Abstract
We have identified a novel mouse gene, Usp2, encoding two ubiquitin-specific proteases (USPs) due to alternate splicing of 5′ exons. Usp2-45 consists of 396 amino acids (45.2 kDa), while Usp2-69 is 619 amino acids (69.5 kDa). Usp2-69 results from the splicing of different combinations of untranslated 5′ exons (1A, 1B, 1C) onto exon 1D and the 40-kDa catalytic core (exons 3–13), while Usp2-45 has exon 2 spliced onto the core. The catalytic core contains the highly conserved motifs of the UBP family of deubiquitinating enzymes. We can find no evidence for a reported 41-kDa isoform (UBP41) in any sequence databases. Usp2-69 is able to form a complex with Usp2-45 and with itself. Antibodies raised against the catalytic core recognized a 69-kDa protein, but did not detect a 45-kDa protein in mouse tissues. Using Northern blot, Western blot, and immunohistochemistry, Usp2 expression was observed in many adult and embryonic tissues including testis, heart, skeletal muscle, diaphragm, brain, kidney, liver, pancreas, lung, and skin. Both Usp2 isoforms were localized to the cytoplasm when overexpressed in COS-7 and NIH3T3 cells. The Usp2 expression pattern indicates that this protein might be involved in specific processes in different types of cells, especially those that are differentiating, and that its function is not restricted to a development of a particular organ.
Keywords: Ubiquitin, Ubiquitin-specific protease, UBP41, Differentiation, Apoptosis
UBIQUITIN is a 76-amino acid protein that serves as a multifunctional covalent “tag” for a wide variety of cellular proteins. Its best understood function is in marking proteins for degradation by the 26S proteasome, but ubiquitin conjugation is also important in diverse pathways including DNA repair, receptor internalization, and signal transduction (11,13,30). Ubiquitin-specific proteases (USPs) are a large family of cysteine proteases, capable of cleaving both linear ubiquitin precursor proteins and posttranslationally formed ubiquitin–protein conjugates, and are the mammalian homologs of the yeast UBP family (4,5,27). These enzymes contain two highly conserved sequence motifs that were discovered during sequence alignment of the first three yeast UBPs (4). These are the “Cys box” and the “His box,” which contain a conserved cysteine and histidine residue, respectively, that were proposed to form part of the active site of these thiol proteases (4), consistent with the results of subsequent mutagenesis (9,10,15,23,32,33) and structural (14) studies. The region between the Cys box and the His box defines the catalytic core responsible for deubiquitinating activity of the enzyme. USPs also contain N-terminal or/and C-terminal extensions of varying length and unique sequence.
Humans contain at least 28 USPs that have been characterized to varying extents (5,6,31), with a total of 50 suggested by analysis of the human genome sequence. The mouse genome would be expected to contain a similar number. It is proposed that the presence of such a large family of USPs in all eukaryotes is not required merely for the cleavage of ubiquitin precursors, and is indicative of other regulatory roles in the ubiquitin pathway, where variable N-terminal extensions might be responsible for specific functions in selecting and binding substrates, and/or mediating localization of the enzyme (6,30,31).
Here we describe a gene structure and tissue expression pattern for a novel member of the mouse USP gene family, Usp2, which exists as two isoforms of 45 and 69.5 kDa, termed Usp2-45 and Usp2-69, respectively. These proteins are highly similar to the chicken UBP41 family of deubiquitinating enzymes, and include a region that is almost identical to a sequence in GenBank reported to be the mouse UBP41 enzyme (AF079565).
Rat orthologs of mouse Usp2-45 and Usp2-69 have been described as testis-specific and developmentally regulated proteins Ubp-t1 and Ubp-t2, with Ubp-t1 being induced in step 16 to 19 spermatids and Ubp-t2 expressed in step 18 to 19 spermatids (19). Ubp-t1 and Ubp-t2 were shown to localize to the nucleus and cytoplasm, respectively, when overexpressed in COS-7 cells as GFP or c-myc N-terminal fusions (19). The same group has shown that the N-terminal extensions change substrate specificity and inhibit the deubiquitinating activity of the catalytic core (20). The same rat proteins, termed UBP45 and UBP69, have also been reported to have a role in muscle differentiation (24). In addition, mRNA expression of rat Usp2 (UBP41) was shown to be selectively upregulated in bone by osteotropic agents parathyroid hormone (PTH), parathyroid hormone related peptide (PTHrP), and prostaglandin E2 (PGE2), possibly via the PKA/cAMP pathway (22). These authors also detected expression of UBP41 mRNA in primary osteoblasts and in osteoblast-like cell lines as well as in rat brain, heart, skeletal muscle, kidney, liver, and testis tissues.
Given these varying reports of Usp2 tissue-specific function, and some confusion as to the occurrence of UBP41, we carried out a systematic study of mouse Usp2 gene structure, mRNA expression, protein expression, and tissue distribution. We conclude that Usp2 expression in mouse might be cell specific but is not confined to a certain tissue or organ and appears to be present mainly in highly differentiated or rapidly differentiating tissues.
MATERIALS AND METHDOS
Computer Methods
Databases were accessed and searched using the BLAST algorithm (1) at the National Centre for Biotechnology Information, USA (http://www.ncbi.nlm.nih.gov/BLAST/). Sequences were also analyzed using BestFit and Isoelectric from the GCG package of programs (Program Manual for the Wisconsin Package Version 8, August 1994, Genetics Computer Group, Madison, Wisconsin, USA), and Clustal W (v1.7) (26), both provided by the Australian National Genomic Information Service (ANGIS; http://www.angis.su.oz.au/).
DNA Manipulation and Cloning
EST cDNA clones BG294999, BG294744 and BG296510 (mouse Usp2-69; IMAGE # 4503992, 4504061, and 4506362, respectively) and AI876889 (mouse Usp2-45; IMAGE # 1922050) were obtained from Genome Systems (USA) and sequenced in full on both strands (ThermoSequenase, Amersham; ABI Big Dye). DNA was amplified by the polymerase chain reaction (PCR) using Expand High Fidelity polymerase (Roche Molecular Biosciences) or Pfu (Promega), and following manufacturer’s instructions.
AI876889 cDNA containing Usp2-45 was used as a template to amplify the catalytic core (CC) of Usp2 (exons 3–13) to clone into pET15-b (Novagen) and pGEX-4T-1 (Amersham) vectors to produce His-fusion and GST-fusion proteins.
BG294999 was used to amplify the 900-bp N-terminal extension of Usp2-69 (the open reading frame of exon 1D) for ligation into pGEX-4T-1 and pET28a vectors for the expression of GST- and His-fusion proteins, respectively.
Full-length Usp2-45 (amplified from AI876889) and full-length human USP2-69 (N-terminal extension was amplified from human genomic DNA and ligated to the catalytic core at EcoRI site) were cloned into pGEX-4T-1 vector to produce GST-fusion proteins for GST pull-down assay.
Full-length Usp2-45 (amplified from AI876889), full-length Usp2-69 (amplified from BG294999), and catalytic core were engineered for mammalian expression using the Gateway cloning system and the pT-Rex-Dest30 vector (Invitrogen). A double flag epitope was added at the C-terminus of each construct by PCR. All manipulations were performed according to the manufacturer’s instructions. Plasmids were purified for transfection using the Maxiprep kit (Qiagen). All PCR products were sequenced in full to reveal no PCR errors. Primer sequences are available on request.
Northern Blot
The following probes were used: the mouse Usp2-69 exon 1A/1D SalI-EcoRI fragment from BG294999, the mouse Usp2 catalytic core (CC) sequence downstream of the EcoRI site from AI876889, and the mouse Usp2-45 exon 2 BamHI-EcoRI fragment from AI876889. Probes were labeled with [32P]dATP by random primer extension using a Megaprime kit (Amersham). Mouse Multiple Tissue Northern blots were purchased from ClonTech and hybridized according to the manufacturer’s instructions.
Protein Purification and Antibody Production
Plasmid constructs for bacterial expression of recombinant proteins were transformed into E. coli BL21(DE3). Overnight liquid culture (10 ml) was grown in 400 ml LB/amp and induced with 0.6 mM IPTG for 5-6 h. Cells were harvested by centrifugation and frozen at -70°C overnight. Tagged proteins were purified on nickel-NTA resin as described by Qiagen or on GSH-agarose (Sigma) according to manufacturer’s instructions. Polyhistidine tagged CC and a GST-fusion of the 29-kDa N-terminal extension of Usp2-69 (N-29) were used to inoculate New Zealand White rabbits (three injections of 200 μg) to produce antibodies by standard methods (12). The antiserum obtained from the first rabbit (termed CC antiserum) was first affinity purified against GST-CC fusion protein immobilized on PVDF membrane. In the same way the antiserum obtained from the second rabbit (termed N-29 antiserum) was purified on a His-tagged 29-kDa N-terminal extension of Usp2-69. For affinity purification, the membrane containing the protein was blocked for 1 h in 5% skim milk/ PBS/0.05% Tween-20, then incubated with 200 μl of antiserum diluted in 800 μl PBS/0.05% Tween-20 for 3 h, washed three times in PBS/0.05% Tween-20 for 20 min each wash, and eluted in 200 μl of 200 mM glycine, pH 3, 0.1% BSA for 20 min with rotation. The antibody solution was neutralized by adding 1/10 volume of 1 M Tris to achieve pH 7-8. The CC antiserum was also affinity purified against a peptide (termed CC2) originating from the CC, CPETLDHL PDEEKGR, using the Sulfolink kit (Pierce) according to the manufacturer’s instructions.
