Abstract
The Forkhead Box (Fox) proteins are an extensive family of transcription factors that shares homology in the winged helix DNA binding domain. Liver regeneration studies with the –3 kb transthyretin (TTR) promoter-driven FoxM1B transgenic (TG) mice demonstrated that premature hepatocyte nuclear localization of the FoxM1B transgene protein at 16 h following partial hepatectomy (PHx) caused an 8-h acceleration in the onset of hepatocyte DNA replication (S-phase) and mitosis by stimulating earlier expression of cell cycle genes. Whether the FoxM1B transgene protein participates in immediate early events during liver regeneration remains to be determined. Here, we found that the FoxM1B transgene protein translocated to hepatocyte nuclei immediately following PHx, that its nuclear staining persisted for the first 6 h after surgery, and that this translocation was associated with an increase in size of regenerating TG hepatocytes. However, regenerating TTR-FoxM1B liver did not exhibit altered expression of proteins that have been implicated in mediating increased cell size, including serum-and-gucocorticoid-inducible protein kinase (SGK), protein kinase-B/Akt, the tumor suppresser gene PTEN (negative regulator of the PI3K/Akt pathway), c-Myc, or peroxisome proliferation. Moreover, we demonstrated that hepatocyte nuclear translocation of the FoxM1B transgene protein was rapidly induced during the hepatic acute phase response, which occurs during the immediate early stages of liver regeneration. Analysis of cDNA expression arrays identified a number of genes such as immediate early transcription factors (ID-3, Stat3, Nur77), matrix metalloproteinase-9 (MMP-9), and several glutathione S-transferase (GST) isoforms and stress response genes, whose expression is elevated in regenerating TTR-FoxM1B TG livers compared with regenerating wild-type (WT) liver. These liver regeneration studies demonstrate that hepatocyte nuclear translo-cation of the FoxM1B transgene protein was sustained for the first 6 h after PHx, and was associated with transient hypertrophy of regenerating TG hepatocytes and increased expression of genes that may enhance hepa-tocyte proliferation.
Keywords: Forkhead Box transcription factor, Liver regeneration, Hypertrophy, FoxM1, Immediate early transcription factors, Winged helix domain, Nuclear localization
THE mammalian liver is one of the few adult organs capable of completely regenerating itself in response to injury (32). Liver regeneration induced by two thirds partial hepatectomy (PHx) results in synchronous induction of hepatocyte DNA replication (S-phase) and mitosis. This proliferative response is initiated by the release of growth factors and cytokines that stimulate expression and nuclear translocation of immediate early transcription factors, which mediate reentry of terminally differentiated hepatocytes into the cell cycle (12,16,32,43). The Forkhead Box (Fox) transcription factors are an extensive family of transcription factors, consisting of more than 50 mammalian proteins (21), that shares homology in the winged helix DNA binding domain (7,30). Its members play important roles in regulating transcription of genes involved in cellular proliferation (46,47,49, 57,58), differentiation (6,9,13,59), and metabolic homeostasis (20,35). The FoxM1B (FoxM1) protein (previously known as HFH-11B, Trident, or WIN) is a proliferation-specific transcriptional activator that is expressed in all replicating cells examined (27,56,58). FoxM1B expression is extinguished in the liver postnatally, but its expression is markedly induced in regenerating liver during the G1/S transition and continues throughout the period of hepatic cell proliferation (58). Premature expression of FoxM1B in regenerating liver of transthyretin (TTR)-FoxM1B transgenic (TG) mice accelerated the onset of hepatocyte DNA replication and mitosis by stimulating earlier expression of cell cycle genes (46,57). Furthermore, maintaining hepatocyte expression of FoxM1B in 12-month-old (old-aged) TTR-FoxM1B TG mice is sufficient to increase regenerating hepatocyte DNA replication and mitosis and reestablish expression of cell cycle regulatory genes to levels found in young regenerating mouse liver (49). These results suggest the hypothesis that Foxm1b controls the transcriptional network of genes essential for progression through the cell cycle.
Consistent with this hypothesis, we used the albumin promoter/enhancer-driven Cre recombinase transgene to mediate conditional deletion of the Foxm1b LoxP/LoxP (fl/fl) allele in adult hepatocytes, and found that Foxm1b deficiency resulted in significant reduction in hepatocyte DNA replication and mitosis following partial hepatectomy (47). Reduced hepatocyte DNA replication was associated with increased protein levels of Cdk inhibitor p21Cip1 and reduced protein expression of Cdc25A phosphatase, leading to decreased Cdk2 activation and progression into S-phase (47). Diminished hepatocyte mitosis was associated with undetectable expression of the Cdc25B phosphatase and delayed accumulation of cyclin B1, which is required for cyclin B-Cdk1 kinase activation and entry into mitosis.
In this study, we compared wild-type (WT) and TTR-FoxM1B transgenic (TG) mice during the initial time points following partial hepatectomy (PHx). The FoxM1B transgene protein was found to translocate immediately to hepatocyte nuclei following PHx, and its nuclear staining was sustained for the first 6 h following PHx. FoxM1B nuclear staining was associated with a significant increase in size of regenerating TG hepatocytes. However, regenerating TTR-FoxM1B liver did not exhibit altered expression of proteins which have been implicated in mediating increase in cell size, including serum-and-gucocorticoid-inducible protein kinase (SGK) (4,5,26,45,50), Akt (8,14, 41,44), the tumor suppressor gene PTEN (2,3,10,18, 28,52), and c-Myc (24). Furthermore, regenerating TG liver displayed WT levels of the peroxisomal membrane protein 70 (PMP 70), indicating that there was no increase in peroxisome proliferation, which is also known to increase hepatocyte size (19,37). These studies demonstrate that the transient nuclear translocation of FoxM1B transgene protein is associated with increase in the size of regenerating TG hepatocytes and utilized pathways that are distinct from SGK, PI3K/Akt, c-Myc, or peroxisome proliferation. Analysis of cDNA expression arrays identified a number of genes whose expression is elevated in regenerating TTR-FoxM1B TG livers compared with regenerating WT liver. These include immediate early transcription factors (ID-3, Stat3, Nur77), matrix metalloproteinase-9 (MMP-9), and several glutathione S-transferase (GST) isoforms and stress response genes. These liver regeneration studies demonstrate that a transient hepatocyte nuclear translocation of the FoxM1B transgene protein immediately following PHx was associated with transient hypertrophy of regenerating TG hepatocytes and increased expression of genes that may enhance cell cycle progression.
MATERIALS AND METHODS
Partial Hepatectomy Surgery, LPS Administration, Immunohistochemical Staining, and Western Blot Analysis
Generation of FoxM1B transgenic CD-1 mice, which used the -3 kb TTR promoter to constitutively express the human FoxM1B cDNA transgene in he-patocytes, has been described previously (57). Two-month-old WT or TTR-FoxM1B TG CD-1 mice were subjected to two thirds PHx to induce liver regeneration as described previously (57). We sacrificed two mice per time point following PHx operation (15 min, 1, 2, 4, 6, and 8 h) using CO2 asphyxiation, and regenerating livers were dissected. To induce the acute phase response, 2-month-old WT or TG CD-1 mice were treated with 100 μg of lipopolysaccharide (LPS; serotype O11B4; Sigma) by IP injection, sacrificed at either 1 or 2 h following LPS administration, and their livers dissected as described previously (38). Dissected mouse liver tissue was divided into two portions: one to isolate nuclear protein extracts as described previously (33); the rest of the liver was paraffin embedded and 5-μm sections were prepared with a Microtome and then processed for immunohistochemical staining as described previously (57). Furthermore, two 8-week-old WT and TTR-FoxM1B TG mice were subjected to PHx, sacrificed at 4 h following surgery, and liver tissue was dissected. These livers were used to prepare total protein extracts and total RNA as described previously (39,57). These mouse experiments were approved by the University of Illinois Animal Welfare committee.
