Abstract
We have identified adapt33 as a multiple stress-responsive gene that is induced under conditions of a cytoprotective “adaptive response.” adapt33 RNA does not contain any appreciable open reading frame nor produce a protein product and is therefore classified as a stress-inducible riboregulator. Although a number of oxidant stress-modulated, protein-encoding genes have been reported and characterized, very few stress-inducible riboregulator RNAs are known. Here we extend previous studies toward understanding the underlying regulation of expression and function of this rare mammalian riboregulator. mRNA stability and transcription studies determined that adapt33 induction by hydrogen peroxide is at the mRNA stability level, and that adapt33 has a very short half-life. Surprisingly, adapt33 mRNA also exhibits altered electrophoretic migration in response to both hydrogen peroxide and cis-platinum treatment. Although no transcriptional modulation in response to hydrogen peroxide was observed, fusion promoter constructs revealed that adapt33 has an unusually strong promoter that is active in both hamster and human cells. Analysis of expression following the stimulation of apoptosis with hydrogen peroxide and staurosporine revealed a strong correlation with apoptosis, suggesting a possible novel, noncoding RNA component of the apoptotic mechanism. We conclude that adapt33 is a stress-inducible, apoptosis-associated RNA with unique structural and gene promoter characteristics.
Keywords: Riboregulator, Adaptive response, Gene expression, Hydrogen peroxide, Oxidative stress, Hamster fibroblasts
HA-1 hamster fibroblasts respond to a minimally toxic “pretreatment” dose of hydrogen peroxide by synthesizing RNAs and proteins that protect them against subsequent exposure to a highly cytotoxic concentration of hydrogen peroxide (5,7,8,28). We have used this model system to identify mRNAs whose levels are modulated during pretreatment (7,9–11,26). These modulated RNAs represent potentially cytoprotective genes that mediate the observed adaptive response and hold potential clinical utility in treating oxidant-related diseases and disorders. We have designated such pretreatment-modulated mRNAs as “adapts.” One of these adapts, adapt78, has already been shown to be cytoprotective (21).
Of the five adapt genes that we have reported to date, two—adapt15 and adapt33—do not encode any discernable protein product. They are therefore classified as riboregulators. Riboregulator RNAs are spliced and polyadenylated RNAs that contain no apparent open reading frame or translational product. Their importance and characterization have been largely ignored due to their infrequent identification and lack of a functional protein product. However, it has become increasingly clear that these RNAs are associated with a wide range of biological activities, suggesting that they are important cellular regulators.
Riboregulators, also referred to noncoding RNAs (ncRNAs) in eukaryotes, have been implicated in transcriptional regulation, RNA processing, tumor suppression, prenatal lethality, chromosome condensation, developmental timing, protein synthesis regulation, and growth arrest (3,4,18,24,25,27). A recent review listed 28 identified riboregulators to date in organisms ranging from bacteria and plant to human (16).
Although adapt15 has been shown to suppress cell growth, the intracellular function of adapt33 is not yet known. Two adapt33 mRNAs species of 1.46 and 0.99 kb have been identified by Northern blot hybridization (26). Both variants are inducible by hydrogen peroxide in a calcium-dependent manner (26). Cell fractionation studies have revealed that a significant proportion of adapt33 RNA is associated with the actively translating polysome region. adapt33 RNA may therefore act directly as a regulatory RNA at the site of actively translating ribosomes during translation. As has been previously speculated for adapt15, it is also possible that adapt33 RNA acts both via RNA structure and a small peptide translated from one of the many small open reading frames. The mRNA levels of both of these adapt riboregulators are strongly induced by hydrogen peroxide, and under adaptive response conditions where cellular protection is observed. This suggests that, whatever the mechanism, these RNAs are involved in protecting cells against the damaging effects of oxidative and perhaps other stress. Together, adapt15 and adapt33 represent the only known mammalian stress-inducible riboregulators. Baterial oxyS and Drosophila hsr[omega] (15,16) are the only other reported stress-inducible riboregulators to date. Here we extend previous studies on adapt33 to gain more insight into the regulation and action of this rare stress-inducible riboregulator.
MATERIALS AND METHODS
Cell Culture and Treatment Conditions
Hamster HA-1 cells, a Chinese hamster ovary fibroblast cell line, were maintained in Eagle’s minimal essential medium (MEM) supplemented with 15% heat-inactivated fetal bovine serum, penicillin (100 U/ml), and streptomycin (100 μg/ml). The cultures were grown in a humidified incubator atmosphere of 95% air and 5% CO2 at 37°C.
Treatment of HA-1 Cells With Hydrogen Peroxide
HA-1 cells were trypsinized and plated at 200,000 cells per 60-mm plate. After 2 days of incubation, cells were divided into two groups, one receiving phosphate-buffered saline only (controls), and the other the appropriate concentration of hydrogen peroxide (H2O2). Cells were then returned to the incubator for the designated period of time.
mRNA Stability
HA-1 cells were pretreated with 4 μmol of hydrogen peroxide per 107 cells as described above. Three hours later, 40 μg/ml of DRB (Sigma, St. Louis, MO) was added to the cell culture to completely block RNA polymerase II transcription. Total RNA was then extracted 0, 2, 4, and 6 h later. The levels of specific mRNAs were analyzed by Northern blot hybridization as described previously (5,7) using cDNA probes. mRNA decay in control and treated cells was normalized to the GAPDH signal.