Western Blot
Mouse tissues were homogenized in lysis buffer (50 mM Tris, pH 7.4, 300 mM sucrose, 0.5% NP-40), frozen and thawed twice, and centrifuged for 20 min at 16,000 × g at 4°C. Supernatant was collected and protein concentration was measured using the DC Protein Assay kit (BioRad). Total protein (100 μg) for each tissue was mixed with 3× protein loading buffer (187.5 mM Tris-HCl, pH 6.8, 6% SDS, 30% glycerol, 0.0075% bromophenol blue, 1 μM β-mercaptoethanol), boiled for 5 min, and resolved on 10% Tris-glycine SDS-PAGE. Proteins were transferred to PVDF membrane; the membrane was blocked in 5% skim milk/PBS/0.05% Tween for 1 h, incubated with primary antibodies (CC antiserum 1:10,000 dilution; N-29 antiserum 1:5,000 dilution), or CC affinity-purified antibodies 1:200 dilution) for 1 h at RT or overnight at +4°C, washed three times, incubated with anti-rabbit HRP-conjugated secondary antibody (DAKO) for 1 h at RT, washed, and visualized with ECL detection reagent (Amersham Pharmacia).
Immunoprecipitation
COS-7 cells were grown in 10% FBS RPMI medium at 37°C, 10% CO2, and passaged routinely. Cells were seeded in 100-mm dishes and transfected at 50% confluence with 2 μg DNA and 5 μl Lipofectamine (Invitrogen) for 5 h in 5 ml of serum-free medium. An equal volume of 20% medium was then added and cells were incubated for 18 h. Cells were harvested by scraping in 500 μl lysis buffer (50 mM Tris, pH 7.4, 300 mM sucrose, 0.5% NP-40), centrifuged for 20 min at 16,000 × g at 4°C, and the supernatant was collected and stored at –70°C until further analysis. Cell lysate (200 μg) was precleared with nonimmune rabbit serum and protein G beads (Pharmacia) for 1 h; the supernatant was removed and mixed with antibody at 1:100 dilution and 30 μl protein G beads and incubated for 3 h at 4°C. Immunocomplexes bound to protein G were washed five times with 1 ml of lysis buffer each time, resus-pended in 30 μl of protein loading buffer, boiled for 5 min, and loaded on the gel or stored at –20°C until analysis.
GST Pull-Down
GST-fusions of mouse USP2 catalytic core, mouse Usp2-45, and human USP2-69 were produced as described above and purified on GSH-agarose (Sigma) according to the manufacturer’s instructions. The proteins were left bound to the beads and an equal molar amount of each was used for GST pull-down. Mouse and human Usp2-69 were transfected into COS-7 cells and harvested as described above. Total protein concentration was quantified using DC Protein Assay (Bio-Rad) and 300 μg was mixed with GST-fusion proteins bound to GSH-agarose. The mixture was incubated for 3 h, washed five times in cells lysis buffer, eluted with protein loading buffer, and resolved on 10% PAGE. Western blot was performed as described, using primary anti-flag M2 monoclonal antibody (Sigma) at dilution 1:3000 and secondary anti-mouse HRP-conjugated antibody (Sigma) at dilution 1:10,000.
Immunofluorescence
Approximately 2 × 104 cells were seeded onto coverslips in 12-well plates and transfected at 50–60% confluence with 1 μg DNA and 1 μl of Lipofectamine (Invitrogen) in 1 ml of serum-free medium for 5 h. An equal volume of 20% medium was then added and cells were incubated for 18 h. Cells were fixed in 4% paraformaldehyde for 20 min, washed twice with PBS, permeabilized in PBS, 1% BSA, 0.1% SDS for 15 min, blocked in PBS, 1% BSA for 45 min, incubated with 20 μl of undiluted primary antibodies for 3 h at RT, washed five times with PBS, incubated with anti-rabbit FITC-conjugated secondary antibody (1:100 dilution) (Jackson Immuno-Research Labs) for 1 h, washed with PBS five times, mounted in 2% propylgallate, and fixed on coverslips using nail polish.
Immunohistochemistry on Embryo Sections
Twenty female Balb/C mice were mated with Balb/C males overnight; females with a vaginal plug were separated next morning and sacrificed at expected 7.5, 9.5, 13.5, and 16.5 days postcoitum (dpc). Embryos were isolated and fixed in 4% paraformal-dehyde for 1–3 days and processed for histological sectioning (4 μm thick). Slides with paraffin-embedded sections were washed in xylene and ethanol, rehydrated, and used for immunohistochemistry. Sections were blocked with 3% hydrogen peroxide for 3 min, washed in running water for 3 min, washed in two changes of Tris-buffered saline, 0.1% Tween (TBST), and blocked with 10% normal bovine serum for 30 min. The primary antibodies were applied overnight (affinity-purified CC dilution 1:5, N-29 and CC2 undiluted). After that sections were washed in two changes of TBST and incubated in anti-rabbit HRP-conjugated antibodies for 30 min (1:50 dilution), sections were washed in TBST twice and visualized using DAB-enhanced liquid substrate system (Sigma) until brown staining appeared but no longer than 5 min. Sections were washed, fixed with in DRX mountant (BDH), and photographed. Interpretation of the images was made referring to the Atlas of Mouse Development (17) and with the kind help of Dr. Stephen Wood.
RESULTS
Identification of Usp2 cDNA Clones
Using the chicken UBP41 cDNA sequence (3), we searched mouse EST databases for orthologous sequences. Given that the human ortholog of chicken UBP41 has been termed USP2, we have named these mouse proteins Usp2 (ubiquitin-specific protease 2), to be consistent with the nomenclature proposed for human ubiquitin-specific proteases [(5); http://www.gene.ucl.ac.uk/nomenclature].
We obtained IMAGE clone 1922050 (GenBank: AI876889) and sequenced it in full to reveal an insert of 2051 bp plus a poly(A) tail (submitted to Gen-Bank; Accession number AY255637). An open reading frame (ORF) of 1191 bp encoded a protein of 396 amino acids with a predicted molecular mass of 45.2 kDa and a predicted pI of 8.92. We have termed this protein Usp2-45. Similarly, we obtained IMAGE clones 4503992, 4504061, and 4506362 (GenBank: BG294999, BG294744, and BG296510, respectively) and sequenced them in full to reveal inserts of ∼2.1 kb including an ORF of 1860 bp encoding a protein of 619 amino acids with a predicted molecular mass of 69.5 kDa and a predicted pI of 9.34. We have termed this protein Usp2-69. These sequences have been submitted to GenBank with accession numbers AY255638, AY255639, and AY255640, respectively. These sequences are not given in isolation, but are shown with the subsequently determined gene sequence in Figure 1. Notably, IMAGE clone 4504061 (BG294744; our sequence AY255639) had a deletion of 18 bp within the open reading frame compared with the other two Usp2-69 cDNAs. Closer inspection revealed two directly repeated copies of this 18-bp sequence, followed by a third imperfect copy with three mismatches, resulting in three copies of R-T-S-D/E-G-Y/F in the protein (Fig. 1). The protein encoded by IMAGE clone 4504061 thus lacks one of these repeats. The mechanism of the deletion is unknown, but could be a natural polymorphism, an artifact due to slipping of reverse transcriptase during cDNA construction, or subsequent deletion in E. coli. No other ESTs have this deletion. We will only consider the Usp2-69 protein with all three copies of the six-residue repeat in subsequent sections.
Figure 1.
