example SCN1A testing in suspected Dravet syndrome. If targeted-WES was performed immediately after the tier at which a diagnosis was suspected, it was considered that the diagnosis would be confirmed on targeted-WES rather than targeted gene testing. For Dravet syndrome, we assumed the diagnosis would be suspected after Tier 1 testing. The cost for Pathway 2 would then be $3,854 (Tier 1 $3,202→ SCN1A sequencing $652) and the cost for Pathway 5 $4,841 (tier 1 $3,202→ Targeted-WES $1,639). The costs of genetic testing were not included where infants had a clearcut etiology (e.g. tuberous sclerosis diagnosed on MRI). The model considered that infants with or without an affected sibling would progress through the pathway in the same manner (Supplementary information).
For each pathway, the number of diagnoses, total cost, cost per diagnosis and cost per patient were calculated. Incremental cost-effectiveness ratio (ICER), defined as the cost required to make one additional diagnosis, was calculated for each pathway relative to Pathway 1 without targeted-WES. The formula used was (total cost Pathway X – total cost Pathway 1)/(number of diagnoses in Pathway X – number of diagnoses in Pathway 1). Willingness to pay was defined as the maximum amount that a funder (e.g. hospital/insurance company) would be willing to pay to achieve an additional diagnosis.
Sensitivity analyses were performed on Model A (for Pathways 1,2,5 and 7), varying the cost of WES (+/−20%), the diagnostic yield of targeted-WES (+/− 4 diagnoses) and the diagnostic yield of the first MRI brain scan (+/− 4 diagnoses) to determine the impact of variability in cost and yield of these investigations on the ICER relative to pathway 1. An additional sensitivity analysis was performed (Model B), using the assumption that infants would only continue beyond step 1 of the pathway if seizures were ongoing (step 2 performed if seizures were ongoing 1 month after presentation, step 3 if ongoing at 3 months, steps 4 and 5 if ongoing at 6 months).
The CHEERS guidelines for reporting economic evaluations were followed (https://www.ispor.org/Health-Economic-Evaluation-Publication-CHEERS-Guidelines.asp).
Study approvals
The study was approved by the Human Research and Ethics Committees of RCH, Monash Children’s Hospital, Royal Women’s Hospital, Mercy Hospital for Women, Austin Health and Geelong Hospital. Written informed consent was obtained for research clinical assessments and genetic testing.
Results
Incidence
114 infants with SEI were born in Victoria during 2011–2013. All were identified through EEG laboratories, 11 from NICU databases (Figure 1) and 43 from neurologist referrals. 107 (94%) infants were identified multiple times, 47 from >1 source and 60 from multiple EEGs. During 2011–2013, there were 222,818 live births in Victoria, yielding an incidence of SEI, adjusted for population migration (Supplementary material), of 54/100,000 live births/year (95% confidence interval 45–65/100,000). Infantile spasms occurred in 74 infants, with an adjusted incidence of 35/100,000 live births/year (95% confidence interval 28–44/100,000).
Etiology
Etiology was identified in 76 (67%) infants (Table 3). Fourteen (12%) had an acquired brain injury, 62 (54%) had genetic or presumed genetic etiologies, and 38 (33%) had unknown etiologies. Brain malformations were identified in 31 (27%), including FCD in 14 (12%). Six (5%) infants had metabolic disorders, nine (8%) had chromosomal abnormalities, and 16 (14%) had single gene disorders (excluding genes for malformation and metabolic disorders), including 11 (10%) with channelopathies.
Table 3.
