Abstract
Septin 3 is a novel member of the septin subfamily of GTPase domain proteins that was recently identified in human neuronal cells. These proteins are involved in vesicle trafficking, neurite outgrowth, and neurofibrillary tangle formation; however, the expression and functional role of septin 3 in normal neuronal tissues and as an etiological agent in neurological disorders is currently unclear. To further characterize these parameters, the present study analyzed the expression of three isoforms of septin 3 (A, B, and C) in fetal and adult human brains and polymorphism of the septin 3 exon 11 microsatellite in control, pure Alzheimer’s disease (AD), Lewy body variant (LBV) of AD, and Parkinson’s disease. Septin 3 mRNAs for isoforms A and B, but not C, were detected in the frontal cortex of fetus and adult human samples, as measured by reverse transcription-coupled polymerase chain reaction. Genotype analyses indicated that polymorphic septin 3 alleles were distributed in two peaks of frequency in both control and disease groups. Categorization of the alleles into short (S) and long (L) types revealed a significant difference between AD patients and controls (p = 0.034 by chi-square test). Furthermore, the S-allele homozygosity was significantly underrepresented in AD compared with control (p = 0.015 by chi-square test). These results suggest that polymorphism in exon 11 of septin 3 may have a determinative role in the pathogenesis of AD.
Key words: Septin 3, Isoforms, Polymorphism, Alzheimer’s disease, Lewy body variant of Alzheimer’s disease, Parkinson’s disease, CYP2D6
SEPTINS are a group of proteins that were first identified in a genetic screen of budding yeast Saccharomyces cerevisiae for mutants defective in cytokinesis (9). Further studies led to the characterization of septins in eurkaryotes (8). Ten homologues have since been identified in mammals (12,15), many of which are present in the brain (14,22). Recently, involvement of neuronal septins in vesicle trafficking (2,10), neurite outgrowth (23), and neurofibrillary tangle formation (13) was reported, and one of the neuronal septins, CDCrel-1, was identified as a substrate of parkin, a ubiquitin-protein ligase that is present as the PARK2 mutant in individuals with autosomal recessive juvenile parkinsonism (26). These findings suggest functional roles of human septins in neuronal networking, survival, and, possibly, degeneration.
Septin 3 was originally cloned as a gene upregulated upon neuronal differentiation of a human teratocarcinoma cell line (16). Its encoding protein, septin 3, belongs to a family of GTPases (40–50 kDa) that are distinct from small Ras-like GTPases or other known GTPases (12). Human septin 3 is highly homologous to mouse Sep3 (24) and rat G-septin (25). The septin 3 gene has been mapped to chromosome 22q13.2 in the vicinity of CYP2D6 gene and contains a microsatellite with dinucleotide repeats in exon 11 (Fig. 1). Although chromosome 22 was not identified as a linkage region in recent genomic screenings for Alzheimer’s disease (AD), previous studies have shown an association between the CYP2D6 polymorphism and Parkinson’s disease (PD) or Lewy body variant (LBV) of AD (1,18,19,21). To further clarify the role of septin 3 in normal and diseased tissues, the present study aimed to assess the expression of septin 3 isoforms in human fetal and adult cerebral cortices and analyze whether microsatellite polymorphism of septin 3 is associated with pure AD, LBV, or PD. In addition, a possible linkage between CYP2D6 polymorphism and septin 3 polymorphism was investigated.
Figure 1.
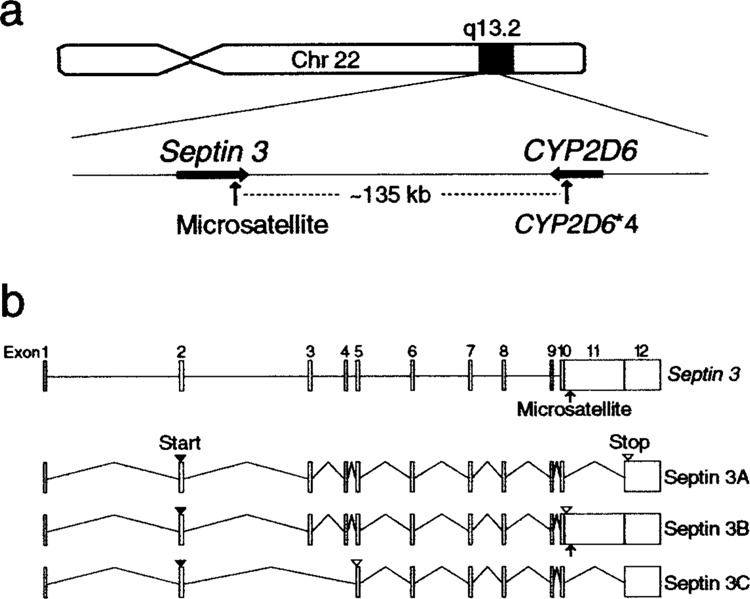
Chromosomal locus and genomic structure of septin 3. (a) Septin 3 gene is mapped to chromosome 22q13.2 in the vicinity of CYP2D6 gene. The distance between the two polymorphic sites, septin 3 exon 11 microsatellite and CYP2D6*4, is approximately 135 kb. (b) Septin 3 gene consists of 12 exons (boxes) and contains a microsatellite in exon 11 (arrow). Septin 3 has three isoforms, A, B, and C, that are produced by alternative splicing of a single transcript. Start and stop codons are indicated by closed and open arrowheads, respectively. Based on Ensembl’s Human Genome Browser (http://www.ensembl.org/Homo_sapiens/) and Methner et al. (16).