For immunohistochemical staining, paraffin wax was removed from liver sections with xylenes, and they were rehydrated with decreasing graded ethanol washes. Microwave retrieval was used to enhance the antigenic activity as described previously (39,57). FoxM1B (HFH-11B) antibody (57,58) was used for immunohistochemical detection using the ABC kit and DAB peroxidase substrate purchased from Vector Laboratories (Burlingame, CA). For Western blot analysis, 200 μg of liver nuclear extract (33) was separated on SDS polyacrylamide gel electrophoresis (PAGE) and transferred to Protran membrane (Schleicher & Schuell, Keene, NH). The primary FoxM1B N-terminal-specific antibody was amplified by biotin conjugated anti-rabbit IgG (Bio-Rad, Hercules, CA), and developed signals with enhanced chemiluminescence (ECL, Amersham Pharmacia Biotech, Piscataway, NJ).
RNase Protection Assays
RNase protection assay (RPA) probe for the mouse c-Myc gene was a gift from Dr. Nissim Hay (Dept. of Molecular Genetics, University of Illinois at Chicago). The RPA GSK probe was generated by RT-PCR of TG liver mRNA isolated at 4 h following PHx using the following primers: 5′-acctcctgtggattcc ttcctctg-3′ and 5′-actgcggggattcctcttagac-3′. RT-PCRs were performed using SUPERSCRIPT™ One-Step RT-PCR with PLATINUM® Taq kit (Invitrogen, CA) according to the manufacturer’s recommendations as described previously (23). The PCR product was cloned into pCR®II vector (TA Cloning® Kit Dual promoter, Invitrogen Life Technologies. Carlsbad, CA) and antisense-labeled RNA probe was synthesized with T7 RNA polymerase and [α-32P]UTP (ICN, Irvine, CA) using BamHI digested pCR®II GSK plasmid. RPAs were performed by hybridizing 40 μg of total liver RNA with [32P]UTP-labeled anti-sense RNA probe followed by RNase one digestion, electrophoresis, and autoradiography as described previously (39,46,57). Quantitation of expression levels was determined with scanned X-ray films or agar-ose gel tiff files by using the BioMax 1D program (Eastman Kodak Co) and were normalized to cyclophilin RNA levels. The mean was determined from two distinct regenerating livers using the Microsoft Excel program.
Analysis of cDNA Arrays
To identify genes whose expression is altered in regenerating TTR-FoxM1B TG liver at 4 h following PHx, we performed gene expression analysis using Clontech Mouse Atlas 1.2 cDNA Expression Array analysis as described previously (22,42,46). We synthesized radioactive cDNA probes from RNA prepared from two distinct regenerating livers isolated from either WT or TTR-FoxM1B TG mice at 4 h after PHx using [α-33P]dATP (ICN, Irvine, CA) as described by the manufacturer’s protocol. Each liver cDNA probe was used to hybridize to a distinct Atlas mouse 1.2 array (Clontech, Palo Alto, CA) as described previously (42,46). Following hybridizing and rinsing, Atlas cDNA arrays were exposed to the phosphor-imaging screens for 1 or 2 days and scanned with a storm 840 Phosphorimager. After subtraction of background, hybridization signals were normalized to two control genes: 40S ribosomal protein S29 (L31609) and cytoplasmic β-actin (M12481). We used the AtlasImage 1.5 program (Clontech) to determine normalized expression levels and used them to determine the fold induction of gene expression in regenerating TTR-FoxM1B TG liver compared with regenerating WT liver.
RESULTS AND DISCUSSION
FoxM1B Transgene Protein Is Immediately Translocated to the Nucleus Following PHx
Our previous studies demonstrated that deletion of the terminal 800 nucleotides from the 3′ end of the human FoxM1B (HFH-11B) cDNA resulted in stabilization of the FoxM1B transgene mRNA in uninjured liver and that the FoxM1B transgene protein was cytoplasmic in quiescent hepatocytes (57). Following PHx, nuclear FoxM1B transgene protein was detected by 16 h following PHx, which was 16 h earlier than in regenerating WT liver (57). This premature nuclear localization of the FoxM1B transgene protein in regenerating TG liver caused an 8-h acceleration of the onset of hepatocyte DNA replication and mitosis by stimulating earlier expression of cell cycle genes (46,57). In this study, we used the same TTR-FoxM1B TG mouse line to determine whether the FoxM1B transgene protein was translocated to the nucleus at early time points following PHx. WT and TTR-FoxM1B TG mice were subjected to PHx, sacrificed at 15 min (resected and remnant liver) and at 1, 2, 4, 6, and 8 h following PHx, and regenerating liver tissue was harvested. Regenerating liver was used to either prepare nuclear protein extracts for Western Blot analysis (33) or paraffin embedded and sectioned for immunohistochemical staining to determine FoxM1B nuclear levels with an N-terminal FoxM1B antibody (57,58). Surprisingly, immunohis-tochemical staining with a FoxM1B antibody showed rapid hepatocyte nuclear translocation of FoxM1B transgene protein within 15 min after beginning PHx (Fig. 1B), whereas FoxM1B protein was cytoplasmic in quiescent hepatocytes without surgery (Fig. 1A). In contrast, only low levels of hepatocyte FoxM1B nuclear staining were found immediately following PHx in WT liver (Fig. 1C–D), but this FoxM1B staining rapidly disappeared within 1 h following PHx (Fig. 2A, F). Furthermore, hepatocyte nuclear staining of the FoxM1B transgene protein persisted for the first 6 h after PHx and significantly diminished by 8 h postsurgery (Fig. 2B–E). In contrast, nuclear staining was undetectable in regenerating WT hepatocytes between 1 and 8 h following PHx (Fig. 2F–J).
Figure 1.
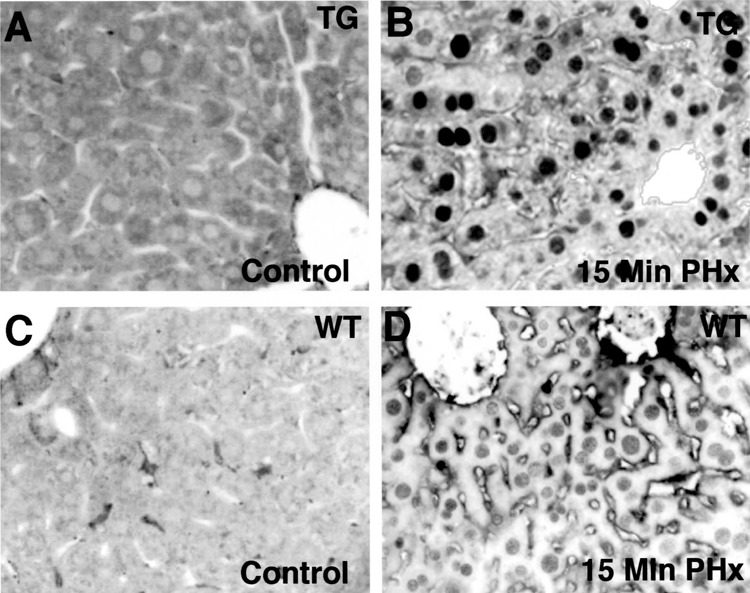
Rapid nuclear localization of FoxM1B transgene protein in regenerating transgenic hepatocytes immediately following partial hepatectomy. Eight-week-old wild-type (WT) and TTR-FoxM1B transgenic (TG) CD1 mice were subjected to two thirds partial hepatectomy (PHx) and regenerating livers were harvested 15 min after beginning the PHx surgery (B and D) or without surgery (control, A and C). Paraffin embedded liver sections were used for immunohistochemical staining with affinity-purified antibody specific for FoxM1B as described previously (48,49). Shown is regenerating or hepatocyte staining of FoxM1B protein in liver sections from either TG (A, B) or WT (C, D) mice. Original magnification 400×.
