Abstract
Alcohol addiction is a serious condition perpetuated by enduring physiological and behavioral adaptations. An important component of these adaptations is the long-term rearrangement of neuronal gene expression in the brain of the addicted individual. Epigenetic histone modifications have recently surfaced as important modulators of the transcriptional adaptation to alcohol as these are thought to represent a form of transcriptional memory that is directly imprinted on the chromosome. Some histone modifications affect transcription by modulating the accessibility of the underlying DNA, whereas others have been proposed to serve as marks read by transcription factors as a “histone code” that helps to specify the expression level of a gene. Although the effects of some epigenetic modifications on the transcriptional activity of genes are well known, the mechanisms by which alcohol consumption produces this rearrangement and leads to lasting changes in behavior remain unresolved. Recent advances using the Drosophila model system have started to unravel the epigenetic modulators underlying functional alcohol neuroadaptations. In this review, we discuss the role of 3 different histone modification systems in Drosophila, which have a direct impact on key alcohol neuroadaptations associated with the addictive process. These systems involve the histone deacetylase Sirt1, the histone acetyltransferase CREB-binding protein (CBP), and a subset of the Drosophila JmjC-Domain histone demethylase family.
Keywords: Alcohol, addiction, Drosophila, epigenetics, histone, Sirtuin, CBP, JmjC domain, KDM, HAT, HDAC
Introduction
The processing of information that underlies specific behavioral traits is ultimately dictated by the particular characteristics of the neuronal circuits in the brain.1 However, as the nervous system is exposed to distinct environmental conditions, the molecular identity of the neurons within these circuits can change. This transformation allows for functional continuity despite an ever-changing environment.2 As a neural depressant, alcohol consumption can trigger a series of compensatory neuroadaptations that may lead to a progressive increase in alcohol tolerance and the emergence of physiological dependence. Both tolerance and dependence have been linked to increased alcohol consumption through the dysregulation of the brain reward system.3–5
In recent years, numerous efforts have been directed to understanding the molecular mechanisms by which alcohol initiates and perpetuates the changes in neural physiology and behavior that drive the alcoholic state. It has become clear that many of these mechanisms involve changes in gene expression that result in a transcriptional reprogramming of the specific neuronal circuits that regulate alcohol use disorders. Studies in animal models as well as in humans have shown that both acute and chronic alcohol consumption can significantly alter the brain transcriptome.6–11 However, while the number of differentially expressed “alcohol genes” identified keeps growing, little is known of the underlying molecular mechanisms that orchestrate these transcriptional neuroadaptations to alcohol. Identifying these mechanisms is crucial for the development of more effective treatments for alcoholism.
To understand these mechanisms, it is important to note that every cell in an organism shares essentially the same genetic sequence. Nevertheless, different cells express distinct sets of genes. It is now well known that the ability to express a particular gene set is dependent on epigenetic modifications that alter chromatin structure and DNA accessibility.12,13 In the 1940s, Conrad Waddington introduced the term epigenetics as “the branch of biology which studies the causal interaction between genes and their products, which brings the phenotype into being.”14 In modern terms, epigenetics refers to the set of nongenetic modifications that determines the gene expression profile of a cell.15 By altering the structure of chromatin, epigenetic modifications can not only allow cells to differentiate from each other developmentally but they can also mediate the transcriptional reprogramming required for adaptation to environmental stimuli. The adaptations emerging from drug exposure are certainly a great example of this reprogramming.16–18
Epigenetic chromatin remodeling is mediated by posttranscriptional modifications of histone tails or through direct chemical modification of DNA. Each modification can have a specific effect on the biophysical structure of chromatin. Together, these chromatin changes regulate the accessibility of the transcriptional machinery to the underlying DNA and can also serve as signals that can influence the timing, speed, and/or volume of transcription.19 The best-studied examples of epigenetic modifications include methylation and acetylation of lysines in histone tails and direct DNA methylation. Acetylation of histone H3 (H3ac) and acetylation of histone H4 (H4ac) are generally associated with processes in which the DNA is made accessible and thus transcriptional active.20,21 Meanwhile, methylation of histones can have a wide range of transcriptional effects depending on the specific histone residue modified. Histone lysine methylation can occur on both the histone H3 and H4 tails, where each lysine residue can be mono- (me1), di- (me2), or trimethylated (me3). Methylation of residues H3K4 and H3K36 is associated with transcriptionally active chromatin. Genome-wide analyses of the different H3K4 methylation states indicate that H3K4me3 is enriched at transcription start sites.22 H3K4 trimethylation, in combination with histone acetylation, accurately reflects promoter activity, whereas H3K4me1 is mostly associated with the location of cis-regulatory elements or enhancers.21,23 The H3K36me3 mark is considered to be a marker for transcriptional elongation as it appears with the passage of RNA polymerase II.24 Monitoring the occurrence of H3K36me3 can, therefore, be regarded as taking a snapshot of recent transcriptional activity. Finally, H3K27me3 and H3K9me 3 are tightly associated with inactive gene promoters and with the inhibition of gene transcription and act in opposition to H3K4me3 and H3 acetylation.25,26 Together, these chromatin marks hold great promise because their involvement in gene regulation is conserved from Drosophila to humans.22,25,27,28
Although epigenetic modifications can remain for the entire lifespan of an organism—or even inherited through the germ line—the process is enzymatically controlled and reversible.29 Chromatin remodeling enzymes can be informally subdivided into “writers” or those that add a modification and “erasers,” which refer to those that remove the modifications. Both writers and erasers are critical mediators of transcriptional reprogramming and often work in concert with transcription factors (aka “readers”) to influence the expression profile of a cell.30 And, because of their reversible nature, these mechanisms have the potential of being exploited as therapeutic interventions.31 In this article, we review the recent advances made in identifying the epigenetic writers and erasers that control alcohol neuroadaptation in Drosophila melanogaster.