RNA Isolation and Analysis
Total RNA was isolated by direct addition of RNA lysis buffer containing guanidinium isothiocyanate (RNAzol, Biotecz, Houston, TX and RNA Isolator, Genosys, Woodlands, TX) to PBS-washed cultures. The RNA was then extracted according to the manufacturer, and the final, semidried pellet resuspended in diethylpyrocarbonate-treated distilled, deionized water. Electrophoresis, Northern blotting, and hybridization were performed as previously described (6,7). Final washed blots were exposed to X-ray film.
Nuclear Run-on Analysis
Transcription of adapt33 gene was assessed by nuclear run-on. Cells were pretreated with hydrogen peroxide as described above. Medium was removed 1.5 and 4.5 h after treatment; cells were rinsed with ice-cold PBS, scraped, and centrifuged for 5 min at 500 × g at 4°C. The cell pellet was resuspended in NP-40 lysis buffer A (10 mM Tris-HCl, pH 7.4, 10 mM NaCl, 3 mM MgCl2, 0.5% v/v NP-40), incubated 5 min on ice, centrifuged as above, and the supernatant removed. The nuclear pellet was resuspended in 4 ml NP-40 lysis buffer A as above, centrifuged as above, and the pellet nuclei resuspended in 50 mM Tris-HCl, pH 8.3, 40% v/v glycerol, 5 mM MgCl2, and 0.1 mM EDTA and stored in liquid nitrogen. For nuclear run-on transcription of nascent RNA transcripts, 170 μl of 2× reaction buffer with nucleotides (10 mM Tris-HCl, pH 8.0, 5 mM MgCl2, 0.3 M KCl, 1 mM of each of ATP, CTP, and GTP, and 5 mM DTT) plus 10 μl of 10 mCi/ml [α-32P]UTP was added to the thawed nuclei and incubated 30 min at 30°C with shaking. Total RNA was then extacted as above and the final pellet resuspended in 10 mM TES, pH 7.4, 10 mM EDTA, and 0.2% w/v SDS. The labeled RNA was then hybridized to cDNAs on each gene of interest slot blotted to nitrocellulose.
Rapid Amplification of cDNA Ends (RACE)
RACE was performed on RNA extracted as above using a RACE kit according to the manufacturer (Gibco Invitrogen, Grand Island, NY). In summary, a 17-mer oligomer specific to the 5′ region of adapt33 cDNA was used as the first gene-specific primer (GSP-1) from reverse transcription from RNA extracted from control and peroxide-treated HA-1 cells. The generated cDNA fragment was PCR amplified by using a second gene-specific primer (GSP-2). The PCR product was then cloned into a TA-vector and subjected to sequencing analysis.
Isolation of adapt33 Genomic Clone
Full-length adapt33 cDNA was used to screen a Chinese hamster ovary genomic library in the λ FIX vector according to the manufacturer (Stratagene, La Jolla, CA). A single positive clone was identified and found to contain a 16-kb insert. To subclone the fragment containing the 5′-flanking region of adapt33 gene, the 16-kb insert was digested with EcoR1, electrophoresed through an agarose gel, and subjected to Southern blot hybridization using a Pvu1/HindIII fragment of the cDNA as a probe corresponding to the 5′ region of the full-length adapt33 cDNA. A 4-kb fragment hybridized and was subcloned into pBlue-script SK vector (Stratagene).
Construction of the CAT–Fusion Plasmid
A 2.5-kb fragment of the 5′-flanking region of the adapt33 gene was generated by PCR using a hamster adapt33 genomic clone we had obtained as template. The upper primer T7 was specific to the vector sequence immediately upstream from the 5′ end of the insert. The lower primer 5′-ACAAACTAGTACA ATGCTCGCCCAAAG-3′ was specific to the cDNA sequence downstream from the determined (see above RACE) transcriptional start site. A subsequent deletion at the 5′ end was made on the 2.5-kb fragment by cutting an internal HindIII site to generate a 1.4-kb fragment. These fragments were ligated into a pCAT vector (Promega, Madison, WI) where the tested promoter region was fused upstream to the coding region of the CAT (chloramphenicol acetyl transferase) reporter gene.
HA-1 Cell Transfection and CAT Assay
Plasmid DNA was transfected into HA-1 cells using lipofectamine (Gibco Invitrogen, Gaithersburg, MD). HA-1 cells were seeded at a density of 1.5 × 105 cells per 60-mm culture dish and incubated at 37°C. After 48 h, lipofectamine–DNA coprecipitate was added according to the manufacturer. After 6 h, the transfection medium was replaced with fresh complete medium and, after 24 h, 4 μmol of hydrogen peroxide was added per 107 cells. The CAT activity from the transfected cells was measured by the diffusion method normalizing to protein mass.
DNA Extraction and Analysis of Internucleosomal DNA Fragmentation
HA-1 control cultures and cells treated with varying concentrations of hydrogen peroxide or staurosporine were collected at the appropriate time points by centrifuging the culture medium with washed monolayer cells. The cells were lysed by exposure to 1% NP-40 in 20 mM EDTA and 50 mM Tris-HCl, pH 7.5, for 10 s and centrifuged. The supernatant containing the lower molecular weight fraction was treated with 1% SDS and RNase (5 μg/μl) followed by Proteinase K (2.5 μg/μl) as described previously (17). After precipitation, the DNA was run on a 1.5% agarose gel and stained with SYBR green.