Figure 2.
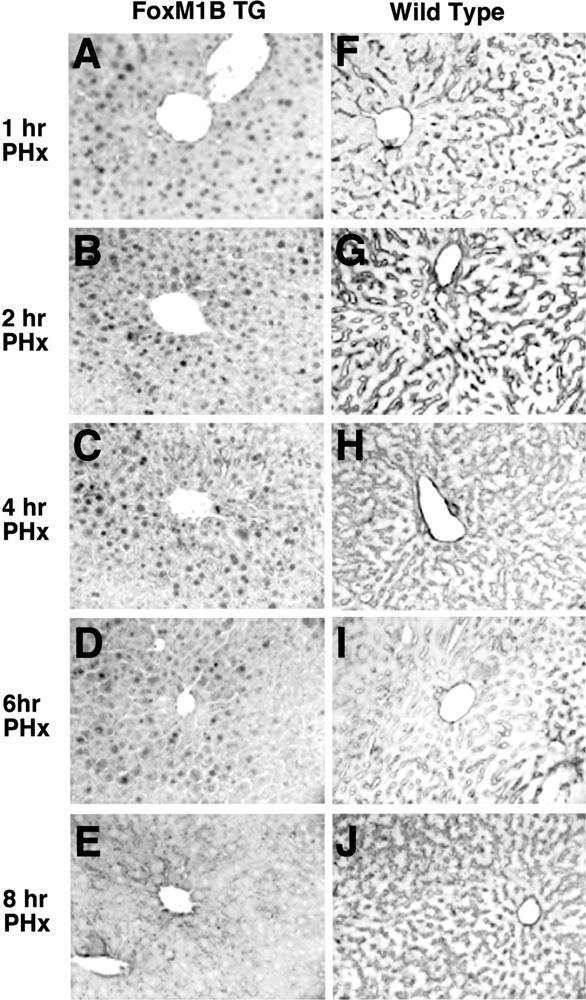
Nuclear translocation of the FoxM1B transgene protein within the first 6 h following partial hepatectomy. Eight-week-old WT (F–J) and TG (A–E) CD1 mice were subjected to two thirds partial hepatectomy (PHx) and regenerating livers were harvested at either 1, 2, 4, 6, or 8 h following PHx and used for immunohistochemical staining with affinity-purified FoxM1B-specific antibody (48,49). Original magnification 200×.
To measure FoxM1B nuclear protein, we used Western blot analysis with nuclear extracts prepared from regenerating liver and an N-terminal-specific FoxM1B antibody as described in Materials and Methods. This Western blot analysis demonstrated increased nuclear levels of the FoxM1B protein in either resected (Res) or remnant (Rem) TTR-FoxM1B TG liver compared with resected WT liver immediately following PHx (Fig. 3A). Furthermore, TG liver expressed the largest amount of nuclear FoxM1B protein within 15 min following PHx compared with regenerating TG and WT liver at 28 and 32 h following PHx (Fig. 3A). Western Blot analysis with nuclear extracts prepared from regenerating TG liver demonstrated abundant levels of FoxM1B trans-gene protein at 1, 2, and 4 h after PHx (Fig. 3B). Consistent with diminished nuclear staining of FoxM1B at 8 h following PHx (Fig. 2E), expression of nuclear FoxM1B protein was significantly diminished by the 8-h time point (Fig. 3B). These studies demonstrated that only regenerating TG liver displayed significant nuclear levels of the FoxM1B protein and that these levels persisted throughout the initial 6 h following PHx.
Figure 3.
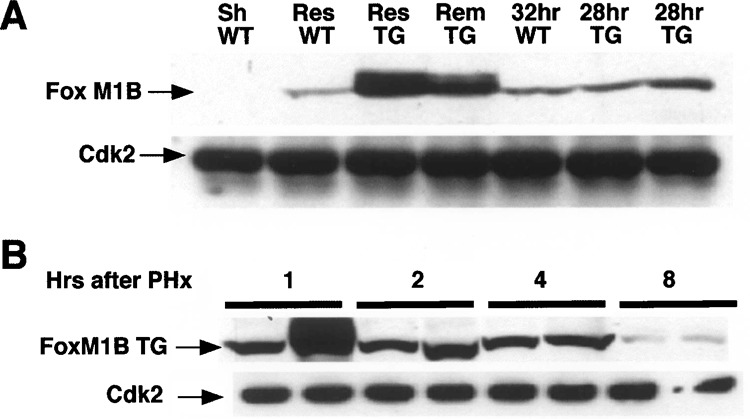
Nuclear FoxM1B transgene protein levels are increased in regenerating liver. Nuclear extracts were prepared from regenerating liver tissue 15 min after PHx, sham-operated (Sh), or regenerating wild-type (WT) and transgenic (TG) liver at the indicated times following PHx, as described previously (33). Lanes marked Res (resected) were nuclear extracts prepared from the liver tissue removed during partial hepatectomy (PHx). Lanes marked Rem (remnant) were nuclear extracts prepared immediately after PHx from the remaining liver tissue. Nuclear protein extracts from regenerating WT liver at 32 h following PHx and duplicate regenerating TG liver at 28 h after PHx were included for comparison. Liver nuclear extract (200 μg) was analyzed by Western blot analysis with polyclonal antibodies against either FoxM1B or Cdk2 (loading control). (A) FoxM1B Western blot analysis of regenerating liver nuclear extracts from WT and TG mice immediately after PHx. (B) Western blot analysis of nuclear extracts prepared from regenerating TG liver prepared from two distinct mice (1, 2, 4, or 8 h following PHx) with FoxM1B antibody.
Acute Phase Response Induces Nuclear Translocation of the FoxM1B Transgene Protein
The initial stages of liver regeneration are dominated by an acute phase response, which involves the release of the cytokines tumor necrosis factor-α (TNF-α), interleukin 6 (IL-6), and IL-1, which are critical for priming hepatocytes to respond to mitogenic signaling (11,51,53–55). We therefore examined whether induction of the hepatic acute phase response caused hepatocyte nuclear translocation of the FoxM1B transgene protein. Bacterial LPS bind to the LPS receptor on macrophages and induce secretion of TNF-α that subsequently stimulates synthesis and secretion of IL-6 and IL-1 cytokines, which bind to their cognate hepatocyte receptors, causing changes in expression of hepatic transcription factors and acute phase response genes (36). Furthermore, LPS treatment has been shown to augment hepatocyte growth factor (HGF)-mediated stimulation of hepatocyte proliferation through activation of the immediate early AP-1 (c-Jun and c-Fos) transcription factors (17). To induce the acute phase response, both TG and WT mice were subjected to IP injections of LPS, mice were sacrificed at either 1 or 2 h after LPS injection, and their livers were isolated as described previously (38). These livers were used to either prepare nuclear extracts for Western blot analysis or processed for immunohistochemical staining to determine FoxM1B nuclear levels with a FoxM1B antibody. Approximately half of the TG hepatocytes displayed nuclear staining of the FoxM1B transgene protein within 1 h of LPS injection (Fig. 4A). By 2 h following LPS injection, nearly all of the TG hepatocytes exhibited nuclear FoxM1B staining (Fig. 4B). In contrast, no detectable FoxM1B nuclear staining was found in WT liver following LPS treatment (Fig. 4C–D). Although nuclear FoxM1B protein was detected by immunohistochemical staining in a subset of TG hepatocytes at 1 h following LPS injection, Western blot analysis was only sensitive enough to detect nuclear FoxM1B transgene protein by 2 h following LPS treatment (Fig. 4E). These studies suggest that acute phase cytokines are sufficient to mediate nuclear translocation of the FoxM1B transgene protein, but only minimal WT hepatocyte expression of nuclear Foxm1b protein was found in response to LPS administration.