Drosophila Paves the Way
Drosophila has proven to be an effective biological model to study alcohol addiction. Fruit flies not only show behavioral responses to alcohol that closely resemble those of humans but will also develop the underlying neuroadaptations.32–34 On repeated exposure, flies display tolerance, increased alcohol preference, and symptoms of withdrawal.35–39 All of these phenotypes are good predictors of alcohol dependence in humans.40 Moreover, the behavioral assays to measure these adaptations in flies are uncomplicated and mostly straightforward and thus lend themselves quite well to genetic screens. It is important to note that dependence to alcohol can also be directly measured in Drosophila as shown by Robinson and colleagues.41,42 In these studies, the authors showed that after withdrawal from chronic alcohol exposure, the performance of Drosophila larvae in a cognitive task (learning) is significantly impaired. Interestingly, on alcohol reinstatement, their learning performance improves significantly. These results demonstrate that larvae can indeed become dependent to the presence of alcohol.
Alcohol tolerance is by far the easiest alcohol neuroadaptation to measure in Drosophila. Tolerance is generally defined as an increase in resistance to the drug in response to repeated exposure. However, tolerance can take different forms depending on the mechanism by which it is manifested. A distinction is often made between functional tolerance and metabolic or dispositional tolerance. The former refers to adaptations elicited to compensate for the disruption caused by alcohol in both behavior and physiology, whereas the latter refers to a change in the ability to metabolize or eliminate the drug. Because adult flies do not display metabolic tolerance to alcohol, they are a great system to study functional tolerance.43
In flies, a single sedating dose of alcohol can elicit tolerance that is evident as early as 4 hours but can still be detected a week after the initial exposure.44,45 Alcohol is often delivered to flies as vapor, and the time to sedation (and/or recovery) is recorded either visually by an observer or through an automated locomotion-tracking device. An identical protocol is used to monitor the knockdown (and recovery) period of flies undergoing a second sedation. These 2 events are directly compared and assessed. Because tolerance is defined as a reduced effect to the drug on repeated exposure to the drug, a significant delay in knockdown (or a speedier recovery) is a clear indicator that the flies have acquired tolerance. Other methods to monitor alcohol neuroadaptations such as increased preference, withdrawal, and cognitive dependence are usually more laborious but just as robust. These methods have been reviewed in detail in the work by Rothenfluh et al.46
The genetic component of alcohol neuroadaptation can be easily studied in Drosophila because screening large numbers of genes for a participatory role is possible and practical. Flies are one of the few animals in which community resources provide access to mutations and RNAi transgenes for almost all genes. Also, their minimal maintenance cost makes the screening of large numbers of genes practical, and their rapid life cycle means that combinations of genetic tools can be quickly created. Furthermore, Drosophila also facilitates the use of powerful tools to study the neural components of behavior. The UAS-Gal4 system championed in flies47 allows one to genetically manipulate individual neural circuits and explore their relevance in different behavioral processes including alcohol neuroadaptation. This technique has been successfully used to map the brain regions important for the rewarding aspects of alcohols in flies.38
Drosophila research translates well to mammals and has provided the keys for understanding important regulatory pathways and neurobiological processes involved in alcohol neuroadaptation. Examples of alcohol genes identified in Drosophila and then used to identify their mammalian counterparts in both sequence and function include the BK-type K+ channel gene slo48 and the LMO protein gene Bx.49 The slo gene contributes to similar neural processes in mammals and flies. In both, slo is required for implementing circadian rhythms,50 and expression of slo BK channels has been shown to underlie physiological alcohol tolerance in both, the rat hypothalamo-neurohypophysial explant system and behavioral alcohol tolerance in the fly.36,51 The Bx gene was first shown to be involved in the response of flies to cocaine and alcohol. This fly result translated well to mammals, as 2 of the mammalian Bx homologues (Lmo4 and Lmo3) were later shown to play similar roles in mammals. While Lmo4 was shown to regulate the response to cocaine in mice,52 Lmo3 was implicated in the response to alcohol.53,54
Over the years, close to a hundred genes covering many different biological pathways have been associated with alcohol neuroadaptation in Drosophila (reviewed in the works by Rodan and Rothenfluh55 and Park et al34). It is now evident that the adaptations elicited during alcohol exposure are very complex and not dependent on single genes. Instead, these responses seem to be choreographed by multigene networks. It has thus become increasingly clear that proper understanding of these adaptations requires the analysis of the molecular mechanisms that coordinate the regulation of these networks. Because of its intricate role in the regulation of gene expression, epigenetic histone modifications have become a major focus of research in this area. In Drosophila, a series of studies have led the way toward understanding the role of epigenetic histone modifications in the control of alcohol-induced transcriptional changes. A wave of histone acetylation around the BK channel gene slo was first detected in response to the anesthetic benzyl alcohol.56 This increase in histone acetylation was directly linked to an increase in expression of the gene, the development of tolerance to this drug, and to the subsequent binding of the transcription factor CREB.57 A similar mechanism was later confirmed for alcohol.58 Moreover, a recent genomic study that exploits genome-wide analysis of epigenetic modifications induced by different drugs identified a network of genes that show common alcohol-induced histone acetylation responses.59 These genes were not only shown to have a direct role in the development of alcohol tolerance but also share highly correlated expression profiles in response to diverse environmental stimuli. These results strongly suggest that the genes in this network are coordinately regulated. Indeed, these genes fall into interconnected categories and encode a set of proteins that are tightly associated with the regulation of synaptic plasticity.
In the past 2 years, 3 alcohol studies from different Drosophila groups have identified a set of histone-modifying enzymes with direct roles in controlling adaptive alcohol responses.60–62 These findings, reviewed below, have strengthened the link between epigenetic modifications and alcohol-induced neuroadaptations and have solidified the importance of the Drosophila model in elucidating the mechanisms of alcohol addiction.
