Abstract
Increasing global temperatures due to climate change have resulted in respective increases in the severity and frequency of epizootics around the globe. Corals in particular have faced rapid declines due to disease outbreaks. Understanding immune responses and associated potential life-history trade-offs is therefore a priority. In the autumn of 2011, a novel disease of octocorals of the genus Eunicea was first documented in the Florida Keys. Termed Eunicea Black Disease (EBD), the disease is easily identified by the dark appearance of affected tissue, caused by a strong melanization response on the part of the host. In order to better understand the response of corals to EBD, we conducted full transcriptome analysis of 3 healthy and 3 diseased specimens of Eunicea calyculata collected from offshore southeast Florida. Differential expression and protein analyses revealed a strong, diverse immune response to EBD characterized by phagocytosis, adhesion and melanization on the part of the host. Furthermore, coexpression network analyses suggested this might come at the cost of reduced cell cycle progression and growth. This is in accordance with past histological studies of naturally infected hard corals, suggesting that potential trade-offs during infection may affect post-outbreak recovery of reef ecosystems by reducing both organismal growth and fecundity. Our findings highlight the importance of considering factors beyond mortality when estimating effects of disease outbreaks on ecosystems.
Keywords: cnidarians, invertebrates, transcriptomics, ecoimmunology, invertebrate immunity, coral ecology
1. Introduction
Increasing global temperatures as well as other anthropogenic stressors have resulted in dramatic increases in the prevalence, frequency and severity of disease outbreaks [1,2]. Increases in disease have necessitated subsequent increases in our understanding of organismal immune systems and the physiological consequences of mounting an immune response [3–6]. These efforts have given rise to the field of ecological immunity, which is primarily concerned with understanding how immune responses vary between individuals and species, the dynamics of life-history–immune response trade-offs, and the consequences of immune response [7–11]. In particular, numerous studies focused on organismal level, reproductive consequences of immune response have been published in recent years [12–15]. Furthermore, many theories regarding the costs of immunity for individuals have also been developed [8,9,16–18]. Still many gaps exist in our knowledge of immunity and the consequences of immune trade-offs on community structures as a whole. Knowledge of the consequences of immune response on individuals and subsequently community structure is key to developing a robust understanding of how disease outbreaks shape community structure.
Marine ecosystems have arguably been the hardest hit by recent increases in epizootic outbreaks [1]. These rises in disease outbreaks in ocean ecosystems have resulted in significant die offs of urchins [19], corals [20] and shellfish [21]. Furthermore, many of these vulnerable invertebrate species are also essential to ecosystem function. Corals in particular form the structural and trophic basis of diverse coral reef ecosystems [22] and provide many important ecosystem services, such as serving as coastal protection and supporting fishing industries [23]. However, despite these grim predictions of increasing disease threat, and the importance of affected species, many gaps still exist in our knowledge of marine diseases. These include lack of information regarding the origins and spread of marine diseases, infectious stages and host ranges, and immune responses of host organisms [24].
Invertebrates possess a robust innate immune system consisting of three main processes: pathogen recognition, signalling pathways and effector responses [6]. Yet, while extensive research has revealed a framework for invertebrate immunity [17,25], many questions of host response, and potential trade-offs still remain. While it is clear that immunity comes at a cost, organisms cannot maintain optimal fecundity or longevity while also maintaining a robust immune system [16], few studies have investigated these trade-offs in vulnerable marine invertebrates. In part this is because the species most affected by disease outbreaks are non-model organisms such as corals [20] and echinoderms [26], for which there are limited existing resources [6,27]. Therefore, it is essential to increase efforts to understand immune function, and trade-offs between immunity and life history in these groups so as to better predict future ecosystem structures under rapid global change.
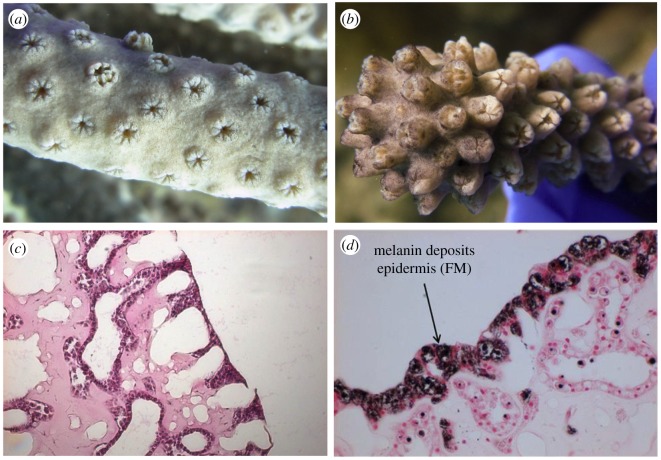
In the autumn of 2011, a novel disease (Eunicea Black Disease; EBD) causing a heavily melanized appearance was observed affecting gorgonian corals of the genus Eunicea off the coast of Florida. To date EBD has been documented along the Florida Reef Tract from the Dry Tortugas National Park to offshore southeast Florida with prevalence ranging from 12 to 86% within the Eunicea community [28]. While the aetiological agent of this disease remains unknown, the disease is easily identifiable due to the darkening of affected tissue resulting from a strong melanization response (figure 1). Additionally, since its first observation, the disease has been reported in multiple other locations throughout the Caribbean (L.E.F. and L.D.M. 2017, personal observation). In order to document the response of these corals to this disease, and any potential trade-offs associated with the disease or immune-stimulated state, we conducted a transcriptional study of naturally affected Eunicea calyculata colonies. Here we document both the strong immune response of individuals to the disease that articulate the late stage or effector responses, and clear trade-offs with growth associated with the disease.
Figure 1.
Macro and histological views of healthy (a,c) and diseased (b,d) Eunicea. (a) Close up of a healthy colony of Eunicea lacking any characteristic black pigmentation associated with the disease. (b) Close up of a colony of Eunicea infected with EBD; note the characteristic black pigmentation of the tissue. (c) Standard haematoxylin and eosin stained histology slides of apparently healthy sample showing good tissue organization and normal staining characteristics (pink staining) and (d) slides of tissue from an infected colony stained with Fontana-Masson showing the epidermis with melanin deposition.
2. Methods
2.1. Sample collection
During October 2013, fragments of Eunicea calyculata were collected from 7 healthy and 7 diseased colonies from Southeast Coral Reef Evaluation and Monitoring Project site DC3, an octocoral reef, near Miami, FL (25° 50.526′ −80° 05.286′). Samples were collected while working with the Florida Fish and Wildlife Conservation Commission in compliance with Chapter 68B-8.016 of the Marine Special Activity License programme. Diseased corals were identified based on the presence of black tissue lesions and 3–5 cm fragments were cut from the affected areas. Fragments of the same size and approximate location were cut from normally pigmented colonies that did not show any signs of disease. Samples were immediately frozen in liquid nitrogen and shipped back to the University of Texas at Arlington on dry ice and stored at −80°C until processing for protein and transcriptomic analysis.
