Abstract
Aim:
Pathway analysis of gastric atrophy to find new molecular prospective of disease.
Background:
Gastric atrophy as a process which is accompanied with “loss of glans” in stomach can be considered as a risk factor of gastric cancer. Here, the correlated biochemical pathways to the disorder have been analyzed via protein-protein interaction (PPI) network analysis.
Methods:
The genes related to gastric atrophy were retrieved by STRING database and organized in a network by Cytoscape. Three significant clusters were determined by ClusterONE plug-in of Cytoscape. The elements of cluster-2 which contained all central nodes of the network were enriched by ClueGO and the biochemical pathways discussed in details.
Results:
The number of seven central nodes (which are included in cluster-2); INS, TP53, IL6, TNF, SRC, MYC, and IL8 were identified. The biochemical pathways related to the elements of cluster-2 were determined and clustered in nine groups. The pathways were discussed in details.
Conclusion:
Pathway analysis indicates that the introduced central genes of the network can be considered as biomarkers of gastric atrophy.
Key Words: Gastric atrophy, Network analysis, Gene, Biochemical pathway, Central node.
Introduction
Gastric atrophy is characterized with progressive decrease of glandular structures accompanied with mucosa changes to atrophic (1). Gastric atrophy is recorded as “loss of glands” in stomach. This process may follow ulceration accompanied with damage of glandular layer. In another case which occurs more frequently; it results from a prolonged inflammation. In additional definition atrophy is considered as “loss of specialized cells” (2). There are evidences that gastric atrophy can be considered as risk factor of cancer. It is an important state which causes pathologically gastric cancer. Gastric atrophy diagnosis is improved by endoscopy (especially by new criteria of this method) (3). Variety of studies revealed molecular aspects of gastric atrophy. The role of different genes such as interleukin-2, cyclooxygenase-2 and several genes in the gastric atrophy especially in Helicobacer pylori inducing disease, is investigated and highlighted (4, 5). Helicobacter pylori infection is mentioned as one of important causes of gastric atrophy (6, 7).
Different diseases are assessed via PPI network analysis to discover new molecular aspect of disorder (8-11). In this approach all reported genes or other molecular reagents such as proteins or metabolites which are correlated to the disease are collected and organized in an interactome which is an integrated and interacted unit (12, 13). Based on diverse roles which the elements play in PPI network, they rank and screen (14). Variety of factors are introduced for screening which central parameters such as degree and betweenness are the important ones (15). In the other hand GO analysis is a useful tool to find terms such as biological processes, molecular function, cellular component, and biochemical pathways related to the query genes (16). The identified terms can be clustered by using statistical parameters (17). In this tactic new molecular prospective of disorder will be represented (18). In the present study the genes related to the gastric atrophy are retrieved from STRING database and organized in the PPI network. The network analyzed and the central genes determined and enriched to determine the related biochemical pathways. The pathways discussed to present new views of gastric atrophy.
Methods
The recorded genes in STRING database up dated at 2017(19) related to gastric atrophy from its disease query were retrieved. The genes interacted by Cytoscape software version 3.6.0 (20) to construct an interactome. The PPI network was analyzed by network analyzer a plug-in of Cytoscape and the topological properties of the network were assessed. Two important central parameters (degree and betweenness centrality) were considered for screening of the nodes. The top nodes based on degree (degree more than mean + 2SD) were selected as hub-nodes and the nodes characterized as top 5% based on betweenness were determined as bottleneck-nodes (12, 17). The common hub and bottleneck nodes were identified as hub-bottleneck nodes. Cluster ONE (21, 22) application of Cytoscape was used to determined significant clusters of the network and P-value less than 5% was considered. The elements of the main cluster enriched via gene ontology by ClueGO (v. 2.5.0) plug-in of Cytoscape (23, 24). Related biochemical pathways to the nodes of main cluster were determined; at least three genes per term and three percent attribution in the term were considered. The terms were grouped based on kappa score (25). For better discussion restricted condition including at least seven genes per term and five percent attribution in the term were considered. The terms were discussed in details.
