Abstract
Sensitive and specific detection of pathogens via nucleic acid amplification is currently constrained to laboratory settings and portable equipment with costly fluorescent detectors. Nucleic acid-detecting lateral flow immunoassay strips (LFIAs) offer a low-cost visual transduction strategy at points of need. Unfortunately, these LFIAs frequently detect amplification byproducts that can yield spurious results which can only be deciphered through statistical analysis. We integrated customizable strand displacement probes into standard loop mediated isothermal amplification (LAMP) assays to prevent byproduct capture on commercial LFIAs. We find that combining strand displacement with LAMP (SD-LAMP) yields LFIA test band intensities that can be unequivocally interpreted by human subjects without additional instrumentation, thereby alleviating the need for a portable reader’s analysis. Using SD-LAMP, we capture target amplicons on commercially available LFIAs from as few as 3.5 Vibrio cholerae and 2 750 Escherichia coli bacteria without false positive or false negative interpretation. Moreover, we demonstrate that LFIA capture of SD-LAMP products remain specific even in the presence of complex sample matrixes, providing a significant step toward reliable instrument-free pathogen detection outside of laboratories.
Isothermal nucleic acid amplification techniques, such as loop mediated amplification (LAMP), are promising methods for point-of-need pathogen identification because they can efficiently amplify targets without the expensive thermal cycling equipment of conventional PCR.1−3 LAMP can amplify 10 ng of nucleic acids within 5–10 min4 and from complex patient,5,6 environmental,7 and food sample matrixes.8 Despite the minimal infrastructure required to amplify a pathogenic gene target via LAMP, additional instrumentation is typically required to both sensitively and specifically visualize amplicons which limits the usage of LAMP outside of a laboratory.
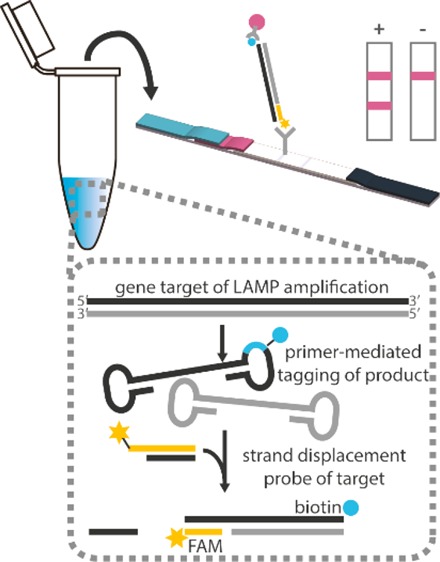
Minimal instrumentation techniques frequently reported for LAMP amplicon visualization, such as intercalating fluorophores, detecting pyrophosphate byproducts,2,9,10 and staining with crystal violet dye11 or pH-sensitive dyes12 all nonspecifically transduce signals from both off-target and target amplicons. Similarly problematic, previously published methods demonstrate amplicon detection by labeling primers for capture on a low-cost lateral flow immunoassay (LFIA), risking capture of primer dimers and spurious amplicons.13−15 The false positive results endemic to these nonspecific signal transduction methods are typically mitigated by redesigning the primer sets or reducing reaction time, which also decreases assay sensitivity. Alternatively, target specific detection of LAMP amplicons can be achieved with hybridization of incorporated primers,16 polymerase-mediated strand displacement of duplexed primers,17 or toehold-mediated strand displacement events,18 but all have required an excited fluorophore and fluorescent reader for signal transduction. Du et al. have recently alleviated the need for a lab-based fluorescent reader by coupling target-specific strand exchange reactions to a home-pregnancy test, an inexpensive and ubiquitous lateral flow immunoassay (LFIA) strip, for visual detection.19 However, multiple additional user steps and reagents to complete this signal transduction complicated the detection and could increase interpretation error. In this work, we leverage the specificity of a strand displacement probe to tag amplified nucleic acids in a one-pot reaction mixture. This probe allows direct capture of the reaction products on a commercial LFIA for visual detection.
To demonstrate specific detection of LAMP amplicons, we integrated tagged strand displacement probes into LAMP assays (which we subsequently call SD-LAMP) for the detection of Escherichia coli and Vibrio cholerae (Figure 1) in human and environmental sample matrixes. A Mastermix of LAMP reagents, strand displacement probes, and bacteria were isothermally heated to simultaneously lyse bacteria and amplify and tag the target DNA. After an hour of heating, reaction products were deposited onto a commercial LFIA which yield colorimetric bands within 30 min to indicate the presence or absence of target DNA. We further evaluated the unaided reading of LFIA results by human subjects, determining LFIA test band intensities required for definitive interpretation. We demonstrated that while LAAMP assays themselves were specific, the LFIA readout of conventional LAMP assays without strand displacement probes yielded results that were difficult to interpret by untrained and even trained users. Rather than redesign the entire primer sets, we found the use of strand displacement probes greatly improved interpretability of LFIAs by increasing the intensity of positive test bands and preventing primer dimerization that yielded false positive test bands. Moreover, the modified amplification and detection assays performed robustly even when diluted in complex matrixes such as pond water and human blood plasma.