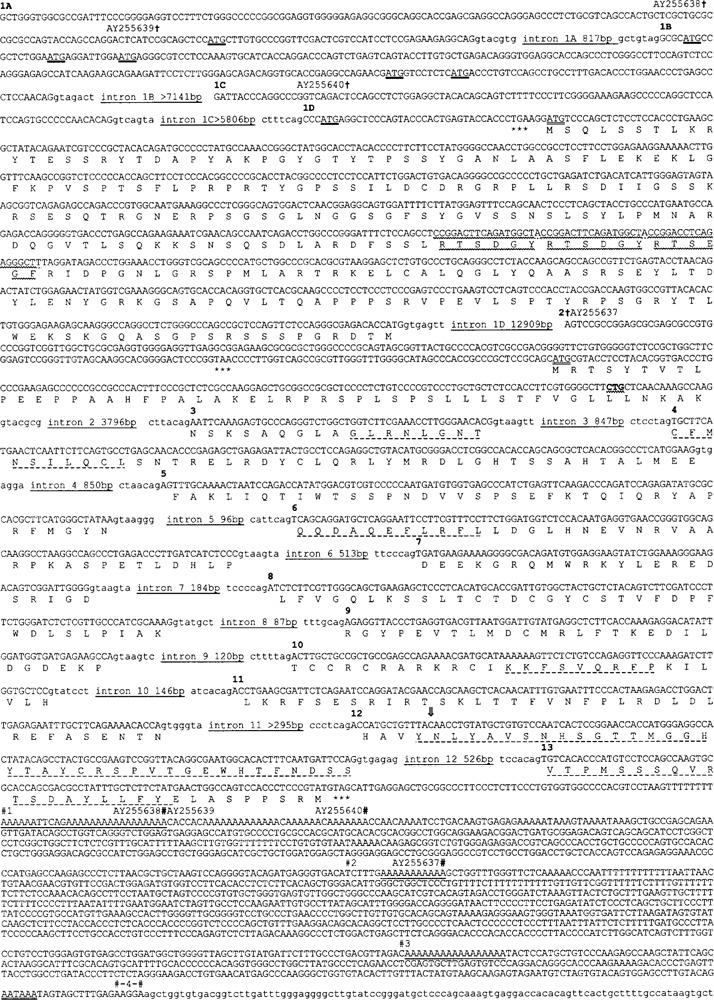
Sequence of Usp2 gene and cDNA clones. Bases 3527763 to 3556612 of GenBank:NW_000352.1 are shown, except that a single “C” has been inserted into exon 12 (open arrow; 14th base of exon 12; after base 3554030 of NW_000352.1; see text). Exons are in upper case; introns in lower case. Exons are numbered above the first base of each exon and intron lengths are indicated. The Usp2-45/Usp2-69 translation is shown below the first base of each codon, with ATG initiation codons doubly underlined. Other ATG codons within 5′ UTR exons are shown with a heavy underline; they form short ORFs that terminate before the bona fide ORFs. Stop codons upstream of, and in-frame with, bona fide ORFs are asterisked (***). An 18-bp direct repeat within exon 1D, one copy of which is deleted from one Usp2-69 cDNA clone, has a wavy underline. Amino acid regions conserved in all UBP sequences have a dashed underline. The AATAAA polyadeny-lation signal in the 3′ UTR is doubly underlined. Polyadenylation sites identified from cDNA or EST sequences are shown # and numbered. Three of these coincide with A-rich sequences in the gene (underlined) and lack AATAAA signals. The reported initiation codon of mouse UBP41, which corresponds to a CTG leucine codon in the gene and all known ESTs and cDNAs, is in bold with a wavy underline (see text; exon 2). The first and last bases of the clones we sequenced are shown † and #, respectively, with their accession numbers.
As there is an in-frame stop codon located 2 co-dons (Usp2-69) or 19 codons (Usp2-45) upstream of each putative initiator methionine codon, we consider these ORFs to be complete (Fig. 1). The Usp2-69 cDNAs all encoded an identical protein (except for the deletion noted above) but had different 5′ UTRs, later determined to be due to alternate splicing (see below).
Usp2-45 and Usp2-69 are identical at the protein/ DNA level over their C-terminal 347 amino acids/ codons. This region contains the catalytic core of the enzymes, bordered by the consensus Cys and His boxes diagnostic of the UBP family of deubiquitinat-ing enzymes, first identified in the yeast Saccharomyces cerevisiae Ubp1p, Ubp2p, and Ubp3p enzymes (4). However, both proteins differ upstream of this catalytic core region, with Usp2-45 having a 49-amino acid N-terminal extension, whereas Usp2-69 has a 272-amino acid N-terminal extension of unrelated sequence. The most likely explanation is alternate splicing of a single gene to produce two proteins, which was confirmed by analysis of the mouse gene sequence.
Usp2 Gene Structure
We searched the mouse genome database (mouse_contig/mgscv3, Posted date: Nov 20, 2002) using the Usp2-45 and Usp2-69 cDNA sequences. All cDNAs produced a single match to the same region of mouse chromosome 9, on Mus musculus WGS supercontig Mm9_WIFeb01_196, a 13.25-megabase contig. The region 3530192 to 3554926 of this contig has been designated NW_000352.1, and annotated to contain the Usp2 and UBP41 gene, although with both transcriptional and translational discrepancies noted during automated annotation. However, this region does not include the full Usp2 gene, as noted below.
Analysis of this ∼29-kb region of chromosome 9 reveals that the Usp2 gene consists of 16 exons in total, spread over a minimum of 29125 bp (Table 1, Fig. 2). The catalytic core of both Usp2 isoforms is encoded by 11 exons (exons 3-13), which contribute 39.9 kDa to the molecular weight. Usp2-45 is formed by splicing exon 2 to exons 3-13 (Table 1, Fig. 2). Usp2-69 is formed by splicing exon 1D (containing the initiation codon) and differing combinations of further upstream exons (1A, 1B, and/or 1C, all purely 5′ UTR) to exons 3-13. The exons range in size from 51 to 857 bp (with the exception of exon 13; see below), while the introns range from 87 to 12,909 bp (the latter includes exon 2 that is not present in Usp2-69). Introns 1C and 11 contain gaps of undefined length (Figs. 1, 2) and thus 29125 bp is a minimum estimate of gene length. All splice junctions confer to consensus sequences (Table 1). There is an apparent sequence error in this version of the mouse genome sequence; exon 11 is missing a single base when compared with our cDNA clones. A search of the mouse EST database using exon 11 revealed that all 38 EST clones that span this region contain the extra base compared to the gene sequence. Given this EST support, and as this base absence would cause a frame-shift in the highly conserved His box, we conclude that the genome sequence is in error, and have provided the corrected sequence in Figure 1.
TABLE 1.
MOUSE Usp2 INTRON/EXON BOUNDARIES
| Exon Number | Intron/EXON . . . EXON/Intron | Exon Length (bp) | Intron Length (bp) | |
|---|---|---|---|---|
| 1A* | GCTGGGT . . . GAGGCAGgtacgtg | 200 | >8216 (to 1D)** | |
| 817 (to 1B) | ||||
| 1B† | gctgtagGCGCATG . . . CCAACAGgtagact | 258 | >7141 (to 1D)** | |
| 1C‡ | GATTACC . .. AACACAGgtcagta | 103 | >5806 (to 1D)** | |
| 1D | ctttcagCCCATGA .. . CACCATGgtgagtt | 857 | 12909 (to 3) | |
| 2§ | AGTCCGC . . . AGCCAAGgtacgcg | 385 | 3796 | |
| 3 | cttacagAATTCAA . .. GAACACGgtaagtt | 51 | 847 | |
| 4 | ctcctagTGCTTCA .. . ATGGAAGgtgagga | 124 | 850 | |
| 5 | ctaacagAGTTTGC . . . GCTATAAgtaaggg | 112 | 96 | |
| 6 | cattcagTCAGCAG . . . ATCTCCCgtaagta | 111 | 513 | |
| 7 | ttcccagTGATGAA . . . ATTGGGGgtaagta | 65 | 184 | |
| 8 | tccccagATCTCTT . .. CGCAAAGgtatgct | 104 | 87 | |
| 9 | tttgcagAGAGGTT .. . GAAGCCAgtaagtc | 81 | 120 | |
| 10 | cttttagACTTGCT . . . GTGCTCCgtatcct | 79 | 146 | |
| 11 | atcacagACCTGAA .. . AACACCAgtgggta | 108 | >295** | |
| 12 | ccctcagACCATGC . . . ATTCCAGgtgagag | 121# | 526 | |
| 13#1¶ | tccacagTGTCACA .. . TTTTTTA | 150 | — | |
| 13#4¶ | tccacagTGTCACA .. . AGAAGGA | 1853 | — | |
| Total | Usp2-68v1 exons 1A/1D/3-13#1 | 2163 | >24789** | |
| Total | Usp2-68v1 exons 1A/1D/3-13#4 | 3865 | >24789** | |
| Total | Usp2-68v2 exons 1C/1D/3-13#1 | 2066 | >22379** | |
| Total | Usp2-68v2 exons 1C/1D/3-13#4 | 3768 | >22379** | |
| Total | Usp2-45v1 exons 2-13#1 | 1491 | >7460** | |
| Total | Usp2-45v1 exons 2-13#4 | 3193 | >7460** |
Exon/intron data were derived from comparison of our cDNA and EST clones to NW_000352.1.
Start site of exon 1A defined from EST BB635565.
Exon 1B is supported by a single EST BB635565.
Start site of exon 1C defined from EST BY097860.
Start site of exon 2 defined from our sequence of AI876889.