Results
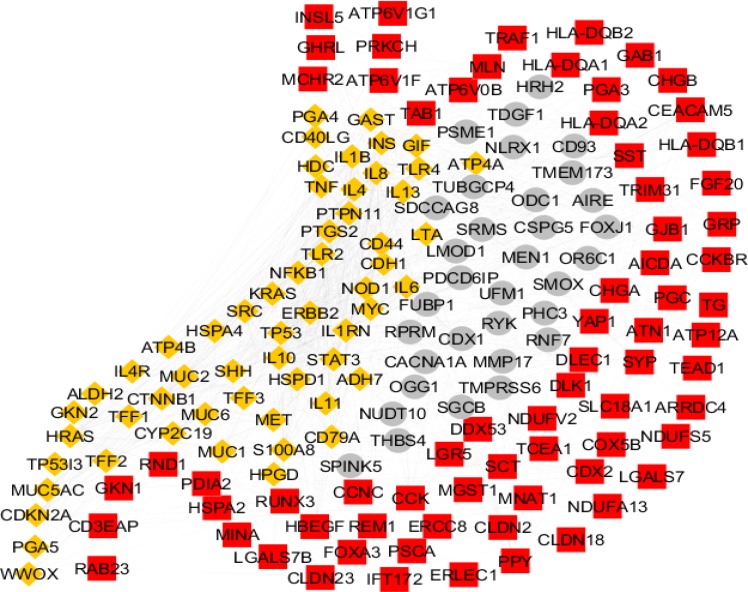
There are 206 genes related to gastric atrophy. As it is shown in the figure 1 the numbers of 161 genes among them are included in the PPI network and the others (45 genes) were isolated. The genes are organized in three clusters including cluster-1 (the red color nodes), cluster-2 (the yellow colored genes), and cluster-3 (the grey colored nodes). The clusters 1-3 contains 71, 57, and 33 nodes, respectively. The network was analyzed and the hub and bottleneck nodes were determined. The common hub and bottleneck nodes including INS, TP53, IL6, TNF, SRC, MYC, and IL8 were identified as hub-bottleneck nodes (see table 1). Degree distribution was fitted in a power law line with y=axb which a and b are equal to 25.745 and -0.840, respectively. Correlation and R-squared (computed on logarithmized values) are 0.922 and 0.749, respectively. Therefore, the network is a scale free network.
Figure 1.
The number of 206 genes related to gastric athrophy were organized in the PPI network. The network was included a main connected component containing 161 nodes and 45 isolated nodes. Confidence score was 0.4. The main connected component was included cluster-1, 2, and 3 which are colored in red, yellow, and grey respectively
Table 1.
Ten hub-nodes of the analyzed network are presented. There are eight bottleneck nodes which seven of them are hub-nodes (see the bold hub-nodes) and another one is GAST. BC, D, and DS are betweenness centrality, degree, and disease score respectively
| R | name | description | BC | D | DS |
|---|---|---|---|---|---|
| 1 | INS | insulin | 0.15 | 62 | 0.67 |
| 2 | TP53 | tumor protein p53 | 0.18 | 59 | 1.57 |
| 3 | IL6 | interleukin 6 (interferon, beta 2) | 0.04 | 51 | 0.59 |
| 4 | TNF | tumor necrosis factor | 0.05 | 48 | 1.11 |
| 5 | SRC | v-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (avian) | 0.09 | 48 | 0.98 |
| 6 | MYC | v-myc myelocytomatosis viral oncogene homolog (avian) | 0.06 | 47 | 0.85 |
| 7 | IL8 | interleukin 8 | 0.05 | 45 | 1.58 |
| 8 | PTGS2 | prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) | 0.04 | 43 | 1.29 |
| 9 | HRAS | v-Ha-ras Harvey rat sarcoma viral oncogene homolog | 0.02 | 39 | 0.56 |
| 10 | NFKB1 | nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 | 0.02 | 38 | 0.63 |
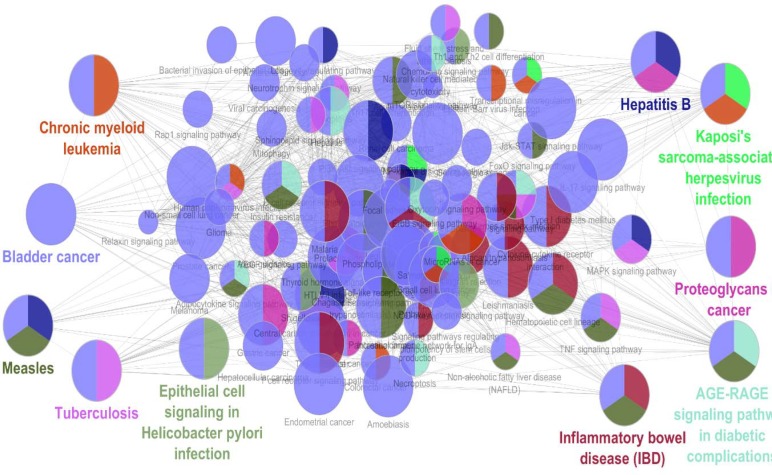