2.2. Protein analyses
Prior to protein processing, two small pieces (approximately 0.5 cm2) were removed from each sample and preserved for later RNA extraction. Proteins were extracted from all 14 samples by grinding the entire remaining sample in liquid nitrogen with a mortar and pestle. A small portion of this frozen powder was placed in a pre-weighed 1.5 ml centrifuge tube and reserved for melanin concentration analysis. Proteins were then extracted from the remaining powder on ice for approximately 45 min using approximately 2 ml of 100 mM sodium phosphate-buffered saline, pH 7.8. The remaining volume was centrifuged for 10 min at 4°C and 3500 r.p.m. in an Eppendorf centrifuge 5810R. The resulting coral extract was divided between 2 ml aliquots, flash frozen in liquid nitrogen, and stored at −80°C [29].
Prophenoloxidase (PPO) activity was estimated by diluting 20 µl of extract in 50 mM phosphate buffer, pH 7.0. Next, samples were incubated for 30 min in 25 µl of trypsin (0.1 mg ml−1). Just prior to the assay, 30 µl of 10 mM L-1,3-dihydroxphenylalanine (L-dopa, Sigma-Aldrich) was added to each sample. Absorbance was then read every 20 min at 490 nm. PPO activity was calculated as the change in absorbance per minute at the steepest part of the curve. A Red660 protein assay (G Biosciences, St Louis, MO) was used to determine total protein concentration in each sample. All colorimetric assays were run in duplicate on 96 well plates using a Synergy 2 multi-detection microplate reader and Gen5 software (Biotek Instruments, Winooski, VT, USA). The results of the PPO activity assay were then standardized by protein concentration as determined using a standard Red660 colorimetric assay [29]. Presented PPO activity results are representative of combined activity of proenzyme (PPO) activated with trypsin and existing active phenoloxidase within a sample.
In addition to PPO activity, total melanin concentration per sample was estimated using the portion of extract reserved for melanin analysis. The aliquot was lyophilized in a tarred 2 ml microcentrifuge tube overnight. At the end of this period, the total dry tissue weight in the aliquot was determined and a full spatula of glass beads (approx. 200 µl in volume) was added to each. Samples were vortexed for 10 s, 400 µl of 10 M NaOH was added to each tube, and the samples were vortexed for another 20 s. Tubes were then incubated for 48 h at room temperature in the dark and vortexed every 24 h. Following incubation, tubes were centrifuged at 1000 r.p.m. at room temperature for 10 min. 40 µl of supernatant was transferred to a ½ well UV plate (Costar, Corning Life Sciences, Lowell, MA, USA) and absorbance was recorded at 410 and 490 nm. Concentrations of melanin were estimated based on a standard curve of melanin dissolved in 10 M NaOH and are presented as mg of melanin per mg of tissue [29].
Assay results were analysed in SPSS software using standard ANOVAs with disease status as a factor. Protein data were tested for normality and outliers prior to statistical analysis. Data from both the PPO and melanin assays were normally distributed and required no transformation. Significance was determined as p < 0.05.
2.3. RNA extraction and sequencing
Total RNA was extracted from approximately 0.5 cm2 pieces of a subset of 3 fragments per group (healthy and diseased). Coral pieces were homogenized in Trizol (ThermoFisher) until all of the polyp tissue was removed from the gorgonin skeleton. Chloroform was added to the solution, incubated for 2–3 min and centrifuged at 12 000 × g at 4°C for 15 min. The aqueous phase was transferred to an RNeasy spin column and the extraction was completed per the RNeasy protocol (Qiagen). RNA quality was assessed using a 2100 BioAnalyzer (Agilent) and samples with RIN values greater than 8.0 were submitted for sequencing. Extracted RNA was submitted to the University of Texas Genome Sequencing Facility that created total RNA libraries using the NEBNext® Ultra™ Directional RNA Library Prep Kit for Illumina® (New England Biolabs, #E7420S) with poly-A tail selection. Libraries were constructed according to the manufacturer's protocol, with modifications to add custom barcodes during the ligation step. Samples were then sequenced at this facility on a single lane using an Illumina HiSeq 2500 platform with 100 base pair paired end reads.
2.4. Transcriptome assembly and differential expression analyses
Using the Trimmomatic software package [30] sequences were trimmed to remove adaptors and low quality reads. All sequences of 36 bp in length or less were discarded. Forward and reverse reads for all samples were concatenated together to create a reference forward and reverse read. Reference reads were fed into Trinity [31] for transcriptome assembly using default parameters. Non-host sequences were filtered out using methods described in [32]. Briefly, symbiont reads were removed by BLAT [33] comparison to a data file consisting of combined genomic and transcriptomic data from multiple species of Symbiodinium. The remaining transcriptome was filtered to obtain host only reads by BLAT comparison against a second meta-data file consisting of multiple cnidarian genomes (O. favolata, Acropora digitifera [34], A. millepora (http://genome.wustl.edu/genomes/detail/acropora-millepora/) and Nematostella vectensis [35]). Transcriptome completeness of the resulting host-only transcriptome was assessed via comparison to the publically available Gorgonia ventalina transcriptome [36]. A tblastx comparison of the two transcriptomes was conducted with a minimum e-value cut-off of 1 × 10−5.
Differentially expressed contigs were identified and normalized read counts generated using the Cufflinks software package with default parameters [37]. Log2fold change values for each contig were generated by comparing average normalized expression values between disease status groups. Significantly differentially expressed contigs were identified as those with an adjusted p < 0.05 between treatments.
2.5. Transcriptome annotation and gene ontology analyses
The full E. calyculata transcriptome generated from collected samples was annotated against the UniProtKB/Swiss-Prot and TrEMBL databases (blastx algorithm, 1.0 × 10−5 e-value threshold). Gene ontology (GO) was first assessed using the online PANTHER database [38] to determine relative abundance of GO terms. Additionally, to identify enriched functional groups that were characteristic of differentially expressed contigs, we conducted GO enrichment analysis using a Fisher's exact test with the R script GO MWU [39].
2.6. Coexpression network analysis
In order to identify groups of coexpressed contigs with potential correlation to disease status, the R package Weighted Gene Correlation Network Analyses [40] was used to analyse the full transcriptome. Normalized read counts for all contigs were log2 transformed. Using these values for all six samples, a single, signed network was built with manual network construction methods (parameters: soft power = 20, minimum module size = 180, deep split = 2, merged cut height = 0.25, minimum verbose = 3, cutHeight = 0.991). Module eigengene values (average expression of all contigs in a module) of each module were correlated to disease status, sample identity and protein analysis data using a bicor correlation. Modules with significant correlations (p-value ≤ 0.05) were selected for further analyses. GO enrichment analysis for each significant module was conducted using a Fisher's exact test with the R script GO MWU [39].