Figure 1.
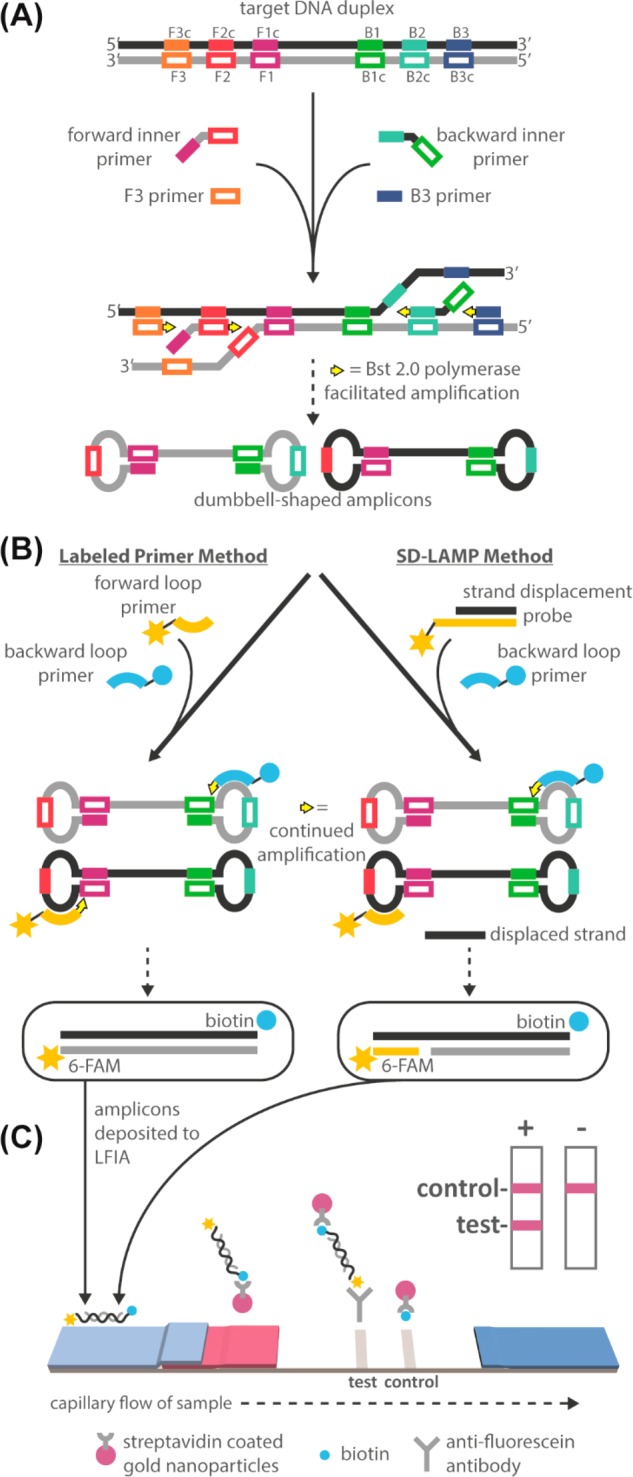
Schematic of standard LAMP and SD-LAMP reactions and their subsequent LFIA detection. (A) The noncyclic step of both reactions in which F3, B3, and inner primers bind to a double-stranded target sequence and polymerase generates dumbbell-shaped products. (B) Dumbbell-shaped products enter the cyclic amplification step during which loop primers accelerate amplification. Products can be labeled by either standard tagging of each loop primer or by SD-LAMP, which uses one labeled loop primer along with a tagged strand displacement probe that hybridizes to the amplicons’ loop region. (C) Labeled amplicons visualized on a standard LFIA strip.
Experimental Section
Bacterial Cell Culture
E. coli strain DH5α (NEB, Ipswich, MA) and V. cholerae strain N16961 (generously donated by Dr. Afsar Ali at the University of Florida) were grown overnight at 37 °C at 350 rpm (Thermo Fisher, Waltham, MA) in lysogeny broth (LB) media. Cultures were diluted in LB media to an OD600 (Ultrospec 10, Biochrom, Cambourne, U.K.) of 1.0, representing 5.5 × 109 cells/mL of E. coli or 7.0 × 109 cells/mL of V. cholerae as determined by fluorescence microscopy of serially diluted cells or counting colony forming units of serially diluted cells, respectively.
Reagents
Oligonucleotide primers and probes were ordered from Integrated DNA Technology (IDT, Coralville, IA) and are summarized in Table S-2. Oligonucleotides were resuspended in DNase/RNase-free DI water. Note that LF and LB primers are tagged with either 6-carboxyfluorescein (FAM) or biotin for capture on commercial lateral flow immunoassay strips. Local pond water was collected and then stored at 4 °C until use. Citrated human whole-blood (catalog no. IPLA-WB1, Innovative Research, Novi, MI) was separated into plasma by centrifugation at 1 000g for 10 min at 4 °C (Eppendorf Centrifuge 5424R, Hamburg, Germany). Whole blood was stored at 4 °C and used within 2 weeks; plasma was stored at −20 °C until use.