Due to presence of all hub-bottleneck nodes in the cluster-2, the elements of cluster-2 were enriched by ClueGO v. 2.5.0. The related chemical pathways were extracted via KEGG and are presented in the figure 2. More restrictions (presence of at least seven genes in term and 5% attribution of genes in the term) were considered and the finding were tabulated in the tables 2 and 3. The number of 31 terms were organized in nine groups including: Kaposis sarcoma associated herpesvirus infection, AGE-RAGE signaling pathway in diabetic complication, Hepatitis B, Proteoglycans in cancer, Chornic myeliod leukemia, Tuberculosis, Herpes simplex infection, Inflammatory bowel disease (IBD), Measles, and bladder cancer.
Figure 2.
The elements of cluster-2 are enriched by ClueGO v. 2.5.0. The biochemical pathways are extracted from KEGG_20.11.2017: 7282. The number of 71 terms are organized in the 10 groups based on kappa score. Presence of at least three genes/term and 3% genes/term were considered. The groups name is shown in different colors
Table 2.
The number of 18 terms among 31 resulted terms from enrichment of elements of cluster-2 which are organized in the eight groups are presented. P-value≤0.05, 7≤genes/term, and 5≤%genes/term. The underlined number refers to repeat of term. The underlined number corresponding to repeat of the term
| R | Terms | Genes/Term | %Genes/Term |
| 1 | Kaposis sarcoma associated herpesvirus infection | 11 | 6 |
| 2 | AGE-RAGE signaling pathway in diabetic complication | 8 | 8 |
| 3 | Toxoplasmosis | 7 | 6 |
| 4 | Hepatitis C | 7 | 5 |
| 5 | Cellular senescence | 8 | 5 |
| 6 | Hepatitis B | 12 | 8 |
| 7 | Measles | 9 | 7 |
| 8 | Hepatitis B_1 | 12 | 8 |
| 9 | Proteoglycans in cancer | 15 | 7 |
| 10 | Gastric cancer | 10 | 7 |
| 11 | Cellular senescence_1 | 8 | 5 |
| 12 | Kaposis sarcoma associated herpesvirus infection_1 | 11 | 6 |
| 13 | Chornic myeliod leukemia | 7 | 9 |
| 14 | Tuberculosis | 9 | 6 |
| 15 | Herpes simplex infection | 8 | 5 |
| 16 | Cytokin-cytokin receptor infection | 12 | 5 |
| 17 | Hematopoietic cell lineage | 7 | 8 |
| 18 | T cell receptor signaling pathway | 7 | 8 |
| 19 | Leishmaniasis | 8 | 12 |
| 20 | Malaria | 9 | 19 |
| 21 | Inflammatory boweldisease (IBD) | 11 | 18 |
| 22 | Hematopoietic cell lineage_1 | 7 | 8 |
| 23 | AGE-RAGE signaling pathway in diabetic complication_1 | 8 | 9 |
| 24 | Measles_1 | 9 | 8 |
| 25 | Herpes simplex infection_1 | 8 | 6 |
| 26 | Inflammatory boweldisease (IBD)_1 | 11 | 18 |
Table 3.