RESULTS
Induction of adapt33 mRNA by Hydrogen Peroxide in HA-1 Cells
As we have previously reported, adapt33 mRNA levels were significantly elevated by exposure of HA-1 cells to 160 μM of hydrogen peroxide (Fig. 1A). The presence and inducibility of two adapt33 variants of 1.5 and 1.0 kb is also apparent from this figure. As a generalization, we noted that adapt33 mRNA levels were lower than for a number of other transcripts and required longer exposure to film or phosphoimaging to obtain a clear signal. Thus, adapt33 appeared to be a low abundant RNA.
adapt33 mRNA Stability
HA-1 cells were treated with the transcriptional inhibitor DRB after hydrogen peroxide pretreatment and RNA levels assessed at various time points. As shown in Figure 1B, the adapt33 RNA transcript had a very short half-life in control cells, dropping to 20% of that before DRB treatment within 2 h. On the other hand, treatment with hydrogen peroxide resulted in a half-life of greater than 6 h. Thus, hydrogen peroxide treatment strongly stabilized adapt33 mRNA. This was observed at both 4 μmol of hydrogen peroxide per 107 cells and especially 10 μmol of hydrogen peroxide per 107 cells.
Figure 1.
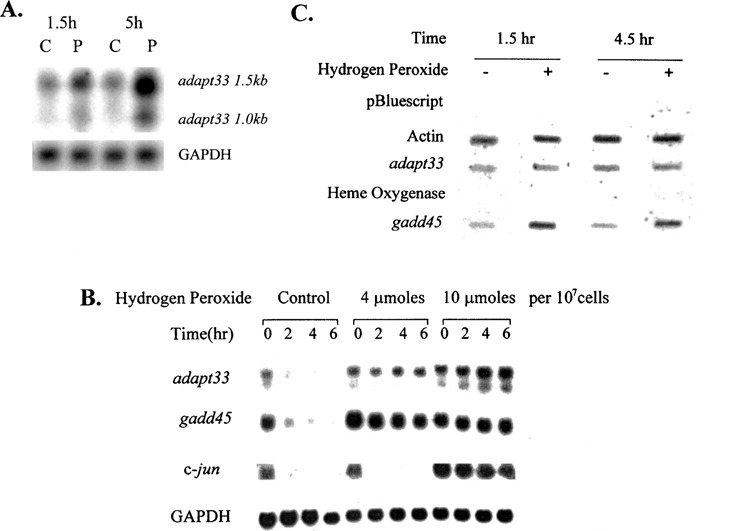
The effect of hydrogen peroxide on adapt33 RNA levels. (A) Northern blot containing RNA from HA-1 cells treated with 160 μM of hydrogen peroxide for 90 min and 5 h was probed with radiolabeled adapt33 cDNA. C, control. P, peroxide treated. The Northern blot was also treated with GAPDH cDNA as a loading control. (B) adapt33 stability. HA-1 cells were treated with 0, 4, or 10 μmol of hydrogen peroxide per 107 cells. Three hours later, all cells were treated with 40 μg/ml of DRB. Total RNA was extracted at indicated times after DRB treatment and subjected to Northern blot analysis. (C) adapt33 transcription. HA-1 cells were treated with or without (control) 4 μmol of hydrogen peroxide per 107 cells. At 1.5 and 4.5 h time points after treatment, cell nuclei were extracted from both control and treated cells and run-on reactions were performed on the nuclei and quantified as described in Materials and Methods.
To determine whether the stabilization of adapt33 by hydrogen peroxide is a general or specific effect, the decay of c-jun, another short-lived message, was also analyzed. It was not stabilized by the pretreatment concentration of hydrogen peroxide (Fig. 1B). Thus, message stabilization is not a general effect of hydrogen peroxide treatment. In addition, gadd45 mRNA was stabilized by hydrogen peroxide, consistent with its known stabilization by other DNA-damaging agents including MMS and UV irradiation (19).
Induction of adapt33 mRNA by Hydrogen Peroxide Is Not at the Transcriptional Level
HA-1 cells were treated with peroxide for 1.5 and 4.5 h and adapt33-specific transcription determined following nuclei isolation. As shown in Figure 1C, adapt33 RNA transcript production did not change in response to peroxide. gadd45, which is known to be induced at the transcriptional level by hydrogen peroxide, was used as a positive control, and actin as a negative control. Combined with the above mRNA stability studies, we conclude that the induction in adapt33 mRNA levels was due to message stabilization.
Exposure of Cells to Stress Agents Leads to Altered Migration of adapt33 mRNA
After exposure of HA-1 cells to hydrogen peroxide and subsequent Northern blot analysis, it was observed that adapt33 mRNA levels were not only induced by peroxide but also exhibited altered migration on agarose gels. Specifically, migration of the adapt33 transcript was moderately accelerated (Fig. 2A, B). This effect did not appear to be specific to peroxide-treated cells, as cis-platinum-treated cells elicited a similar effect, albeit more modestly (see 4-h treatment with 25 mg/ml CisPt, Fig. 2C). However, it was specific to adapt33 as a stress-inducible mammalian riboregulator, because the only other known such riboregulator, adapt15, did not share this altered migration (Fig. 2A). No other transcript was observed to exhibit this altered migration, including GAPDH (Fig. 2).