Exon 13#1 and 13#4 refer to the first and fourth polyadenylation sites, respectively; see text.
Gene sequence NW_000352 is missing 1 bp.
Gap of 100 ns is not included in intron size.
Figure 2.
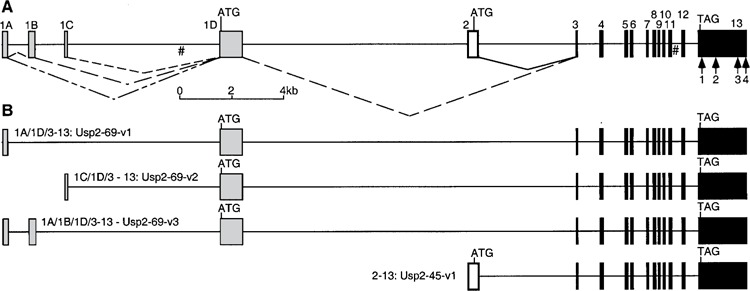
Usp2 gene structure and alternate splicing. (A) Gene structure. Genomic DNA is shown as a solid line, with exons as filled boxes, numbered above the boxes. Different combinations of exons 1A, 1B, and 1C are spliced onto exon 1D (shaded boxes) and exons 3–13 (black boxes; catalytic core) to form Usp2-69 (dotted carets), while exon 2 (white box) is spliced onto exons 3–13 to form Usp2-45 (solid caret). Start (ATG) and stop (TAG) codons are indicated. Polyadenylation sites within exon 13 are shown as numbered arrows; some may be artifacts (see text). Gaps of 100 ns in the mouse genome sequence (NW_000352.1) are indicated #. (B) Known alternately spliced Usp2 products derived from EST and cDNA clones. See text for accession numbers.
Splice Variants of Usp2-69
Two of the clones we sequenced (BG294999 = AY255638 and BG294744 = AY255639) represented cDNAs of the composition exon 1A plus 1D spliced onto exons 3-13, with an exon 1A length of at least 82 bp (Fig. 2). We term this splice variant Usp2-69-v1. A search of the mouse EST database revealed several more sequences that supported this splice variant (BG294915, BBB618107, BB617020, BY324974, BY232473). The longest clone, BB617020, revealed an exon 1A length of 200 bp that perfectly matched the mouse genome sequence, which we have depicted in Figures 1 and 2. Notably, the 18-bp deletion in BG294744 compared with other cDNAs and the gene does not correspond to a splice junction, but is roughly in the middle of exon 1D, and thus presumably does not represent a splice error.
The third clone we sequenced, BG296510 (our sequence AY255640), had a different 5′ UTR sequence upstream of the coding exon 1D, suggesting another splice variant arising from exon 1C spliced onto 1D, that we term Usp2-69-v2. A search of the mouse EST database revealed several more sequences that supported this splice variant (BY097860, BY134117, AA073417, and also BC017517 in the “nr” database). The longest clone, BY097860, revealed an exon 1C length of 103 bp that perfectly matched the mouse genome sequence, and thus we have depicted exon 1C being 103 bp.
One further EST was identified during these searches that contained a novel sequence of 258 bp between exons 1A and 1D. This 258-bp region perfectly matched the gene sequence in a location between exons 1A and 1C, and was flanked by consensus splice acceptor and splice donor sites. We have named this exon 1B, and it thus appears to be a third splice variant, termed Usp2-69-v3, comprising exons 1A/1B/1D/3–13. However, it is only supported by a single EST clone, whereas v1 and v2 are supported by multiple cDNA/EST clones.
While the Usp2-69 protein encoded by all three splice variants is identical (except for the 6-residue deletion in one clone noted above), the different combinations of 5′ UTR exons may represent different transcriptional or translational tissue-specific regulatory mechanisms.
Usp2 Expression and Multiple Polyadenylation Sites
Northern blots using the catalytic core of the mouse Usp2-45 cDNA as a probe identified up to four mRNA species in adult tissues (Fig. 3A). Testis had the strongest expression levels, with mRNAs of 4.0, 3.4, 2.4, and 1.6 kb. The 4.0- and 3.4-kb bands were also present in brain, heart, and skeletal muscle, while bands of 3.4 and 1.6 kb were present in liver and kidney. mRNAs were very weak or absent from spleen and lung.
Figure 3.
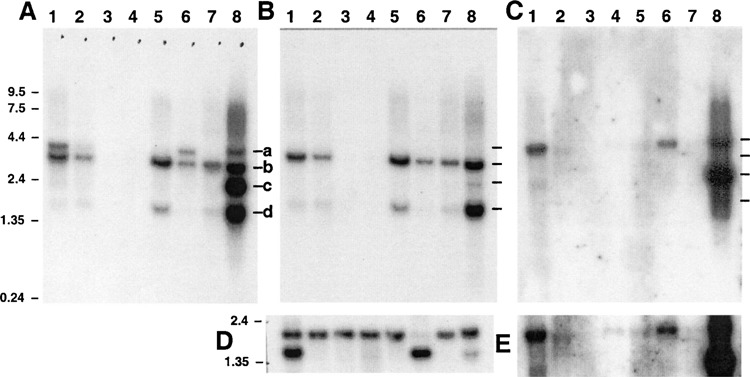
Northern blot analysis. A commercially available mouse Northern blot (ClonTech) was hybridized with (A) catalytic core probe, (B) Usp2-45-specific probe (exon 2), (C) Usp2-69-specific probe (exon 1D), or (D) β-actin probe as a loading control. A longer exposure of part of (C) is shown in (E). Lane 1: heart; lane 2: brain; lane 3: spleen; lane 4: lung; lane 5: liver; lane 6: skeletal muscle; lane 7: kidney; lane 8: testis. Markers in kb are shown on the left. Marks labeled a, b, c, and d identify mRNAs of ∼4.0, 3.4, 2.4, and 1.6 kb, respectively.
Given that the Usp2-45 and Usp2-69 cDNAs that we sequenced were all ∼2.1 kb in length, the identity of the multiple mRNAs was unclear. We therefore used a probe specific for Usp2-45, spanning exon 2 to the EcoRI site at the exon 2/3 boundary. This probe hybridized only to the 3.4- and 1.6-kb mRNA species observed with the catalytic core probe, suggesting that they correspond to Usp2-45 (Fig. 3B).
Conversely, a Usp2-69-specific probe from exon 1D hybridized to the 4.0- and 2.4-kb mRNAs in testis, and the 4.0-kb band in skeletal muscle and heart, and weakly to 4.0-kb bands in other tissues except spleen and lung (Fig. 3C, E). Notably, the 2.4-kb Usp2-69 mRNA is testis specific.
A search of the EST database using the 3′ region of the gene sequence revealed at least four apparent polyadenylation sites (Figs. 1 and 2). The majority of ESTs (43 in total) were apparently polyadenylated at site #1 (Fig. 1); however, this coincides with a long A-rich stretch in the gene sequence and is not preceded by an AATAAA polyadenylation signal, and thus may represent an oligo-dT primer annealing within a longer mRNA. The same may apply for A-rich sites #2 and #3, which are used in 3 and 14 ESTs, respectively. Site #4 appears to be bona fide, in that an AATAAA signal precedes by ∼15 bp two closely spaced polyadenylation sites used by 5 ESTs (Fig. 1). The three Usp2-69 cDNA clones that we sequenced were primed from site #1, whereas the Usp2-45 cDNA was primed from site #2.
The predicted lengths of Usp2-69v1 or v2 mRNA to polyadenylation sites #1 and #4 would be ∼2.2 and ∼3.8 kb, respectively (Table 1), corresponding to the observed 2.4 and 4.0 bands hybridizing to the Usp2-69-specific probe (Fig. 3C). Likewise, predicted Usp2-45 mRNAs to polyadenylation sites #1 and #4 would be ∼1.5 and ∼3.2 kb, respectively (Table 1), corresponding to the observed 1.6- and 3.4-kb Usp2-45-specific bands (Fig. 3B). Thus, although site #1 lacks a consensus AATAAA signal, usage of sites #1 and #4 would explain the four mRNA sizes we observe. Site #1 in Usp2-69 is only used in testis, whereas it is used in Usp2-45 in other tissues as well (Fig. 3B).
No Evidence for the Reported Mouse UBP41 cDNA
There is a GenBank sequence reporting a mouse UBP41 cDNA/protein, that is essentially the catalytic core of Usp2 (AF079565), with an ATG codon six codons upstream of exon 3. However, we could not find this ATG codon in any of our Usp2 cDNAs or gene sequence. The reported mouse UBP41 DNA sequence is practically identical to Usp2-45, but begins with an ATG codon in place of the Usp2-45 Leu44 codon (CTG) (Fig. 4). We searched the mouse EST database with the Usp2-45 cDNA sequence. Of the 41 ESTs that cover the ATG in question, 38 had the Leu codon CTG at this position, while another two had an alternate Leu codon, TTG, which may represent allelic variation or sequence errors. One EST sequence, AA572506, begins with CATG and then matches the Usp2-45/UBP41 sequence (Fig. 4), which does support the UBP41 ATG codon. We obtained this clone (IMAGE #987337) and sequenced it to reveal a completely unrelated sequence, suggesting that an arraying or picking error had been made. We thus cannot confirm the sequence of AA572506, but we believe it highly likely that the reported mouse UBP41 sequence (AF079565) arose from a sequencing artifact, and that there is no 41-kDa-sized protein (corresponding to the catalytic core only) expressed from the Usp2 gene. The same situation is true for available human and rat genome/EST sequences (data not shown).