Etiologies of severe epilepsy in 114 infants
| Etiology | N | Gene mutations identified |
|---|---|---|
| Acquired | 14 (12%) | N/A |
| Perinatal/neonatal hypoxic ischemic encephalopathy | 5 | N/A |
| Periventricular leukomalacia | 3 | N/A |
| Complicated meningitis | 2 | N/A |
| Perinatal hypoxic ischemic encephalopathy and hypoglycaemia | 2 | N/A |
| Perinatal stroke | 2 | N/A |
| Genetic/presumed genetic | 62 (54%) | 39 (50 tested) |
| Structural | 31 | 9 (19 tested) |
| Focal cortical dysplasia | 14 | BRAF (1), DEPDC5 (1), NPRL3 (1), NF (5), NT (6) |
| Tuberous sclerosis | 5 | TSC2 (3), TSC1 (1), NT (1) |
| Malformation of cortical development (other) | 3 | NF (2), NT (1) |
| Polymicrogyria | 3 | NF (1), NT (2) |
| Lissencephaly | 2 | LIS1 (1), NT (1) |
| Other: achondroplasia, Aicardi syndrome, pontocerebellar hypoplasia, Sturge-Weber syndrome | 1 each | FGFR3 (1), NF (2), NT (1) |
| Metabolic | 6 | 5 |
| Mitochondrial disorder | 3 | NDUFAF6 (1), FARS2* (1), NF (1) |
| Other: molybdenum cofactor deficiency, PNPO deficiency, Tay-Sachs disease | 1 each | MOCS2 (1), PNPO (1), HEXA (1) |
| Chromosomal | 9 | 9 |
| Trisomy 21 | 5 | See left |
| Other: chromosome 2q24.3 deletion (incl. SCN1A and SCN2A genes), chromosome 15q21.3q22.2 deletion, isodicentric chromosome 15, Wolf-Hirschhorn syndrome | 4 | See left |
| Single gene | 16 | 16 |
| Channelopathies | 11 | SCN1A (3), KCNQ2 (3), SCN2A (2), SCN8A (2), KCNT1 (1) |
| Other: Aicardi-Goutieres syndrome, SMC1A mutation, Sotos syndrome, SYNGAP1 mutation, TBC1D24 mutation | 1 each | RNASEH2B (1), SMC1A (1), NSD1 (1), SYNGAP1 (1), TBC1D24 (1) |
| Unknown | 38 (33%) | N/A |
N/A= not applicable, NF = not found, NT= not tested,
Potentially pathogenic
The etiology was known prior to epilepsy onset in 28 (25%) infants, including all 14 with acquired etiologies. 37/86 (32%) infants with unknown etiology prior to epilepsy onset, had a diagnosis subsequently made on clinical evaluation (including non-research genetic testing in five and research imaging review in four). 44/49 infants with unknown etiology consented to research genetic testing, the etiology being identified in 11 (25%) (Table S6).
Among infants with unknown etiology prior to epilepsy onset, the highest diagnostic yield investigations were (non-chromosomal) genetic testing (16/49 (32%)) and brain MRI (26/85 (31%)). The genetic tests yielding a diagnosis were single gene testing (5), 4-gene panel (1), MIPS (3), targeted-WES (6) and WGS (1) (Table S7). Nine infants had a variant of unknown significance (VUS) identified on targeted-WES. Chromosomal microarray was diagnostic in 4/74 (5%) infants.
Genetic diagnoses informed reproductive counselling in all infants, led to management change in one (SCN2A mutation with sodium channel blocking AEDs used15) and informed prognostic counselling in most. A significant recurrence risk was identified in five families; two with somatic mosaicism in an unaffected parent (submitted), one with two affected children and presumed parental mosaicism, and two with heterozygous carrier parents for a recessive disorder.
Economic modelling
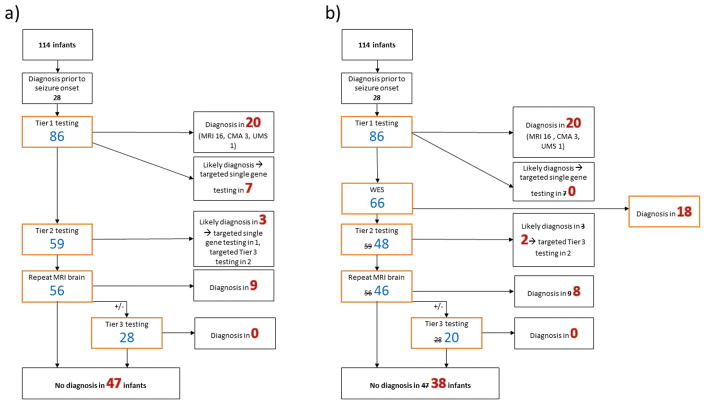
Using Model A (Table 2, Figure 2), Pathway 1 without targeted-WES resulted in a diagnosis in 39/86 (45%) infants with unknown etiology at epilepsy onset, at a total cost of $661,103USD and an average cost per diagnosis of $16,951USD. The addition of targeted-WES after Tier 3 investigations (Pathway 2) increased yield to 48/86 (56%), with a higher total cost ($738,136USD) but lower cost per diagnosis ($15,378USD). The diagnostic yield of adding targeted-WES after Tier 1 investigations (Pathway 5) was the same as Pathway 2, with lower total cost ($677,081USD). The total cost of Pathway 5 was comparable to Pathway 1, with lower cost per diagnosis ($14,106USD).