The number of 13 terms among 31 resulted terms from enrichment of elements of cluster-2 which are organized in one group are presented. P-value≤0.05, 7≤genes/term, and 5≤%genes/term. The underlined number refers to repeat of term. The repeated terms are shown with underlined numbers
| R | Terms | Genes/Term | %Genes/Term |
|---|---|---|---|
| 1 | NF-Kappa B signaling pathway | 8 | 8 |
| 2 | Cellular senescence_2 | 8 | 5 |
| 3 | Toll-like receptor signaling pathway | 7 | 7 |
| 4 | Jak-STAT signaling pathway | 10 | 6 |
| 5 | Hematopoietic cell lineage_2 | 7 | 7 |
| 6 | IL-17 signaling pathway | 10 | 11 |
| 7 | T cell receptor signaling pathway_1 | 7 | 7 |
| 8 | AGE-RAGE signaling pathway in diabetic complication_2 | 8 | 8 |
| 9 | Pertussis | 8 | 11 |
| 10 | Legionellosis | 8 | 14 |
| 11 | Leishmaniasis_1 | 8 | 11 |
| 12 | Chagas disease (American trypanosomiasis) | 8 | 8 |
| 13 | Malaria_1 | 9 | 17 |
| 14 | Toxoplasmosis_1 | 7 | 6 |
| 15 | Amoebiasis | 9 | 9 |
| 16 | Tuberculosis_1 | 9 | 5 |
| 17 | Hepatitis C_1 | 7 | 5 |
| 18 | Hepatitis B_2 | 12 | 8 |
| 19 | Measles_2 | 9 | 7 |
| 20 | Kaposis sarcoma associated herpes virus infection_2 | 11 | 6 |
| 21 | Proteoglycans in cancer_1 | 15 | 7 |
| 22 | Endometrial cancer | 7 | 12 |
| 23 | Prostate cancer | 7 | 7 |
| 24 | Bladder cancer | 9 | 22 |
| 25 | Chronic myeloid leukemia | 7 | 9 |
| 26 | Gastric cancer_1 | 10 | 6 |
| 27 | Inflammatory bowel disease (IBD)_2 | 11 | 17 |
| 28 | Rheumatoid arthritis | 7 | 8 |
Discussion
As it is shown in the figure 1 the numbers of 161 genes are organized in a scale free network. It is possible that several genes among them play critical roles in the network. Presence of limited numbers of crucial genes in network (the seven hub-bottleneck nodes which are shown in table 1) is corresponded to successful screening of the genes by network analysis. Surprisingly, all of the critical nodes are presented in the cluster-2. Since this cluster includes 57 nodes, it seems these genes play central role in the network. There is evidence that the elements of a cluster are related to each other and manage similar functions. Based on this object the mentioned 57 genes were enriched and the biochemical pathways were identified. The results of enrichment and more restricted analysis are shown in the figure 2 and tables 2 and 3, respectively. The 31 terms which organized in the nine groups were introduced. Previous studies indicate that understanding of the related biochemical pathways is useful to know mechanism of disorder. In the following part, the roles of these highlighted groups in gastric atrophy are discussed.