Standard LAMP Reaction of E. coli
In total, 5 μL of E. coli culture diluted in LB media, pond water, or plasma was added to 20 μL reaction mixtures containing 0.3 μM each F3 and B3 primers, 2.4 μM each FIP and BIP primers, 1.2 μM each LF and LB primers, 1100 mM betaine, 1.4 mM dNTPs, 1× Isothermal Buffer, and 8U Bst 2.0 polymerase. Reactions were incubated at 65 °C for 1 h followed by a 1 min 95 °C denaturing step to prevent further amplification (7500 RT-PCR System, Applied Biosystems, Foster City, CA) and then kept at 4 °C until analyzed.
Standard LAMP Reaction of V. cholerae
In total, 5 μL of V. cholerae culture diluted in LB media or pond water was added to 20 μL reaction mixtures containing 0.2 μM each F3 and B3 primers, 1.6 μM each FIP, BIP, LF, and LB primers, 800 mM betaine, 1.4 mM dNTPs, 1× Isothermal Buffer, and 8U Bst 2.0 polymerase. Reactions were incubated at 65 °C for 1 h followed by a 1 min 95 °C denaturing step to prevent further amplification using the 7500 RT-PCR System (Applied Biosystems, Foster City, CA) and then kept at 4 °C until analyzed.
SD-LAMP Probe Design
Strand displacement probes were designed to bind to the forward loop region of the malB and ctxA gene targets following previously published design considerations,18 in order to detect only the target amplification products (Table S-2). We aimed for a free energy change of −18 kcal/mol at the probes’ duplex region, which constrained both probes’ binding sites to the location of the forward loop primers. Probes were tagged with 6-FAM and preannealed to shorter complementary strands tagged with a quencher for real-time amplification verification (data not shown). All 3′-OH groups were blocked against polymerization by including a 3′ inverted dT group. The short, single stranded regions in the FAM-labeled probe strands were designed to act as a toehold to initiate strand displacement (Figure 1A) and bind to the amplicon target. The tagged amplicon can then be visualized on an LFIA (Figure 1B).
SD-LAMP Reaction of E. coli
A stock solution of strand displacement probe was prepared by annealing 1 μM of FAM tagged oligonucleotide to 5 μM of quencher tagged oligonucleotide in Isothermal Buffer at 95 °C for 1 min followed by 0.1 °C/s cooling to room temperature. In total, 5 μL of E. coli culture diluted in LB media, pond water, or plasma and 1.25 μL of prepared probe was added to 18.75 μL reaction mixtures containing 0.3 μM each F3 and B3 primers, 2.4 μM each FIP and BIP primers, 1.2 μM LB primers, 1100 mM betaine, 1.4 mM dNTPs, 1× Isothermal Buffer, and 8U Bst 2.0 polymerase. Reactions were incubated at 65 °C for 1 h followed by a 1 min 95 °C denaturing step kept at 4 °C until analysis to prevent further amplification.
SD-LAMP Reaction of V. cholerae
A stock solution of probe was prepared by annealing 1 μM of FAM tagged oligonucleotide to 5 μM of quencher tagged oligonucleotide in Isothermal Buffer at 95 °C for 1 min followed by 0.1 °C/s cooling to room temperature. In total, 5 μL of V. cholerae culture diluted in LB media or pond water and 2.5 μL of prepared probe was added to 17.5 μL reaction mixtures containing 0.2 μM each F3 and B3 primers, 1.6 μM each FIP, BIP, and LB primers, 800 mM betaine, 1.4 mM dNTPs, 1× Isothermal Buffer, and 8U Bst 2.0 polymerase. Reactions were incubated at 65 °C for 1 h followed by a 1 min 95 °C denaturing step to prevent further amplification and then kept at 4 °C until analyzed.
Gel Electrophoresis Characterization and LFIA Detection
In total, 10 μL of reaction mixtures were loaded with an additional 2 μL of 6× Purple Loading Dye (NEB, Ipswich, MA) into a 2% agarose gel containing ethidium bromide and were run at 100–115 V for 45 min. A total of 10 μL of FastDNA ladder was used as a size standard. The resulting gels were imaged with a 15 s exposure (c400, Azure Biosystems, Dublin, CA). In total, 10 μL of reaction mixtures were mixed with 40 μL of commercial LFIA wash buffer and pipetted onto a fluorescein capturing lateral flow immunoassay (catalog no. D003-03, USTAR Biotechnologies, Hangzhou, China). The FAM and biotin tagged nucleic acid amplification products are expected to complex with the streptavidin-conjugated nanoparticles present in the LFIA’s conjugate pad. This complex flows downstream to the test lines where it will be captured by immobilized antifluorescein antibodies. Remaining streptavidin-conjugated nanoparticles will be captured by immobilized antistreptavidin antibodies. The accumulation of visible nanoparticles at the test and control lines will therefore indicate a positive (2 lines) or negative (1 line) result.