Figure 2.
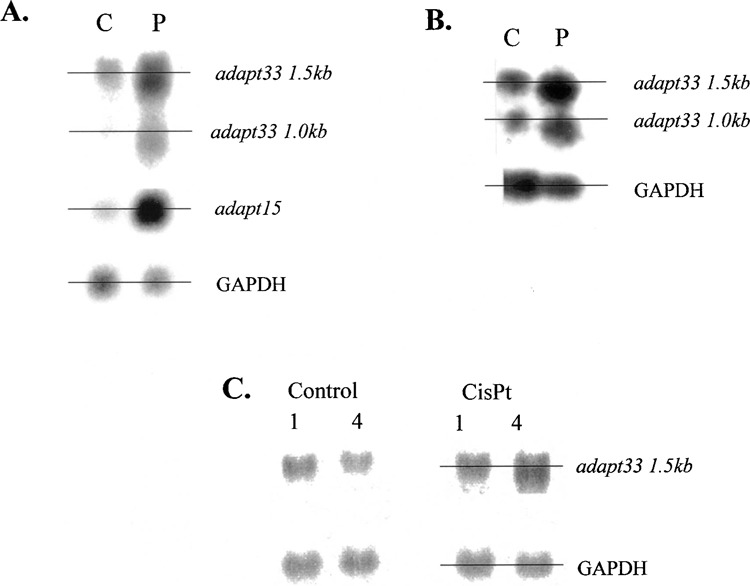
The altered migration of adapt33 mRNA. HA-1 cells were exposed to 4 μmol of hydrogen peroxide per 107 cells. RNA was extracted 5 h later and subjected to Northern blot analysis as described above. Horizontal lines were drawn through the middle of adjacent lanes to more easily assess the accelerated migration observed for adapt33 in the peroxide (P) lanes compared with control (C). (A) Single blot probed successively with radiolabeled cDNA probes for adapt33, adapt15, and GAPDH. (B) Single blot probed successively with radiolabeled cDNA probes for adapt33 and GAPDH. (C) The effect of the DNA-damaging agent cis-platinum (CisPt; 25 mg/ml) on adapt33 expression analyzed as described above.
adapt33 Promoter Constructs
To better understand the regulation of adapt33 gene expression and further test the transcriptional response of the adapt33 gene to peroxide exposure, we performed promoter fusions studies. To establish the promoter region of adapt33, it was necessary to first identify the transcriptional start site. This was done by rapid amplification of the 5′ cDNA ends (RACE). The results are shown in Figure 3A. At the same time, subcloning of a 16-kb adapt33 genomic clone was carried out as described in Materials and Methods. These subclones were probed with a fragment of the adapt33 DNA as a probe corresponding to the 5′ region of the full-length adapt33 cDNA. A 4-kb genomic clone hybridized and was therefore most likely to contain the promoter region of adapt33. The fragment was sublcloned into pBluescript SK (Fig. 3B). Partial sequencing of the subclone revealed that the genomic insert contained the 5′ end of the cDNA and 2.5 kb of 5′-flanking region, using the above RACE information to identify the transcriptional start site. Sequence analysis of the upstream region from the transcription start site indicated that adapt33 has a TATA-less promoter with the upstream region containing a number of known transcriptional elements including NF-κB, Sp-1, and ATF sites (data not shown).
Figure 3.
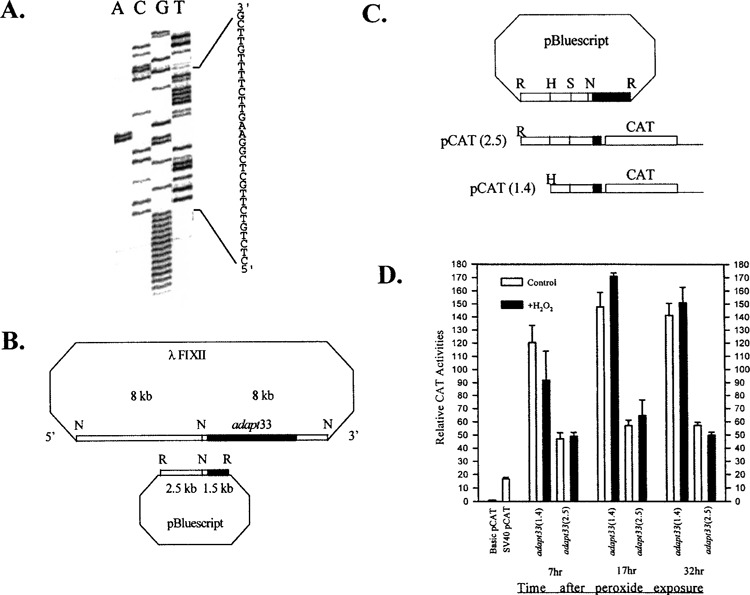
adapt33 promoter studies. (A) 5′ RACE was used to identify the transcription start site of the adapt33 gene using gene-specific primers. The major PCR product was cloned and sequenced. The sequence shown represents the 5′ end of the cDNA. (B) adapt33 genomic clone and subcloning into the pBluescript vector. The shaded box represents the genomic sequence that is overlapping with the 5′ end of adapt33 cDNA. N, Not1 site; R, EcoR1 site. (C) adapt33 promoter–CAT fusion constructs. The top figure shows the pBluescript/adapt33 genomic subclone. A 2.5-kb fragment containing the 5′ end of adapt33 cDNA and the upstream region was generated from this genomic subclone by PCR and ligated upstream of a reporter CAT gene in a promoter-less pCAT vector. A subsequent 5′ deletion at an internal HindIII site generated a construct containing the 1.4-kb adapt33 insert. The 5′ end of adapt33 cDNA is shaded. R, EcoR1 site; H, HindIII site; N, Not1 site; S, Sac1 site. (D) After 6-h transfection, cells were allowed to recover for 24 h and then treated with 4 μmol of hydrogen peroxide per 107 cells. Cell extracts from control and treated cells were prepared as described in Materials and Methods at indicated time points after treatment.