The biggest group which includes 28 terms is represented as bladder cancer cluster. This collection contains 9o% of the identified total terms. It is reported that in the case of gastritis due to Helicobacter pylori infection NF-κB activity increases in the epithelial cells (26). Based on literature; inhibition of FoxM1 hints to cellular Senescence. FoxM1 expression increases in gastric cancer (27). The role of Toll-like receptors in gastric cancer and gastric atrophy is discussed in details (28). Assessment of role of inflammatory factor IL-17 by T helper cells and Jak-STAT signaling pathway in gastritis indicates that these two agents are related to gastric tumourigenesis (29). Ohnishi et al. reported occurrence of gastrointestinal and hematopoietic neoplasms after infection with Helicobacter pylori (30). It is reported that after gastric ulcer healing RAGE mRNA expression is increased (31). As it is presented in the figure 2, this pathway is related to the bladder cancer and measles. Role of Helicobacter pylori in inflammatory bowel disease is confirmed (32). In the other hand, this microorganism is the main mediator of gastric atrophy. A relationship between Bordetella pertussis toxin secretion protein and Helicobacter pylori secretion is found (33). There is evidence that sever malaria infection leads to increased gastroduodenal permeability band hepatitis C virus infection is detected and discussed (34). As it is reported, glypcian-3 which is a tumor suppressor gene belongs to proteoglycan family. Down-regulation of this gene in gastric cancer is described (35, 36). Frequent connections between the introduced pathways in figure 2, indicate that all of them are related directly or indirectly to the gastric atrophy. If this statement be accepted, it can be concluded that cluster-2 is a right core of the main genes which are involved in the disease. As it is mentioned in the result section, this cluster contains the seven central genes. Therefore it is a logical conclusion that the central genes are a suitable biomarker panel for gastric atrophy. Presence of IL-6 and IL-8 as two inflammatory reagents beside TP53 and TNF as two well-known tumor indicators in the introduced biomarker panel, are corresponding to the complex nature of gastric atrophy.
Biochemical pathway analysis of gastric atrophy represents a new prospective of molecular mechanism of disorder. The related biomarkers were proposed which are corresponded to the roles of inflammation and cancerous pathways in this disease. Based on the findings more investigation can lead to introduce safe diagnostic method for the disease instead of endoscopy.
Acknowledgment
This project is supported by Shahid Beheshti University of Medical Sciences.
Conflict of interests
The authors declare that they have no conflict of interest.
References
- 1.Genta RM. Review article: Gastric atrophy and atrophic gastritis—nebulous concepts in search of a definition. Aliment Pharmacol Ther. 1998;12S:17–23. doi: 10.1111/j.1365-2036.1998.00003.x. [DOI] [PubMed] [Google Scholar]
- 2.Dixon MF. Prospects for intervention in gastric carcinogenesis: reversibility of gastric atrophy and intestinal metaplasia. Gut. 2001;49:2–4. doi: 10.1136/gut.49.1.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nomura S, Ida K, Terao S, Adachi K, Kato T, Watanabe H, et al. Research Group for Establishment of Endoscopic Diagnosis of Chronic Gastritis Endoscopic diagnosis of gastric mucosal atrophy: multicenter prospective study. Dig Endosc. 2014;26:709–19. doi: 10.1111/den.12286. [DOI] [PubMed] [Google Scholar]
- 4.Togawa S, Joh T, Itoh M, Katsuda N, Ito H, Matsuo K, et al. Interleukin-2 gene polymorphisms associated with increased risk of gastric atrophy from Helicobacter pylori infection. Helicobacter. 2005;10:172–8. doi: 10.1111/j.1523-5378.2005.00308.x. [DOI] [PubMed] [Google Scholar]