Statistical Analysis and Quantification
All strips were scanned at least 30 min after initial sample application on an Epson V850 Pro scanner (Long Beach, CA) to allow tests to dry. Test band intensities were analyzed with a custom MATLAB script that averages the gray scale pixel intensity (to maximize signal-to-noise) and subtracts out average background signal 25 pixels below the test band. The resultant “background subtracted test line intensity” was then calculated as
where 0 represents the lowest possible pixel intensity (i.e., black). Computationally, this yields a much greater dynamic range than physically possible given the saturation limitation of nanoparticles.20 To determine the limit of detection of E. coli and V. cholerae, 4 test strip replicates from each solution’s starting dilution were analyzed for statistical significance (Figures S1–S3 and S5 contain the replicate gels and LFIA strips). The test band intensities were compared to the averaged LFIA experimental negative control test band intensities using a Dunnett’s test with an alpha set to 0.05.
Human Interpretation of LFIA Test Bands
To determine LFIA test bands’ visible threshold for positive interpretation (independent of experimental negative control test band intensities), 18 volunteers between the ages of 18 and 44 years of age with no self-described vision impairments were asked to evaluate 10 test strips of varying test band intensities (Figure 3), three times each and in varying lighting conditions (indoors, outdoors in sun, and outdoors in shade, amounting to 54 distinct observations). The evaluation study was performed in accordance to Purdue University’s Human Research Protection Program. We performed a two-way ANOVA to evaluate confounding effects of test strip or and lighting conditions on test strip order. A Fisher’s Exact Test was used to determine whether previous instruction on reading LFIAs (e.g., in a classroom or previous research setting) affected volunteers’ interpretation.
Figure 3.
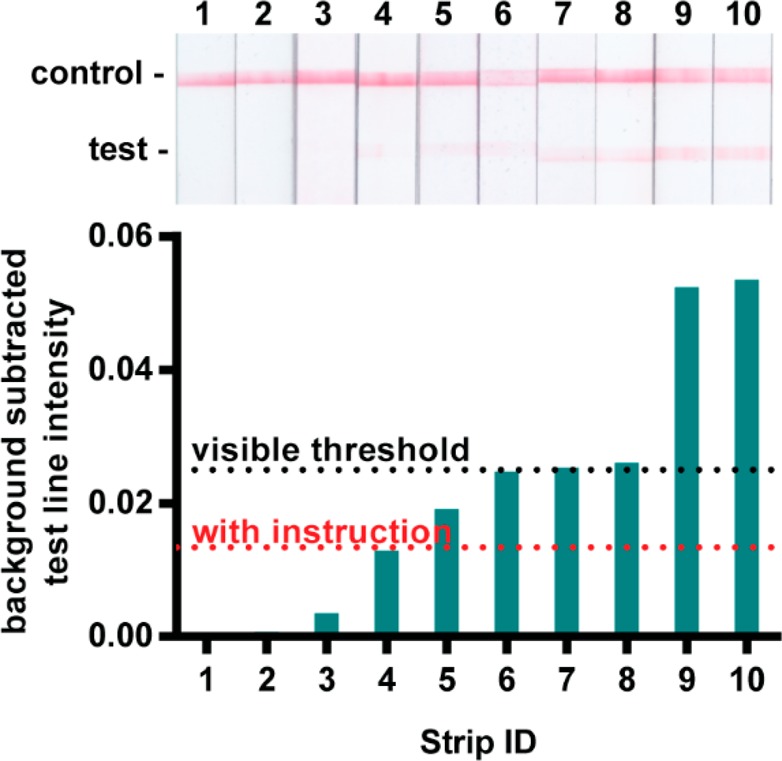
A total of 98 percent of test strips with a background subtracted test intensity of 0.020 are interpreted as positive by unaided human subjects.
Results and Discussion
Detection of Standard LAMP Amplicons on LFIAs
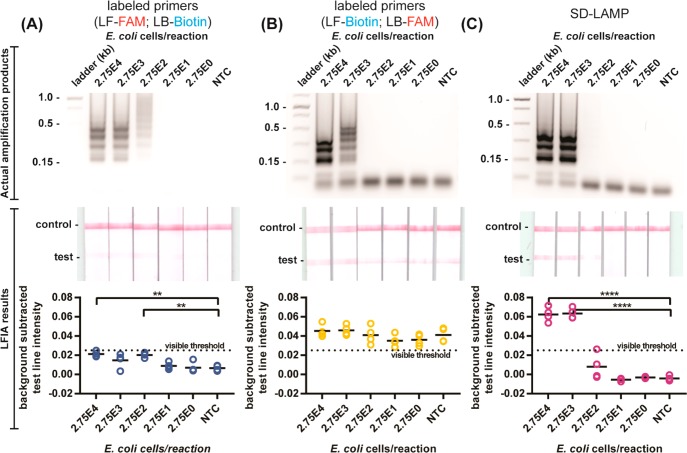
Our initial attempt to capture labeled LAMP amplicons on LFIA strips followed previously published methods for detection of other LAMP amplicon targets using labeled primers but yielded both true positives that had only faint positive test bands as well as false positive results. We tagged the loop primers (forward loop primer with FAM; backward loop primer with biotin) of previously reported LAMP primers designed to amplify the malB gene of E. coli (Table S-1).21 We heated the amplification reactions with serial dilutions of E. coli to simultaneously lyse the bacteria and amplify its DNA. After 1 h of heating, we verified amplification by observing a characteristic ladder-like banding pattern in electrophoresis gels and deposited the reaction products onto LFIA strips. After 30 min, we expected to observe a pink test band, indicating a direct sandwich of the tagged amplicons between streptavidin coated gold nanoparticles and immobilized antifluorescein antibody at the LFIA’s test line (Figure 1B). However, despite observing abundant amplification products by gel electrophoresis from reactions with as few as 275 E. coli cells, we observed only faintly positive test bands in the corresponding LFIA strips (Figure 2A). Only by quantitatively measuring the test band intensity of scanned strips and performing a Dunnett’s multicomparison test were we able to discern positive test bands of reactions with 275 E. coli cells from negative control reactions. Reliance on a computer algorithm to interpret the LFIAs would greatly limit their utility at points of need outside of a laboratory. Therefore, we sought to improve the assay results for instrument-free interpretation.