Figure 2. Yield of modelled diagnostic pathway a) without whole exome sequencing (Pathway 1) and b) with early whole exome sequencing (Pathway 5).
These models assumed that infants suspected clinically to have occult brain malformations did not undergo Tier 3 testing. CMA = chromosomal microarray, UMS= urine metabolic screen, WES = targeted WES
46/86 (53%) infants had a diagnosis made with Pathway 7 in which targeted-WES was performed after Tier 1 investigation, and Tiers 2 and 3 testing was not performed. Both total cost ($455,596USD) and cost per diagnosis ($9,904USD) were considerably reduced in this pathway compared with Pathway 1.
Pathways 6 and 7, in which targeted-WES was used earliest in the diagnostic pathway (after Tier 1 investigations) with Tiers 3 +/− 2 investigations omitted were the most cost-effective relative to Pathway 1. They produced an incremental cost-effectiveness ratio which was ‘dominant’, that is, was cheaper and more effective than Pathway 1. Pathways 3, 4 and 5 had incremental ratios of $1,775–$3,650USD per additional diagnosis achieved. These could be considered cost-effective if the willingness to pay was at least $3,650USD per additional diagnosis made. Pathway 2, in which targeted-WES was placed last in the diagnostic pathway, was the least cost-effective, with an incremental ratio of $8,559USD per additional diagnosis achieved relative to Pathway 1.
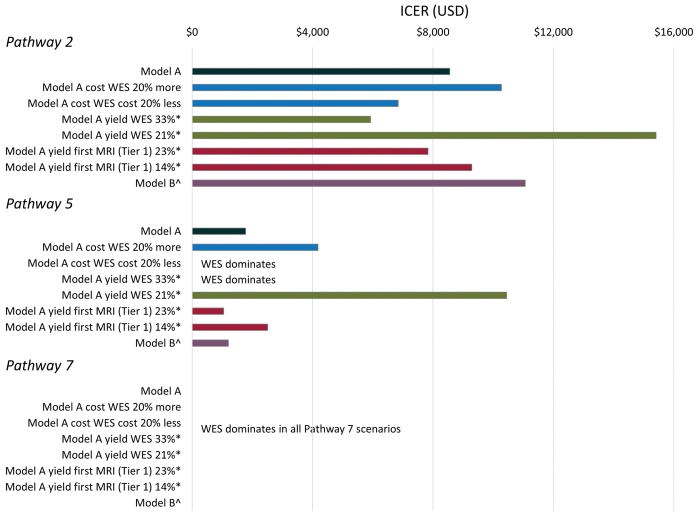
We varied the cost of targeted-WES and the yield of targeted-WES and MRI in Model A, and obtained similar results for most scenarios (Figure 3). The only scenarios in which the pattern of results differed from the base case analysis (Model A) were in Pathway 5, in which WES became dominant when the diagnostic yield was increased (4 additional diagnoses, diagnostic rate 33%) or the cost reduced (by 20%). For Pathways 2 and 5, the sensitivity analyses showed that economic modelling was more sensitive to (varied more with) assumptions about WES yield than WES cost (Pathway 2, base case $8,559, reduced WES yield $15,407, increased WES cost $10,271. Pathway 5, base case $1,775, reduced WES yield $10,442, increased WES cost $4,179). The additional sensitivity analysis (Model B) showed the same pattern of results as Model A (Figure S1, Table S8).