- 5.Pereira C, Sousa H, Ferreira P, Fragoso M, Moreira-Dias L, Lopes C, et al. -765G > C COX-2 polymorphism may be a susceptibility marker for gastric adenocarcinoma in patients with atrophy or intestinal metaplasia. World J Gastroenterol. 2006;12:5473–8. doi: 10.3748/wjg.v12.i34.5473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ye W, Held M, Lagergren J, Engstrand L, Blot WJ, McLaughlin JK, et al. Helicobacter pylori infection and gastric atrophy: risk of adenocarcinoma and squamous-cell carcinoma of the esophagus and adenocarcinoma of the gastric cardia. J Natl Cancer Inst. 2004;96:388–96. doi: 10.1093/jnci/djh057. [DOI] [PubMed] [Google Scholar]
- 7.Beales IL, Crabtree JE, Scunes D, Covacci A, Calam J. Antibodies to CagA protein are associated with gastric atrophy in Helicobacter pylori infection. Eur J Gastroenterol Hepatol. 1996;8:645–9. [PubMed] [Google Scholar]
- 8.Zamanian-Azodi M, Rezaei-Tavirani M, Rahmati-Rad S, Hasanzadeh H, Rezaei Tavirani M, Seyyedi SS. Protein-Protein Interaction Network could reveal the relationship between the breast and colon cancer. Gastroenterol Hepatol Bed Bench. 2015;8:215–24. [PMC free article] [PubMed] [Google Scholar]
- 9.Zali H, Rezaei Tavirani M. Meningioma protein-protein interaction network. Arch Iran Med. 2014;17:262–72. [PubMed] [Google Scholar]
- 10.Safaei A, Rezaei Tavirani M, Arefi Oskouei A, Zamanian Azodi M, Mohebbi SR, Nikzamir AR. Protein-protein interaction network analysis of cirrhosis liver disease. Gastroenterol Hepatol Bed Bench. 2016;9:114–23. [PMC free article] [PubMed] [Google Scholar]
- 11.Goehler H, Lalowski M, Stelzl U, Waelter S, Stroedicke M, Worm U, et al. A protein interaction network links GIT1, an enhancer of huntingtin aggregation, to Huntington's disease. Mol Cell. 2004;15:853–65. doi: 10.1016/j.molcel.2004.09.016. [DOI] [PubMed] [Google Scholar]
- 12.Safari-Alighiarloo N, Taghizadeh M, Tabatabaei SM, Shahsavari S, Namaki S, Khodakarim S, et al. Identification of new key genes for type 1 diabetes through construction and analysis of protein-protein interaction networks based on blood and pancreatic islet transcriptomes. J Diabetes. 2017;9:764–77. doi: 10.1111/1753-0407.12483. [DOI] [PubMed] [Google Scholar]
- 13.Barabási AL, Gulbahce N, Loscalzo J. Network medicine: a network-based approach to human disease. Nat Rev Genet. 2011;12:56–68. doi: 10.1038/nrg2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rezaei-Tavirani M, Zamanian-Azodi M, Rajabi S, Masoudi-Nejad A, Rostami-Nejad M, Rahmatirad S. Protein Clustering and Interactome Analysis in Parkinson and Alzheimer's Diseases. Arch Iran Med. 2016;19:101–9. [PubMed] [Google Scholar]
- 15.Abbaszadeh HA, Peyvandi AA, Sadeghi Y, Safaei A, Zamanian-Azodi M, Khoramgah MS, et al. Er:YAG Laser and Cyclosporin A Effect on Cell Cycle Regulation of Human Gingival Fibroblast Cells. J Lasers Med Sci. 2017;8:143–9. doi: 10.15171/jlms.2017.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rezaei-Tavirani M, Rezaei-Tavirani M, Mansouri V, Mahdavi SM, Valizadeh R, Rostami-Nejad M, et al. Introducing crucial protein panel of gastric adenocarcinoma disease. Gastroenterol Hepatol Bed Bench. 2017;10:21–8. [PMC free article] [PubMed] [Google Scholar]
- 17.Safari-Alighiarloo N, Rezaei-Tavirani M, Taghizadeh M, Tabatabaei SM, Namaki S. Network-based analysis of differentially expressed genes in cerebrospinal fluid (CSF) and blood reveals new candidate genes for multiple sclerosis. PeerJ. 2016;4:e2775. doi: 10.7717/peerj.2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen L, Zhang YH, Zheng M, Huang T, Cai YD. Identification of compound-protein interactions through the analysis of gene ontology, KEGG enrichment for proteins and molecular fragments of compounds. Mol Genet Genomics. 2016;291:2065–79. doi: 10.1007/s00438-016-1240-x. [DOI] [PubMed] [Google Scholar]