Figure 2.
Detection of standard LAMP and SD-LAMP reactions in pure water. Electrophoresis gels verifying amplification (top), LFIA test results (middle), and LFIA test line quantification (bottom). (A) Standard LAMP reaction products for E. coli labeled with primers (LF-FAM and LB-Biotin) yield LFIA results too faint for visual interpretation. (B) LAMP products labeled with reconfigured primers (LF-Biotin and LB-FAM) yield false positive LFIA results in low concentration samples and no template control (NTC) reactions. (C) Probed strand displacement LAMP reactions yield no false positive LFIA results for E. coli. n = 4, replicates indicated by each circle. **** indicates p ≤ 0.0001; ** indicates p ≤ 0.01.
Human Subjects’ Visual Interpretation of LFIAs
To inform assay design specifications that would permit human interpretable LFIAs, we studied 18 human subjects’ visual interpretation of LFIA strips. Half of the participants reported having previous instruction on how to read a lateral flow test (e.g., in a classroom or research setting) and the other half of participants reported having no previous instruction. All participants evaluated test strips and recorded whether they interpreted the LFIA to be positive, negative, or invalid when referencing a provided schematic (Figure S-9). A two-way ANOVA test confirmed that neither the order of test strip presentation nor the lighting conditions had confounding effects on test result interpretation. Test strips with a background-subtracted test band intensity above 0.020 were interpreted as positive more than 98% of the time by participants with and without previous instruction. However, just over half of participants repeatedly interpreted the next faintest test band (strip 4 in Figure 3 with a 0.013 background-subtracted intensity) as negative (Figure S-8), suggesting that there was a different visible threshold for a portion of participants. By using a Fisher’s exact test to evaluate the effect of participants’ previous instruction on their interpretation of test strip 4, we determined that users with prior instruction more frequently interpret this strip as positive while those without prior instruction interpreted the strip as negative (Table 1). For maximum usability by both trained and untrained LFIA readers, we therefore aimed for LFIAs to have a test band intensity of at least 0.020 for positive interpretation and well below 0.013 for negative interpretation.
Table 1. Contingency Table of 54 Observations Indicates a Statistically Significant Interpretation of LFIA Test When Participant Does or Does Not Have Previous Instruction (p-Value <0.01).
| interpret test strip 4 as positive | interpret test strip 4 as negative | total | |
|---|---|---|---|
| previous instruction | 18 | 9 | 27 |
| no previous instruction | 7 | 20 | 27 |
| total | 25 | 29 | 54 |
Noting that all of the LFIA test bands resulting from conventional E. coli LAMP reaction product capture were at, or below, the visible threshold of 0.020, we aimed to improve positive test band intensity. We hypothesized that steric hindrance had prevented efficient capture of the amplicons using LF-FAM and LB-biotin primers and therefore repeated the amplification reactions with LF-biotin and LB-FAM primers (Figure 2B). The swapped primer tag configuration still yielded a limit of detection in agarose gels of 275 E. coli cells but resulted in positive test bands in all LFIAs, including negative controls. Not only were true positive LFIAs indistinguishable from negative control LFIAs by computer analysis, but all LFIAs would be interpreted positive by the human eye. We observed similar nonspecific capture of reaction products when performing a standard LAMP amplification of the V. cholerae ctxA gene using labeled primers (Figure 4A, Table S-1).22
Figure 4.