The Histone Deacetylase Sir2/Sirt1
The Silence Information Regulator 2 or Sir2 is a histone deacetylase protein (HDAC) that is conserved in almost every domain of life. It was first discovered in Saccharomyces cerevisiae for its effect on important mating type loci63 and later associated with the regulation of DNA repair and DNA recombination (reviewed in the work by Loo and Rine64). Molecularly, Sir2 is directly involved in gene silencing through the removal of acetyl groups from histones and other proteins. Sir2 belongs to a family of NAD-dependent protein deacetylases known as Sirtuins.65 In Drosophila, Sir2 is encoded by the gene Sirt1 and has previously been linked to the regulation of longevity and the maintenance of metabolic health and feeding behavior,66,67 for which it has received significant attention.
Drosophila Sirt1 was first linked to alcohol responses through a genome-wide survey of alcohol-induced transcriptional responses using RNA microarrays.6 Sirt1 was significantly downregulated after acute alcohol exposure. This was later confirmed by a second genome-wide study from a different group.68 In the latter study, the authors followed up with behavioral analysis of a Sirt1 mutant and demonstrated the involvement of Sirt1 in functional alcohol tolerance. More recently, however, a study by Engel et al60 have expanded this analysis and found that Drosophila Sirt1 is an integral part of a transcriptional program to alter presynaptic properties and neural responses essential for the development of alcohol tolerance, preference, and reward.
In this most recent article, the authors showed that Sirt1 mutants were both less sensitive to the sedating effects of acute alcohol and showed a significant decrease in alcohol tolerance. These strong behavioral effects were shown to be neurally specific, as RNAi knockdown of Sirt1 in neurons, but not in other tissues, showed the same effects. Moreover, through a more exhaustive mapping of the circuitry controlling this behavior, the authors narrowed the requirement of Sirt1 to the mushroom bodies, a brain neuropil that has been previously implicated in learning and memory, as well as in alcohol preference and reward.38,69,70 Interestingly, the Sirt1 mutant effect extended also to the rewarding aspects of alcohol-induced behaviors. The authors showed that when given a choice between regular food and alcohol supplemented food, Sirt1 mutant flies preferred the alcohol-containing food even without being previously exposed to alcohol. This is in marked contrast to wild-type flies, which are initially uninterested in alcohol but develop a slight preference over time. Similarly, flies that lacked Sirt1 presented a reduction in alcohol-conditioned odor preference, which is indicative of a loss of the alcohol rewarding effects.
Because Sirt1 expression is reduced following alcohol exposure and because its activity is closely linked to the epigenetic regulation of gene expression, the authors hypothesized that Sirt1 sets up a transcriptional program that regulates neural activity. Indeed, when they searched for possible transcriptional targets of Sirt1, they found a small group of neural proteins that were differentially regulated in response to alcohol in Sirt1 mutants. They use RNA-Seq to detect changes in expression induced by ethanol exposure in either Sirt1 mutants or wild-type controls. Of the 492 genes upregulated in wild-type flies in response to alcohol, only 52 were also upregulated in Sirt1 mutant flies, evidencing a very distinct transcriptional profile between the 2 lines. One gene, in particular, Syn, which encodes Synapsin—a protein associated with maintenance of the synaptic vesicle reserve pool71—showed a marked difference in response to alcohol in Sirt1 mutants vs control. While Syn expression was reduced in response to alcohol exposure in wild-type animals, it increased in the Sirt1 mutant. Syn has been previously linked to altered alcohol tolerance in Drosophila72 and its involvement was confirmed here. Furthermore, the authors show that alcohol may also regulate 2 other genes in a Sir2-dependent manner: cac and Cdk5. These genes encode a presynaptic calcium channel (cac) and a cyclin-dependent kinase involved in presynaptic function (Cdk5). However, while Syn decreased, cac and Cdk5 increased. The authors thus propose that acute alcohol exposure leads to presynaptic adaptations that increase neurotransmitter release through the action of Sirt1.
Overall, this study demonstrates that Sirt1 is required to promote alcohol sensitivity and facilitate the development of functional alcohol tolerance. Sirt1 is likely acting through the direct regulation of genes involved in synaptic modulation. Sirtuins, such as Sirt1, target different histone marks, including H4K16Ac, H3K9Ac, H3K56Ac, and H3K18Ac, and non-histone components of the chromatin machinery, such as enzymes and structural proteins.65 It is thus expected that the regulation of alcohol-responsive genes will be mediated through an overall change in acetylation of these targets.
The Histone Acetyltransferase CREB-Binding Protein
The CREB-binding protein (CBP) is one of the best-studied histone acetyltransferases. It acetylates several nuclear proteins, including histone H3 on K14, K18, and K27, and H4 on K5 and K8. It is a member of the bromodomain-like superfamily and contains the highly conserved CBP/p300-type histone acetyltransferase domain.26,73 Through its direct role in transcriptional activation, CBP has been linked to cell proliferation, cell signaling, and differentiation, and in developmental patterning (reviewed in the works by Janknecht74 and Dancy and Cole75). Interestingly, in mammals, CBP has also been linked to alcohol behaviors, as increased levels of CBP and histone acetylation were observed in the amygdaloid brain regions of rats.76
In Drosophila, CBP is encoded by the gene nej and it was recently linked to the development of functional alcohol tolerance.61,77 In this article, Ghezzi and colleagues first showed that a mutation in nej significantly reduces tolerance to alcohol sedation. Moreover, when nej was artificially induced using a heat-inducible transgene, the experimenters were able to induce an increase in alcohol resistance that resembled the wild-type tolerance phenotype. These findings suggested that CBP was indeed necessary and sufficient for the acquisition of functional tolerance.