3. Results
3.1. Protein analyses
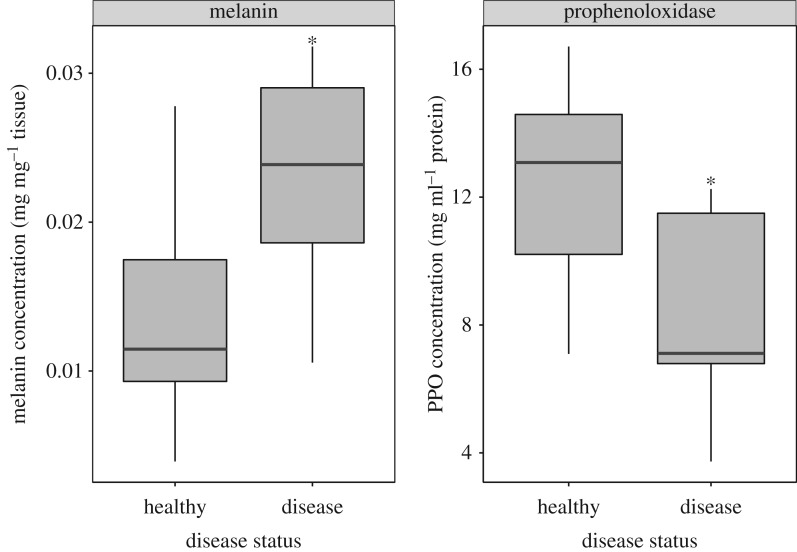
PPO activity was significantly reduced in diseased samples compared to those from healthy colonies (ANOVA, p = 0.040). In contrast, melanin concentration was significantly higher in diseased samples (ANOVA, p = 0.031, figure 2).
Figure 2.
Results of biochemical assays measuring melanin concentration per mg tissue and prophenoloxidase protein activity (standardized by protein concentration). Infected samples had significantly higher melanin concentration (p = 0.031) and significantly lower prophenoloxidase activity (p = 0.040).
3.2. Transcriptome assembly
A total of 37 936 228 paired end reads were obtained from sequencing of all six samples (three healthy and three diseased). Full reads are available for download from NCBI (SRA PRJNA407366). Reads were assembled into a de novo transcriptome consisting of 95 665 contigs with an N50 value of 1364. Comparison of the full transcriptome to the UniProtKB/Swiss-Prot database resulted in annotation of 29 409 (approx. 31%) contigs.
Assessment of transcriptome completeness by comparison to the available G. ventalina transcriptome [36] suggested a robust transcriptome assembly. Blast results yielded 50 498 (52.8%) significant matches between the two transcriptomes. Furthermore, the two transcriptomes are relatively comparable in size, with the G. ventalina transcriptome consisting of 90 230 contigs, while our E. calyculata transcriptome was composed of 95 665 contigs. Finally both transcriptome assemblies had a similar N50 value, 1149 and 1364, for G. ventalina and E. calyculata respectively.
3.3. Differentially expressed contigs
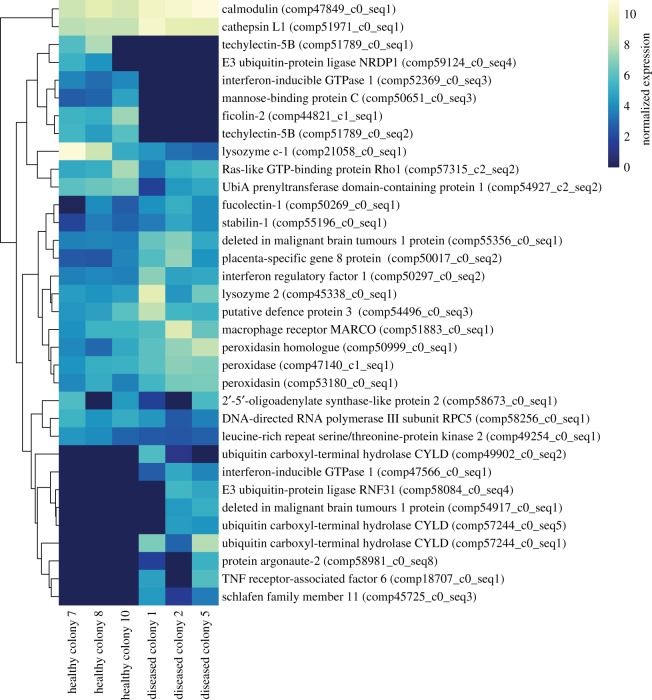
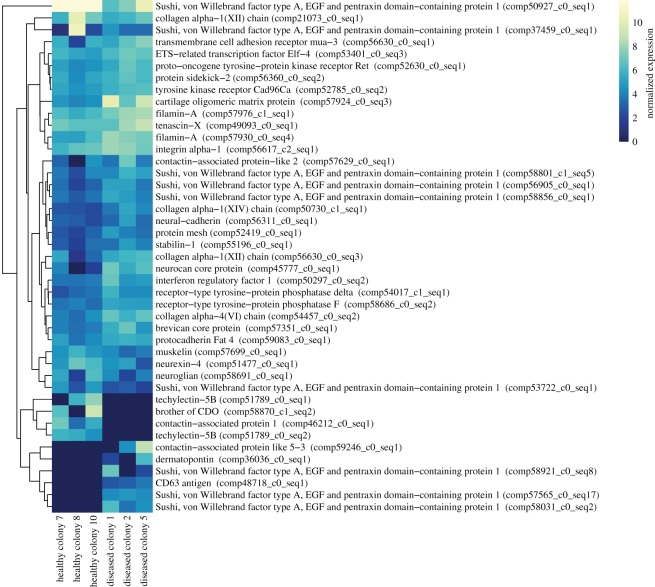
Principal component analysis of patterns of expression among all six samples showed a clear division between healthy and diseased samples along the secondary principal component, and little division among the first principal component between samples. Disease status rather than colony identity appeared to be the primary factor contributing to sample variance in gene expression (electronic supplementary material, figure S1). Differential expression analyses comparing diseased to healthy samples found 1662 differentially expressed contigs: 1124 (approx. 68%) upregulated and 538 (approx. 32%) downregulated. 554 (approx. 33%) of the differentially expressed contigs were annotated (electronic supplementary material, file S1). A total of 37 contigs putatively involved in innate immune processes could be identified from this list (figure 3; electronic supplementary material, file S1). This included contigs involved in multiple different processes such as Toll-like receptor (TLR) signalling, complement cascade, antiviral responses, and the encapsulation and destruction of pathogens.
Figure 3.
Heatmap of expression of all significantly differentially expressed immune contigs. Log2 transformed expression values are displayed for each sample. Colonies are ordered by treatment and contigs are clustered based on similarity in expression patterns. Contigs that were only expressed in one sample not shown.