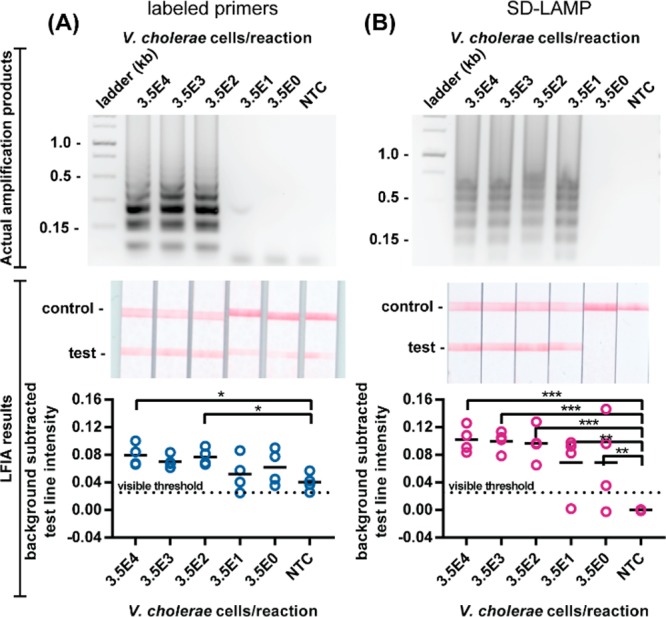
Detection of standard LAMP and SD-LAMP reactions in pure water. Electrophoresis gels verifying amplification (top), LFIA test results (middle), and LFIA test line quantification (bottom). (A) Standard LAMP reaction products for V. cholerae labeled with primers yield false positive LFIA results in low concentration samples and no template control (NTC) reactions. (B) SD-LAMP reactions yield no false positive LFIA results for V. cholerae. n = 4, replicates indicated by each circle. *** indicates p ≤ 0.001; ** indicates p ≤ 0.01; * indicates p ≤ 0.05.
Modification of LAMP with Strand Displacement Probe
Suspecting that the false positive LFIA results were caused by LB and LF primer hybridization, as observed by other researchers,18 we decided to replace the LF primer with a strand displacement probe tagged with FAM and perform SD-LAMP. The probe binds to the targeted loop region of the LAMP products by toehold-mediated strand exchange, a process that has been demonstrated to be exquisitely sequence specific due to the thermodynamic penalties of initiating branch migration.23 Even single nucleotide mismatches prevent strand displacement of the probe. Since the probe does not prime amplification and spurious amplification products caused by LAMP primers’ hybridization lack the probes’ binding site, the probe is expected to hybridize with only true target amplification products. Therefore, no spurious SD-LAMP products are expected to contain both the biotin and FAM labels that facilitate capture on LFIAs and could otherwise cause false positive results. As expected, SD-LAMP reaction products from both E. coli and V. cholerae were deposited on LFIAs and yielded no false positive test bands. As seen in Figure 2C, positive test bands from as few as 2 750 E. coli cells are well above the visible threshold intensity. Moreover, LFIAs from negative control reactions display as true negatives. The order of magnitude decrease in sensitivity compared to the conventional LAMP reaction is likely a result of there being only 1 loop primer instead of the usual 2 that accelerate amplification.24 In the V. cholerae LFIAs (Figure 4B), we did not observe a decrease in sensitivity and consistently visualized SD-LAMP product from as few as 350 cells (and several times as few as 3.5 V. cholera cells), likely because the primers bound more efficiently than the malB primers. However, as with SD-LAMP E. coli products, we observed only intense true positive LFIA test bands and no bands from true negative SD-LAMP V. cholerae products. Notably, the test band intensity (or lack thereof) resulting from SD-LAMP negative control reactions are narrowly distributed and well below the 0.013 threshold indicating that they can be easily interpreted as negative. Conversely, the test band intensities of negative control reactions using labeled primers were broadly distributed which caused user confusion during interpretation. Adding the strand displacement probes to the LAMP protocol is a simpler modification than redesigning primer sets or painstakingly optimizing reaction times. This essential modification mitigates spurious amplification and nonspecific detection in easy-to-use LFIAs, improving LAMP’s utility outside of a laboratory.
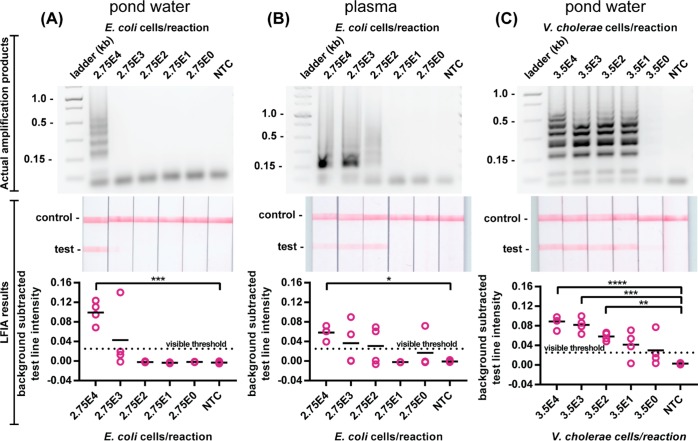
SD-LAMP of Complex Sample Matrixes
Both enteric pathogenic V. cholerae and E. coli have been found in environmental water sources and are significant public health concerns.25,26 Furthermore, E. coli is a common cause of sepsis by infecting the bloodstream of neonates and others with impaired immune systems.27 The rapid, sensitive, and accurate detection of these pathogenic bacteria from complex matrixes such as pond water and blood would aid in the control of waterborne and bloodborne infectious diseases. Given that complex matrixes have been found to inhibit amplification assays such as PCR, we investigated whether pond water or human blood would similarly limit the utility of SD-LAMP visualization on LFIAs (Figure 5). We found LFIAs yielded positive test bands from SD-LAMP reactions performed with 50% pond water (the maximum possible ratio given that the reagents are prehydrated) and as few as 27 500 E. coli and 350 V. cholerae cells. LFIAs yielded positive test bands from SD-LAMP reactions performed with 10% human blood plasma and as few as 27 500 E. coli cells. The decrease in sensitivity caused by pond water and blood plasma is a result of a decrease in LAMP reaction efficiency in these matrixes (as seen in Figure S-4 and also previously reported).28 Because the LFIA results match the agarose gel outputs, we can confirm that these are not due to a decrease in the hybridization efficiency of the strand displacement probes or capture efficiency of the LFIAs. The positive test bands remained well above the 0.02 visible threshold intensity and the negative test bands remained well below the 0.013 threshold. Our combined SD-LAMP and LFIA assay detects E. coli 100 times more sensitively than commercially available LFIA tests29 that rely on surface antigen capture. Moreover, our combined SD-LAMP and LFIA assay can detect 1000 fewer V. cholerae cells than existing commercial dipstick tests (taking into account that the existing dipstick assay requires 200 μL of sample while SD-LAMP is a 25 μL reaction).30
Figure 5.