The authors also linked CBP with the increased acetylation and subsequent transcriptional upregulation of known alcohol tolerance genes. One of which is the BK channel gene slo. The role of slo in drug-related behaviors is well-documented and has been shown to be an important player in the development of alcohol tolerance in both flies and mammals.5,78 In flies, its involvement in the alcohol response is directly mediated by an increase in histone H4 acetylation.58 Artificially induction of the heat-inducible nej transgene failed to increase alcohol resistance in the slo mutant background. In contrast, alcohol-induced expression of the slo gene, as well as 5 other alcohol response genes, was effectively blocked in a nej mutant background. Together, these results directly link CBP activity with the transcriptional regulation of 6 different genes involved in synaptic regulation. And, because CBP was found to bind the transcriptional control regions of these genes directly, it was proposed that CBP orchestrates the expression of these synaptic genes during the adaptation to alcohol.
CBP is well known for (and, in fact, it is named after) its interaction with the transcription factor CREB. The authors thus postulate that CBP-regulated alcohol response genes involve the recruitment of CREB to their transcriptional control regions. This is based on the observation that Drosophila CREB mutants can also block the development of alcohol tolerance while inhibiting the alcohol-induced histone acetylation profiles of the slo gene.56,57 Interestingly, CBP is also known to interact with the Sirt2,79 another chromatin remodeler that, as discussed above, has also been recently linked to presynaptic changes associated with the development of alcohol tolerance and preference.60 It is thus foreseeable that the coordinate induction of CBP and suppression of Sir2 by alcohol can dramatically increase acetylation of histones and significantly reshape the chromatin structure.
The JmjC-domain Histone Demethylases
The most recent epigenetic system implicated in alcohol use disorders is composed of a set of JmjC domain–containing demethylases (JmjC-KDM). These enzymes represent the largest class of histone demethylases. They functionally bind and modify chromatin by removing methyl groups from lysines of histone tails. In Drosophila, there are at least 13 JmjC-KDMs each with different substrate specificity.80 In a recent study, Pinzón et al62 systematically tested the role of all known Drosophila JmjC-KDM in alcohol-induced responses. They focused on 2 main alcohol-related behaviors: innate sensitivity and rapid tolerance. In addition, they tested these behaviors under 3 different doses. Out of the 13 genes known to encode JmjC-KDMs in the fly, 4 induced significant and reproducible impairments in functional alcohol tolerance and/or sensitivity to sedation when mutated. These genes are KDM3, lid, NO66, and HSPBAP1.
KDM3, which is also known in Drosophila by JDHM2, catalyzes the removal of methyl groups from Histone H3 on lysine 9 and thereby promotes an open chromatin structure.81,82 It was first found in the meiotic and postmeiotic male cells and originally described as a coactivator of the androgen receptor with a direct role in primordial germ cell development.81 Lid or little imaginal discs (named for its mutant phenotype) is a trimethyl H3K4 histone demethylase that regulates transcription through both demethylase-dependent and demethylase-independent mechanisms, as it has also been shown to contribute to histone acetyltransferase activity (H3K9 specific).80,83 In flies, Lid has been associated with the regulation of cell growth, circadian rhythm, stress resistance, hematopoiesis, and fertility.84–87 The lid gene interacts with various molecular pathways such as the Notch and the JAK-STAT signaling pathways.85,88,89 Finally, NO66 is a histone demethylase that is so far very understudied. Substrate specificity to H3K4me and H3K36me has only been inferred from UniProt Gene Ontology curation based on sequence similarities.90
Interestingly, mutants of each of the 4 JmjC-KDMs genes have relatively different alcohol behaviors. Knockout of NO66, KDM3, and lid resulted in increased sensitivity in all doses. However, mutations in HSPBAP1 gene resulted in less sensitivity only when flies were exposed to low doses. Furthermore, NO66 had no significant difference in tolerance except in low doses, whereas KDM3 had less tolerance in mid and high doses and no significant difference in low doses. Similarly, HSBAP1 resulted less tolerant in low and high doses and no significant difference in mid dose. In contrast, lid had a strikingly different phenotype. Although it shows more sensitivity to alcohol (such as NO66 and KDM3), it was the only one that showed enhanced tolerance in every dose. Together, these results suggest that different JmjC-KDM genes have different roles in controlling the distinct aspects of alcohol-induced responses, but most importantly, it genetically separated innate sensitivity from the capacity to acquire tolerance.
To confirm that KDM3, NO66, and lid were indeed involved in functional alcohol tolerance, the authors performed transgenic rescue experiments in an attempt to reverse the mutant phenotype. For this, wild-type genomic constructs for each of the 3 JmjC-KDM genes that affected tolerance were recombined with their respective mutant background. While the KDM3 and NO66 genomic rescues were able to restore to wild-type phenotype, the lid rescue failed. However, instead, the authors were able to validate the involvement of lid in alcohol tolerance using a lid-RNAi line that mimicked the lid mutant phenotype and thus ruling out nonspecific effects. In this case, however, the lid RNAi transgene was expressed exclusively in neurons, and yet was still able to recapitualte the mutant phenotype, which indicates that lid is required in the brain for normal responses to alcohol. Similar results were observed for KDM3 and NO66. RNAi knockdown of KDM3 and NO66 using panneuronal promoters also photocopied the mutant tolerance phenotypes, suggesting that all 3 JmjC-KDMs are important for alcohol tolerance specifically in the nervous system.
Altogether, these results demonstrate that JmjC-KDMs serve critical roles in the neuroadaptive behavior to alcohol. Furthermore, the authors speculate that these JmjC-KDMs interact with other components of the chromatin remodeling complex such as Snr1 (a SET domain protein associated with histone lysine methylation) to affect changes in alcohol-induced behaviors. Both Lid and NO66 suppress the expression of the Snr1 gene, whereas KDM3 enhances the expression of the Snr1 gene.91 Snr1 expression is not only regulated by 3 of these 4 JmjC-KDMs but also is influenced by the SWI/SNF chromatin remodeling complex that regulate a vast number of genes and has been associated with alcohol-induced behaviors.91,92
Final Remarks
The brain is one of the most complex tissues in higher organism. Its ability to adapt to environmental changes is remarkable. Not only is this important because it can maintain a constant level of global activity but at the same time it allows selected modifications induced by activity itself to occur. This plasticity is not only crucial for proper functioning of innate behaviors but it is also tightly involved in higher order processing and cognition. Alcohol, similar to many other psychoactive drugs, is an environmental stressor that can directly affect the carefully controlled balance of activity and inhibition within the brain. It is thus not surprising that in response to its consumption, the brain elicits a series of neuroadaptive changes to compensate its effects.