- 19.Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:D362–8. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhi J, Sun J, Wang Z, Ding W. Support vector machine classifier for prediction of the metastasis of colorectal cancer. Int J Mol Med. 2018;41:1419–26. doi: 10.3892/ijmm.2018.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheng L, Liu P, Leung KS. SMILE: a novel procedure for subcellular module identification with localisation expansion. IET Syst Biol. 2018;12:55–61. doi: 10.1049/iet-syb.2017.0085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maddi AMA, Eslahchi C. Discovering overlapped protein complexes from weighted PPI networks by removing inter-module hubs. Sci Rep. 2017;7:3247. doi: 10.1038/s41598-017-03268-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, et al. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25:1091–3. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mlecnik B, Galon J, Bindea G. Comprehensive functional analysis of large lists of genes and proteins. J Proteomics. 2018;171:2–10. doi: 10.1016/j.jprot.2017.03.016. [DOI] [PubMed] [Google Scholar]
- 25.Rezaei-Tavirani M, Rezaei-Tavirani S, Ahmadi N, Naderi N, Abdi S. Pancreatic adenocarcinoma protein-protein interaction network analysis. Gastroenterol Hepatol Bed Bench. 2017;10:S85–92. [PMC free article] [PubMed] [Google Scholar]
- 26.Tak PP, Firestein GS. NF-kappaB: a key role in inflammatory diseases. J Clin Invest. 2001;107:7–11. doi: 10.1172/JCI11830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zeng J, Wang L, Li Q, Li W, Björkholm M, Jia J, et al. FoxM1 is up-regulated in gastric cancer and its inhibition leads to cellular senescence, partially dependent on p27 kip1. J Pathol. 2009;218:419–27. doi: 10.1002/path.2530. [DOI] [PubMed] [Google Scholar]
- 28.Hishida A, Matsuo K, Goto Y, Mitsuda Y, Hiraki A, Naito M, et al. Toll-like receptor 4 +3725 G/C polymorphism, Helicobacter pylori seropositivity, and the risk of gastric atrophy and gastric cancer in Japanese. Helicobacter. 2009;14:47–53. doi: 10.1111/j.1523-5378.2009.00659.x. [DOI] [PubMed] [Google Scholar]
- 29.Kennedy CL, Najdovska M, Jones GW, McLeod L, Hughes NR, Allison C, et al. The molecular pathogenesis of STAT3-driven gastric tumourigenesis in mice is independent of IL-17. J Pathol. 2011;225:255–64. doi: 10.1002/path.2933. [DOI] [PubMed] [Google Scholar]
- 30.Ohnishi N, Yuasa H, Tanaka S, Sawa H, Miura M, Matsui A, et al. Transgenic expression of Helicobacter pylori CagA induces gastrointestinal and hematopoietic neoplasms in mouse. Proc Natl Acad Sci U S A. 2008;105:1003–8. doi: 10.1073/pnas.0711183105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Litwinoff E, Hurtado Del Pozo C, Ramasamy R, Schmidt AM. Emerging Targets for Therapeutic Development in Diabetes and Its Complications: The RAGE Signaling Pathway. Clin Pharmacol Ther. 2015;98:135–44. doi: 10.1002/cpt.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luther J, Dave M, Higgins PD, Kao JY. Association between Helicobacter pylori infection and inflammatory bowel disease: a meta-analysis and systematic review of the literature. Inflamm Bowel Dis. 2010;16:1077–84. doi: 10.1002/ibd.21116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tummuru MK, Sharma SA, Blaser MJ. Helicobacter pylori picB, a homologue of the Bordetella pertussis toxin secretion protein, is required for induction of IL-8 in gastric epithelial cells. Mol Microbiol. 1995;18:867–76. doi: 10.1111/j.1365-2958.1995.18050867.x. [DOI] [PubMed] [Google Scholar]
- 34.Wilairatana P, Meddings JB, Ho M, Vannaphan S, Looareesuwan S. Increased gastrointestinal permeability in patients with Plasmodium falciparum malaria. Clin Infect Dis. 1997;24:430–5. doi: 10.1093/clinids/24.3.430. [DOI] [PubMed] [Google Scholar]
- 35.Rahbari M, Pecqueux M, Reissfelder C, Welsch T, Weitz J, Rahbari NN, et al. Exosomal Glypican-3 is a diagnostic and prognostic biomarker in gastric cancer. Z Gastroenterol. 2017;55:e57–299. [Google Scholar]
- 36.Wang S, Wu Z, Zhou M, Liao W. Effect of GPC1 on epithelial-to-mesenchymal transition and stemness and interaction with ITGB1 in gastric cancer. J Clin Oncol. 2017;35:e15580. [Google Scholar]