Figure 4.
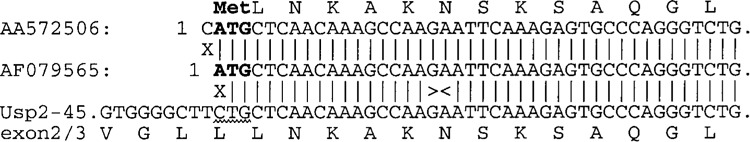
Origin of the reported mouse UBP41 sequence. The 5′ 42 bp of the reported mouse UBP41 cDNA sequence (GenBank: AF079565; center line) were aligned with our mouse Usp2-45 cDNA sequence (AY255637; bottom line) flanking the exon 2/exon3 junction (shown ><), and with the single mouse EST sequence that supports the UBP41 ATG codon AA572506; top line). Translations are given above or below the sequences. The proposed ATG codon of UBP41 (bold) aligns with the codon for Leu-45 of Usp2-45 (wavy underline). Vertical lines indicate sequence identity, and an X indicates a mismatch.
Usp2 Protein Expression
Rabbit polyclonal antibodies were raised against the Usp2 CC and the Usp-2-69 N-terminal extension. They were found to be specific for Usp2. The CC antiserum and affinity-purified CC antibody could recognize recombinant CC, Usp2-45, and Usp2-69, while both N-29 antiserum and affinity-purified N-29 antibodies could only recognize recombinant Usp2-69 (data not shown). In addition, affinity-purified CC antibody could immunoprecipitate Usp2-CC, Usp2-45, and Usp2-69 isoforms at the expected size, when overexpressed in mammalian cells (Fig. 5A).
Figure 5.
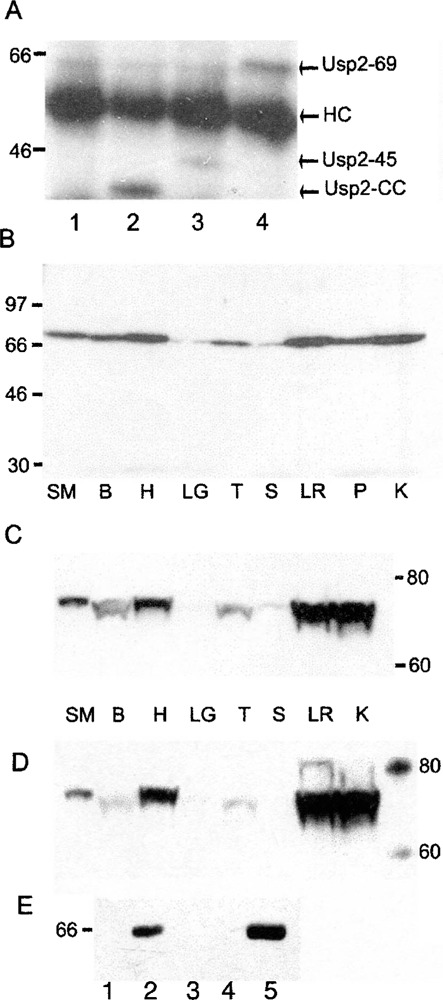
Specificity of affinity-purified Usp2 CC antibody, expression of Usp2 protein in adult mouse tissues and interaction of Usp2-45 and Usp2-69. (A) Lysates of untransfected COS-7 cells (lane 1) or COS-7 cells transiently transfected with plasmids expressing Usp2-CC (lane 2), Usp2-45 (lane 3), or Usp2-69 (lane 4) were immunoprecipitated with affinity-purified CC antibody, resolved by 10% SDS-PAGE, transferred to a PVDF membrane, and detected with affinity-purified CC antibody, followed by an HRP-conjugated secondary antibody and detection with ECL reagent. (B-D) Lysates of adult mouse tissues were resolved and processed as above for Western blot with either crude CC antiserum (B, C) or N-29 antiserum (D). (B) Samples were resolved by 10% SDS-PAGE, while electrophoresis in (C) and (D) was for an extended time in a 7% Trisacetate gel to better resolve larger proteins. For (D), the membrane from (C) was stripped in 62.5 mM Tris-HCl, pH 6.7, 2% SDS, 100 mM β-mercaptoethanol for 30 min at 65°C and probed with N-29 antiserum. Samples were: skeletal muscle (SM), brain (B), heart (H), lung (LG), testis (T), spleen (S), liver (LR), pancreas (P), and kidney (K). (E) GST pull-down assay, where free GST (lanes 1, 3), human Usp2-69 (lane 2), mouse Usp2-CC (lane 4), and mouse Usp2-45 (lane 5) were immobilized on GSH-agarose, mixed with 300 μg of COS-7 cell lysate with overexpressed flag epitope-tagged human Usp2-69 (lanes 1, 2) or mouse Usp2-69 (lanes 3, 4, and 5), incubated for 3 h, washed five times in cell lysis buffer, and resolved on 10% PAGE, followed by the Western blotting.
Using the CC antiserum, the highest level of Usp2 protein expression was detected in adult heart, liver, kidney, and pancreas, with a little less in skeletal muscle and brain, and yet less in testis, while very little (almost no) expression was detected in spleen and lung (Fig. 5B). This protein expression pattern was different from the RNA expression pattern obtained by Northern blot, where Usp2-69 mRNA was more abundant in testis, heart, and skeletal muscle, but could be detected in other tissues after long exposure (Fig. 3C, E).
We could not detect any protein at the 45-kDa size, in spite of abundant Usp2-45 mRNA expression in all tissues (except lung and spleen). To more accurately measure the size of the identified protein, a Western blot of a lower percentage Tris-acetate gel was probed with CC antiserum (Fig. 5C). The protein size was approximately 71 kDa, which is close to the predicted size of 69.5 kDa. Stripping the membrane and reprobing with N-29 antiserum detected the same single band at 71 kDa (Fig. 5D). This confirms that the identified protein is Usp2-69.
Usp2-45 and Usp2-69 Interact With Each Other
Usp2-69 was found to interact with itself and with Usp2-45, as shown by GST pull-down, where Usp2-69 overexpressed with a C-terminal flag epitope in COS-7 cells was able to bind to either Usp2-45 or Usp2-69 immobilized on GSH-agarose as GST-fusions (Fig. 5E). The binding was not mediated by the catalytic core domain, as no interaction was detected between Usp2-69 and the catalytic core construct alone. The interaction may be direct or mediated through a third partner present in COS-7 cells. The functional significance of this interaction is unknown, but its existence indicates that Usp2-45 and Usp2-69 are able to form a complex, where they may function together.
Tissue Distribution of Usp2
Immunohistochemistry performed on embryos at 7.5, 9.5, 13.5, and 16.5 dpc (Fig. 6) revealed that Usp2 does not have a stage-dependent developmental pattern of expression. At the early head-fold stage 7.5 dpc (Fig. 6A) Usp2 protein was already expressed throughout the developing embryo, most strongly in allantois (Fig. 6A1), at the site of embryo attachment (Fig. 6A2), in the endometrial tissue (Fig. 6A3), in the outer zone of the decidual reaction (Fig. 6A4), and in the inner circular layer of myometrial smooth muscle (Fig. 6A5).
Figure 6.