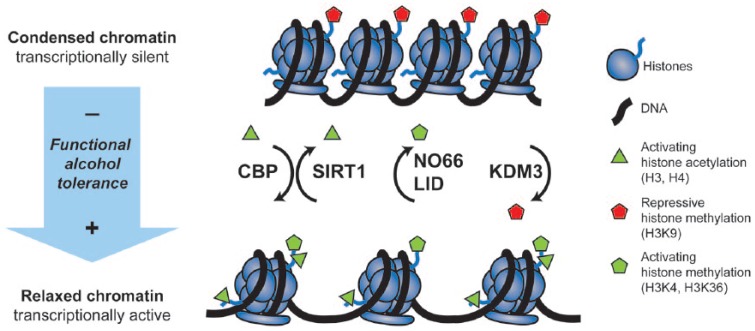
As demonstrated in flies, the responses elicited by alcohol are not short-lived events, but rather adaptations that persist for a relatively extended period of time. A single exposure to alcohol results in a prolonged increase in alcohol resistance that extends over several days.45 This enduring tolerance phenotype is accompanied by an equally long increase in susceptibility to seizures—a common symptom of alcohol withdrawal in humans.39 The persistent nature of drug-induced adaptations suggests that the mechanisms behind them involve long-lasting changes in gene expression and include the epigenetic restructuring of chromosomal regions that perpetuate them. Because the process of gene transcription is dependent on chromatin state,19 enzymes with the ability to remodel chromatin structure have the potential to reprogram the expression pattern of a cell. Here, we reviewed 3 independent epigenetic systems that are not only known to affect chromatin structure but are all linked to alcohol (see Table 1 for a summary of the enzymes implicated). Figure 1 depicts the role each of these enzymes play in chromatin remodeling and its effect on functional tolerance. It is expected that the interplay between these writers and erasers of histone modifications results in the optimal combination of histone marks that produces the adapted cellular state.
Table 1.
Summary of the chromatin remodelers in Drosophila melanogaster known to affect alcohol-induced neuroadaptation.
| Protein name | Gene symbol | Molecular function | Substrate specificity | Mutant phenotype | Human homologue | References |
|---|---|---|---|---|---|---|
| Sir2/Sirt1 | Sirt1 | Sirtuin, histone deacetylase activity | H3 | Enhanced tolerance | SIRT1 | Engel et al60 |
| CBP | Nej | Histone acetyltransferase activity | H3, H4 | Reduced tolerance | CREBBP | Ghezzi et al61 |
| NO66 | NO66 | Histone demethylase activity | H3K4, H3K36 | Enhanced tolerance | RIOX1 | Pinzón et al62 |
| KDM3 | JHDM2 | Histone demethylase activity | H3K9 | Reduced tolerance | KDM3 | Pinzón et al62 |
| LID | Lid | Histone demethylase activity | H3K4 | Enhanced tolerance | KDM5 | Pinzón et al62 |
| HSPBAP1 | CG12879 | Histone demethylase activity | H3K4, H3K36 | Reduced tolerance | HSPBAP1 | Pinzón et al62 |
Shown are their molecular functions, substrate specificity, mutant alcohol phenotype, their human homologues, and the publications demonstrating their role in the alcohol phenotypes in Drosophila.
Figure 1.
Schematic model of the role of epigenetic modifiers in functional tolerance. The figure shows the transition from a transcriptionally silent, condensed chromatin state (top) to a transcriptionally active, relaxed chromatin state (bottom) during the development of functional alcohol tolerance. (1) The histone acetyltransferase CBP, which is induced by alcohol, catalyzes the addition of acetyl groups to the tails of histone H3 and H4, resulting in the relaxation of chromatin and promoting alcohol tolerance. Mutations in CBP reduce alcohol tolerance. (2) The histone deacetylase SIRT1, which is suppressed by alcohol, catalyzes the removal of acetyl groups from the tails of histone H3, resulting in chromatin condensation and reduces alcohol resistance. Mutations in SIRT1 enhance alcohol tolerance. (3) The histone demethylases NO66 and LID catalyze the removal of activating methyl groups from the tails of histone H3 residues K4 and K36, resulting in chromatin condensation and reduce alcohol resistance. Mutations in NO66 and LID enhance alcohol tolerance. (4) The histone demethylase KDM3 catalyzes the addition of repressive methyl groups from the tails of histone H3 residue K9, resulting in the relaxation of chromatin and favoring alcohol tolerance. Mutations in KDM3 reduce alcohol tolerance.
Many of the enzymes implicated so far can interact with each other—either directly through protein-protein interactions or indirectly by modulating the same histone substrates. It is thus expected that together these enzymes maintain a tightly controlled balance between acetylation/deacetylation and methylation/demethylation of chromatin regions (Figure 1). By doing so, they can fine-tune neuronal excitability. Even small changes in chromatin state can have a significant impact on the transcriptional profile of individual cells of the nervous system.
An important problem in medical research today is that it is difficult to extrapolate from any single model system to humans. The strong evolutionary conservation of the mechanistic response between distantly related species such as Drosophila and humans is a good predictor that response will continue to humans. Histones are among the most highly conserved proteins in eukaryotes, so it is extremely likely that the study of how alcohol modifies these proteins will translate directly to humans.
Footnotes
Funding:The author(s) disclosed receipt of the following financial support for the research, authorship and/or publication of this article: The authors received funding from the “Fondo Institucional para la Investigación” (FIPI) from the University of Puerto Rico–Rio Piedras and from the National Science Foundation Award #1736026.