MATERIALS AND METHODS
Subjects
Seventy-four control subjects (mean age ± SD, 74.8 ± 8.8 years), 99 patients with AD (78.7 ± 8.8 years), 58 patients with LBV (78.5 ± 8.0 years), and 45 patients with PD (74.2 ± 6.1 years) were studied. The diagnosis of the diseases was pathologically confirmed at autopsy. The presence of Lewy bodies in neocortical and subcortical regions served as a criterion for differentiating LBV from AD. The control group contained 28 subjects whose brains exhibited no neuropathological changes characteristic of AD, LBV, or PD, and 46 subjects who had been clinically diagnosed not to have dementia or parkinsonism. The latter subjects were included in the control group to supplement a limited number of control brains available for the allelic association study. All subjects were Caucasian. Brain samples were obtained from the University of California (UCSD; San Diego, CA) Alzheimer’s Disease Research Center (ADRC), the University of Pennsylvania Medical Center (Philadelphia, PA), and the National Neurological Research Specimen Bank in Los Angeles (CA). Blood samples of the control group were obtained from UCSD-ADRC. Informed consent was obtained for all samples.
Reverse Transcription-Coupled Polymerase Chain Reaction (RT-PCR)
Total RNA was extracted from the frontal cortex of a control subject by the guanidinium/CsCl method (6). First-strand cDNA was synthesized from 3 μg total RNA with oligo(dT)20 primer (Toyobo Co., Ltd., Osaka, Japan) using 100 units of Moloney murine leukemia virus reverse transcriptase ReverTra Ace (Toyobo Co., Ltd.) in a total volume of 20 μl. The cDNA thus formed served as a template for polymerase chain reaction (PCR). The following primers were used to detect mRNA of septin 3 isoforms A (GenBank, Bethesda, MD; septin 3A, Accession No. AF285107), B (septin 3B, AF285109), and C (septin 3C, AF285108): 5′-GAT TCA TGT CCA AAG GGC TC-3′ (septin 3A, 3B, and 3C; forward), 5′-ACA GCA CTA AAA AGG GCT TGG-3′(3A, reverse), 5′-CTT ACA AGG GAC TCT CCA GG-3′ (3B, reverse), and 5′-TTC AAG CTC CTT GCT GAG GT-3′ (3C, reverse). PCR was performed with 0.65 unit of Pyrobest DNA polymerase (Takara Bio Inc., Otsu, Japan) in 20 μl of 1× PCR buffer containing 0.4 μl cDNA prepared as above, 0.2 mM each dNTP mixture, 1.0 mM MgCl2, and 0.2 μM each of the primers. The mixture was subjected to 35 cycles of denaturation at 95°C for 30 s, annealing at 55°C for 30 s, and extension at 72°C for 1.5 min, followed by a final extension at 72°C for 2 min. The Human Fetal Brain Large-Insert cDNA library (BD Biosciences Clontech, Palo Alto, CA) was also subjected to PCR. The PCR products were electrophoresed on 1.5% agarose gel containing ethidium bromide.
Analysis of Septin 3 Polymorphism
Genomic DNA was extracted from frozen brain samples or leukocyte preparations by the phenol/chloroform method. Polymorphism of septin 3 was analyzed by PCR with primers flanking the polymorphic region. The sequence of the forward primer was 5′-GTC TGG TAT TTG TGG AGC ATC-3′, corresponding to the sequence from nt 1170 to 1190 in exon 11 of septin 3B mRNA (GenBank, Accession No. AF285109), and that of the reverse primer was 5′-CTT ACA AGG GAC TCT CCA GG-3′, complementary to the sequence from nt 1318 to 1299 in exon 11. PCR was performed with 1.0 unit of rTaq DNA polymerase (Toyobo Co., Ltd.) in 20 μl of 1× PCR buffer containing 0.1 μg genomic DNA, 0.2 mM each dNTP mixture, 1.5 mM MgCl2, and 0.2 μM each of the primers. [α-32P]dCTP 1.5 (μCi was added to this mixture. The mixture was subjected to 32 cycles of denaturation at 95°C for 1 min, annealing at 55°C for 1 min, and extension at 75°C for 30 s, followed by a final extension at 75°C for 5 min. The PCR products were denatured and separated on a 6% polyacrylamide gel containing 8.0 M urea. The alleles were identified by autoradiography. An M13mp18 DNA sequencing ladder was used as a size marker.