3.4. Gene ontology enrichment analyses
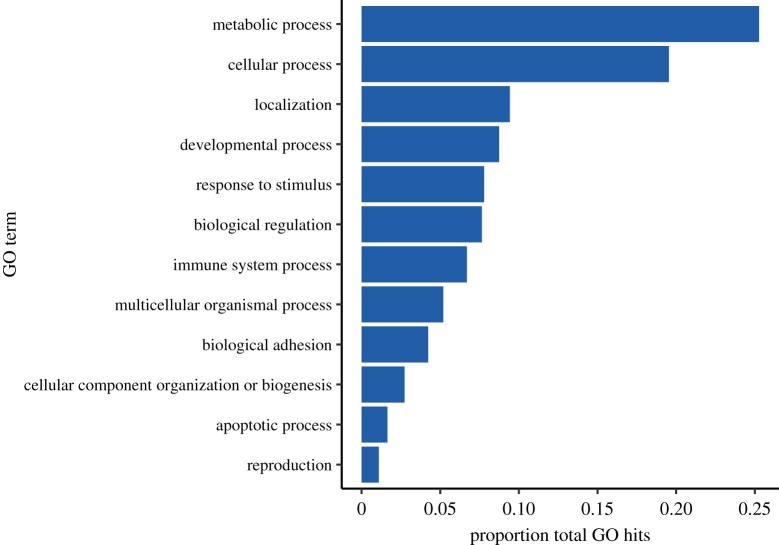
Top tier GO breakdown of the 554 differentially expressed contigs revealed a large portion of contigs involved in response to stimulus, immune response and biological adhesion processes (figure 4). GO enrichment analyses of differentially expressed contigs found one significantly enriched term: biological adhesion. A total of 50 differentially expressed contigs were included in the biological adhesion GO term (figure 5). This included contigs with potential immune function such as those related to interferon responses, techylectins and integrins.
Figure 4.
Top tier gene ontology breakdown of all annotated differentially expressed contigs. Shown as ratio of hits to total hits within a given category.
Figure 5.
Heatmap of differentially expressed contigs that are involved in biological adhesion. Log2 transformed expression values are displayed for each sample. Colonies are ordered by treatment and contigs are clustered based on similarity in expression patterns. Contigs that were only expressed in one sample not shown.
3.5. Coexpression analyses
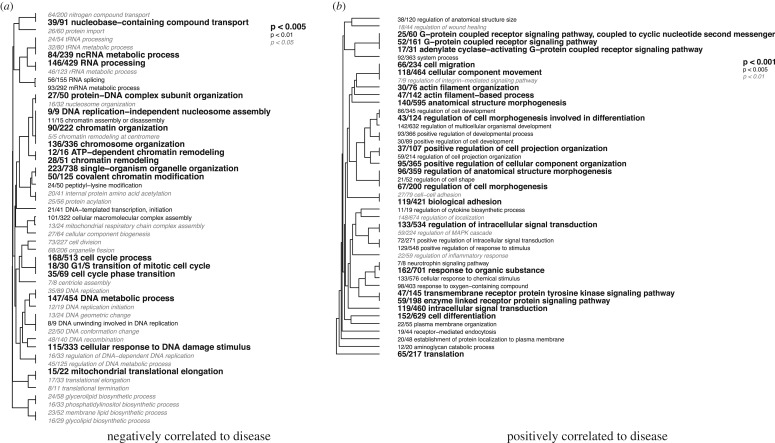
Coexpression analyses of all expressed contigs using WGCNA resulted in a network composed of 29 modules ranging in size from 190 to 3252 contigs each. Of these 29 modules, 2 were significantly correlated to disease status, modules 4 (r = −0.93, p = 0.008) and 19 (r = 0.98, p < 0.001) (electronic supplementary material, figure S2). These two modules were significantly enriched for multiple biological process GO terms, 50 terms for module 4 and 114 terms for module 19 (p < 0.05; figure 6, electronic supplementary material, file S3). Module 4, which was negatively correlated to disease status, was significantly enriched with terms involved in RNA and DNA processing, as well as cell cycle progression (electronic supplementary material, figure S3). Module 19, which was positively correlated to disease status, was significantly enriched with immune terms such as inflammatory response, regulation of MAPK cascade, receptor mediated endocytosis and regulation of wound healing. Additionally this module was significantly enriched for terms involved in cell signalling such as G-protein coupled receptor signalling and regulation of intracellular signalling, as well as terms involved in cell adhesion such as cell–cell adhesion and biological adhesion (electronic supplementary material, figure S4).
Figure 6.
Hierarchical clustering of significantly enriched biological process gene ontology enrichment of the two modules correlated to disease status: (a) module 4 (negatively correlated) and (b) module 19 (positively correlated).
An additional four modules were correlated with either melanin concentration or PPO activity (electronic supplementary material, figure S2). Module 9 was significantly negatively correlated to melanin activity (p = 0.03, r = −0.83). This module was not significantly enriched for any biological process GO terms. Three modules were significantly correlated to PPO activity: module 12 (p = 0.03, r = 0.86), module 14 (p = 0.02, r = 0.89) and module 21 (p = 0.04, r = −0.84). Module 14 was also significantly correlated to healthy colony 8. Again, none of these modules were significantly enriched for any biological process GO terms.
4. Discussion
Coastal ecosystems such as coral reefs have experienced rapid declines due to increasing global temperatures and other anthropogenic stressors [1,2]. This outbreak of yet another novel coral disease is further evidence of rapidly declining ocean conditions and coral health. This disease in particular is clearly characterized by darkly pigmented tissue resulting from a strong immune response dominated by melanin deposition (figure 1). The disease phenotype is in fact the result of a melanization response as evidenced by the results of our assays (figure 2). Furthermore, patterns of gene expression among samples were predominately driven by disease status rather than colony identity (electronic supplementary material, figure S1). Differential expression and GO analyses reveal significant changes in numerous immune and adhesion contigs underlying these responses. Finally, coexpression analyses suggest that this response may come at the cost of reduced cell division and growth. Here we further examine the immune response and associated fitness trade-offs of the host E. calyculata using transcriptional analyses.
4.1. Infected E. calyculata mount a strong immune response characterized by melanization to an unknown pathogen
Black pigmented diseased tissue E. calyculata demonstrated a clear immune response as evidenced by both protein and gene expression data. Biochemical analysis of protein samples revealed that infected tissue had significantly lower PPO activity than apparently healthy tissue. Consequently, infected tissue also had higher melanin concentration. This is as would be expected for a late-stage immune response. Melanization is the end product of the PPO cascade in invertebrates [41,42] and has been previously documented as an important component of the cnidarian immune response [43,44]. Therefore, the observed response is likely indicative of previous rapid melanization and subsequent reduction in PPO availability or activity. These findings here add to existing ecological observations and histology results that suggest that melanization is a key component of the response of E. calyculata to EBD.
Supporting our protein results, we found contigs representative of various components of innate immunity to be differentially expressed, including components of the TLR signalling pathway, complement cascade, antiviral response, phagocytosis and antioxidant activity. TLR signalling is an important mechanism by which hosts detect pathogens and activate immune responses [45]. Here we document 8 differentially expressed contigs involved in or downstream of this pathway, including two copies of the signalling protein deleted in malignant brain tumour protein 1 (DMBT1) which is regulated by TLR signalling and has roles in pathogen recognition [46]. This protein has previously been shown to be an important part of the coral immune response in multiple studies and has been linked to host production of antimicrobial compounds [47,48]. Other TLR-related contigs we observed to be differentially expressed included tumour necrosis factor receptor-associated factor 6 [49,50], and contigs involved in the negative regulation of TLR signalling, E3 ubiquitin-protein ligase RNF31 [51] and 4 copies of ubiquitin carboxyl-terminal hydrolase CYLD [52]. Additionally, we observed upregulation of the protein argonaute 2, an antiviral protein that has previously been described as part of the response of acroporid corals to white diseases [53].