Figure 4.
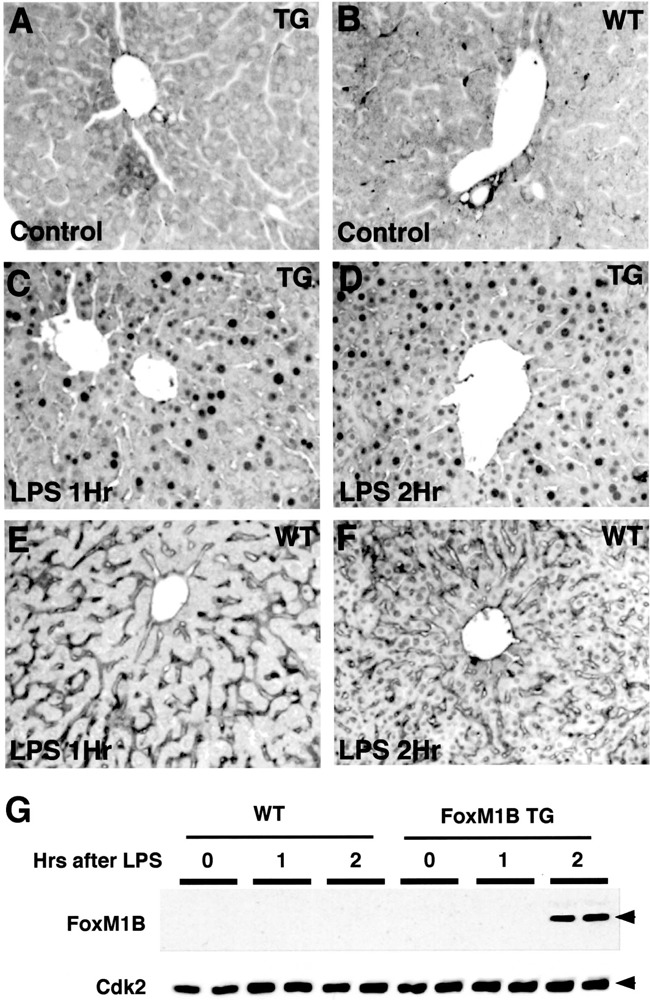
Increased hepatocyte nuclear staining of the FoxM1B transgene protein during the hepatic acute phase response. Eight-week-old WT (B, E, F) and TG (A, C, D) CD1 mice were subjected to lipopolysaccharide (LPS) injection and livers were harvested at either 0 (control; A, B), 1 h (C, E), or 2 h (D, F) following LPS treatment and used for immunohistochemical staining with affinity-purified FoxM1B antibody (48,49). (G) Western blot analysis of nuclear extracts prepared from WT and TG liver following LPS treatment with FoxM1B antibody. Nuclear extracts were prepared from WT and TG liver isolated at either 0, 1, or 2 h following LPS treatment and used for Western blot analysis with either FoxM1B or Cdk2 (loading control) specific antibodies. The numbers above the panels refer to the hours following LPS treatment. Original magnification 200×.
Hepatocyte Nuclear Staining of the FoxM1B Transgene Protein Is Associated With Increase in Regenerating Hepatocyte Size
Hematoxylin and eosin (H&E) staining of regenerating liver sections from TG and WT mice revealed an increase in regenerating TG hepatocytes size (Fig. 5A–F) compared with regenerating WT hepatocytes (Fig. 5G–L). To quantitate this increase in size, we counted the number of hepatocyte nuclei per 200× field from H&E-stained WT and TG regenerating liver sections and plotted these data for each of the time points following PHx (Fig. 6). By 1 h following PHx, TG hepatocytes displayed an increase in hepatocyte size as evidenced by a decrease in the number of hepatocyte nuclei per field (Fig. 6). These results suggested that nuclear translocation of FoxM1B protein was mediating an increase in hepatocyte size. This increase in TG hepatocyte size continued until 6 h following PHx, and the greatest size difference between WT and TG hepatocytes was observed at 4 h after surgery (Fig. 6). Interestingly, hepatocyte size began to decrease at 8 h following PHx (Fig. 6), which coincided with our inability to detect FoxM1B nuclear levels (Fig. 2). These results suggested that the transient nuclear localization of the FoxM1B transgene protein is associated with increase in regenerating TG hepatocyte size during the immediate early period of hepatocyte proliferation.
Figure 5.
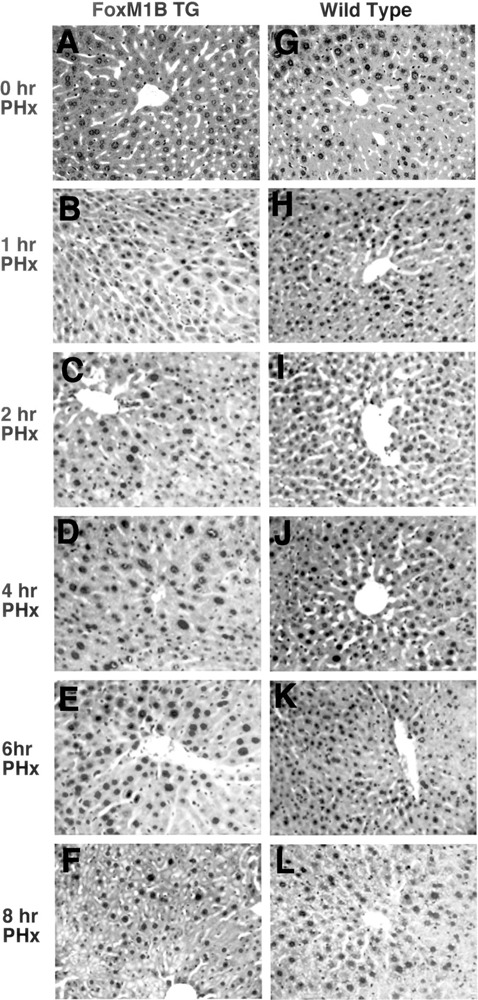
Transient increase in regenerating transgenic hepatocyte size. Eight-week-old WT (F–J) and TG (A–E) CD1 mice were subjected to PHx, and regenerating livers were harvested at either 0, 1, 2, 4, 6, or 8 h following PHx and then stained with hematoxylin and eosin (H&E) to visualize hepatocyte nuclei. Note that TG hepatocytes display an increase in size between 1 and 6 h following PHx. Original magnification 200×.
Figure 6.