Figure 3. Economic evaluation one-way sensitivity analyses for Model A (base case analysis) comparing Pathways 2, 5 and 7 with Pathway 1.
ICER = Incremental cost-effectiveness ratio
*Yield of targeted-WES and MRI was increased or decreased by 4 diagnoses from that of the base case analysis.
^Model B, infants continued through the diagnostic pathway until an etiology was identified only if seizures were ongoing.
’Dominates’ refers to an economic evaluation result where the intervention/pathway is cheaper and more effective (i.e. greater yield) than the comparison.
Discussion
Understanding the epidemiology and etiologies of SEI is essential for optimisation of diagnostic strategies. The approach taken in this study is pertinent to many human disorders with heterogeneous etiologies, providing a basis for estimating the burden of disease and potential avenues for precision medicine. We found that SEI have an incidence of 1/2000 live births, greater than Duchenne muscular dystrophy (1/4000) and neurofibromatosis type 1 (1/2700).16,17 The incidence of infantile spasms in this study (approximately 1/3000) was similar to previous reports, providing some validation of the study methodology.18 The health burden of SEI accounts for a significant proportion of pediatric inpatient costs, with huge psychosocial and economic impacts on families and society.19,20
We identified the etiology in two-thirds of infants with SEI, with research genetic testing and expert review of MRI. This high yield carries critical prognostic and genetic counselling implications for most patients, and therapeutic implications in some. Acquired causes such as hypoxic-ischemic encephalopathy accounted for only 12% of infants; however, perinatal causes will be more common in developing countries.21
A key finding with management implications was that brain malformations comprised the most common etiological group (27%). FCD predominated (12% patients), and represents a critical group for early diagnosis, as surgery may control seizures, potentially enabling developmental acceleration.10 MRI sequences that increase sensitivity to identify FCD (e.g. double inversion recovery sequences)22, together with expert review, and repeat brain imaging later in infancy, facilitate detection of occult FCD; advances in imaging promise to further improve detection.
Single gene disorders (excluding those associated with malformations or metabolic disorders) were identified in 14% of cases, including 32% of infants in whom MRI, metabolic and chromosomal testing were non-diagnostic, similar to gene panel yield in non-population-based cohorts.23,24 We may have underestimated the yield of targeted-WES as the parents of five infants declined genetic testing. Given the genetic heterogeneity of SEI, our numbers were too small to determine which genes are most common, however, ion channel gene mutations were found in 11/16 cases. One-third of infants remain without a diagnosis, most despite targeted-WES.
Our economic modelling established at a population level that targeted-WES increases diagnostic yield. Diagnostic costs are lower when targeted-WES is performed early in the diagnostic evaluation, and are comparable to the cost of investigation without targeted-WES. Early targeted-WES becomes overwhelmingly cost-effective when the low yield Tiers 2 and 3 metabolic testing are also removed from the diagnostic pathway.
Our model likely underestimates the diagnostic yield and cost-effectiveness of WES as we did not interrogate genes other than those in our panel. Conversely, we did not include the cost of investigating VUS. We also did not consider indirect cost benefits of making an etiologic diagnosis, an important area for future research. The potential benefits of early diagnosis, including optimizing treatments, recognition of comorbidities and accurate reproductive counselling will improve health and economic outcomes.25
Implementation of WES into clinical practice is currently a major focus in many areas of medicine. WES has improved rates of etiologic diagnosis in conditions with high genetic heterogeneity and phenotypic overlap, such as epilepsy. However, optimal use of WES depends not only on diagnostic yield, but also on cost-effectiveness relative to either not using WES or using it late in the diagnostic pathway. This is the first economic study of the utility of WES in epilepsy, with few reported for other conditions.26,27 Most economic evaluations have been performed on retrospective cohorts in tertiary settings, featuring only patients who remain without diagnoses following extensive and expensive investigation. A recent, prospective clinic-based study of 80 infants with suspected monogenic disorders found that WES early in diagnostic pathways was cost-effective compared with standard care without WES28; however, it was not an epidemiological sample and therefore not representative of total costs to the healthcare system. Studies to date have not assessed the role of WES within diagnostic pathways that utilize multiple investigations, such as brain imaging, which is not replaceable by WES. Our population-based study addressed these limitations and strongly supports early use of targeted-WES. Modelling showed that use of targeted-WES after exhaustive investigation, the usual practice for implementation of new technologies, was the least cost-effective strategy. Our findings likely have corollaries in other genetically-heterogeneous neurological conditions in which WES has high yield, such as leukodystrophies and neuropathies. 29,30 Early WES will reduce use of non-genetic, low-yield and often repeated or invasive investigations.