adapt33 Transcriptional Activity Using CAT Gene Reporter Constructs
adapt33 transcriptional activity was further studied using CAT gene reporter fusion constructs. A 2.5-kb and a 1.4-kb fragment of the 5′-flanking region of the adapt33 gene were generated by PCR using the above hamster adapt33 genomic clone as a template and fused to the CAT gene as shown in Figure 3C. Transfection was then performed as described in Materials and Methods and transcription assessed. Consistent with the above nuclear run-on analyses, no additional promoter activity was observed in cells treated with hydrogen peroxide, as shown in Figure 3D. This lack of a peroxide effect was observed under all conditions tested including 7, 17, and 32 h after peroxide exposure and for both promoter constructs. However, adapt33 promoter showed unusually strong constitutive activity. Both promoters exhibited significantly higher activity than the popular SV40 promoter/enhancer and, maximally, was almost 10-fold higher than the SV40 promoter in HA-1 cells in the 1.4-kb adapt33 promoter construct. The promoter activity was also tested in a HT1080 human cell line and showed a similar result. These results indicate that adapt33 constitutive promoter activity is very high. It also indicates that adapt33 mRNA is low abundance despite a strong promoter, apparently due to its very short half-life (described above).
adapt33 Is Associated With Apoptosis
adapt33 mRNA induction is maximal in cells exposed to a concentration of 10 μmol hydrogen peroxide per 107 cells (Fig. 4A). This concentration of peroxide also elicits the maximum apoptosis response in these cells as assessed by DNA fragmentation (Fig. 4B). Thus, adapt33 induction is closely associated with apoptosis in HA-1 cells. This correlation was exact by 5 h in that, at a higher concentration of peroxide (25 μmol hydrogen peroxide per 107 cells), adapt33 levels decreased in parallel with apoptosis as apparent necrosis increased (compare Fig. 4A and B). To further assess the correlation and possible involvement of adapt33 expression with apoptosis, we treated cells with another apoptosis inducer, the protein kinase C inhibitor staurosporine. As shown in Figure 5A and B, the cellular level of GAPDH, actin, and even the proapoptotic bax decreased with time in the presence of the staurosporine. This is probably due to an inhibiting effect of staurosporine on general mRNA transcription. In contrast, adapt33 mRNA levels did not decrease but increased at the later time points in the treated cells (Fig. 5A, B, C). We also observed that adapt33 RNA is induced by cis-platinum (Fig. 2C), a known apoptotic agent that is used chemotherapeutically (13,22). These data provide further evidence that adapt33 is an apoptosis-associated gene.
Figure 5.
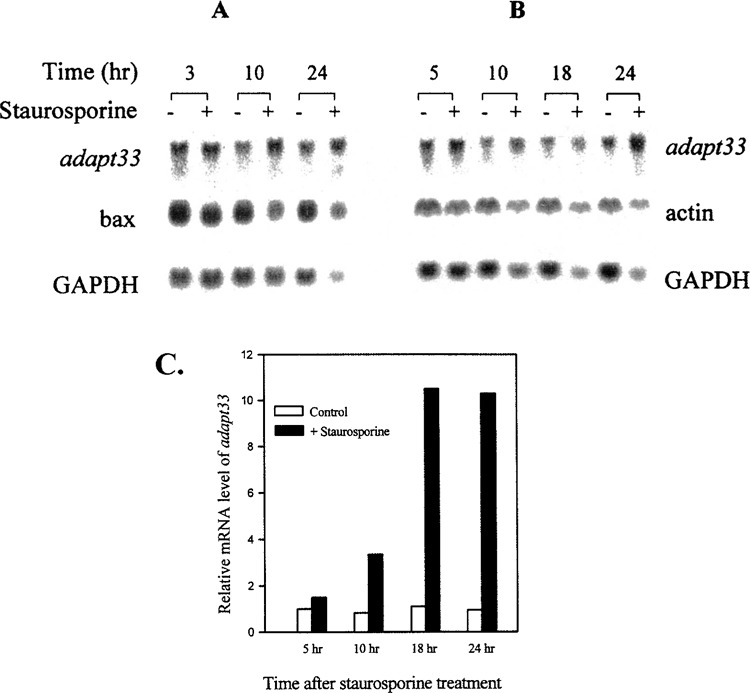
The effect of staurosporine on adapt33 mRNA levels. HA-1 cells were exposed to 200 μM staurosporine and incubated for the indicated times prior to RNA extraction. (A, B) Northern blots containing total RNA from two independent experiments. The blots were probed with radiolabeled cDNAs to (A) adapt33, bax, and GAPDH, and (B) adapt33, actin, and GAPDH, respectively. (C) Densitometric quantification of induction of adapt33 by staurosporine. Relative mRNA levels of adapt33 were normalized to that of GAPDH.