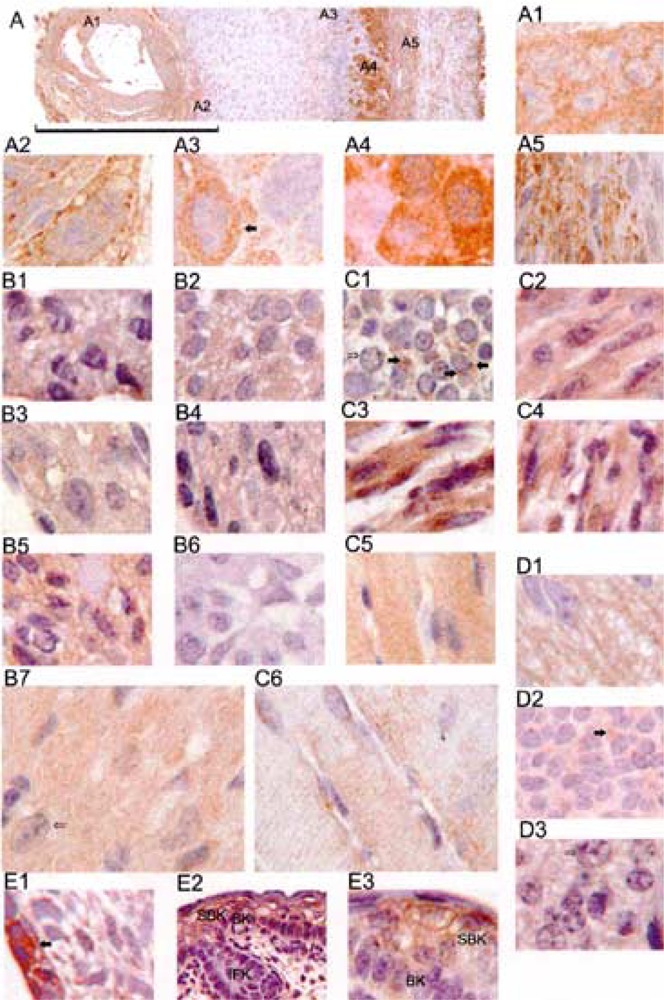
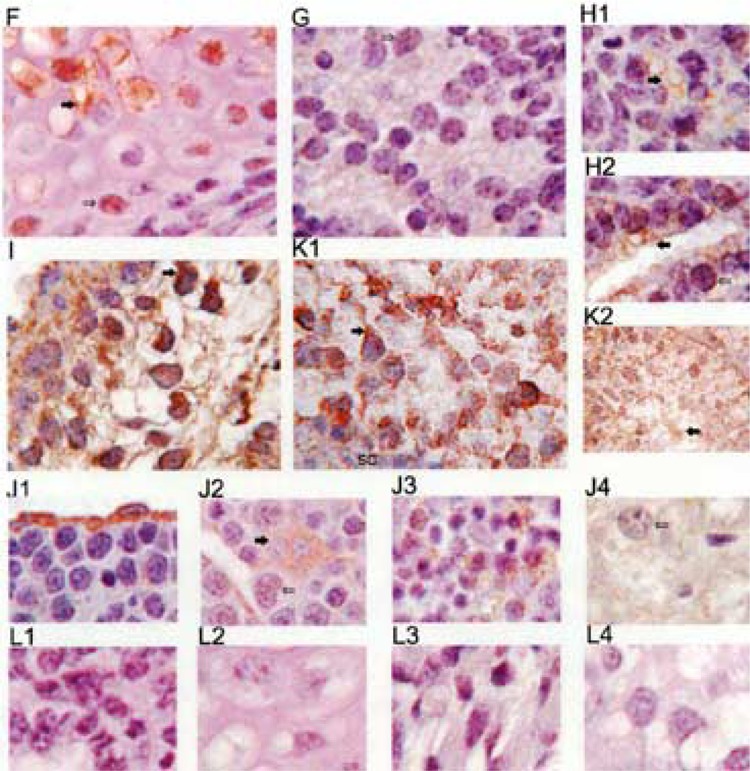
Usp2 expression in tissues of 7.5, 9.5, 14.5, and 16.5 dpc embryos. Embryo sections were stained with affinity-purified CC antibody (unless indicated otherwise), followed by an HRP-conjugated secondary antibody, and visualized with DAB (brown color). Nuclei were stained with hematoxylin (blue color). (A) 7.5 dpc embryo and part of the uterus, scale bar is 0.4 mm; (A1) allantois; (A2) embryo attachment site; (A3) endomentrial tissue; (A4) outer zone of the decidual reaction; (A5) myometrium; (B1) 9.5 dpc embryo heart, stained with peptide CC2 antibody; (B2) 13.5 dpc embryo heart; (B3) 13.5 dpc embryo heart, stained with peptide CC2 antibody; (B4) 16.5 dpc embryo heart, stained with N-29 antibody; (B5) 16.5 dpc embryo heart; (B6) control 13.5 dpc embryo heart, where primary antibody was omitted; (B7) adult mouse heart; (C1) 13.5 dpc embryo tongue; (C2) 13.5 dpc tongue; (C3) 16.5 dpc skeletal muscle; (C4) 16.5 dpc diaphragm; (C5) adult mouse diaphragm; (C6) adult heart diaphragm; (D1) 13.5 dpc medulla oblongata; (D2) 13.5 dpc brain; (D3) 16.5 dpc brain; (E1) 13.5 dpc epidermis; (E2 and E3) 16.5 dpc skin; (F) 16.5 dpc long bone; (G) 16.5 dpc eye; (H1 and H2) 16.5 dpc lung; (I) 13.5 dpc placenta; (J1) 13.5 dpc liver; (J2) 16.5 dpc liver; (J3) adult liver; (K1 and K2) adult testis; (L1) control 16.5 dpc lung; (L2) control 16.5 dpc bone; (L3) control 16.5 dpc skeletal muscle. The scale bar is 30 μm for (E2) and (K2) and 10 μm for all images, except A (which has its own scale bar).
We found Usp2 protein to be strongly and consistently expressed in heart throughout development: in 9.5 dpc embryo, when the heart is already fully developed (Fig. 6B1), in 13.5 dpc (Fig. 6B2, B3), and in 16.5 dpc (Fig. 6B4, B5) embryos. No staining was observed when primary antibody was omitted (Fig. 6B6). Likewise, there was very strong expression of Usp2 protein in mouse adult heart (Fig. 6B7). The expression of Usp2 was also very strong in muscles. In 13.5 dpc embryo tongue (which does not yet have fully formed muscle fibers) Usp2 was concentrated in the cytoplasm of cells in distinct areas around the nuclei (Fig. 6C1, filled arrows). Other muscles such as 16.5 dpc embryo tongue (Fig. 6C2), skeletal muscle (Fig. 6C3), and diaphragm (Fig. 6C4) as well as adult diaphragm (Fig. 6C5) and adult skeletal muscle (Fig. 6C6) show uniform staining along the fibers. Staining along the striations of muscle fibers was well defined in the section of adult skeletal muscle (Fig. 6C6).
At 13.5 dpc, strong staining was observed in the inner part of the medulla oblongata, which is comprised of neuronal axons (Fig. 6D1), and also in the cytoplasm of 13.5 dpc brain cells (Fig. 6D2, filled arrow) and 16.5 dpc brain cells (Fig. 6D3).
Usp2 was strongly expressed in epidermal cells of the 13.5 dpc embryo (Fig. 6E1) and in forming skin of 16.5 dpc embryo (Fig. 6E2), in particular, in basal and supra-basal keratinocytes of the epidermis of skin (Fig. 6E3), but not in the interfollicular keratinocytes (Fig. 6E2).
We observed Usp2 protein in some cells of 16.5 dpc embryo developing bone (Fig. 6F), with increasing amount of protein in cells towards the most differentiated middle of the bone (filled arrow). Likewise, Usp2 was detected in forming primitive lens of developing eye of 16.5 dpc embryo (Fig. 6G), in lung epithelial cells that are forming a new airway (Fig. 6H1, filled arrow), or more strongly where the airway is well defined (Fig. 6H2, filled arrow).
Additionally, Usp2 was expressed very strongly in the placenta of 13.5 dpc embryo (Fig. 6I), in the cytoplasm of some liver epithelial cells of 13.5 dpc (Fig. 6J1) and hepatocytes (Fig. 6J2, filled arrow), in 16.5 dpc embryonic liver (Fig. 6J3), and weakly throughout adult liver (Fig. 6J4). In testis, strong perinuclear staining was observed in spermatogonia (Fig. 6K1, filled arrow), continuing through all stages of spermatogenesis, including mature spermatozoa in the middle of the seminiferous tubule (Fig. 6K2, filled arrow). In contrast, Usp2 was not visible in Sertoli cells that surround each seminiferous tubule (Fig. 6K1).
Sample controls, where primary antibody were omitted, are presented in Figure 6L: 16.5 dpc lung (Fig. 6L1), 16.5 dpc bone (Fig. 6L2), 16.5 dpc skeletal muscle (Fig. 6L3), and adult testis (Fig. 6L4). Controls were observed for all embryos and tissues.
To ensure that the CC antibody did not cross-react with other Usps with similar catalytic cores, we affinity purified CC antiserum against a synthetic peptide from the Usp2 catalytic core CPETLDHLPDEEKGR (named CC2). This peptide was predicted to be on the surface of Usp2 (18) and had a unique sequence according to EMBL protein database searches. Pep-tide CC2-purified antibody produced an identical immunohistochemical staining pattern to the CC affinity-purified antibody, as shown in 9.5 dpc embryo (Fig. 6B1) and 13.5 dpc embryo (Fig. 6B3) heart sections, compared with the 13.5 dpc embryo heart section, stained with CC antibody (Fig. 6B2). Thus, the CC antibodies are specific in recognizing Usp2 protein by immunohistochemistry. Affinity-purified N-29 antibody gave the same staining pattern as the affinity-purified CC antibody in all tissues analyzed, as exemplified by the 16.5 dpc embryo heart section (Fig. 6B4). This indicates that the protein detected by immunohistochemistry is Usp2-69. We did not find any difference between the staining pattern obtained with the CC antibody common to both Usp2-45 and Usp2-69, and the antibody against the unique N-terminal extension of Usp2-69. This might indicate that Usp2-45 is not expressed at detectable levels, or that it is colocalized with Usp2-69.