In addition to differential expression of conventional immune contigs, the biological process GO term biological adhesion was significantly enriched in our dataset. The enrichment of biological adhesion as a response to a pathogen is not surprising, as cellular adhesion is a basic component of multiple innate and adaptive immune responses [54–57]. Specifically, cellular adhesion is an important characteristic of the PPO activating system that results in the formation of melanin [25,56] that can be used to encapsulate pathogens for later phagocytosis [58]. Biological adhesion is also often associated with phagocytic response [25,55,56]. Interestingly, the majority of immune-related differentially expressed contigs were in fact related to phagocytosis and antioxidant responses. This included several contigs involved in recognition and agglutination of pathogens for later phagocytosis such as MARCO, techylectin-5B, stabilin-1, fucolectin-1 and DMBT1 [46,59–62], as well as lysozyme and peroxidase contigs which can be associated with phagocytosis [63]. Upregulation of antioxidants and lysozymes, which are frequently associated with degradation of phagocytized particles, is further suggestion of ongoing phagocytic processes [64–67]. Together these patterns of expression strongly support our hypothesis that diseased samples are undergoing a late-stage immune response characterized by phagocytosis of melanized particles and pathogens. Therefore, despite our limited sample size, our combination of protein data and robust transcriptomic analyses provides a clear and unified picture of late-stage immune responses in cnidarians.
Finally, it should be noted that the transcriptomic responses documented here share a great deal of overlap with a number of other transcriptomic studies of cnidarian immune responses [47,48,53,68,69]. Many of these studies document changes in a core subset of contigs that are shared across cnidarian species in response to both pathogenic challenge and immune stimulation. This suggests a common, conserved role of a robust innate immune response characterized by pathways such as the TLR signalling pathway in the cnidarian immune response.
4.2. Eunicea calyculata undergo a fitness trade-off during infection
GO enrichment analyses of the two modules that were significantly correlated to disease status demonstrated a trade-off associated with immune response. The module positively correlated to disease status was enriched with immune system and signalling processes, while the one negatively correlated to disease status was enriched with processes associated with cell cycle progression, including DNA and RNA processes. These findings were corroborated by examination of patterns of expression of contigs included in these two modules (figure 6 and electronic supplementary material, figures S3 and S4), suggesting that cellular division and growth may be impacted during a pathogenic infection of E. calyculata. Decreases in cell cycle and other like processes have the potential to detrimentally impact growth of the organism as a whole. As colonial organisms, octocorals such as E. calyculata undergo modular, iterative growth via replication of polyps through asexual budding [70,71]. Furthermore, asexual reproduction is dependent on proliferative cells; therefore decreases in cellular growth may have impacts on the growth, size, and ultimately fitness of the whole organism [72]. However the effects of decreased cell cycle progression may not only result in decreased size of the organism, but also potentially organism fecundity. Growth and size are an important determinant of fecundity, and therefore fitness, in colonial organisms such as gorgonian corals. Many gorgonian corals must maintain a certain colony size to reproduce [73,74], and larger colonies have larger reproductive outputs [74]. Subsequently, in theory, reduction in growth during disease would correspond to reduced long-term fecundity and fitness. While this is the first documentation of transcriptomic evidence of the potential consequences of disease on reproductive output in a cnidarian, it is in accordance with past observations reporting reduced fecundity (measured as amount of gravid polyps) in a hard coral infected with disease [75]. Therefore, our findings here provide new molecular context to previously published findings that disease comes at the cost of fecundity in corals.
It should be noted that samples used for transcriptomic analysis of diseased colonies were collected from heavily melanized lesion locations. This leaves the potential that observed signals are the result of either death response or the cellular stress response. However, we observed little differential expression of apoptotic, death or autophagic pathways in our analysis. Similarly we saw little evidence of the cellular stress response, particularly in comparison to other similar studies that document this as part of the cnidarian disease response [69]. Additionally these lesions often affect the majority of diseased colonies. Therefore, we are confident that the trends discussed here are relevant and have important potential ecological consequences.
Our findings documented here point to an ecologically relevant potential trade-off. Increasing sea surface temperatures as a result of climate change have resulted in increases in the prevalence of coral disease [76]. During active infection, our findings suggest that E. calyculata may reduce cell cycle progression, growth and fecundity. Therefore, as disease outbreaks become more common and individuals are more frequently infected, E. calyculata growth, and fecundity, may be impacted. This could have implications beyond individual corals as self-seeding is one of the most important sources of recruitment on a recovering reef [73,77]. Regardless, as Eunicea corals are relatively slow growing under normal conditions [78,79], recovery times will be slow, and even slower still should our findings here regarding reduced expression of growth related processes following disease be corroborated by further investigation. Therefore, mortality of Eunicea and reduced growth of remaining colonies may lead to displacement of the species by faster growing species such as those from the genus Antillogorgia [80].
5. Conclusion
The results presented here are some of the first transcriptomic evidence to corroborate past field and histological observations of the growth and fecundity consequences of disease in cnidarians. Furthermore, our documentation of the response of E. calyculata to disease joins a growing body of literature reporting shared transcriptomic responses of cnidarians to pathogenic infection and immune stimulation. The severity of this disease, robustness of the associated immune response, and potential related fitness trade-offs cast a grim outlook for the future of Eunicea octocorals on Caribbean reefs. As climate change effects increase and with it the frequency of disease outbreaks affecting numerous species [81], resilience of these communities may decrease, resulting in decreases or shifts in octocoral communities on reefs [82]. Therefore, it is essential to continue to explore our findings regarding the consequences of disease on growth and fecundity of individual corals. Increasing understanding of these potential trade-offs will improve our ability to understand species and community level effects of disease beyond mortality. Our findings open the door to future research that will increase our ability to predict coral community trajectories under increasing pressure from disease and climate change.
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
The authors would like to thank the Florida Department of Environmental Protection for boat support in the field. Additionally we would like to thank Esther Peters, Jan Landsberg, Yasu Kiryu, Noretta Perry and Yvonne Waters for assistance in obtaining the histopathology pictures shown in figure 1. Finally, we would like to thank the staff of the Genetics Core Facility (UTA) for assistance with laboratory work and anonymous reviewers for edits to improve the manuscript. The analyses were run on a server provided by the UTA Office of Information Technology.
Ethics
Samples were collected while working with the Florida Fish and Wildlife Conservation Commission in compliance with Chapter 68B-8.016 of the Marine Special Activity License programme.
Data accessibility
Raw RNAseq reads are available on NCBI GenBank (SRA PRJNA407366).
Authors' contributions
L.D.M., W.T.M. and V.B. planned collections and collected samples from reefs in the Florida Keys. W.T.M. and L.R.J. processed samples for RNA extraction and sequencing. W.T.M. assembled the transcriptome. L.E.F. and W.T.M. conducted differential expression analyses. L.E.F. conducted gene ontology and WGCNA analyses and constructed all figures. L.E.F. and L.D.M. wrote the manuscript with editorial assistance from W.T.M. and V.B.
Competing interests
The authors declare no competing interests.
Funding
Funding was provided by NSF grant no. IOS #1017458 to L.D.M. This material is based upon work supported by the National Science Foundation Graduate Research Fellowship under grant no. 1144240 to L.E.F.