Figure 4.
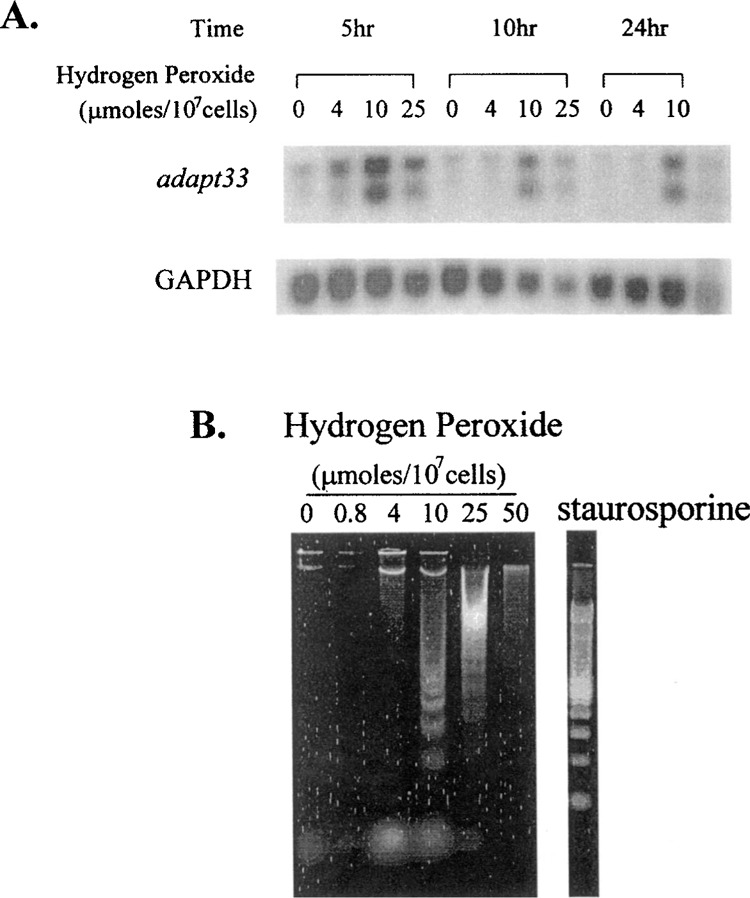
adapt33 RNA expression and apoptosis. (A) HA-1 cells were exposed to 0, 4, 10, or 25 μmol of hydrogen peroxide per 107 cells and incubated for the indicated times prior to RNA extraction. A Northern blot containing total RNA from these sample was then probed with a cDNA to adapt33. This blot was also probed with GAPDH cDNA as a loading control. (B) HA-1 cells treated with 0, 0.8, 4, 10, 25, or 50 μmol of hydrogen peroxide per 107 cells and staurosporine for 24 h were collected, lysed, centrifuged, and treated. After precipitation, the DNA was run out on a 1.5% agarose gel and stained with SYBR green.
DISCUSSION
Previously, we have described that adapt33 is a novel gene induced by hydrogen peroxide under conditions where adaptive response occurs. Here we extend these studies to better understand the expression and function of this gene. In addition, it provides insight into a possibly novel mammalian system that combines the actions of untranslated riboregulator adapt RNAs (adapt15 and adapt33) with other known protein stress-response mediators. If true, it would resemble the bacterial OxyR system where both non-coding oxyS RNA and other protein-encoding RNAs are induced following exposure to hydrogen peroxide; and the Drosophila heat shock response where both hsr[omega] noncoding RNAs are induced along with other RNAs encoding heat shock proteins, although the latter are induced independently (15,17). Clearly, there are a number of similarities between adapt33 and adapt15, including inducibility by hydrogen peroxide under conditions of adaptive response, their similar peroxide-induction kinetics, the role of calcium in these inductions, and their lack of large open reading frames (7,8,26). It is therefore possible that these two adapts belong to the same family. We have identified adapt15/gadd7 as a growth arrest- and DNA damage-inducible sequence (8,12). Thus, adapt15 and adapt33 appear to belong to the same family as gadd45 and gadd153. This suggests that adapt15 and adapt33 RNAs act as riboregulators in concert with Gadd45 and Gadd153 proteins to bring about growth arrest and perhaps protect against DNA damage.
The short half-life of adapt33 indicates that it probably has a regulatory role during cellular response. We have previously reported that the induction of adapt33 mRNA levels is calcium dependent. It is possible that calcium-dependent protein kinase pathways are involved in the RNA stability control. This regulation must be specific because other shortlived transcripts such as c-jun are not stabilized by the same concentration of hydrogen peroxide.