Gene panel analysis (non-WES-based) is an alternative clinical NGS technology. The utility of targeted-WES over gene panels was not compared. Although there is considerable variability, gene panel analyses are typically cheaper and more rapidly analysed. A major advantage of targeted-WES, is that, when negative, data can be reanalysed for new genes as they emerge, or exome-wide analysis can be performed. Thus, additional sequencing costs are not incurred (apart from scientist’s time for data reanalysis). In our modelling, 18 diagnoses were made on WES. Two commercially-available epilepsy gene panels include the causative genes in 14/18 (GeneDx, www.genedx.com) and 16/18 (Ambry Genetics, www.ambrygen.com) patients. Our sensitivity analysis showed that early targeted-WES and limited metabolic testing remained cost-effective even when the yield of WES was reduced to 14 instead of 18 diagnoses. Given this, gene panel analysis would also likely be cost-effective relative to investigation without another NGS technology, although the subsequent need for WES when the panel is negative would add significantly to the total cost. In future, newer molecular techniques, such as WGS, will likely supersede current technologies, although their utility in SEI is not yet known.
In SEI, there is an argument for limiting second- and third-tier metabolic testing at the group level given its low yield, invasiveness and high cost. Metabolic disorders are genetically-determined, such that WES may obviate the need for complex biochemical testing (with the exception of some mitochondrial disorders as WES does not detect mutations in the mitochondrial genome). However, in clinically-suspected treatable metabolic conditions, the turn-around-time for metabolic testing may be faster, and therefore warranted until genomic testing is more rapid. Although the simulated diagnostic pathway in which second- and third-tier metabolic testing was removed yielded two fewer diagnoses, these diagnoses were strongly suspected on clinical grounds and were diagnosed with targeted metabolic testing. Targeted testing should be performed in infants in whom a metabolic diagnosis is strongly suspected and in subgroups with higher likelihood of treatable metabolic conditions, such as neonates and parental consanguinity.6 Although not modelled here, the yield of first-tier metabolic testing and chromosomal microarray probably warrant their continued use.31,32
Our cohort is population-based, but relatively small. Thus, an important consideration is whether our findings are generalizable to similar (high income, low perinatal morbidity) populations. Given the heterogeneity of SEI, there will undoubtedly be differences in etiologies between similar populations. However, it is likely that the proportion of patients in each etiologic group (malformative vs metabolic vs genetic) will vary considerably less. Supporting this assumption are Canadian and US studies of infantile epilepsies showing similarly low rates of metabolic disorders to our study.31,33 Thus, there is likely to be less difference between the yield of each diagnostic test than actual etiologies. Our sensitivity analyses, in which the yield of MRI or WES was increased or reduced as a proxy for variability in the proportion of patients with malformative and single gene disorders, showed that early WES is cost-effective across the range of diagnostic yields modelled.
The cost of WES and other diagnostic investigations may vary between countries. For example, the cost of singleton WES reported in health economic studies from the United States is $1,060–2,471USD.34 While we did not specifically model the costs of all diagnostic investigations in other countries, our sensitivity analysis showed that early targeted-WES is cost-effective across the range of WES prices modelled ($1,639USD +/− 20%), which is within the aforementioned range of US prices. There may be different challenges in obtaining funding for WES in all healthcare settings, be they ‘public’ (government pays) or ‘private’ (insurance company or patient pays). However, where the funder is also responsible for the costs of other diagnostic investigations, the cost-savings achieved through preventing costly metabolic and re-imaging investigations is likely to provide sufficient incentive for performing early targeted-WES.Early targeted-WES in SEI where initial MRI, chromosomal microarray and Tier 1 metabolic investigations are negative is both clinically- and cost-effective. Not performing targeted-WES, or performing it as a final investigation after exhaustive metabolic and specific genetic testing, are suboptimal diagnostic approaches. Early diagnosis of the etiology of a child’s severe epilepsy may carry time-critical genetic counselling and management implications.