Identification of CYP2D6*4
Analysis of the CYP2D6*4 allele was carried out by the PCR-BstNI restriction fragment length polymorphism technique (19). The sequence of the forward primer was 5′-GCC TTC GCC AAC CAC TCC G-3′, corresponding to the sequence from nt 3359 to 3377 in exon 3 of CYP2D6 gene (GenBank, Accession No. M33388), and that of the reverse primer was 5′-AAA TCC TGC TCT TCC GAG GC-3′, complementary to the sequence from nt 3713 to 3694 in intron 4. PCR amplification was performed as described above for septin 3 polymorphism, except for omission of [α-32P]dCTP and annealing at 60°C. The PCR product of 355 bp was digested with BstNI restriction endonuclease. The CYP2D6*4 allele missing the recognition site gave a 355-bp band, whereas the wild-type allele was digested to give 105- and 250-bp bands.
Statistical Analysis
Genetic association was examined using the contingency tables and incorporated the use of Kruskal-Wallis and Wilcoxon rank sum statistics. Odds ratio (OD) and 95% confidence interval (CI) were generated from subset 2 × 2 tables, with simple tests for association calculated using the chi-square test or Fisher’s exact test. The latter test was applied when at least one expected value was smaller than five. These statistical analyses were carried out by using InStat, version 2.03 (GraphPad Software, San Diego, CA) and SAS, version 8.2 (SAS Statistical, Software Inc., Cary, NC).
RESULTS
Detection of Septin 3 mRNAs by RT-PCR
Expression of three isoforms of septin 3 was analyzed in fetal and adult human brains by RT-PCR. Septin 3A and 3B mRNAs were clearly detected as the bands of expected sizes in both fetal and adult human brains, whereas septin 3C mRNA was not detectable in either tissue (Fig. 2).
Figure 2.
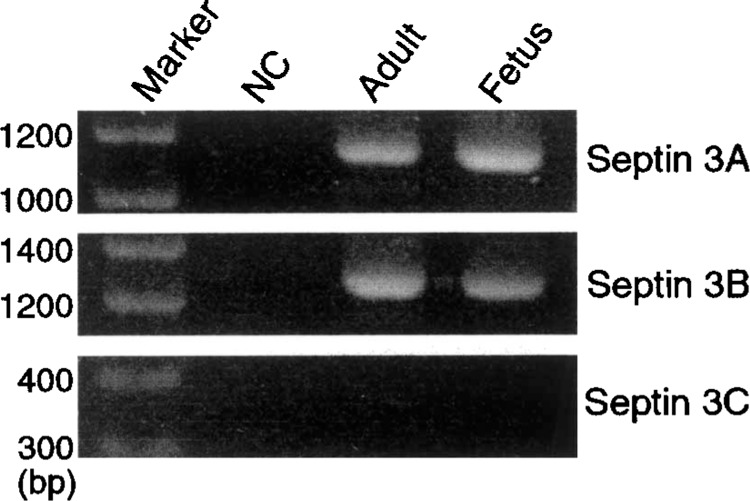
Detection of septin 3 mRNAs in human brains. cDNA synthesized from mRNA isolated from fetal and adult human brains was used as templates for PCR. Three sets of primers were used to detect alternatively spliced forms of septin 3 mRNA. The expected sizes of septin 3A, 3B, and 3C mRNAs were 1160, 1308, and 397 bp, respectively. Septin 3A and 3B, but not 3C, mRNAs were detected in both fetal and adult brains. Sterilized water was used as a negative control (NC) for PCR.