Declaration of conflicting interests:The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Author Contributions: All authors critically reviewed the research articles, and contributed discussions to the final manuscript. AG wrote the manuscript with support from MR and CB.
ORCID iD: Alfredo Ghezzi  https://orcid.org/0000-0001-9176-1320
https://orcid.org/0000-0001-9176-1320
References
- 1. Wolfram V, Baines RA. Blurring the boundaries: developmental and activity-dependent determinants of neural circuits. Trends Neurosci. 2013;36:610–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. McClung CA, Nestler EJ. Neuroplasticity mediated by altered gene expression. Neuropsychopharmacology. 2008;33:3–17. [DOI] [PubMed] [Google Scholar]
- 3. Littleton J. Neurochemical mechanisms underlying alcohol withdrawal. Alcohol Health Res World. 1998;22:13–24. [PMC free article] [PubMed] [Google Scholar]
- 4. Koob GF Le Moal M. Review. Neurobiological mechanisms for opponent motivational processes in addiction. Philos Trans R Soc Lond B Biol Sci. 2008;363:3113–3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ghezzi A, Atkinson NS. Homeostatic control of neural activity: a Drosophila model for drug tolerance and dependence. Int Rev Neurobiol. 2011;99:23–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Morozova TV, Anholt RR, Mackay TF. Transcriptional response to alcohol exposure in Drosophila melanogaster. Genome Biol. 2006;7:R95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mulligan MK, Ponomarev I, Hitzemann RJ, et al. Toward understanding the genetics of alcohol drinking through transcriptome meta-analysis. Proc Natl Acad Sci U S A. 2006;103:6368–6373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Morozova TV, Mackay TF, Anholt RR. Transcriptional networks for alcohol sensitivity in Drosophila melanogaster. Genetics. 2011;187:1193–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ponomarev I, Wang S, Zhang L, Harris RA, Mayfield RD. Gene coexpression networks in human brain identify epigenetic modifications in alcohol dependence. J Neurosci. 2012;32:1884–1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Osterndorff-Kahanek EA, Becker HC, Lopez MF, et al. Chronic ethanol exposure produces time- and brain region-dependent changes in gene coexpression networks. PLoS ONE. 2015;10:e0121522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Marballi K, Genabai NK, Blednov YA, Harris RA, Ponomarev I. Alcohol consumption induces global gene expression changes in VTA dopaminergic neurons. Genes Brain Behav. 2016;15:318–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bonn S, Zinzen RP, Girardot C, et al. Tissue-specific analysis of chromatin state identifies temporal signatures of enhancer activity during embryonic development. Nat Genet. 2012;44:148–156. [DOI] [PubMed] [Google Scholar]
- 13. Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. [DOI] [PubMed] [Google Scholar]
- 14. Waddington CH. Canalization of development and the inheritance of acquired characters. Nature. 1942;150:563–565. [DOI] [PubMed] [Google Scholar]
- 15. Allis CD, Jenuwein T, Reinberg D, Caparros M-L. Epigenetics. Woodbury, NY: Cold Spring Harbor Laboratory Press; 2007. [Google Scholar]
- 16. Maze I, Nestler EJ. The epigenetic landscape of addiction. Ann N Y Acad Sci. 2011;1216:99–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Moonat S, Pandey SC. Stress, epigenetics, and alcoholism. Alcohol Res. 2012;34:495–505. [PMC free article] [PubMed] [Google Scholar]
- 18. Ponomarev I. Epigenetic control of gene expression in the alcoholic brain. Alcohol Res. 2013;35:69–76. [PMC free article] [PubMed] [Google Scholar]
- 19. Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21:381–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Roh TY, Cuddapah S, Zhao K. Active chromatin domains are defined by acetylation islands revealed by genome-wide mapping. Genes Dev. 2005;19:542–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Heintzman ND, Hon GC, Hawkins RD, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Heintzman ND, Stuart RK, Hon G, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–318. [DOI] [PubMed] [Google Scholar]
- 23. Bernstein BE, Kamal M, Lindblad-Toh K, et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. [DOI] [PubMed] [Google Scholar]
- 24. Joshi AA, Struhl K. Eaf3 chromodomain interaction with methylated H3-K36 links histone deacetylation to Pol II elongation. Mol Cell. 2005;20:971–978. [DOI] [PubMed] [Google Scholar]
- 25. Barski A, Cuddapah S, Cui K, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. [DOI] [PubMed] [Google Scholar]
- 26. Tie F, Banerjee R, Stratton CA, et al. CBP-mediated acetylation of histone H3 lysine 27 antagonizes Drosophila Polycomb silencing. Development. 2009;136:3131–3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kharchenko PV, Alekseyenko AA, Schwartz YB, et al. Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature. 2011;471:480–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yin H, Sweeney S, Raha D, Snyder M, Lin H. A high-resolution whole-genome map of key chromatin modifications in the adult Drosophila melanogaster. PLoS Genet. 2011;7:e1002380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33:245–254. [DOI] [PubMed] [Google Scholar]
- 30. Goldberg AD, Allis CD, Bernstein E. Epigenetics: a landscape takes shape. Cell. 2007;128:635–638. [DOI] [PubMed] [Google Scholar]
- 31. Arrowsmith CH, Bountra C, Fish PV, Lee K, Schapira M. Epigenetic protein families: a new frontier for drug discovery. Nat Rev Drug Discov. 2012;11:384–400. [DOI] [PubMed] [Google Scholar]
- 32. Guarnieri DJ, Heberlein U. Drosophila melanogaster, a genetic model system for alcohol research. Int Rev Neurobiol. 2003;54:199–228. [DOI] [PubMed] [Google Scholar]