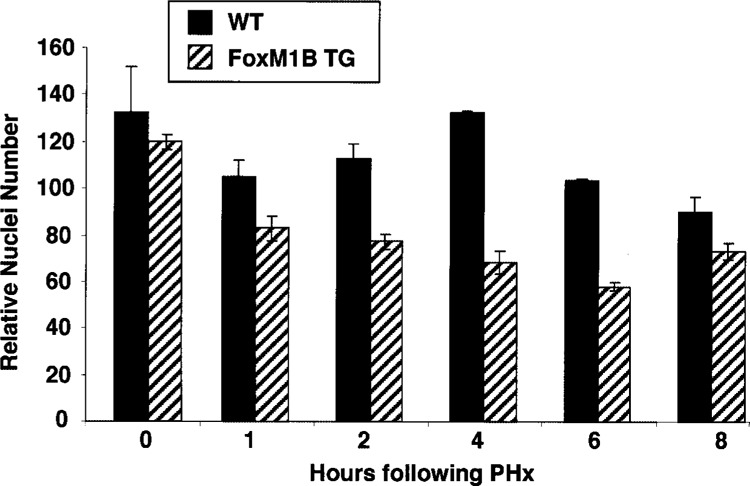
Quantitation of the increase in regenerating transgenic hepatocyte size. We counted and calculated the mean number of hepatocyte nuclei (±SD) per 200× in hematoxylin and eosin (H&E)-stained regenerating WT and TG liver section fields (five separate 200× fields from two distinct regenerating livers). Note that increase in regenerating hepatocyte size is indicated by a decrease in the number of hepatocyte nuclei per 200× field.
Regenerating TG Liver Displayed no Changes in Expression of Proteins Known to Increase Cell Size
Total RNA and protein extracts were prepared from regenerating WT and TTR-FoxM1B TG livers at 4 h following PHx because this was the time point at which we observed the maximal size difference between regenerating WT and TG hepatocytes (Figs. 5 and 6). We used these hepatic RNA samples and protein extracts to examine for expression of genes or proteins that are known to regulate cell size. Published reports describe a rapid increase in SGK expression, which stimulates activity of several potassium/chloride ion channels critical for rapid increase of cell size (4,5,26,45,50). RNase protection assays demonstrated that regenerating liver displayed an increase in Akt-related SGK expression, but no significant difference was found in SGK levels between WT and TG liver at 4 h following PHx (Fig. 7A). Because in vivo adenovirus-mediated delivery of c-myc to mouse liver caused a rapid increase in hepatocyte size (24), we used RPA to determine that c-Myc mRNA levels were unchanged in regenerating TG liver compared with regenerating WT liver (Fig. 7A). Increased phosphatidylinositol 3′-kinase (PI3K) signaling activates protein kinase B/Akt protein, which has been implicated in mediating an increase in cell size (8,14,41,44). We therefore performed Western blot analysis with total protein extracts from regenerating liver and either an antibody that recognizes the Akt protein or an antibody specific to the Akt residues surrounding phosphoserine 473, which is phosphorylated during activation of the Akt kinase. This Western blot analysis demonstrated that neither Akt protein levels nor Akt activity were altered in regenerating TTR-FoxM1B TG liver. Diminished expression of the tumor suppressor gene PTEN, which negatively regulates the PI3K pathway and inhibits activation of Akt, has been shown to increase size (2,3,10,18,28,52). Furthermore, stimulation of hepatocyte peroxisome proliferation is known to increase hepatocyte size (19,37). Western blot analysis demonstrated that neither Pten nor peroxisomal membrane protein 70 (PMP 70) levels were altered in regenerating TG livers. These liver regeneration studies demonstrate that nuclear hepatocyte translocation of the FoxM1B transgene protein persisted for the first 6 h following PHx and that this was associated with a transient increase in size of regenerating TG hepatocytes. Our data suggest that nuclear expression of the FoxM1B transgene protein mediated increase in size of TG he-patocytes through pathways other than SGK, PI3K/ Akt, c-Myc, or peroxisome proliferation.
Figure 7.
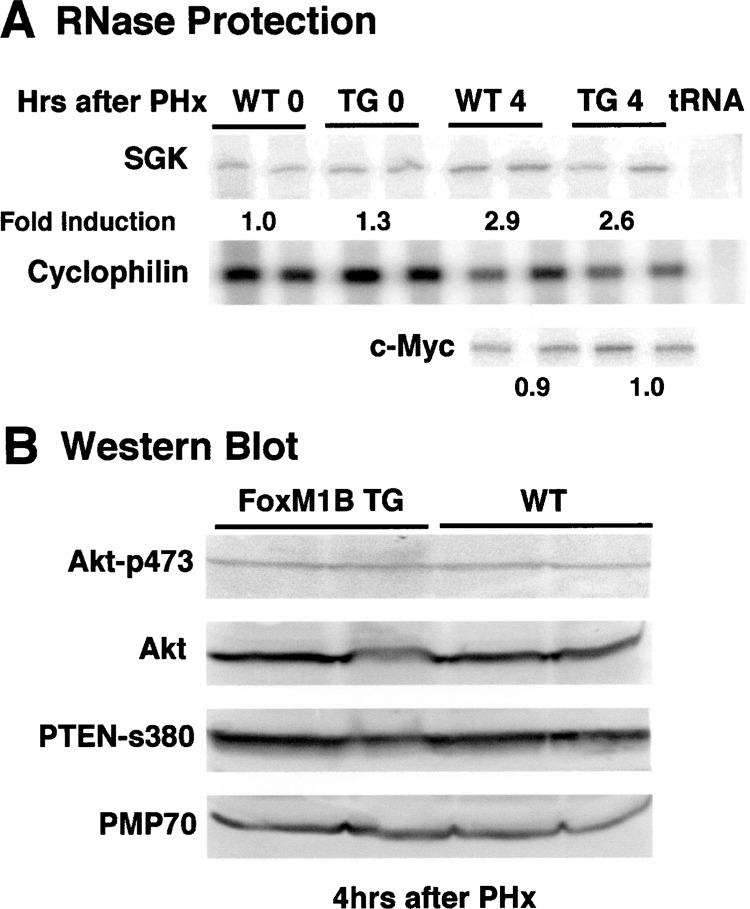
Regenerating transgenic liver displayed no changes in expression of proteins known to increase cell size. (A) Total liver RNA was isolated from two WT or TG mice at 4 h following PHx or prior to PHx and used for RNase protection assays to measure mRNA levels for serum-and-gucocorticoid-inducible protein kinase (SGK), c-myc, and cyclophilin as described in Materials and Methods. (B) Total protein extracts from regenerating TTR-FoxM1B TG liver displayed no changes in Akt, Pten, or PMP70 protein levels. Total protein extracts were prepared from regenerating WT and TG liver isolated at the indicated times following PHx and used for Western blot analysis with antibody specific to phosphoserine 473, whose phosphorylation is required to stimulate Akt kinase activity. The same Western blot was stripped and then reacted with antibody specific to either Akt, activated phosphorylated PTEN (s380), or peroxisomal membrane protein 70 (PMP 70).