References
- 1.Burge CA, et al. 2014. Climate change influences on marine infectious diseases: implications for management and society. Ann. Rev.Mar. Sci. 6, 249–277. (doi:10.1146/annurev-marine-010213-135029) [DOI] [PubMed] [Google Scholar]
- 2.Hoegh-Guldberg O, Bruno JF. 2010. The impact of climate change on the world's marine ecosystems. Science 328, 1523–1528. (doi:10.1126/science.1189930) [DOI] [PubMed] [Google Scholar]
- 3.Daszak P, Cunningham AA, Hyatt AD. 2001. Anthropogenic environmental change and the emergence of infectious diseases in wildlife. Acta Trop. 78, 103–116. (doi:10.1016/S0001-706X(00)00179-0) [DOI] [PubMed] [Google Scholar]
- 4.Ghosh J, Man Lun C, Majeske AJ, Sacchi S, Schrankel CS, Smith LC. 2011. Invertebrate immune diversity. Dev. Comp. Immunol. 35, 959–974. (doi:10.1016/j.dci.2010.12.009) [DOI] [PubMed] [Google Scholar]
- 5.Magnadóttir B. 2006. Innate immunity of fish (overview). Fish Shellfish Immunol. 20, 137–151. (doi:10.1016/j.fsi.2004.09.006) [DOI] [PubMed] [Google Scholar]
- 6.Palmer CV, Traylor-Knowles N. 2012. Towards an integrated network of coral immune mechanisms. Proc. R. Soc. B 279, 4106–4114. (doi:10.1098/rspb.2012.1477) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brock PM, Murdock CC, Martin LB. 2014. The history of ecoimmunology and its integration with disease ecology. Integr. Comp. Biol. 54, 353–362. (doi:10.1093/icb/icu046) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.French SS, Moore MC, Demas GE. 2009. Ecological immunology: the organism in context. Integr. Comp. Biol. 49, 246–253. (doi:10.1093/icb/icp032) [DOI] [PubMed] [Google Scholar]
- 9.Graham AL, Shuker DM, Pollitt LC, Auld SKJR, Wilson AJ, Little TJ. 2011. Fitness consequences of immune responses: strengthening the empirical framework for ecoimmunology. Funct. Ecol. 25, 5–17. (doi:10.1111/j.1365-2435.2010.01777.x) [Google Scholar]
- 10.Hawley DM, Altizer SM. 2011. Disease ecology meets ecological immunology: understanding the links between organismal immunity and infection dynamics in natural populations. Funct. Ecol. 25, 48–60. (doi:10.1111/j.1365-2435.2010.01753.x) [Google Scholar]
- 11.Martin LB, Hawley DM, Ardia DR. 2011. An introduction to ecological immunology. Funct. Ecol. 25, 1–4. (doi:10.1111/j.1365-2435.2010.01820.x) [Google Scholar]
- 12.Adamo SA, Jensen M, Younger M. 2001. Changes in lifetime immunocompetence in male and female Gryllus texensis (formerly G. integer): trade-offs between immunity and reproduction. Anim. Behav. 62, 417–425. (doi:10.1006/anbe.2001.1786) [Google Scholar]
- 13.Bascunan-Garcia AP, Lara C, Cordoba-Aguilar A. 2010. Immune investment impairs growth, female reproduction and survival in the house cricket, Acheta domesticus. J. Insect Physiol. 56, 204–211. (doi:10.1016/j.jinsphys.2009.10.005) [DOI] [PubMed] [Google Scholar]
- 14.Carlton ED, Cooper CL, Demas GE. 2014. Metabolic stressors and signals differentially affect energy allocation between reproduction and immune function. Gen. Comp. Endocrinol. 208, 21–29. (doi:10.1016/j.ygcen.2014.08.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gwynn D, Callaghan A, Gorham J, Walters KFA, Fellowes MDE. 2005. Resistance is costly: trade-offs between immunity, fecundity, and survival in the pea aphid. Proc. R. Soc. B 272, 1803–1808. (doi:10.1098/rspb.2005.3089) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee KA. 2006. Linking immune defenses and life history at the levels of the individual and the species. Integr. Comp. Biol. 46, 1000–1015. (doi:10.1093/icb/icl049) [DOI] [PubMed] [Google Scholar]
- 17.Rolff J, Siva-Jothy MT. 2003. Invertebrate ecological immunology. Science 301, 472–475. (doi:10.1126/science.1080623) [DOI] [PubMed] [Google Scholar]
- 18.Sadd BM, Schmid-Hempel P. 2009. Principles of ecological immunology. Evol. Appl. 2, 113–121. (doi:10.1111/j.1752-4571.2008.00057.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hughes TP. 1994. Catastrophes, phase shifts, and large-scale degradation of a Caribbean coral reef. Science 265, 1547–1551. (doi:10.1126/science.265.5178.1547) [DOI] [PubMed] [Google Scholar]
- 20.Burge CA, Douglas NL, Conti-Jerpe I, Weil E, Roberts S, Friedman CS, Harvell CD. 2012. Friend or foe: the association of Labyrinthulomycetes with the Caribbean sea fan Gorgonia ventalina. Dis. Aquat. Organ. 101, 1–12. (doi:10.3354/dao02487) [DOI] [PubMed] [Google Scholar]
- 21.Mann R, Harding JM, Southworth MJ. 2009. Reconstructing pre-colonial oyster demographics in the Chesapeake Bay, USA. Estuar. Coast. Shelf Sci. 85, 217–222. (doi:10.1016/j.ecss.2009.08.004) [Google Scholar]
- 22.Sebens KP. 1994. Biodiversity of coral-reefs—what are we losing and why. Am. Zool. 34, 115–133. (doi:10.1093/icb/34.1.115) [Google Scholar]
- 23.Spurgeon JPG. 1992. The economic valuation of coral reefs. Mar. Pollut. Bull. 24, 529–536. (doi:10.1016/0025-326X(92)90704-A) [Google Scholar]
- 24.Harvell D, et al. 2004. The rising tide of ocean diseases: unsolved problems and research priorities. Front. Ecol. Environ. 2, 375–382. (doi:10.1890/1540-9295(2004)002[0375:TRTOOD]2.0.CO;2) [Google Scholar]
- 25.Iwanaga S, Lee BL. 2005. Recent advances in the innate immunity of invertebrate animals. J. Biochem. Mol. Biol. 38, 128–150. [DOI] [PubMed] [Google Scholar]
- 26.Fuess LE, et al. 2015. Up in arms: immune and nervous system response to sea star wasting disease. PLoS ONE 10, e0133053 (doi:10.1371/journal.pone.0133053) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dheilly NM, Raftos DA, Haynes PA, Smith LC, Nair SV. 2013. Shotgun proteomics of coelomic fluid from the purple sea urchin, Strongylocentrotus purpuratus. Dev. Comp. Immunol. 40, 35–50. (doi:10.1016/j.dci.2013.01.007) [DOI] [PubMed] [Google Scholar]
- 28.Brinkhuis V, Lunz K. 2015. Investigation of a previously undescribed octocoral disease affecting the Florida reef tract. Florida State Wildlife Grant. [Google Scholar]
- 29.Mydlarz LD, Palmer CV. 2011. The presence of multiple phenoloxidases in Caribbean reef-building corals. Comp. Biochem. Physiol. A 159, 372–378. (doi:10.1016/j.cbpa.2011.03.029) [DOI] [PubMed] [Google Scholar]