Cell Localization of Usp2 Isoforms
Immunohistochemistry experiments showed that the localization of the protein was cytoplasmic in cells of 7.5 dpc embryos (Fig. 6A1–A5), embryonic heart (Fig. 6B1–B5, embryonic and adult muscle (Fig. 6C2–C6), neurons (Fig. 6E1–E2), epithelial cells of skin (Fig. 6E1–E3), eye (Fig. 6G), some cells of lung (Fig. 6H1–H2), liver (Fig. 6J1–J4), placenta (Fig. 6I), and spermatogonia of testis (Fig. 6K1–K2), with strong concentration around the nuclei in some of the cells (Fig. 6A3, I, K1, filled arrows). However, it was not clear from some sections (Fig. 6C1, B7, E3, F, G, H2, J2, J4, open arrows) whether some cells had nuclear localization of Usp2 or whether the brown staining seen was still perinuclear and the nuclei were located below the focal plane.
To investigate this matter, we overexpressed Usp2 CC, Usp2-45, and Usp2-69 isoforms with in COS-7 cells and in NIH3T3 fibroblasts (Fig. 7), and detected expression using affinity-purified CC antibody. Usp2 CC was highly expressed throughout the cells except the nucleus (Fig. 7A, B), whereas the Usp2-45 iso-form was localized to the cytoplasm (Fig. 7C, D). Usp2-69 was mainly concentrated in a particular area near the nucleus, which could be Golgi, and was also observed near the plasma membrane (Fig. 7E, F).
Figure 7.
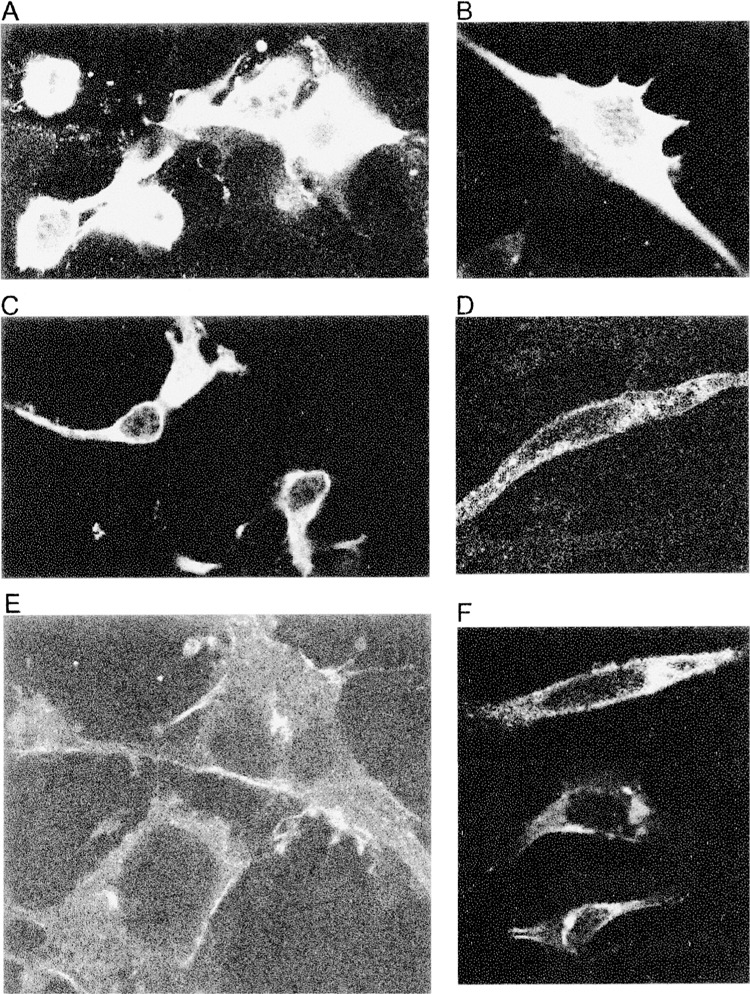
Localization of Usp2 isoforms in transfected cells. Usp2 CC (A, B), Usp2-45 (C, D), and Usp2-69 (E, F) were overexpressed in COS-7 cells (A, C, E) or NIH3T3 cells (B, D, F) growing on coverslips, fixed, detected with affinity-purified CC antibody (A, C, D, E), or peptide CC2 antibody (B, F), followed by anti-rabbit FITC-conjugated secondary antibody, and mounted on slides for confocal microscopy.
DISCUSSION
The Usp2 protein was first described as a 41-kDa protein from chicken, followed by cDNA sequences reported for mouse and human. Here we have identified the mouse Usp2 gene, which is alternatively spliced to give two isoforms, Usp2-45 and Usp2-69. Usp2-69 has at least three splice variants due to alternate splicing of exons at the 5′ end, which may be a regulatory mechanism as it does not affect the Usp2-69 protein. The previous description of one or both Usp2 isoforms as UBP41 has led to some confusion. Several reports describe a 41-kDa UBP41 as an independent protein in mammals, presumably referring to the single mouse (AF079565) and human (AF079564) GenBank entries, but we can find no evidence for its existence. We conclude that UBP41 is indeed the catalytic core of Usp2, but it cannot be expressed in vivo, as we have shown here there is no appropriate initiation codon.
When comparing the Northern and Western blotting results, it is evident that there is a discrepancy between the level of Usp2-69 mRNA expression and the level of Usp2-69 protein translation in the tissues analyzed. Indeed, the protein appears to be expressed well in most of the tissues with some degree of variation, while the RNA is highly expressed only in testis, muscle, and heart, and very weakly in other tissues. The protein expression in heart seems to be quite high, while it corresponds to only moderate levels of mRNA detected in this tissue. Interestingly, testis has very low protein level and the highest level of mRNA, compared with other tissues. As it is established for many testis proteins, gene transcription leads to mRNAs that are stored for several days before transcription ceases and cells start to differentiate, followed by delayed and highly regulated translation to ensure protein production late in germ cell development (7,25,28). Usp2 bears some hallmarks of this process: discrepancy between mRNA and protein levels; alternate splicing of 5′ UTR exons; and alternate and testis-specific polyadenylation sites. Thus, Usp2 may be subject to a similar, complex regulation of its transcription and translation in the testis, as well as in other tissues. In this respect, there are also numerous small ORFs within the 5′ UTRs of the different alternatively spliced Usp2-69 mRNAs, primarily in exon 1B, but also in exons 1A and 1D.
Such ORFs are often found in mRNAs encoding regulatory proteins and are known to regulate expression of the main ORF at the translational level (21). Interestingly, other USPs including Usp4 and Usp15 also have small upstream ORFs (2,5).
Notably, mRNA specific for Usp2-45 is present in all tissues except lung and spleen, while no 45-kDa protein was detected in any tissue by Western blotting. This is surprising, given that our antibody could detect ectopically expressed Usp2-45. We suggest that Usp2-45 might be expressed below the detection level or may not be constitutively translated at all and only be induced in the cell upon a specific signal or at a specific time. In connection to this, it is very interesting that Usp2-69 interacts with Usp2-45, which gives an indication that they may form a complex in vivo and function together. It is also very interesting that Usp2-69 is able to interact with itself, indicating that more than one molecule of Usp2-69 is involved in forming a complex, and that Usp2 may exist as homo- and/or heterodimer.
In rats, Usp2 was originally described as a testis-specific protein termed Ubp-t1 and Ubp-t2 (19). While our Northern blots reveal that Usp2 expression at the mRNA level is strongest in mouse testis, substantial expression was also observed in other tissues, which we have also confirmed at the protein level, at least for Usp2-69. As mentioned above, other studies have revealed that rat Usp2 mRNA isoforms are also expressed in muscle and have been proposed to have a role in muscle differentiation (24), while Miles et al. have shown wide tissue expression by Northern blot in rat (22), which is identical to the Northern blot results that we obtained in mouse with the catalytic core probe.
In our immunohistochemistry experiments Usp2 was detected at all stages of embryonic development and in different tissue types, from which we conclude that it does not regulate the development of particular organs as it has been suggested before, but it functions in many tissues and in different types of cells. However, Usp2 was very well expressed throughout heart, in muscle tissues and testis, which is consistent with our Northern blot results and previous findings.