Septin 3 Polymorphism in AD, LBV, and PD
Microsatellite polymorphism in the septin 3 gene was investigated for a possible association with AD, LBV, or PD. The size of PCR products carrying septin 3 polymorphism ranged from 135 to 157 bp. Septin 3 alleles were distinguished based on the size of their PCR products. Due to the low allele frequency on either extreme of the distribution, the tails were recategorized as ≤145 and ≥155 for analysis. Heterozygosity of this microsatellite polymorphism among control subjects was 0.757 (56 out of 74), and the polymorphic information content (3) was 0.688. Apparently, in both control and disease groups, alleles were distributed in two peak frequencies: a major peak at allele 147 and a minor peak at allele 153 (Table 1). The statistical difference was observed upon comparison of AD distribution to that of controls using the Wilcoxon rank sum test (test statistic = 5.504, p = 0.019). Analysis of the frequency of each allele indicated that septin 3 allele ≤145 was underrepresented in AD and LBV, compared with controls (AD vs. control, OD = 0.32, 95% CI = 0.11–0.95, p = 0.039 by Fisher’s exact test; LBV vs. control, OD = 0.22, 95% CI = 0.05–1.01, p = 0.044 by Fisher’s exact test). Septin 3 allele 153 was overrepresented in AD (AD vs. control, OD = 1.63, 95% CI = 1.01–2.62, p = 0.044 by chi-square test). However, these differences were not statistically different after Bonferroni’s correction (critical p value for 6 alleles = 0.0083).
TABLE 1.
DISTRIBUTION OF SEPTIN 3 ALLELES IN CONTROL AND DISEASE GROUPS
| Group | n | ≤145 | 147 | 149 | 151 | 153 | ≥155 |
|---|---|---|---|---|---|---|---|
| Control | 148 | 11 (7.4) | 58 (39.2) | 32 (21.6) | 5 (3.4) | 36 (24.3) | 6 (4.1) |
| AD | 198 | 5 (2.5) | 68 (34.3) | 40 (20.2) | 8 (4.0) | 68 (34.3) | 9 (4.5) |
| LBV | 116 | 2 (1.7) | 54 (46.6) | 20 (17.2) | 3 (2.6) | 32 (27.6) | 5 (4.3) |
| PD | 90 | 2 (2.2) | 37 (41.1) | 19 (21.1) | 1 (1.1) | 28 (31.1) | 3 (3.3) |
n: number of chromosomes analyzed. Number in parentheses indicates percentage.
Overall statistic (Kruskal-Wallis test) = 6.062, df = 3, p = 0.109.
Wilcoxon rank sum statistic:
AD vs. controls: test statistic = 5.504, df = 1, p = 0.019.
LBV vs. controls: test statistic = 0.355, df = 1, p = 0.551.
PD vs. controls: test statistic = 0.854, df = 1, p = 0.355.
Based on the allele distribution in two high frequency peaks, septin 3 polymorphic alleles were dichotomized. The alleles of 149 and shorter were termed short-type (S) allele, and those of 151 and longer were termed long-type (L) allele in this study. After this categorization, a significant difference was found between control and AD (AD vs. control, p = 0.034 by chi-square test) (Table 2). In LBV and PD, the allelic distribution of septin 3 polymorphism did not differ significantly from control. The proportions of three septin 3 genotypes, S/S, S/L, and L/L, in control and disease groups are summarized in Table 3. The progression from S/S to L/L was assumed to be ordinal for statistical analysis. There was no significant difference in overall genotypic distribution between the control and disease groups (Kruskal-Wallis statistic = 5.087, p = 0.167); however, a statistically significant difference in genotype distribution was noted between AD patients and controls (Wilcoxon rank sum statistic = 4.706, p = 0.030). Genotype S/S was found to be underrepresented in AD compared with controls (AD vs. control, OD = 0.46, 95% CI = 0.25–0.87, p = 0.015 by chi-square test) (Fig. 3). Conversely, AD patients carrying at least one L allele (S/L, L/L) were significantly more abundant than controls. This difference was significant even after Bonferroni’s correction (critical p value for 3 genotypes = 0.017). The frequency of genotypes S/L or L/L did not differ significantly in any disease groups from control.
TABLE 2.
FREQUENCY OF SEPTIN 3 ALLELES IN CONTROL AND DISEASE GROUPS
| Group | N | S | L | χ2 | p Value |
|---|---|---|---|---|---|
| Control | 148 | 101 (68.2) | 47 (31.8) | ||
| AD | 198 | 113 (57.1) | 85 (42.9) | 4.480 | 0.034 |
| LBV | 116 | 76 (65.5) | 40 (34.5) | 0.219 | 0.640 |
| PD | 90 | 58 (64.4) | 32 (35.6) | 0.364 | 0.546 |
n: number of chromosomes analyzed. S: short-type allele (≤149). L: long-type allele (≥151). Number in parentheses indicates percentage.
Overall statistic (Kruskal-Wallis test) = 5.120, df = 3, p = 0.163.
TABLE 3.