Supplementary Material
Key Points.
Severe epilepsies of infancy have an incidence of 1:2000, greater than that of neurofibromatosis type 1 and Duchenne muscular dystrophy.
Etiology can be determined in two-thirds of infants with current diagnostic technologies.
Focal cortical dysplasias were the most common cause, and are a critical group for early diagnosis as surgery may improve outcomes.
Early use of targeted whole exome sequencing, with omission of some metabolic testing, yields more diagnoses for lower cost.
Acknowledgments
We thank the patients and their families who participated in this study; senior EEG scientists and neonatal unit database managers at all sites for assistance with database searches; A/Prof Susan Donath for advice on epidemiological analysis; and Li Huang, Malathi Jeremiah and Justin Green for assistance with the health economic analysis.
Footnotes
Author contributions
The Victorian Severe Epilepsy of Infancy Study Group is Katherine B. Howell (primary investigator), A. Simon Harvey (senior investigator), Ingrid E Scheffer (senior investigator), John Archer, Samuel F. Berkovic, Gemma L. Carvill, Eunice K. Chan, Mark Corbett, Gabriel Dabscheck, Kim Dalziel, Stefanie Eggers, Michael Fahey, Jeremy L. Freeman, Jozef Gecz, Michael Hayman, James Holberton, Rod W. Hunt, Sue Jacobs, Andrew J. Kornberg, Richard J. Leventer, Mark T. Mackay, Simone Mandelstam, Jacinta M McMahon, Heather C. Mefford, Candace T. Myers, Julie Panetta, Jessica Riseley, Victoria Rodriguez-Casero, Monique M. Ryan, Amy Schneider, Lindsay Smith, Flora Wong and Eppie M. Yiu.
KBH, ASH, IES, SFB, and KD conceived of and designed the study. KBH, ASH, IES, JA, GLC, EC, MC, GD, SE, MF, JLF, JG, MH, JH, RWH, SJ, AJK, RJL, MTM, SM, JMM, HCM, CTM, JP, JR, VRC, MMR, AS, LS, FW and EMY acquired data. KBH, ASH, IES, KD, SE, GLC, SM, HM, CM and JR analysed and interpreted the data. All authors critically reviewed the manuscript, have given approval for publication and agree to be accountable for all aspects of the work.
Ethical publication statement
We confirm that we have read the Journal’s position on issues involved in ethical publication and affirm that this report is consistent with those guidelines.
Declaration of interests
Dr Howell was supported by the Gustav Nossal National Health and Medical Research Council (NHMRC) Postgraduate Scholarship and the Clifford PhD Scholarship. Dr Mefford is support by National Institutes of Health (NINDS R01 NS069605). Prof Scheffer is supported by a NHMRC Program Grant and Practitioner Fellowship. None of the authors has any conflicts of interest to disclose.