Analysis of cDNA Arrays Revealed Increased Expression of Immediate Early Transcription Factors, GST, and Oxidative Stress Genes in Regenerating TG Liver
To identify genes whose expression is altered in regenerating TTR-FoxM1B TG liver at 4 h following PHx, we performed gene expression analysis using Clontech Mouse Atlas 1.2 cDNA Expression Array analysis (Table 1). Radioactive cDNA probes were made to RNA prepared from two distinct regenerating livers isolated from either WT or TTR-FoxM1B TG mice at 4 h after PHx. These probes hybridized to the Mouse Atlas 1.2 cDNA Expression Array Blots (Clontech) as described in Materials and Methods. Gene expression levels were determined using Atlas-Image program version 1.5 (Clontech) and expressed as fold induction levels of TG versus WT regenerating liver as described in Materials and Methods. This cDNA expression array analysis (Table 1) demonstrated that regenerating TTR-FoxM1B TG livers displayed increased expression of several immediate early transcription factor genes (ID3, Stat3, Nur77; Table 1), which are required to mediate hepatocyte proliferation during liver regeneration 9 gene, which functions to proteolytically activate growth factors during liver regeneration (25). We also found an increase in hepatic expression of GST isoforms and stress response genes, which are likely to be critical to prevention of oxidative stress during hepatocyte DNA replication in regenerating liver. These studies demonstrate that hepatocyte nuclear translocation of the FoxM1B transgene protein was associated with transient TG hepatocyte hypertrophy and increased expression of genes that may enhance hepatocyte proliferation.
TABLE 1.
CHANGES IN EXPRESSION OF REGENERATING TG LIVER GENES INVOLVED IN PROLIFERATION
| Gene Name | Genbank | TG/WT Fold Induction |
|---|---|---|
| Immediate early transcription factors and proteases | ||
| ID-3; inhibitor of basic helix–loop–helix (bHLH) dimerization 3 | M60523 | 7.0 |
| Stat3; signal transducer and activator of transcription 3 | U06922 | 4.0 |
| Nur77; immediate early response nuclear hormone receptor | J04113 | 4.0 |
| MMP-9; matrix metalloproteinase-9 | X72795 | 4.0 |
| Glutathione S-transferase and stress response | ||
| GST A; glutathione S-transferase A | J03958 | 7.1 |
| Microsomal GST1 | J03752 | 5.8 |
| GST M5-5; GST mu | J04696 | 6.8 |
| GST theta 1 | X98055 | 3.8 |
| HSP60; heat shock protein 60-kDa mitochondrial matrix protein P1 | X53584 | 7.0 |
| HO-2; heme oxygenase 2 | AF029874 | 7.0 |
| ERp72; endoplasmic reticulum stress protein | J05186 | 4.6 |
Radioactive cDNA probes were made to RNA prepared from regenerating wild-type (WT) liver and transthyretin (TTR) FoxM1B transgenic (TG) liver at 4 h after partial hepatectomy (PHx). Each of these cDNA probes was separately hybridized to the Mouse Atlas 1.2 cDNA Expression Array Blots (Clontech), rinsed, and then exposed for phosphorimager analysis. Gene expression levels were determined by using AtlasImage program version 1.5 (Clontech) following normalization to 40S ribosomal protein S29 (L31609) and cytoplasmic β-actin levels (M12481) and expressed as fold induction of gene levels in regenerating TTR-FoxM1B TG liver compared with regenerating WT liver.
Significance of Rapid Nuclear Translocation of FoxM1B Transgene in Regenerating Liver
In this study, we showed that the transient nuclear localization of the FoxM1B transgene protein in hepatocytes during the immediate early period following PHx is associated with a transient increase in regenerating TG hepatocyte size. Hepatocyte hypertrophy plays an important role in restoring liver mass and function during liver regeneration following PHx (34). Furthermore, swelling of primary rat hepatocytes causes the activation of signal transducers and activators of transcription 1 and Stat3 proteins (31), the latter of which is an immediate early transcription factor that is critical for regenerating hepatocyte DNA replication (29). Consistent with this finding, regenerating TTR-FoxM1B TG livers exhibited increased expression of Stat3 mRNA, suggesting that hepatocyte hypertrophy may participate in priming cellular proliferation through stimulating immediate early transcription factors such as the Stat3 protein. Furthermore, we found that expression of the nuclear receptor Nur77, which is known to be induced as an immediately early gene during liver regeneration (40), was elevated in regenerating TTR-FoxM1B TG liver as was ID3, which is known to stimulate cellular proliferation (1). Moreover, ectopic expression of the c-Myc transgene protein is known to stimulate earlier onset of hepatocyte DNA replication during liver regeneration (15) and in vivo adenovirus-mediated hepatic delivery of the c-Myc protein caused hepatocyte hypertrophy (24). These results suggested that hepa-tocyte hypertrophy precedes hepatocyte DNA replication and is required to stimulate hepatocyte entry into S-phase. Based on the above studies, we speculate that premature hepatocyte nuclear localization of the FoxM1B transgene protein immediately following PHx mediates increase in hepatocyte size, which may enhance activation of immediate early transcription factors. This induction of immediate early transcription factors may in turn play a role in priming TTR-FoxM1B TG hepatocytes for earlier entry into S-phase following liver injury (57). Furthermore, FoxM1B transgene protein-mediated hepatocyte hypertrophy may also play a role in compensation for the loss of liver mass immediately following surgery (this study) or at later times following PHx (16 h post-PHx), when the FoxM1B transgene protein reenters the nucleus (57).
ACKNOWLEDGMENTS
This study was supported by the US Public Health Service grant R01 DK54687-05 from NIDDK (R.H.C.). We thank members of the Costa laboratory and Nissim Hay for helpful discussions.