GENOTYPIC DISTRIBUTION OF SEPTIN 3 POLYMORPHISM IN CONTROL AND DISEASE GROUPS
| Group | N | S/S | S/L | L/L |
|---|---|---|---|---|
| Control | 74 | 35 (47.3) | 31 (41.9) | 8 (10.8) |
| AD | 99 | 29 (29.3) | 55 (55.6) | 15 (15.2) |
| LBV | 58 | 25 (43.1) | 26 (44.8) | 7 (12.1) |
| PD | 45 | 21 (46.7) | 16 (35.6) | 8 (17.8) |
N: number of subjects analyzed. S: short-type allele (≤149). L: long-type allele (≥151). Number in parentheses indicates percentage.
Overall statistic (Kruskal-Wallis test) = 5.087, df = 3, p = 0.165.
Wilcoxon rank sum statistic:
AD vs. controls: test statistic = 4.706, df = 1, p = 0.030.
LBV vs. controls: test statistic = 0.212, df = 1, p = 0.645.
PD vs. controls: test statistic = 0.325, df = 1, p = 0.568.
Figure 3.
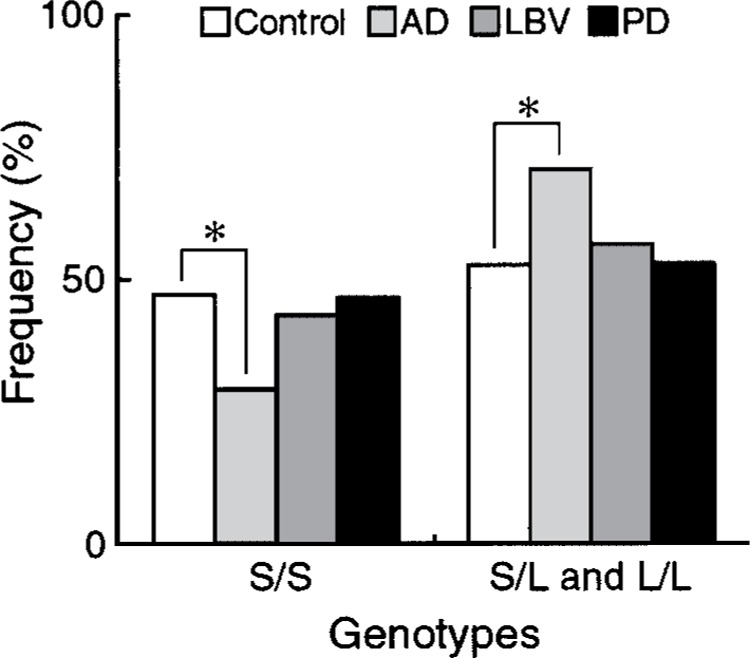
Frequency of septin 3 genotypes with L allele (S/L, L/L) or without L allele (S/S) in control and disease groups. *χ2 (1) = 5.142, p = 0.015 by chi-square test for AD versus control.
Septin 3 Polymorphism and CYP2D6 Polymorphism
A possible linkage between CYP2D6 polymorphism and septin 3 polymorphism due to their proximity was investigated. CYP2D6*4 (formerly designated as CYP2D6 B mutation) allele was used as a marker of CYP2D6 polymorphism. Analysis of the allele frequency showed a strong linkage between septin 3 allele 147 and the CYP2D6*4 allele (W/*4 vs. W/W, OD = 2.88, 95% CI = 1.95–4.27, p < 0.0001 by chi-square test; *4/*4 vs. W/W, OD = 18.9, 95% CI = 5.56–64.5, p < 0.0001 by Fisher’s exact test) (Table 4). In addition, a significant difference was observed in the overall allelic distribution of the septin 3 locus between W/W genotype and W/*4 genotype (Wilcoxon rank sum statistic = 10.647, p = 0.001).
TABLE 4.
ASSOCIATION OF SEPTIN 3 POLYMORPHISM WITH CYP2D6 POLYMORPHISM
| CYP2D6 Genotype | n | Septin 3 Allele | |||||
|---|---|---|---|---|---|---|---|
| ≤145 | 147 | 149 | 151 | 153 | ≥155 | ||
| W/W | 340 | 14 (4.1) | 98 (28.8) | 82 (24.1) | 12 (3.5) | 118 (34.7) | 16 (4.7) |
| W/*4 | 156 | 4 (2.6) | 84 (53.8)* | 19 (12.2) | 5 (3.2) | 38 (24.4) | 6 (3.8) |
| *4/*4 | 26 | 0 (0.0) | 23 (88.5)† | 3 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
n: number of chromosomes analyzed. W: wild-type allele of CYP2D6. *4: CYP2D6*4 allele. Number in parentheses indicates percentage.
Wilcoxon rank sum statistic (W/W vs. W/*4) = 10.647, df = 1, p = 0.001.
χ2(1) = 28.82, p < 0.0001 (chi-square test for W/*4 vs. W/W), critical p value = 0.0083.