Adenylate/uridylate-rich elements represent a common determinant of RNA stability in mammalian cells and are found in the 3′ untranslated regions of many mRNAs, including those that code for proto-oncogenes, nuclear transcription factors, cytokines, and gadd genes (1,19). The adapt33 transcript has two adenylate/uridylate-rich elements, which probably explains its very short constitutive half-life. Importantly, many molecules that mediate rapid cell response to environmental change are capable of rapid turnover. On the other hand, structural genes are long-lived so that constitutive, high-level transcription of these genes is not required to supply the basic components of cellular machinery.
We have also determined here that adapt33 has an unusually strong promoter that is active in both hamster and human cells. In both cases, its strength was compared with that of the SV40 promoter. SV40 is commonly considered a strong promoter and used to drive the expression of other genes in functional studies. The adapt33 promoter was eight- and sixfold stronger than SV40 promoter in hamster HA-1 cells and human HT1080 cells, respectively. Such a high activity is rarely seen in mammalian promoters. It may be used as a tool in studying other gene’s functions. Despite its strong promoter, adapt33 has a very short half-life. It seems that while the cell is always generating large amounts of adapt33 transcripts, it is also rapidly degrading them. This pattern is reminiscent of some genes that are involved in only emergency response. Because adapt33 is a riboregulator that probably functions mainly, if not exclusively, at the RNA level, increased RNA stability during stress would lead to the ready availability of large amounts of functional RNA molecules.
The alteration in adapt33 RNA migration following the exposure of cells to hydrogen peroxide or cis-platinum is curious. We speculate that this altered migrating form may represent an “activated” form of adapt33 involving altered primary or secondary structure in response to cellular stress. Because, as a riboregulator, it is believed that adapt33 acts primarily if not exclusively at the level of RNA, this molecular alteration may better enable adapt33 to perform its stress response role, whatever it is. Several possibilities for this role include cytoprotection (due to the induction of adapt33 and other adapt mRNAs during adaptive response when an overall cytoprotection is observed), the regulation of protein translation due to its fractionation with the actively translating fraction of HA-1 cells (26), and, at higher concentrations of peroxide, a role in apoptosis. A similar alteration in adapt15 RNA migration was not observed, indicating that this effect is not a general one (Fig. 2). To our knowledge, adapt15 and adapt33 are the first and only reported mammalian stress-inducible riboregulators. Because adapt33 mRNA stability is increased in response to peroxide, it is unlikely that the change in migration represents a partial degradation of adapt33 RNA, although we cannot absolutely rule this out such as in the form of poly(A) tail truncation. It is also possible that a chemical modification has occurred that alters migration, such the loss of a chemical moiety(s).
The association of adapt33 with apoptosis is seemingly paradoxical because it is induced under conditions of a protective adaptive response. However, a number of genes have been reported that are involved in both recovery of sublethally damaged cells and apoptosis of extensively damaged cells. These genes include p53, RB phosphatase, p21, and c-Abl protein tyrosine kinase (2,14,20,23). Although the exact mechanisms that determine how these genes decide on recovery or suicide is not known, it is known that growth arrest plays an important role. Growth arrest, on the one hand, allows time for a cell to repair damage and is thus a protective response. On the other hand, it is also observed prior to apoptosis in extensively stress-damaged cells. adapt33 may also possess a similar dual type activity.
We conclude that adapt33 is a stress-inducible, apoptosis-associated riboregulator RNA with unique structural and gene promoter characteristics and represents a unique marker of cellular oxidative stress.
ACKNOWLEDGMENTS
We thank Drs. Al Fornace, Nikki Holbrook, Rex Tyrrell, and L. F. Lau for use of the gadd45, gadd153, heme oxygenase, and CH134 cDNA probes.
REFERENCES
- 1. Bakheet T. M.; Frevel B. R.; Williams W. G.; Khabar K. S. ARED: Human AU-rich element-containing mRNA database reveals an unexpectedly diverse functional repertoire of encoded proteins. Nucleic Acids Res. 29:246–254; 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bargonetti J.; Manfredi J. J. Multiple roles of the tumor suppressor p53. Curr. Opin. Oncol. 14:86–91; 2002. [DOI] [PubMed] [Google Scholar]
- 3. Brockdorff N. A.; Ashworth G. F.; Kay V. M.; McCabe D. P.; Norris P. J.; Cooper S. S.; Rastan S. The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell 71:515–526; 1992. [DOI] [PubMed] [Google Scholar]