REFERENCES
- 1. Asp J.; Thornemo M.; Inerot S.; Lindahl A. The helix-loop-helix transcription factors Id1 and Id3 have a functional role in control of cell division in human normal and neoplastic chondrocytes. FEBS Lett. 438:85–90; 1998. [DOI] [PubMed] [Google Scholar]
- 2. Backman S.; Stambolic V.; Mak T. PTEN function in mammalian cell size regulation. Curr. Opin. Neurobiol. 12:516; 2002. [DOI] [PubMed] [Google Scholar]
- 3. Backman S. A.; Stambolic V.; Suzuki A.; Haight J.; Elia A.; Pretorius J.; Tsao M. S.; Shannon P.; Bolon B.; Ivy G. O.; Mak T. W. Deletion of Pten in mouse brain causes seizures, ataxia and defects in soma size resembling Lhermitte-Duclos disease. Nat. Genet. 29:396–403; 2001. [DOI] [PubMed] [Google Scholar]
- 4. Bohmer C.; Wagner C. A.; Beck S.; Moschen I.; Melzig J.; Werner A.; Lin J. T.; Lang F.; Wehner F. The shrinkage-activated Na(+) conductance of rat hepatocytes and its possible correlation to rENaC. Cell Physiol. Biochem. 10:187–194; 2000. [DOI] [PubMed] [Google Scholar]
- 5. Buse P.; Tran S. H.; Luther E.; Phu P. T.; Aponte G. W.; Firestone G. L. Cell cycle and hormonal control of nuclear-cytoplasmic localization of the serum-and glucocorticoid-inducible protein kinase, Sgk, in mammary tumor cells. A novel convergence point of anti-proliferative and proliferative cell signaling pathways. J. Biol. Chem. 274:7253–7263; 1999. [DOI] [PubMed] [Google Scholar]
- 6. Carlsson P.; Mahlapuu M. Forkhead transcription factors: Key players in development and metabolism. Dev. Biol. 250:1–23; 2002. [DOI] [PubMed] [Google Scholar]
- 7. Clark K. L.; Halay E. D.; Lai E.; Burley S. K. Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. Nature 364:412–420; 1993. [DOI] [PubMed] [Google Scholar]
- 8. Condorelli G.; Drusco A.; Stassi G.; Bellacosa A.; Roncarati R.; Iaccarino G.; Russo M. A.; Gu Y.; Dalton N.; Chung C.; Latronico M. V.; Napoli C.; Sadoshima J.; Croce C. M.; Ross J. Jr. Akt induces enhanced myocardial contractility and cell size in vivo in transgenic mice. Proc. Natl. Acad. Sci. USA 99:12333–12338; 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Costa R. H.; Kalinichenko V. V.; Lim L. Transcription factors in mouse lung development and function. Am. J. Physiol. Lung Cell Mol. Physiol. 280:L823–L838; 2001. [DOI] [PubMed] [Google Scholar]
- 10. Crackower M. A.; Oudit G. Y.; Kozieradzki I.; Sarao R.; Sun H.; Sasaki T.; Hirsch E.; Suzuki A.; Shioi T.; Irie-Sasaki J.; Sah R.; Cheng H. Y.; Rybin V. O.; Lembo G.; Fratta L.; Oliveira-dos-Santos A. J.; Benovic J. L.; Kahn C. R.; Izumo S.; Steinberg S. F.; Wymann M. P.; Backx P. H.; Penninger J. M. Regulation of myocardial contractility and cell size by distinct PI3K-PTEN signaling pathways. Cell 110:737–749; 2002. [DOI] [PubMed] [Google Scholar]
- 11. Cressman D. E.; Greenbaum L. E.; DeAngelis R. A.; Ciliberto G.; Furth E. E.; Poli V.; Taub R. Liver failure and defective hepatocyte regeneration in interleukin-6-deficient mice. Science 274:1379–1383; 1996. [DOI] [PubMed] [Google Scholar]
- 12. Diehl A. M. Cytokine regulation of liver injury and repair. Immunol. Rev. 174:160–171; 2000. [DOI] [PubMed] [Google Scholar]
- 13. Duncan S. A. Transcriptional regulation of liver development. Dev. Dyn. 219:131–142; 2000. [DOI] [PubMed] [Google Scholar]
- 14. Edinger A. L.; Thompson C. B. Akt maintains cell size and survival by increasing mTOR-dependent nutrient uptake. Mol. Biol. Cell 13:2276–2288; 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Factor V. M.; Kiss A.; Woitach J. T.; Wirth P. J.; Thorgeirsson S. S. Disruption of redox homeostasis in the transforming growth factor-α/c-myc transgenic mouse model of accelerated hepatocarcinogenesis. J. Biol. Chem. 273:15846–15853; 1998. [DOI] [PubMed] [Google Scholar]
- 16. Fausto N. Liver regeneration. J. Hepatol. 32:19–31; 2000. [DOI] [PubMed] [Google Scholar]
- 17. Gao C.; Jokerst R.; Gondipalli P.; Cai S. R.; Kennedy S.; Flye M. W.; Ponder K. P. Lipopolysaccharide potentiates the effect of hepatocyte growth factor on hepatocyte replication in rats by augmenting AP-1 activity. Hepatology 30:1405–1416; 1999. [DOI] [PubMed] [Google Scholar]
- 18. Gao X.; Neufeld T. P.; Pan D. Drosophila PTEN regulates cell growth and proliferation through PI3K-dependent and -independent pathways. Dev. Biol. 221:404–418; 2000. [DOI] [PubMed] [Google Scholar]
- 19. Green S. PPAR: A mediator of peroxisome proliferator action. Mutat. Res. 333:101–109; 1995. [DOI] [PubMed] [Google Scholar]
- 20. Kaestner K. H. The hepatocyte nuclear factor 3 (HNF3 or FOXA) family in metabolism. Trends Endocrinol. Metab. 11:281–285; 2000. [DOI] [PubMed] [Google Scholar]
- 21. Kaestner K. H.; Knochel W.; Martinez D. E. Unified nomenclature for the winged helix/forkhead transcription factors. Genes Dev. 14:142–146; 2000. [PubMed] [Google Scholar]
- 22. Kalinichenko V. V.; Lim L.; Beer-Stoltz D.; Shin B.; Rausa F. M.; Clark J.; Whitsett J. A.; Watkins S. C.; Costa R. H. Defects in pulmonary vasculature and perinatal lung hemorrhage in mice heterozygous null for the Forkhead box f1 transcription factor. Dev. Biol. 235:489–506; 2001. [DOI] [PubMed] [Google Scholar]
- 23. Kalinichenko V. V.; Zhou Y.; Bhattacharyya D.; Kim W.; Shin B.; Bambal K.; Costa R. H. Haploin-sufficiency of the mouse Forkhead box f1 gene causes defects in gall bladder development. J. Biol. Chem. 277:12369–12374; 2002. [DOI] [PubMed] [Google Scholar]
- 24. Kim S.; Li Q.; Dang C. V.; Lee L. A. Induction of ribosomal genes and hepatocyte hypertrophy by adenovirus-mediated expression of c-Myc in vivo. Proc. Natl. Acad. Sci. USA 97:11198–11202; 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kim T. H.; Mars W. M.; Stolz D. B.; Michalopoulos G. K. Expression and activation of pro-MMP-2 and pro-MMP-9 during rat liver regeneration. Hepatology 31:75–82; 2000. [DOI] [PubMed] [Google Scholar]
- 26. Klingel K.; Warntges S.; Bock J.; Wagner C. A.; Sauter M.; Waldegger S.; Kandolf R.; Lang F. Expression of cell volume-regulated kinase h-sgk in pancreatic tissue. Am. J. Physiol. Gastrointest. Liver Physiol. 279:G998–G1002; 2000. [DOI] [PubMed] [Google Scholar]
- 27. Korver W.; Roose J.; Clevers H. The winged-helix transcription factor Trident is expressed in cycling cells. Nucleic Acids Res. 25:1715–1719; 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kwon C. H.; Zhu X.; Zhang J.; Knoop L. L.; Tharp R.; Smeyne R. J.; Eberhart C. G.; Burger P. C.; Baker S. J. Pten regulates neuronal soma size: A mouse model of Lhermitte-Duclos disease. Nat. Genet. 29:404–411; 2001. [DOI] [PubMed] [Google Scholar]
- 29. Li W.; Liang X.; Kellendonk C.; Poli V.; Taub R. STAT3 contributes to the mitogenic response of hepatocytes during liver regeneration. J. Biol. Chem. 277:28411–28417; 2002. [DOI] [PubMed] [Google Scholar]
- 30. Marsden I.; Jin C.; Liao X. Structural changes in the region directly adjacent to the DNA-binding helix highlight a possible mechanism to explain the observed changes in the sequence-specific binding of winged helix proteins. J. Mol. Biol. 278:293–299; 1998. [DOI] [PubMed] [Google Scholar]