- 33. Devineni AV, Heberlein U. The evolution of Drosophila melanogaster as a model for alcohol research. Annu Rev Neurosci. 2013;36:121–138. [DOI] [PubMed] [Google Scholar]
- 34. Park A, Ghezzi A, Wijesekera TP, Atkinson NS. Genetics and genomics of alcohol responses in Drosophila. Neuropharmacology. 2017;122:22–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Scholz H, Franz M, Heberlein U. The hangover gene defines a stress pathway required for ethanol tolerance development. Nature. 2005;436:845–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cowmeadow RB, Krishnan HR, Ghezzi A, Al’Hasan YM, Wang YZ, Atkinson NS. Ethanol tolerance caused by slowpoke induction in Drosophila. Alcohol Clin Exp Res. 2006;30:745–753. [DOI] [PubMed] [Google Scholar]
- 37. Devineni AV, Heberlein U. Preferential ethanol consumption in Drosophila models features of addiction. Curr Biol. 2009;19:2126–2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kaun KR, Azanchi R, Maung Z, Hirsh J, Heberlein U. A Drosophila model for alcohol reward. Nat Neurosci. 2011;14:612–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ghezzi A, Krishnan HR, Atkinson NS. Susceptibility to ethanol withdrawal seizures is produced by BK channel gene expression. Addict Biol. 2014;19:332–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders: DSM-5. 5th ed. Washington, DC: American Psychiatric Association; 2013. [Google Scholar]
- 41. Robinson BG, Khurana S, Kuperman A, Atkinson NS. Neural adaptation leads to cognitive ethanol dependence. Curr Biol. 2012;22:2338–2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Robinson BG, Khurana S, Atkinson NS. Drosophila larvae as a model to study physiological alcohol dependence. Commun Integr Biol. 2013;6:e23501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Atkinson NS. Tolerance in Drosophila. J Neurogenet. 2009;23:293–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Scholz H, Ramond J, Singh CM, Heberlein U. Functional ethanol tolerance in Drosophila. Neuron. 2000;28:261–271. [DOI] [PubMed] [Google Scholar]
- 45. Cowmeadow RB, Krishnan HR, Atkinson NS. The slowpoke gene is necessary for rapid ethanol tolerance in Drosophila. Alcohol Clin Exp Res. 2005;29:1777–1786. [DOI] [PubMed] [Google Scholar]
- 46. Rothenfluh A, Troutwine BR, Ghezzi A, Atkinson NS. The genetics of alcohol responses of invertebrate model systems. In: Noronha ABC, Cui C, Harris RA, Crabbe JC, eds. Neurobiology of Alcohol Dependence. Amsterdam, The Netherlands: Elsevier; 2014:463–491. [Google Scholar]
- 47. Duffy JB. GAL4 system in Drosophila: a fly geneticist’s Swiss army knife. Genesis. 2002;34:1–15. [DOI] [PubMed] [Google Scholar]
- 48. Atkinson NS, Robertson GA, Ganetzky B. A component of calcium-activated potassium channels encoded by the Drosophila slo locus. Science. 1991;253:551–555. [DOI] [PubMed] [Google Scholar]
- 49. Milán M, Diaz-Benjumea FJ, Cohen SM. Beadex encodes an LMO protein that regulates Apterous LIM-homeodomain activity in Drosophila wing development: a model for LMO oncogene function. Genes Dev. 1998;12:2912–2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Meredith AL, Wiler SW, Miller BH, et al. BK calcium-activated potassium channels regulate circadian behavioral rhythms and pacemaker output. Nat Neurosci. 2006;9:1041–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Pietrzykowski AZ, Friesen RM, Martin GE, et al. Posttranscriptional regulation of BK channel splice variant stability by miR-9 underlies neuroadaptation to alcohol. Neuron. 2008;59:274–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lasek AW, Kapfhamer D, Kharazia V, Gesch J, Giorgetti F, Heberlein U. Lmo4 in the nucleus accumbens regulates cocaine sensitivity. Genes Brain Behav. 2010;9:817–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lasek AW, Giorgetti F, Berger KH, Tayor S, Heberlein U. Lmo genes regulate behavioral responses to ethanol in Drosophila melanogaster and the mouse. Alcohol Clin Exp Res. 2011;35:1600–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Savarese A, Zou ME, Kharazia V, Maiya R, Lasek AW. Increased behavioral responses to ethanol in Lmo3 knockout mice. Genes Brain Behav. 2014;13:777–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Rodan AR, Rothenfluh A. The genetics of behavioral alcohol responses in Drosophila. Int Rev Neurobiol. 2010;91:25–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wang Y, Krishnan HR, Ghezzi A, Yin JC, Atkinson NS. Drug-induced epigenetic changes produce drug tolerance. PLoS Biol. 2007;5:e265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wang Y, Ghezzi A, Yin JC, Atkinson NS. CREB regulation of BK channel gene expression underlies rapid drug tolerance. Genes Brain Behav. 2009;8:369–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Krishnan HR, Li X, Ghezzi A, Atkinson NS. A DNA element in the slo gene modulates ethanol tolerance. Alcohol. 2016;51:37–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ghezzi A, Krishnan HR, Lew L, Prado FJ, Ong DS, Atkinson NS. Alcohol-induced histone acetylation reveals a gene network involved in alcohol tolerance. PLoS Genet. 2013;9:e1003986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Engel GL, Marella S, Kaun KR, et al. Sir2/Sirt1 links acute inebriation to presynaptic changes and the development of alcohol tolerance, preference, and reward. J Neurosci. 2016;36:5241–5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ghezzi A, Li X, Lew LK, Wijesekera TP, Atkinson NS. Alcohol-induced neuroadaptation is orchestrated by the histone acetyltransferase CBP. Front Mol Neurosci. 2017;10:103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Pinzón JH, Reed AR, Shalaby NA, Buszczak M, Rodan AR, Rothenfluh A. Alcohol-induced behaviors require a subset of Drosophila JmjC-domain histone demethylases in the nervous system. Alcohol Clin Exp Res. 2017;41:2015–2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Klar AJ, Fogel S, Macleod K. MAR1—a regulator of the HMa and HMalpha Loci in Saccharomyces Cerevisiae. Genetics. 1979;93:37–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Loo S, Rine J. Silencing and heritable domains of gene expression. Annu Rev Cell Dev Biol. 1995;11:519–548. [DOI] [PubMed] [Google Scholar]