- 30.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. (doi:10.1093/bioinformatics/btu170) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grabherr MG, et al. 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644 (doi:10.1038/nbt.1883) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pinzon JH, Kamel B, Burge CA, Harvell CD, Medina M, Weil E, Mydlarz LD. 2015. Whole transcriptome analysis reveals changes in expression of immune-related genes during and after bleaching in a reef-building coral. R. Soc. open sci. 2, 140214 (doi:10.1098/rsos.140214) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kent WJ. 2002. BLATL: the BLAST-like alignment tool. Genome Res. 12, 656–664. (doi:10.1101/gr.229202) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shinzato C, et al. 2011. Using the Acropora digitifera genome to understand coral responses to environmental change. Nature 476, 320–382. (doi:10.1038/nature10249) [DOI] [PubMed] [Google Scholar]
- 35.Putnam NH, et al. 2007. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science 317, 86–94. (doi:10.1126/science.1139158) [DOI] [PubMed] [Google Scholar]
- 36.Burge CA, Mouchka ME, Harvell CD, Roberts S. 2013. Immune response of the Caribbean sea fan, Gorgonia ventalina, exposed to an Aplanochytrium parasite as revealed by transcriptome sequencing. Front. Physiol. 4, 180 (doi:10.3389/fphys.2013.00180) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L. 2013. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 31, 46–53. (doi:10.1038/nbt.2450) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thomas PD, et al. 2003. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 13, 2129–2141. (doi:10.1101/gr.772403) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wright RM, Aglyamova GV, Meyer E, Matz MV. 2015. Gene expression associated with white syndromes in a reef building coral, Acropora hyacinthus. BMC Genomics 16, 371 (doi:10.1186/s12864-015-1540-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Langfelder P, Horvath S. 2008. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9, 559 (doi:10.1186/1471-2105-9-559) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cerenius L, Kawabata SI, Lee BL, Nonaka M, Soderhall K. 2010. Proteolytic cascades and their involvement in invertebrate immunity. Trends Biochem. Sci. 35, 575–583. (doi:10.1016/j.tibs.2010.04.006) [DOI] [PubMed] [Google Scholar]
- 42.Cerenius L, Lee BL, Soderhall K. 2008. The proPO-system: pros and cons for its role in invertebrate immunity. Trends Immunol. 29, 263–271. (doi:10.1016/j.it.2008.02.009) [DOI] [PubMed] [Google Scholar]
- 43.Mydlarz LD, Holthouse SF, Peters EC, Harvell CD. 2008. Cellular responses in sea fan corals: granular amoebocytes react to pathogen and climate stressors. PLoS ONE 3, e1811 (doi:10.1371/journal.pone.0001811) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Palmer CV, Mydlarz LD, Willis BL. 2008. Evidence of an inflammatory-like response in non-normally pigmented tissues of two scleractinian corals. Proc. R. Soc. B 275, 2687–2693. (doi:10.1098/rspb.2008.0335) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Akira S, Takeda K. 2004. Toll-like receptor signalling. Nat. Rev. Immunol. 4, 499–511. (doi:10.1038/nri1391) [DOI] [PubMed] [Google Scholar]
- 46.Rosenstiel P, et al. 2007. Regulation of DMBT1 via NOD2 and TLR4 in intestinal epithelial cells modulates bacterial recognition and invasion. J. Immunol. 178, 8203–8211. (doi:10.4049/jimmunol.178.12.8203) [DOI] [PubMed] [Google Scholar]
- 47.Fuess LE, Pinzon CJ, Weil E, Mydlarz LD. 2016. Associations between transcriptional changes and protein phenotypes provide insights into immune regulation in corals. Dev. Comp. Immunol. 62, 17–28. (doi:10.1016/j.dci.2016.04.017) [DOI] [PubMed] [Google Scholar]
- 48.Wright RM, Kenkel CD, Dunn CE, Shilling EN, Bay LK, Matz MV. 2017. Intraspecific differences in molecular stress responses and coral pathobiome contribute to mortality under bacterial challenge in Acropora millepora. Sci. Rep. 7, 2609 (doi:10.1038/s41598-017-02685-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kawai T, et al. 2004. Interferon-alpha induction through Toll-like receptors involves a direct interaction of IRF7 with MyD88 and TRAF6. Nat. Immunol. 5, 1061–1068. (doi:10.1038/ni1118) [DOI] [PubMed] [Google Scholar]
- 50.Medzhitov R, Preston-Hurlburt P, Kopp E, Stadlen A, Chen C, Ghosh S, Janeway CA. 1998. MyD88 is an adaptor protein in the hToll/IL-1 receptor family signaling pathways. Mol. Cell 2, 253–258. (doi:10.1016/S1097-2765(00)80136-7) [DOI] [PubMed] [Google Scholar]
- 51.Bowman J, Rodgers MA, Shi M, Amatya R, Hostager B, Iwai K, Gao S-J, Jung JU. 2015. Posttranslational modification of HOIP blocks toll-like receptor 4-mediated linear-ubiquitin-chain formation. MBio 6, e01777-15 (doi:10.1128/mBio.01777-15) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yoshida H, Jono H, Kai H, Li J. 2005. The tumor suppressor cylindromatosis (CYLD) acts as a negative regulator for toll-like receptor 2 signaling via negative cross-talk with TRAF6 AND TRAF7. J. Biol. Chem. 280, 41 111–41 121. (doi:10.1074/jbc.M509526200) [DOI] [PubMed] [Google Scholar]
- 53.Libro S, Kaluziak ST, Vollmer SV. 2013. RNA-seq profiles of immune related genes in the staghorn coral Acropora cervicornis infected with white band disease. PLoS ONE 8, e81821 (doi:10.1371/journal.pone.0081821) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Evans R, Patzak I, Svensson L, De Filippo K, Jones K, Mcdowall A, Hogg N. 2009. Integrins in immunity. J. Cell Sci. 122, 215–225. (doi:10.1242/jcs.019117) [DOI] [PubMed] [Google Scholar]
- 55.Lavine MD, Strand MR. 2002. Insect hemocytes and their role in immunity. Insect Biochem. Mol. Biol. 32, 1295–1309. (doi:10.1016/S0965-1748(02)00092-9) [DOI] [PubMed] [Google Scholar]
- 56.Liu H, et al. 2007. Phenoloxidase is an important component of the defense against Aeromonas hydrophila infection in a crustacean, Pacifastacus leniusculus. J. Biol. Chem. 282, 33 593–33 598. [DOI] [PubMed] [Google Scholar]