- 31. Meisse D.; Dusanter-Fourt I.; Husson A.; Lavoinne A. Cell swelling activates STAT1 and STAT3 proteins in cultured rat hepatocytes. FEBS Lett. 463:360–364; 1999. [DOI] [PubMed] [Google Scholar]
- 32. Michalopoulos G. K.; DeFrances M. C. Liver regeneration. Science 276:60–66; 1997. [DOI] [PubMed] [Google Scholar]
- 33. Nag A.; Datta A.; Yoo K.; Bhattacharyya D.; Chakrabortty A.; Wang X.; Slagle B. L.; Costa R. H.; Raychaudhuri P. DDB2 induces nuclear accumulation of the hepatitis B virus X protein independently of binding to DDB1. J. Virol. 75:10383–10392; 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nagy P.; Teramoto T.; Factor V. M.; Sanchez A.; Schnur J.; Paku S.; Thorgeirsson S. S. Reconstitution of liver mass via cellular hypertrophy in the rat. Hepatology 33:339–345; 2001. [DOI] [PubMed] [Google Scholar]
- 35. Nakae J.; Kitamura T.; Silver D. L.; Accili D. The forkhead transcription factor Foxo1 (Fkhr) confers insulin sensitivity onto glucose-6-phosphatase expression. J. Clin. Invest. 108:1359–1367; 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Poli V. The role of C/EBP isoforms in the control of inflammatory and native immunity functions. J. Biol. Chem. 273:29279–29282; 1998. [DOI] [PubMed] [Google Scholar]
- 37. Qi C.; Zhu Y.; Reddy J. K. Peroxisome proliferator-activated receptors, coactivators, and downstream targets. Cell Biochem. Biophys. 32:187–204; 2000. [DOI] [PubMed] [Google Scholar]
- 38. Qian X.; Samadani U.; Porcella A.; Costa R. H. Decreased expression of hepatocyte nuclear factor 3a during the acute-phase response influences transthyretin gene transcription. Mol. Cell. Biol. 15:1364–1376; 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rausa F. M.; Tan Y.; Zhou H.; Yoo K.; Stolz D. B.; Watkins S.; Franks R. R.; Unterman T. G.; Costa R. H. Elevated Levels of HNF-3β in mouse hepatocytes influence expression of genes involved in bile acid and glucose homeostasis. Mol. Cell. Biol. 20:8264–8282; 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Scearce L. M.; Laz T. M.; Hazel T. G.; Lau L. F.; Taub R. RNR-1, a nuclear receptor in the NGFI-B/ Nur77 family that is rapidly induced in regenerating liver. J. Biol. Chem. 268:8855–8861; 1993. [PubMed] [Google Scholar]
- 41. Shioi T.; McMullen J. R.; Kang P. M.; Douglas P. S.; Obata T.; Franke T. F.; Cantley L. C.; Izumo S. Akt/protein kinase B promotes organ growth in transgenic mice. Mol. Cell. Biol. 22:2799–2809; 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tan Y.; Hughes D. E.; Wang X.; Costa R. H. Adenovirus-mediated hepatic increase in HNF-3β or HNF-3α shows differences in levels of liver glycogen and gene expression. Hepatology 35:30–39; 2002. [DOI] [PubMed] [Google Scholar]
- 43. Taub R.; Greenbaum L. E.; Peng Y. Transcriptional regulatory signals define cytokine-dependent and -independent pathways in liver regeneration. Semin. Liver Dis. 19:117–127; 1999. [DOI] [PubMed] [Google Scholar]
- 44. Verdu J.; Buratovich M. A.; Wilder E. L.; Birnbaum M. J. Cell-autonomous regulation of cell and organ growth in Drosophila by Akt/PKB. Nat. Cell Biol. 1:500–506; 1999. [DOI] [PubMed] [Google Scholar]
- 45. Waldegger S.; Barth P.; Raber G.; Lang F. Cloning and characterization of a putative human serine/threonine protein kinase transcriptionally modified during anisotonic and isotonic alterations of cell volume. Proc. Natl. Acad. Sci. USA 94:4440–4445; 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wang X.; Hung N.-J.; Costa R. H. Earlier expression of the transcription factor HFH 11B (FOXM1B) diminishes induction of p21CIP1/WAF1 levels and accelerates mouse hepatocyte entry into S-phase following carbon tetrachloride liver injury. Hepatology 33:1404–1414; 2001. [DOI] [PubMed] [Google Scholar]
- 47. Wang X.; Kiyokawa H.; Dennewitz M. B.; Costa R. H. The Forkhead box m1b transcription factor is essential for hepatocyte DNA replication and mitosis during mouse liver regeneration. Proc. Natl. Acad. Sci. USA 99:16881–16886; 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wang X.; Krupczak-Hollis K.; Tan Y.; Dennewitz M. B.; Adami G. R.; Costa R. H. Increased hepatic Forkhead box M1B (FoxM1B) levels in old-aged mice stimulated liver regeneration through diminished p27Kip1 protein levels and increased Cdc25B expression. J. Biol. Chem. 277: 44310–44316; 2002. [DOI] [PubMed] [Google Scholar]
- 49. Wang X.; Quail E.; Hung N.-J.; Tan Y.; Ye H.; Costa R. H. Increased levels of Forkhead box M1B transcription factor in transgenic mouse hepatocytes prevents age-related proliferation defects in regenerating liver. Proc. Natl. Acad. Sci. USA 98:11468–11473; 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Warntges S.; Grone H. J.; Capasso G.; Lang F. Cell volume regulatory mechanisms in progression of renal disease. J. Nephrol. 14:319–326; 2001. [PubMed] [Google Scholar]
- 51. Webber E. M.; Bruix J.; Pierce R. H.; Fausto N. Tumor necrosis factor primes hepatocytes for DNA replication in the rat. Hepatology 28:1226–1234; 1998. [DOI] [PubMed] [Google Scholar]
- 52. Xu Z.; Stokoe D.; Kane L. P.; Weiss A. The inducible expression of the tumor suppressor gene PTEN promotes apoptosis and decreases cell size by inhibiting the PI3K/Akt pathway in Jurkat T cells. Cell Growth Differ. 13:285–296; 2002. [PubMed] [Google Scholar]
- 53. Yamada Y.; Fausto N. Deficient liver regeneration after carbon tetrachloride injury in mice lacking type 1 but not type 2 tumor necrosis factor receptor. Am. J. Pathol. 152:1577–1589; 1998. [PMC free article] [PubMed] [Google Scholar]
- 54. Yamada Y.; Kirillova I.; Peschon J. J.; Fausto N. Initiation of liver growth by tumor necrosis factor: Deficient liver regeneration in mice lacking type I tumor necrosis factor receptor. Proc. Natl. Acad. Sci. USA 94:1441–1446; 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yamada Y.; Webber E. M.; Kirillova I.; Peschon J. J.; Fausto N. Analysis of liver regeneration in mice lacking type 1 or type 2 tumor necrosis factor receptor: Requirement for type 1 but not type 2 receptor. Hepatology 28:959–970; 1998. [DOI] [PubMed] [Google Scholar]
- 56. Yao K. M.; Sha M.; Lu Z.; Wong G. G. Molecular analysis of a novel winged helix protein, WIN. Expression pattern, DNA binding property, and alternative splicing within the DNA binding domain. J. Biol. Chem. 272:19827–19836; 1997. [DOI] [PubMed] [Google Scholar]
- 57. Ye H.; Holterman A.; Yoo K. W.; Franks R. R.; Costa R. H. Premature expression of the winged helix transcription factor HFH-11B in regenerating mouse liver accelerates hepatocyte entry into S-phase. Mol. Cell. Biol. 19:8570–8580; 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ye H.; Kelly T. F.; Samadani U.; Lim L.; Rubio S.; Overdier D. G.; Roebuck K. A.; Costa R. H. Hepatocyte nuclear factor 3/fork head homolog 11 is expressed in proliferating epithelial and mesenchymal cells of embryonic and adult tissues. Mol. Cell. Biol. 17:1626–1641; 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zaret K. S. Regulatory phases of early liver development: Paradigms of organogenesis. Nat. Rev. Genet. 3:499–512; 2002. [DOI] [PubMed] [Google Scholar]