References
- 1.Organisation WH. Neurological disorders: Public Health Challenges. 2006. [Google Scholar]
- 2.Berg AT, Berkovic SF, Brodie MJ, et al. Revised terminology and concepts for organization of seizures and epilepsies: report of the ILAE Commission on Classification and Terminology, 2005–2009. Epilepsia. 2010;51:676–85. doi: 10.1111/j.1528-1167.2010.02522.x. [DOI] [PubMed] [Google Scholar]
- 3.McTague A, Howell KB, Cross JH, Kurian MA, Scheffer IE. The genetic landscape of the epileptic encephalopathies of infancy and childhood. Lancet Neurol. 2016;15:304–16. doi: 10.1016/S1474-4422(15)00250-1. [DOI] [PubMed] [Google Scholar]
- 4.Bureau M, Thomas P, Genton P. Epileptic syndromes in infancy, childhood and adolescence. p. 682. Fifth edition with video sequences. ed:1 online resource. [Google Scholar]
- 5.Wilmshurst JM, Gaillard WD, Vinayan KP, et al. Summary of recommendations for the management of infantile seizures: Task Force Report for the ILAE Commission of Pediatrics. Epilepsia. 2015;56:1185–97. doi: 10.1111/epi.13057. [DOI] [PubMed] [Google Scholar]
- 6.Van Hove JL, Lohr NJ. Metabolic and monogenic causes of seizures in neonates and young infants. Mol Genet Metab. 2011;104:214–30. doi: 10.1016/j.ymgme.2011.04.020. [DOI] [PubMed] [Google Scholar]
- 7.Consortium EK; Allen AS, et al. Epilepsy Phenome/Genome P. De novo mutations in epileptic encephalopathies. Nature. 2013;501:217–21. doi: 10.1038/nature12439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dyment DA, Tetreault M, Beaulieu CL, et al. Whole-exome sequencing broadens the phenotypic spectrum of rare pediatric epilepsy: a retrospective study. Clin Genet. 2015;88:34–40. doi: 10.1111/cge.12464. [DOI] [PubMed] [Google Scholar]
- 9.Berg AT, Loddenkemper T, Baca CB. Diagnostic delays in children with early onset epilepsy: impact, reasons, and opportunities to improve care. Epilepsia. 2014;55:123–32. doi: 10.1111/epi.12479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Loddenkemper T, Holland KD, Stanford LD, Kotagal P, Bingaman W, Wyllie E. Developmental outcome after epilepsy surgery in infancy. Pediatrics. 2007;119:930–5. doi: 10.1542/peds.2006-2530. [DOI] [PubMed] [Google Scholar]
- 11.Feigin VL, Lawes CM, Bennett DA, Anderson CS. Stroke epidemiology: a review of population-based studies of incidence, prevalence, and case-fatality in the late 20th century. Lancet Neurol. 2003;2:43–53. doi: 10.1016/s1474-4422(03)00266-7. [DOI] [PubMed] [Google Scholar]
- 12.O’Roak BJ, Vives L, Fu W, et al. Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science. 2012;338:1619–22. doi: 10.1126/science.1227764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sadedin SP, Dashnow H, James PA, et al. Cpipe: a shared variant detection pipeline designed for diagnostic settings. Genome Med. 2015;7:68. doi: 10.1186/s13073-015-0191-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Drummond MFSM, Torrance GW, O’Brien BJ, Stoddart GL. Methods for the Economic Evaluation of Health Care Programmes Third Edition. United States: Oxford University Press; 2005. [Google Scholar]
- 15.Howell KB, McMahon JM, Carvill GL, et al. SCN2A encephalopathy: A major cause of epilepsy of infancy with migrating focal seizures. Neurology. 2015;85:958–66. doi: 10.1212/WNL.0000000000001926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Evans DG, Howard E, Giblin C, et al. Birth incidence and prevalence of tumor-prone syndromes: estimates from a UK family genetic register service. Am J Med Genet A. 2010;152A:327–32. doi: 10.1002/ajmg.a.33139. [DOI] [PubMed] [Google Scholar]
- 17.Mendell JR, Shilling C, Leslie ND, et al. Evidence-based path to newborn screening for Duchenne muscular dystrophy. Ann Neurol. 2012;71:304–13. doi: 10.1002/ana.23528. [DOI] [PubMed] [Google Scholar]
- 18.Cowan LD, Hudson LS. The epidemiology and natural history of infantile spasms. J Child Neurol. 1991;6:355–64. doi: 10.1177/088307389100600412. [DOI] [PubMed] [Google Scholar]