χ2(1) = 38.82, p < 0.0001 (Fisher’s exact test for *4/*4 vs. W/W), critical p value = 0.0083.
CYP2D6 Polymorphism in AD, LBV, and PD
Genotypic distribution and allelic frequency of CYP2D6*4 in control and disease groups are summarized in Table 5. Overall genotype distribution by groups was not statistically significant (Kruskal-Wallis statistic = 7.352, p = 0.061); however, a statistically significant difference was found between LBV patients and controls (Wilcoxon rank sum statistic = 5.238, p = 0.022). Analysis of the frequency of individual genotypes indicated that genotype W/W was underrepresented and W/*4 was overrepresented in LBV compared with controls (W/W, OD = 0.37, 95% CI = 0.17–0.80, p = 0.011 by chi-square test; W/*4, OD = 2.64, 95% CI = 1.18–5.92, p = 0.017 by chi-square test). These differences were statistically significant after Bonferroni’s correction (critical p value for 3 genotypes = 0.017). The CYP2D6*4 allele was significantly overrepresented in LBV (28.3%) compared with controls (15.5%) (OD = 2.15, 95% CI = 1.13–4.09, p = 0.018 by chi-square test).
TABLE 5.
GENOTYPIC DISTRIBUTION AND ALLELIC FREQUENCY OF CYP2D6*4 BY GROUP
| Group | Genotype | Allele | |||||
|---|---|---|---|---|---|---|---|
| N | W/W | W/*4 | *4/*4 | n | W | *4 | |
| Control | 71 | 52 (73.2) | 16 (22.5) | 3 (4.2) | 142 | 120 (84.5) | 22 (15.5) |
| AD | 99 | 68 (68.7) | 28 (28.3) | 3 (3.0) | 198 | 164 (82.8) | 34 (17.2) |
| LBV | 46 | 23 (50.0) | 20 (43.5) | 3 (6.5) | 92 | 66 (71.7) | 26 (28.3) |
| PD | 45 | 27 (60.0) | 14 (31.34) | 4 (8.9) | 90 | 68 (75.6) | 22 (24.4) |
N: number of subjects analyzed. n: number of chromosomes analyzed. W: wild-type allele. CYP2D6*4: CYP2D6*4 allele. Number in parentheses indicates percentage.
Genotype Distribution:
Overall statistic (Kruskal-Wallis test) = 7.352, df = 3, p = 0.061.
Wilcoxon rank sum statistic:
AD vs. controls: test statistic = 0.159, df = 1, p = 0.690.
LBV vs. controls: test statistic = 5.238, df = 1, p = 0.022.
PD vs. controls: test statistic = 2.457, df = 1, p = 0.117.
Allele Distribution:
Overall statistic (Kruskal-Wallis test) = 7.833, df = 3, p = 0.050.
Wilcoxon rank sum statistic:
AD vs. controls: test statistic = 0.169, df = 1, p = 0.681.
LBV vs. controls: test statistic = 5.582, df = 1, p = 0.018.
PD vs. controls: test statistic = 2.872, df = 1, p = 0.090.
DISCUSSION
Septin 3 has been shown to occur in three alternatively spliced forms, septin 3A, 3B, and 3C [Fig. 1b and (16)]. Septin 3A and 3B differ in the absence or presence of exon 11, respectively. The omission of exon 11 in septin 3A leads to translation of C-terminal amino acids from exon 12, whereas the presence of exon 11 in septin 3B leads to translation of 13 amino acids followed by a stop codon and an additional 3′ untranslated region (3′ UTR). Septin 3C is an atypical member lacking a highly conserved central domain, which is produced by omission of exons 3 and 4 leading to a frameshift and early stop. In a previous study, only septin 3B was reported as detectable in adult human brain by Northern blotting (16). In the present study, the expression of both septin 3A and 3B mRNAs was demonstrated in fetal and adult human brains, a difference that may be due to primer selection, RNA purity, etc.
Human septin 3 gene is highly homologous to mouse Sep3 and rat G-septin genes. Mouse Sep3 is upregulated in neuronal tissues during development (24), while rat G-septin was identified as a specific substrate for type I cGMP-dependent protein kinase (PKG) and detected as a phosphoprotein in nerve termini (25). Therefore, septin 3 and its modification by PKG may play an important role in brain functions.