- 65. North BJ, Verdin E. Sirtuins: Sir2-related NAD-dependent protein deacetylases. Genome Biol. 2004;5:224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Rogina B, Helfand SL. Sir2 mediates longevity in the fly through a pathway related to calorie restriction. Proc Natl Acad Sci U S A. 2004;101:15998–16003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kusama S, Ueda R, Suda T, Nishihara S, Matsuura ET. Involvement of Drosophila Sir2-like genes in the regulation of life span. Genes Genet Syst. 2006;81:341–348. [DOI] [PubMed] [Google Scholar]
- 68. Kong EC, Allouche L, Chapot PA, et al. Ethanol-regulated genes that contribute to ethanol sensitivity and rapid tolerance in Drosophila. Alcohol Clin Exp Res. 2010;34:302–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Akalal DB, Wilson CF, Zong L, Tanaka NK, Ito K, Davis RL. Roles for Drosophila mushroom body neurons in olfactory learning and memory. Learn Mem. 2006;13:659–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Ojelade SA, Jia T, Rodan AR, et al. Rsu1 regulates ethanol consumption in Drosophila and humans. Proc Natl Acad Sci U S A. 2015;112:E4085–4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Bykhovskaia M. Synapsin regulation of vesicle organization and functional pools. Semin Cell Dev Biol. 2011;22:387–392. [DOI] [PubMed] [Google Scholar]
- 72. Godenschwege TA, Reisch D, Diegelmann S, et al. Flies lacking all synapsins are unexpectedly healthy but are impaired in complex behaviour. Eur J Neurosci. 2004;20:611–622. [DOI] [PubMed] [Google Scholar]
- 73. Crump NT, Hazzalin CA, Bowers EM, Alani RM, Cole PA, Mahadevan LC. Dynamic acetylation of all lysine-4 trimethylated histone H3 is evolutionarily conserved and mediated by p300/CBP. Proc Natl Acad Sci U S A. 2011;108:7814–7819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Janknecht R. The versatile functions of the transcriptional coactivators p300 and CBP and their roles in disease. Histol Histopathol. 2002;17:657–668. [DOI] [PubMed] [Google Scholar]
- 75. Dancy BM, Cole PA. Protein lysine acetylation by p300/CBP. Chem Rev. 2015;115:2419–2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Pandey SC, Ugale R, Zhang H, Tang L, Prakash A. Brain chromatin remodeling: a novel mechanism of alcoholism. J Neurosci. 2008;28:3729–3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Akimaru H, Chen Y, Dai P, et al. Drosophila CBP is a co-activator of cubitus interruptus in hedgehog signalling. Nature. 1997;386:735–738. [DOI] [PubMed] [Google Scholar]
- 78. Treistman SN, Martin GE. BK Channels: mediators and models for alcohol tolerance. Trends Neurosci. 2009;32:629–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Smolik SM. Heterochromatin-mediated gene silencing is not affected by Drosophila CBP activity. J Hered. 2009;100:465–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Lloret-Llinares M, Carre C, Vaquero A, de Olano N, Azorin F. Characterization of Drosophila melanogaster JmjC+N histone demethylases. Nucleic Acids Res. 2008;36:2852–2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Yamane K, Toumazou C, Tsukada Y, et al. JHDM2A, a JmjC-containing H3K9 demethylase, facilitates transcription activation by androgen receptor. Cell. 2006;125:483–495. [DOI] [PubMed] [Google Scholar]
- 82. Herz HM, Morgan M, Gao X, et al. Histone H3 lysine-to-methionine mutants as a paradigm to study chromatin signaling. Science. 2014;345:1065–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Secombe J, Li L, Carlos L, Eisenman RN. The Trithorax group protein Lid is a trimethyl histone H3K4 demethylase required for dMyc-induced cell growth. Genes Dev. 2007;21:537–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. DiTacchio L, Le HD, Vollmers C, et al. Histone lysine demethylase JARID1a activates CLOCK-BMAL1 and influences the circadian clock. Science. 2011;333:1881–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Tarayrah L, Li Y, Gan Q, Chen X. Epigenetic regulator Lid maintains germline stem cells through regulating JAK-STAT signaling pathway activity. Biol Open. 2015;4:1518–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Morán T, Bernués J, Azorín F. The Drosophila histone demethylase dKDM5/LID regulates hematopoietic development. Dev Biol. 2015;405:260–268. [DOI] [PubMed] [Google Scholar]
- 87. Li L, Greer C, Eisenman RN, Secombe J. Essential functions of the histone demethylase lid. Plos Genet. 2010;6:e1001221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Di Stefano L, Walker JA, Burgio G, et al. Functional antagonism between histone H3K4 demethylases in vivo. Genes Dev. 2011;25:17–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Liefke R, Oswald F, Alvarado C, et al. Histone demethylase KDM5A is an integral part of the core Notch-RBP-J repressor complex. Genes Dev. 2010;24:590–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. The UniProt Consortium. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2018;46:2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Curtis BJ, Zraly CB, Marenda DR, Dingwall AK. Histone lysine demethylases function as co-repressors of SWI/SNF remodeling activities during Drosophila wing development. Dev Biol. 2011;350:534–547. [DOI] [PubMed] [Google Scholar]
- 92. Mathies LD, Blackwell GG, Austin MK, et al. SWI/SNF chromatin remodeling regulates alcohol response behaviors in Caenorhabditis elegans and is associated with alcohol dependence in humans. Proc Natl Acad Sci U S A. 2015;112:3032–3037. [DOI] [PMC free article] [PubMed] [Google Scholar]