- 19.Berry JG, Poduri A, Bonkowsky JL, et al. Trends in resource utilization by children with neurological impairment in the United States inpatient health care system: a repeat cross-sectional study. PLoS Med. 2012;9:e1001158. doi: 10.1371/journal.pmed.1001158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Riechmann J, Strzelczyk A, Reese JP, et al. Costs of epilepsy and cost-driving factors in children, adolescents, and their caregivers in Germany. Epilepsia. 2015;56:1388–97. doi: 10.1111/epi.13089. [DOI] [PubMed] [Google Scholar]
- 21.Gulati S, Jain P, Kannan L, Sehgal R, Chakrabarty B. The Clinical Characteristics and Treatment Response in Children with West Syndrome in a Developing Country: A Retrospective Case Record Analysis. J Child Neurol. 2015;30:1440–7. doi: 10.1177/0883073815569304. [DOI] [PubMed] [Google Scholar]
- 22.Wong-Kisiel LC, Britton JW, Witte RJ, et al. Double Inversion Recovery Magnetic Resonance Imaging in Identifying Focal Cortical Dysplasia. Pediatr Neurol. 2016;61:87–93. doi: 10.1016/j.pediatrneurol.2016.04.013. [DOI] [PubMed] [Google Scholar]
- 23.Lemke JR, Riesch E, Scheurenbrand T, et al. Targeted next generation sequencing as a diagnostic tool in epileptic disorders. Epilepsia. 2012;53:1387–98. doi: 10.1111/j.1528-1167.2012.03516.x. [DOI] [PubMed] [Google Scholar]
- 24.Allen NM, Conroy J, Shahwan A, et al. Unexplained early onset epileptic encephalopathy: Exome screening and phenotype expansion. Epilepsia. 2016;57:e12–7. doi: 10.1111/epi.13250. [DOI] [PubMed] [Google Scholar]
- 25.Might M, Wilsey M. The shifting model in clinical diagnostics: how next-generation sequencing and families are altering the way rare diseases are discovered, studied, and treated. Genet Med. 2014;16:736–7. doi: 10.1038/gim.2014.23. [DOI] [PubMed] [Google Scholar]
- 26.Soden SE, Saunders CJ, Willig LK, et al. Effectiveness of exome and genome sequencing guided by acuity of illness for diagnosis of neurodevelopmental disorders. Sci Transl Med. 2014;6:265ra168. doi: 10.1126/scitranslmed.3010076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Monroe GR, Frederix GW, Savelberg SM, et al. Effectiveness of whole-exome sequencing and costs of the traditional diagnostic trajectory in children with intellectual disability. Genet Med. 2016;18:949–56. doi: 10.1038/gim.2015.200. [DOI] [PubMed] [Google Scholar]
- 28.Stark Z, Schofield D, Alam K, et al. Prospective comparison of the cost-effectiveness of clinical whole-exome sequencing with that of usual care overwhelmingly supports early use and reimbursement. Genet Med. 2017 doi: 10.1038/gim.2016.221. [DOI] [PubMed] [Google Scholar]
- 29.Vanderver A, Simons C, Helman G, et al. Whole exome sequencing in patients with white matter abnormalities. Ann Neurol. 2016;79:1031–7. doi: 10.1002/ana.24650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Walsh M, Bell KM, Chong B, et al. Diagnostic and cost utility of whole exome sequencing in peripheral neuropathy. Ann Clin Transl Neurol. 2017;4:318–25. doi: 10.1002/acn3.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wirrell EC, Shellhaas RA, Joshi C, et al. How should children with West syndrome be efficiently and accurately investigated? Results from the National Infantile Spasms Consortium. Epilepsia. 2015;56:617–25. doi: 10.1111/epi.12951. [DOI] [PubMed] [Google Scholar]
- 32.Allen NM, Conroy J, Shahwan A, et al. Chromosomal microarray in unexplained severe early onset epilepsy - A single centre cohort. Eur J Paediatr Neurol. 2015;19:390–4. doi: 10.1016/j.ejpn.2015.03.010. [DOI] [PubMed] [Google Scholar]
- 33.Mercimek-Mahmutoglu S, Patel J, Cordeiro D, et al. Diagnostic yield of genetic testing in epileptic encephalopathy in childhood. Epilepsia. 2015;56:707–16. doi: 10.1111/epi.12954. [DOI] [PubMed] [Google Scholar]
- 34.Schwarze K, Buchanan J, Taylor JC, Wordsworth S. Are whole-exome and whole-genome sequencing approaches cost-effective? A systematic review of the literature. Genet Med. 2018 doi: 10.1038/gim.2017.247. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.