Detection of SD-LAMP reactions in complex matrixes. Electrophoresis gels verifying amplification (top), LFIA test results (middle), and LFIA test line quantification (bottom). SD-LAMP reactions yield no false positive LFIA results for E. coli diluted in (A) pond water and (B) human plasma. SD-LAMP reactions yield no false positive LFIA results for (C) V. cholerae diluted in pond water. n = 4, replicates indicated by each circle. **** indicates p ≤ 0.0001; *** indicates p ≤ 0.001; ** indicates p ≤ 0.01; * indicates p ≤ 0.05.
Until now, the utility of LAMP isothermal amplification outside of a laboratory has been restricted by the lack of signal transduction that is easy-to-use, low-cost, and specific. Strand displacement probes are a simple improvement to LAMP and provide instrument-free nucleic acid detection on low-cost LFIAs without requiring additional user steps. Future research will explore the combination of strand displacement probes with other isothermal amplification assays for instrument-free detection.
Acknowledgments
This work was funded by Purdue University’s Innovations in International Development Lab, Purdue Institute for Inflammation, Immunology and Infectious Disease (PI4D), Purdue Research Foundation, the National Science Foundation Graduate Research Fellowship Program (Grant DGE-1333468), and the Grand Challenges Explorations Program (Grant OPP1150806), an initiative of the Bill & Melinda Gates Foundation.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.analchem.8b00269.
Additional materials and methods and data (PDF)
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Niemz A.; Ferguson T. M.; Boyle D. S. Trends Biotechnol. 2011, 29, 240–250. 10.1016/j.tibtech.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parida M.; Sannarangaiah S.; Dash P. K.; Rao P. V.; Morita K. Rev. Med. Virol. 2008, 18, 407–421. 10.1002/rmv.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L.; Zhou J.; Zheng Y.; Gamson A. S.; Roembke B. T.; Nakayama S.; Sintim H. O. Mol. BioSyst. 2014, 10, 970–1003. 10.1039/c3mb70304e. [DOI] [PubMed] [Google Scholar]
- Tanner N. A.; Evans T. C. In Current Protocols in Molecular Biology; John Wiley & Sons, Inc., 2014; pp 1–14. [Google Scholar]
- Modak S. S.; Barber C. A.; Geva E.; Abrams W. R.; Malamud D.; Ongagna Y. S. Y. Infect. Dis.: Res. Treat. 2016, 9, IDRT.S32162. 10.4137/IDRT.S32162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francois P.; Tangomo M.; Hibbs J.; Bonetti E.-J.; Boehme C. C.; Notomi T.; Perkins M. D.; Schrenzel J. FEMS Immunol. Med. Microbiol. 2011, 62, 41–48. 10.1111/j.1574-695X.2011.00785.x. [DOI] [PubMed] [Google Scholar]
- Sotiriadou I.; Karanis P. Diagn. Microbiol. Infect. Dis. 2008, 62, 357–365. 10.1016/j.diagmicrobio.2008.07.009. [DOI] [PubMed] [Google Scholar]
- Kokkinos P. A.; Ziros P. G.; Bellou M.; Vantarakis A. Food Analytical Methods 2014, 7, 512–526. 10.1007/s12161-013-9748-8. [DOI] [Google Scholar]
- Xu G.; Nolder D.; Reboud J.; Oguike M. C.; van Schalkwyk D. A.; Sutherland C. J.; Cooper J. M. Angew. Chem., Int. Ed. 2016, 55, 15250–15253. 10.1002/anie.201606060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safavieh M.; Kaul V.; Khetani S.; Singh A.; Dhingra K.; Kanakasabapathy M. K.; Draz M. S.; Memic A.; Kuritzkes D. R.; Shafiee H. Nanoscale 2017, 9, 1852–1861. 10.1039/C6NR06417E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S.; Mohd-Naim N. F.; Safavieh M.; Ahmed M. U. ACS Sens 2017, 2, 1713–1720. 10.1021/acssensors.7b00671. [DOI] [PubMed] [Google Scholar]