Identification of three septins (Nedd5, H5, and Diff6) in neurofibrillary tangles, neuropil threads, and dystrophic neurites in AD brains suggests their involvement in neurological disorders (13). To test whether septin 3 interacts with any of these septins, we prepared and analyzed HeLa cells in which septin 3 and Nedd5 were transiently coexpressed. Our preliminary analysis revealed that septin 3 was coimmunoprecipitated with Nedd5, suggesting their interaction and possible involvement of septin 3 in neurodegenerative processes (unpublished data). In this context, noteworthy is our present result that homozygous septin 3 S allele was underrepresented among AD patients, which suggests a recessively protective effect of S allele on AD pathogenesis or a dominantly causative effect of L allele.
The septin 3 microsatellite is located within the 3′ UTR in exon 11, which is retained only in septin 3B mRNA. Elements of 3′ UTR, such as the poly(A) tail and AU-rich sequences, modulate reportedly mRNA translation and stability (4,5,11). In other genes, dinucleotide or trinucleotide repeats within the 3′ UTR have been identified as binding sites for RNA binding proteins that regulate localization, stability, and translation of mRNA (7,17,20). It seems possible that the septin 3 polymorphism act as cis-acting regulatory element(s), directly affecting posttranscriptional regulation of septin 3B mRNA by action of some RNA-binding protein(s).
The septin 3 gene is located in close proximity to the CYP2D6 gene (Fig. 1). A strong linkage was found between septin 3 allele 147 and CYP2D6*4 allele. Overrepresentation of CYP2D6*4 allele in LBV, as revealed in the present study, confirmed our previous observation (18,21). Septin 3 allele 147 was also overrepresented in LBV, although the difference did not reach a significant level. This trend is probably due to the linkage between septin 3 allele 147 and pathogenic CYP2D6*4 allele.
In summary, our present findings suggest that the polymorphism of septin 3 gene is associated with the susceptibility of the brain to neurodegenerative changes specific for AD, but not LBV or PD. Further investigation of septin 3 polymorphism in different ethnic populations and their relevance to changes in gene expression are required to confirm this notion.
ACKOWLEDGMENTS
We thank Drs. R. Katzman, L. J. Thal (USDA), and J. Q. Trojanowski (University of Pennsylvania) for providing brain samples. Some of the brain specimens were obtained from the National Neurological Research Specimen Bank (Los Angels, CA), which is sponsored by NINDS/NIMH, National Multiple Sclerosis Society, Hereditary Disease Foundation, Comprehensive Epilepsy Program, Tourette Syndrome Association, Dystonia Medical Research Foundation, and Veterans Health Services and Research Administration, Department of Veterans Affairs. This work was supported by grants from Japan Foundation for Applied Enzymology and Yamanouchi Foundation for Research on Metabolic Disorders (K.U.) and NIH grant AG5131 (E.M.), and a research fellowship from the 21st Century COE Program, Kyoto University Alliance for Chemistry (M.T.).
REFERENCES
- 1. Armstrong M.; Daly A. K.; Cholerton S.; Bateman D. N.; Idle J. R. Mutant debrisoquine hydroxylation genes in Parkinson’s disease. Lancet 339:1017–1018; 1992. [DOI] [PubMed] [Google Scholar]
- 2. Beites C. L.; Xie H.; Bowser R.; Trimble W. S. The septin CDCrel-1 binds syntaxin and inhibits exocytosis. Nat. Neurosci. 2:434–439; 1999. [DOI] [PubMed] [Google Scholar]
- 3. Botstein D.; White R. L.; Skolnick M.; Davis R. W. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am. J. Hum. Genet. 32:314–331; 1980. [PMC free article] [PubMed] [Google Scholar]
- 4. Brennan C. M.; Steitz J. A. HuR and mRNA stability. Cell. Mol. Life Sci. 58:266–277; 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chen C. Y.; Shyu A. B. AU-rich elements: Characterization and importance in mRNA degradation. Trends Biochem. Sci. 20:465–470; 1995. [DOI] [PubMed] [Google Scholar]
- 6. Chirgwin J. M.; Przybyla A. E.; MacDonald R. J.; Rutter W. J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry 18:5294–5299; 1979. [DOI] [PubMed] [Google Scholar]