- 57.Tsirogianni AK, Moutsopoulos NM, Moutsopoulos HM. 2006. Wound healing: immunological aspects. Injury 37(Suppl 1), S5–S12. (doi:10.1016/j.injury.2006.02.035) [DOI] [PubMed] [Google Scholar]
- 58.Nappi AJ, Christensen BM. 2005. Melanogenesis and associated cytotoxic reactions: applications to insect innate immunity. Insect Biochem. Mol. Biol. 35, 443–459. (doi:10.1016/j.ibmb.2005.01.014) [DOI] [PubMed] [Google Scholar]
- 59.Kawabata S, Iwanaga S. 1999. Role of lectins in the innate immunity of horseshoe crab. Dev. Comp. Immunol. 23, 391–400. (doi:10.1016/S0145-305X(99)00019-1) [DOI] [PubMed] [Google Scholar]
- 60.Multerer KA, Smith LC. 2004. Two cDNAs from the purple sea urchin, Strongylocentrotus purpuratus, encoding mosaic proteins with domains found in factor H, factor I, and complement components C6 and C7. Immunogenetics 56, 89–106. (doi:10.1007/s00251-004-0665-2) [DOI] [PubMed] [Google Scholar]
- 61.Park SY, Bae DJ, Kim MJ, Piao ML, Kim IS. 2012. Extracellular low pH modulates phosphatidylserine-dependent phagocytosis in macrophages by increasing stabilin-1 expression. J. Biol. Chem. 287, 11 261–11 271. (doi:10.1074/jbc.M111.310953) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.van der Laan LJ, et al. 1999. Regulation and functional involvement of macrophage scavenger receptor MARCO in clearance of bacteria in vivo. J. Immunol. 162, 939–947. [PubMed] [Google Scholar]
- 63.Nauseef WM. 2007. How human neutrophils kill and degrade microbes: an integrated view. Immunol. Rev. 219, 88–102. (doi:10.1111/j.1600-065X.2007.00550.x) [DOI] [PubMed] [Google Scholar]
- 64.Becker K, Tilley L, Vennerstrom JL, Roberts D, Rogerson S, Ginsburg H. 2004. Oxidative stress in malaria parasite-infected erythrocytes: host-parasite interactions. Int. J. Parasitol. 34, 163–189. (doi:10.1016/j.ijpara.2003.09.011) [DOI] [PubMed] [Google Scholar]
- 65.Cheng TC, Rodrick GE, Foley DA, Koehler SA. 1975. Release of lysozyme from hemolymph cells of Mercenaria mercenaria during phagocytosis. J. Invertebr. Pathol. 25, 261–265. (doi:10.1016/0022-2011(75)90076-2) [DOI] [PubMed] [Google Scholar]
- 66.Forman HJ, Torres M. 2002. Reactive oxygen species and cell signaling: respiratory burst in macrophage signaling. Am. J. Respir. Crit. Care Med. 166, S4–S8. (doi:10.1164/rccm.2206007) [DOI] [PubMed] [Google Scholar]
- 67.Shimada T, et al. 2010. Staphylococcus aureus evades lysozyme-based peptidoglycan digestion that links phagocytosis, inflammasome activation, and IL-1beta secretion. Cell Host Microbe 7, 38–49. (doi:10.1016/j.chom.2009.12.008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fuess LE, Pinzon CJ, Weil E, Grinshpon RD, Mydlarz LD. 2017. Life or death: disease-tolerant coral species activate autophagy following immune challenge. Proc. R. Soc. B 284, 20170771 (doi:10.1098/rspb.2017.0771) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.van de Water JAJM, Chaib De Mares M, Dixon GB, Raina J-B, Willis BL, Bourne DG, van Oppen MJH. 2018. Antimicrobial and stress responses to increased temperature and bacterial pathogen challenge in the holobiont of a reef-building coral. Mol. Ecol. 27, 1065–1080. (doi:10.1111/mec.14489) [DOI] [PubMed] [Google Scholar]
- 70.Lasker HR, Sanchez JA. 2002. Allometry and astogeny of modular organisms. In Reproductive biology of invertebrates (ed. Hughes RN.), pp. 207–253. New York, NY: Wiley. [Google Scholar]
- 71.Sanchez JA, Lasker HR. 2003. Patterns of morphological integration in marine modular organisms: supra-module organization in branching octocoral colonies. Proc. R. Soc. Lond. B 270, 2039–2044. (doi:10.1098/rspb.2003.2471) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Campagna D, Gasparini F, Franchi N, Vitulo N, Ballin F, Manni L, Valle G, Ballarin L. 2016. Transcriptome dynamics in the asexual cycle of the chordate Botryllus schlosseri. BMC Genomics 17, 275 (doi:10.1186/s12864-016-2598-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bruno JF, Ellner SP, Vu I, Kim K, Harvell CD. 2011. Impacts of aspergillosis on sea fan coral demography: modeling a moving target. Ecol. Monogr. 81, 123–139. (doi:10.1890/09-1178.1) [Google Scholar]
- 74.Kapela W, Lasker HR. 1999. Size-dependent reproduction in the Caribbean gorgonian Pseudoplexaura porosa. Mar. Biol. 135, 107–114. (doi:10.1007/s002270050607) [Google Scholar]
- 75.Weil E, Croquer A, Urreiztieta I. 2009. Yellow band disease compromises the reproductive output of the Caribbean reef-building coral Montastraea faveolata (Anthozoa, Scleractinia). Dis. Aquat. Organ. 87, 45–55. (doi:10.3354/dao02103) [DOI] [PubMed] [Google Scholar]
- 76.Weil E, Rogers C. 2011. Coral reef diseases in the Atlantic-Caribbean. In Coral reefs: an ecosystem in transition (eds Dubinsky Z, Stambler N), pp. 465–491. Dordrecht, The Netherlands: Springer. [Google Scholar]
- 77.Nishikawa A, Katoh M, Sakai K. 2003. Larval settlement rates and gene flow of broadcast-spawning (Acropora tenuis) and planula-brooding (Stylophora pistillata) corals. Mar. Ecol. Prog. Ser. 256, 87–97. (doi:10.3354/meps256087) [Google Scholar]
- 78.Brinkhuis VIP. 2009. Assessment of gorgonian transplantation techniques offshore southeast Florida. Fort Lauderdale, FL: Nova Southeastern University Oceanographic Center. [Google Scholar]
- 79.Cary LR. 1914. Observation on the growth rate and oecology of gorgonians. Carn. Inst. Washington Publ. 43, 79–90. [Google Scholar]
- 80.Yoshioka PM. 1994. Size-specific life history pattern of a shallow-water gorgonian. J. Exp. Mar. Biol. Ecol. 184, 111–122. (doi:10.1016/0022-0981(94)90169-4) [Google Scholar]
- 81.Bruno JF, Selig ER, Casey KS, Page CA, Willis BL, Harvell CD, Sweatman H, Melendy AM. 2007. Thermal stress and coral cover as drivers of coral disease outbreaks. PLoS Biol. 5, 1220–1227. (doi:10.1371/journal.pbio.0050124) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Linares C, Coma R, Garrabou J, Diaz D, Zabala M. 2008. Size distribution, density and disturbance in two Mediterranean gorgonians: Paramuricea clavata and Eunicella singularis. J. Appl. Ecol. 45, 688–699. (doi:10.1111/j.1365-2664.2007.01419.x) [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw RNAseq reads are available on NCBI GenBank (SRA PRJNA407366).