- Tanner N. A.; Zhang Y.; Evans T. C. BioTechniques 2015, 58, 59–68. 10.2144/000114253. [DOI] [PubMed] [Google Scholar]
- Kiatpathomchai W.; Jaroenram W.; Arunrut N.; Jitrapakdee S.; Flegel T. W. J. Virol. Methods 2008, 153, 214–217. 10.1016/j.jviromet.2008.06.025. [DOI] [PubMed] [Google Scholar]
- Singleton J.; Osborn J. L.; Lillis L.; Hawkins K.; Guelig D.; Price W.; Johns R.; Ebels K.; Boyle D.; Weigl B.; LaBarre P. PLoS One 2014, 9, e113693. 10.1371/journal.pone.0113693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez N. M.; Linnes J. C.; Fan A.; Ellenson C. K.; Pollock N. R.; Klapperich C. M. Anal. Chem. 2015, 87, 7872–7879. 10.1021/acs.analchem.5b01594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball C. S.; Light Y. K.; Koh C.-Y.; Wheeler S. S.; Coffey L. L.; Meagher R. J. Anal. Chem. 2016, 88, 3562–3568. 10.1021/acs.analchem.5b04054. [DOI] [PubMed] [Google Scholar]
- Tanner N. A.; Zhang Y.; Evans T. C. Jr. BioTechniques 2012, 53, 81–89. 10.2144/0000113902. [DOI] [PubMed] [Google Scholar]
- Jiang Y. S.; Bhadra S.; Li B.; Wu Y. R.; Milligan J. N.; Ellington A. D. Anal. Chem. 2015, 87, 3314–3320. 10.1021/ac504387c. [DOI] [PubMed] [Google Scholar]
- Du Y.; Pothukuchy A.; Gollihar J. D.; Nourani A.; Li B.; Ellington A. D. Angew. Chem., Int. Ed. 2017, 56, 992–996. 10.1002/anie.201609108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holstein C. A.Development of a Novel Paper-Based Flu Test for Improved Diagnosis at the Point of Care. University of Washington, Seattle, WA, 2015. [Google Scholar]
- Hill J.; Beriwal S.; Chandra I.; Paul V. K.; Kapil A.; Singh T.; Wadowsky R. M.; Singh V.; Goyal A.; Jahnukainen T.; Johnson J. R.; Tarr P. I.; Vats A. Journal of Clinical Microbiology 2008, 46, 2800–2804. 10.1128/JCM.00152-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada K.; Chantaroj S.; Taniguchi T.; Suzuki Y.; Roobthaisong A.; Puiprom O.; Honda T.; Sawanpanyalert P. Diagn. Microbiol. Infect. Dis. 2010, 66, 135–139. 10.1016/j.diagmicrobio.2009.09.004. [DOI] [PubMed] [Google Scholar]
- Srinivas N.; Ouldridge T. E.; Šulc P.; Schaeffer J. M.; Yurke B.; Louis A. A.; Doye J. P. K.; Winfree E. Nucleic Acids Res. 2013, 41, 10641–10658. 10.1093/nar/gkt801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagamine K.; Hase T.; Notomi T. Mol. Cell. Probes 2002, 16, 223–229. 10.1006/mcpr.2002.0415. [DOI] [PubMed] [Google Scholar]
- Jang J.; Hur H. G.; Sadowsky M. J.; Byappanahalli M. N.; Yan T.; Ishii S. J. Appl. Microbiol. 2017, 123, 570–581. 10.1111/jam.13468. [DOI] [PubMed] [Google Scholar]
- Alam M. T.; Weppelmann T. A.; Weber C. D.; Johnson J. A.; Rashid M. H.; Birch C. S.; Brumback B. A.; Beau de Rochars V. E. M.; Glenn J.; Ali A. Emerging Infect. Dis. 2014, 20, 356–363. 10.3201/eid2003.131293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonsen K. A.; Anderson-Berry A. L.; Delair S. F.; Davies H. D. Clin. Microbiol. Rev. 2014, 27, 21–47. 10.1128/CMR.00031-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connelly J. T.; Rolland J. P.; Whitesides G. M. Anal. Chem. 2015, 87, 7595–7601. 10.1021/acs.analchem.5b00411. [DOI] [PubMed] [Google Scholar]
- Phillips E. A.; Shen R.; Zhao S.; Linnes J. C. Lab Chip 2016, 16, 4230–4236. 10.1039/C6LC00945J. [DOI] [PubMed] [Google Scholar]
- Mukherjee P.; Ghosh S.; Ramamurthy T.; Bhattacharya M. K.; Nandy R. K.; Takeda Y.; Nair G. B.; Mukhopadhyay A. K. Jpn. J. Infect. Dis. 2010, 63, 234–238. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.