- 7. Clerch L. B. A 3′ untranslated region of catalase mRNA composed of a stem-loop and dinucleotide repeat elements binds a 69-kDa redox-sensitive protein. Arch. Biochem. Biophys. 317:267–274; 1995. [DOI] [PubMed] [Google Scholar]
- 8. Field C. M.; Kellogg D. Septins: Cytoskeletal polymers or signalling GTPases? Trends Cell Biol. 9:387–394; 1999. [DOI] [PubMed] [Google Scholar]
- 9. Hartwell L. H. Genetic control of the cell division cycle in yeast. IV. Genes controlling bud emergence and cytokinesis. Exp. Cell Res. 69:265–276; 1971. [DOI] [PubMed] [Google Scholar]
- 10. Hsu S. C.; Hazuka C. D.; Roth R.; Foletti D. L.; Heuser J.; Scheller R. H. Subunit composition, protein interactions, and structures of the mammalian brain sec6/8 complex and septin filaments. Neuron 20: 1111–1122; 1998. [DOI] [PubMed] [Google Scholar]
- 11. Jackson R. J. Cytoplasmic regulation of mRNA function: The importance of the 3′ untranslated region. Cell 74:9–14; 1993. [DOI] [PubMed] [Google Scholar]
- 12. Kartmann B.; Roth D. Novel roles for mammalian septins: From vesicle trafficking to oncogenesis. J. Cell Sci. 114:839–844; 2001. [DOI] [PubMed] [Google Scholar]
- 13. Kinoshita A.; Kinoshita M.; Akiyama H.; Tomimoto H.; Akiguchi I.; Kumar S.; et al. Identification of septins in neurofibrillary tangles in Alzheimer’s disease. Am. J. Pathol. 153:1551–1560; 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kinoshita A.; Noda M.; Kinoshita M. Differential localization of septins in the mouse brain. J. Comp. Neurol. 428:223–239; 2000. [DOI] [PubMed] [Google Scholar]
- 15. Macara I. G.; Baldarelli R.; Field C. M.; Glotzer M.; Hayashi Y.; Hsu S. C.; et al. Mammalian septins nomenclature. Mol. Biol. Cell 13:4111–4113; 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Methner A.; Leypoldt F.; Joost P.; Lewerenz J. Human septin 3 on chromosome 22q13.2 is upregulated by neuronal differentiation. Biochem. Biophys. Res. Commun. 283:48–56; 2001. [DOI] [PubMed] [Google Scholar]
- 17. Philips A. V.; Timchenko L. T.; Cooper T. A. Disruption of splicing regulated by a CUG-binding protein in myotonic dystrophy. Science 280:737–741; 1998. [DOI] [PubMed] [Google Scholar]
- 18. Saitoh T.; Xia Y.; Chen X.; Masliah E.; Galasko D.; Shults C.; et al. The CYP2D6B mutant allele is overrepresented in the Lewy body variant of Alzheimer’s disease. Ann. Neurol. 37:110–112; 1995. [DOI] [PubMed] [Google Scholar]
- 19. Smith C. A.; Gough A. C.; Leigh P. N.; Summers B. A.; Harding A. E.; Maraganore D. M.; et al. Debrisoquine hydroxylase gene polymorphism and susceptibility to Parkinson’s disease. Lancet 339:1375–1377; 1992. [DOI] [PubMed] [Google Scholar]
- 20. Takahashi N.; Sasagawa N.; Suzuki K.; Ishiura S. The CUG-binding protein binds specifically to UG dinucleotide repeats in a yeast three-hybrid system. Biochem. Biophys. Res. Commun. 277:518–523; 2000. [DOI] [PubMed] [Google Scholar]
- 21. Tanaka S.; Chen X.; Xia Y.; Kang D. E.; Matoh N.; Sundsmo M.; et al. Association of CYP2D microsatellite polymorphism with Lewy body variant of Alzheimer’s disease. Neurology 50:1556–1562; 1998. [DOI] [PubMed] [Google Scholar]
- 22. Trimble W. S. Septins: A highly conserved family of membrane-associated GTPases with functions in cell division and beyond. J. Membr. Biol. 169:75–81; 1999. [DOI] [PubMed] [Google Scholar]
- 23. Vega I. E.; Hsu S. C. The septin protein Nedd5 associates with both the exocyst complex and microtubules and disruption of its GTPase activity promotes aberrant neurite sprouting in PC12 cells. Neuroreport 14:31–37; 2003. [DOI] [PubMed] [Google Scholar]
- 24. Xiong J. W.; Leahy A.; Stuhlmann H. Retroviral promoter-trap insertion into a novel mammalian septin gene expressed during mouse neuronal development. Mech. Dev. 86:183–191; 1999. [DOI] [PubMed] [Google Scholar]
- 25. Xue J.; Wang X.; Malladi C. S.; Kinoshita M.; Milburn P. J.; Lengyel I.; et al. Phosphorylation of a new brain-specific septin, G-septin, by cGMP-dependent protein kinase. J. Biol. Chem. 275:10047–10056; 2000. [DOI] [PubMed] [Google Scholar]
- 26. Zhang Y.; Gao J.; Chung K. K.; Huang H.; Dawson V. L.; Dawson T. M. Parkin functions as an E2-dependent ubiquitin-protein ligase and promotes the degradation of the synaptic vesicle-associated protein, CD-Crel-1. Proc. Natl. Acad. Sci. USA 97:13354–13359; 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]