- 4. Brunkow M. E.; Tilghman S. M. Ectopic expression of the H19 gene in mice causes prenatal lethality. Genes Dev. 5:1092–1101; 1991. [DOI] [PubMed] [Google Scholar]
- 5. Crawford D. R.; Davies K. J. A. Adaptive response and oxidative stress. Environ. Health Perspect. 102:25–28; 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Crawford D. R.; Edbauer-Nechamen C. A.; Lowry C. V.; Salmon S. L.; Kim Y. K.; Davies J. M. S.; Davies K. J. A. Assessing gene-expression during oxidative stress. Methods Enzymol. 234:175–217; 1994. [DOI] [PubMed] [Google Scholar]
- 7. Crawford D. R.; Schools G. P.; Salmon S. L.; Davies K. J. A. Hydrogen peroxide induces the expression of adapt15, a novel RNA associated with polysomes in hamster HA-1 cells. Arch. Biochem. Biophys. 325:256–264; 1996. [DOI] [PubMed] [Google Scholar]
- 8. Crawford D. R.; Schools G. P.; Davies K. J. A. Oxidant-inducible adapt15 RNA is associated with growth arrest- and DNA damage-inducible gadd153 and gadd45. Arch. Biochem. Biophys. 329:137–144; 1996. [DOI] [PubMed] [Google Scholar]
- 9. Crawford D. R.; Leahy K. P.; Wang Y.; Schools G. P.; Kochheiser J. C.; Davies K. J. Oxidative stress induces the levels of a MafG homolog in hamster HA-1 cells. Free Radic. Biol. Med. 21:521–525; 1996. [DOI] [PubMed] [Google Scholar]
- 10. Crawford D. R.; Davies K. J. A. Modulation of a cardiogenic shock-inducible RNA by chemical stress: adapt73/PigHep3. Surgery 121:581–587; 1997. [DOI] [PubMed] [Google Scholar]
- 11. Crawford D. R.; Leahy K. P.; Abramova N.; Lan L.; Wang Y.; Davies K. J. A. Hamster adapt78 mRNA is a Down syndrome critical region-homologue that is inducible by oxidative stress. Arch. Biochem. Biophys. 342:6–12; 1997. [DOI] [PubMed] [Google Scholar]
- 12. Crawford D. R. Regulation of gene expression by reactive oxygen species. In: Gilbert D. L.; Colton C. , eds. Reactive oxygen species in biological systems: An interdisciplinary approach. New York: Plenum Publishing; 1999:155–171. [Google Scholar]
- 13. Cummings B. S.; Schnellmann R. G. Cisplatin-induced renal cell apoptosis: Caspase 3-dependent and -independent pathways. J. Pharmacol. Exp. Ther. 302:8–17; 2002. [DOI] [PubMed] [Google Scholar]
- 14. Dou Q. P.; An B.; Will P. L. Induction of a retinoblastoma phosphatase activity by anticancer drugs accompanies p53-independent G1 arrest and apoptosis. Proc. Natl. Acad. Sci. USA 92:9019–9023; 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Erdmann V. A.; Szymanski M.; Hochberg A.; de Groot N.; Barciszewski J. Collection of mRNA-like non-coding RNAs. Nucleic Acids Res. 27:192–195; 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Erdmann V. A.; Barciszewska M. Z.; Szymanski M.; Hochberg A.; de Groot N.; Barciszewski J. The non-coding RNAs as riboregulators. Nucleic Acids Res. 29:189–193; 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Herrmann M.; Lorenz H. M.; Voll R.; Grunke M.; Woith W.; Kalden J. R. A rapid and simple method for the isolation of apoptotic DNA fragments. Nucleic Acids Res. 22:5506–5507; 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hollander M. C.; Alamo I.; Fornace A. J. A novel DNA damage-inducible transcript gadd7, inhibits cell growth, but lacks a protein product. Nucleic Acids Res. 24:1589–1593; 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jackman J.; Alamo I.; Fornace A. J. Genotoxic stress confers preferential and coordinate messenger RNA stability on the five gadd genes. Cancer Res. 54:5656–5662; 1994. [PubMed] [Google Scholar]
- 20. Kharbanda S.; Yuan Z. M.; Weichselbaum R.; Kufe D. Determination of cell fate by c-Abl activation in the response to DNA damage. Oncogene 17:3309–3318; 1998. [DOI] [PubMed] [Google Scholar]
- 21. Leahy K. P.; Crawford D. R. adapt78 protects cells against stress damage and suppresses cell growth. Arch. Biochem. Biophys. 379:221–228; 2000. [DOI] [PubMed] [Google Scholar]
- 22. Park M. S.; De Leon M.; Devarajan P. Cisplatin induces apoptosis in LLC-PK1 cells via activation of mitochondrial pathways. J. Am. Soc. Nephrol. 13:858–865; 2002. [DOI] [PubMed] [Google Scholar]
- 23. Roninson I. B. Oncogenic functions of tumour suppressor p21(Waf1/Cip1/Sdi1): Association with cell senescence and tumour-promoting activities of stromal fibroblasts. Cancer Lett. 179:1–14; 2002. [DOI] [PubMed] [Google Scholar]
- 24. Ruvkun G. Molecular biology—glimpses of a tiny RNA world. Science 294:797–799; 2002. [DOI] [PubMed] [Google Scholar]
- 25. Storz G. An expanding universe of noncoding RNAs. Science 296:1260–1263; 2002. [DOI] [PubMed] [Google Scholar]
- 26. Wang Y.; Crawford D. R.; Davies K. J. A. adapt33, a novel oxidant-inducible RNA from hamster HA-1 cells. Arch. Biochem. Biophys. 332:255–260; 1996. [DOI] [PubMed] [Google Scholar]
- 27. Wickens M.; Takayama K. RNA. Deviants—or emissaries. Nature 367:17–18; 1994. [DOI] [PubMed] [Google Scholar]
- 28. Wiese A. G.; Pacific R. E.; Davies K. J. A. Transient adaptation to oxidative stress in mammalian cells. Arch. Biochem. Biophys 318:231–240; 1995. [DOI] [PubMed] [Google Scholar]