We found an interesting expression pattern in forming bone of 16.5 dpc embryo, where Usp2 was found only in proliferating cells of growing bone that were committing to differentiate to form future bone tissue. Recently, rat Usp2 (described as Ubp41) has been implicated in bone formation (22). It was found to be rapidly and transiently upregulated at the mRNA level in bone and in osteoblasts in response to bone metabolism modulator PTH and other osteotropic agents. Presumably, one or both of the Usp2 isoforms are involved in the cascade of events mediating differentiation of osteoblasts.
We were also intrigued by the very high expression of Usp2 protein in many epithelial cells and in skin—a tissue that has not been tested before either by Northern or Western blot. In skin Usp2 protein is located mainly in the supra-basal keratinocytes of epidermis, which are postmitotic, starting to differentiate and would undergo maturation and apoptotic death in the adult animal. Likewise, in the eye Usp2 expression was concentrated mainly in the area where cells were differentiating and forming primitive lens IP: 103.62.30.226 On: We fibers. In testis, expression was found in differentiating spermatogonia and mature spermatozoa, which implies that Usp2 participates the processes of differentiation in testis, and this is in agreement with the data reported by Lin et al. (19).
Our immunohistochemistry results demonstrated Usp2 to be mainly a cytoplasmic protein with some perinuclear concentration in some tissues. We conclude that the tissue sections where brown staining appears to be nuclear were a sectioning artifact, because some nuclei would be below the focal plane, but could appear brown because of the presence of cytoplasm above them, or perinuclear localization of the protein. We confirmed the cytoplasmic localization of Usp2 isoforms using confocal microscopy on mammalian cells with overexpressed Usp2 isoforms. Our data indicate that both Usp2 isoforms are localized to the cytoplasm. Because Usp2-69 was observed around the nucleus, as well as at the plasma membrane, we suggest that Usp2-69 is able to traffic to different subcellular compartments in the cells, and experiments are under way to study the trafficking of Usp2 more precisely. Although there is no inconsistency in the localization of Usp2-69, the localization of Usp2-45 is different to the previously reported nuclear localization for its rat orthologue Ubp-t1 (19). The difference may be due to the position of the epitope tag, which was C-terminal in our case and N-terminal in the rat protein. However, we do not exclude the possibility that Usp2-45, when being expressed at low levels and under certain stimulus, is able to travel to the nucleus at certain stages or in certain cells.
Although we could not identify a specific single cell type that Usp2 may function in, a common theme can be observed in the reported cases of involvement of Usp2 in differentiation of testis, skeletal muscle, and bone. Presumably Usp2 does not define the future tissue type of the differentiating cells, but might be involved in cell cycle exit or early events initiating differentiation. These events could be common for differentiation of these highly specialized tissues. We also observed Usp2-69 in some proliferating cells and fully differentiated tissues, suggesting that it may have some additional functions. It might also be involved in apoptotic events that are an essential part of differentiation processes in tissues. Recently, Park et al. have described the induction of rat Usp2 iso-form mRNAs during first 4 days of rat myoblast differentiation (24). This period of differentiation is known to incorporate associated apoptotic events (16,29). It still remains unknown whether the expression of Usp2 changes during further differentiation and if it is involved in formation of myotubes, but its expression during the first 4 days of differentiation may support the involvement of Usp2 in apoptosis. Interestingly, overexpression of the Usp2 catalytic core alone (described as Ubp41) has been found to be proapoptotic (8). Given our data on the gene structure of Usp2 and the apparent absence of the template for translation of Ubp41 protein, it remains to be determined whether either of the bona fide Usp2-45 or Usp2-69 proteins are also apoptotic. In this respect it is notable that the N-terminal extensions of Usp2-45 and Usp2-69 appear to inhibit the activity of the catalytic core, at least in vitro (20). It is possible to speculate that either Usp2-45 or Usp2-69 or both are indeed involved in apoptosis, because removal of the regulatory N-terminal extension could have accelerated the activity of the enzyme. In conclusion, we have described the complete gene structure, RNA, protein expression, and tissue distribution of Usp2, which gives a basis for a future understanding of the function of Usp2.
ACKNOWLEDGMENTS
Supported in part by a grant from the Australian Research Council (Queen Elizabeth II Fellowship) to R.T.B. Natalia Gousseva is the recipient of an Eccles PhD scholarship (JCSMR). We thank Ann-Maree Ca-tanzariti for purifying Usp2 catalytic core protein used for antibody generation; Xiao-Wen Wang for assistance with one Northern blot; Elissa Sutcliffe for DNA sequencing; Ann Prins for all her help with im-munohistochemistry; Dr. Klaus Matthaei for providing mouse embryos; and Drs. Stephen Wood, Simon Hogan, and Michael Crouch for consultations on interpreting immunohistochemistry images.
REFERENCES
- 1. Altschul S. F.; Madden T. L.; Schèffer A. A.; Zhang J.; Zhang Z.; Miller W.; Lipman D. J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402; 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Angelats C.; Wang X.-W.; Jermiin L. S.; Copeland N. G.; Jenkins N. A.; Baker R. T. Isolation and characterization of the mouse ubiquitin-specific protease Usp15. Mamm. Genome 14:31–46; 2003. [DOI] [PubMed] [Google Scholar]
- 3. Baek S. H.; Choi K. S.; Yoo Y. J.; Cho J. M.; Baker R. T.; Tanaka K.; Chung C.-H. Molecular cloning of a novel ubiquitin-specific protease, UBP41, with isopeptidase activity in chick skeletal muscle. J. Biol. Chem. 272:25560–25565; 1997. [DOI] [PubMed] [Google Scholar]
- 4. Baker R. T.; Tobias J. T.; Varshavsky A. Ubiquitin-specific proteases of Saccharomyces cerevisiae . J. Biol. Chem. 267:23364–23375; 1992. [PubMed] [Google Scholar]
- 5. Baker R. T.; Wang X.-W.; Woollatt E.; White J.; Sutherland G. R. Identification, functional characterization and chromosomal localisation of USP15, a novel human ubiquitin-specific protease related to the Unp/USP4 oncoprotein, and a systematic nomenclature proposal for human ubiquitin-specific proteases. Genomics 59:264–274; 1999. [DOI] [PubMed] [Google Scholar]
- 6. Baker R. T. Deubiquitinating enzymes and the regulation of proteolysis. In: Wolf D.; Hilt W., eds. Proteasomes: The world of regulatory proteolysis. Austin, TX: Landes Bioscience Co.; 2000:238–255. [Google Scholar]
- 7. Braun R. E. Temporal control of protein synthesis during spermatogenesis. Int. J. Androl. 23(Suppl. 2):92–94; 2000. [DOI] [PubMed] [Google Scholar]
- 8. Gewies A.; Grimm S. UBP41 is a proapoptotic ubiquitin-specific protease. Cancer Res. 63:682–688; 2003. [PubMed] [Google Scholar]
- 9. Gilchrist C. A.; Gray D. A.; Baker R. T. A ubiquitin-specific protease that efficiently cleaves the ubiquitin-proline bond. J. Biol. Chem. 272:32280–32285; 1997. [DOI] [PubMed] [Google Scholar]
- 10. Gilchrist C. A.; Baker R. T. Characterization of the ubiquitin-specific protease activity of the mouse/human Unp/Unph oncoprotein. Biochem. Biophys. Acta 1481:297–309; 2000. [DOI] [PubMed] [Google Scholar]
- 11. Glickman M. H.; Ciechanover A. The ubiquitin-proteasome proteolytic pathway: Destruction for the sake of construction. Physiol. Rev. 82:373–428; 2002. [DOI] [PubMed] [Google Scholar]
- 12. Harlow E.; Lane D. Antibodies: A laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- 13. Hicke L. Protein regulation by monoubiquitin. Nat. Rev. Mol. Cell. Biol. 2:195–201; 2001. [DOI] [PubMed] [Google Scholar]
- 14. Hu M.; Li P.; Li M.; Li W.; Yao T.; Wu J. W.; Gu W.; Cohen R. E.; Shi Y. Crystal structure of a UBP-family deubiquitinating enzyme in isolation and in complex with ubiquitin aldehyde. Cell 111:1041–1054; 2002. [DOI] [PubMed] [Google Scholar]
- 15. Huang Y.; Baker R. T.; Fischer-Vize J. Control of cell fate by a deubiquitinating enzyme encoded by the fat facets gene. Science 270:1828–1831; 1995. [DOI] [PubMed] [Google Scholar]
- 16. Huppertz B.; Tews D. S.; Kaufmann P. Apoptosis and syncytial fusion in human placental trophoblast and skeletal muscle. Int. Rev. Cytol. 205:215–253; 2001. [DOI] [PubMed] [Google Scholar]
- 17. Kaufman M. The atlas of mouse development. London: Academic Press Limited; 1995. [Google Scholar]
- 18. Kyte J.; Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157:105–132; 1982. [DOI] [PubMed] [Google Scholar]
- 19. Lin H.; Keriel A.; Morales C. R.; Bedard N.; Zhao Q.; Hingamp P.; Lefrancois S.; Combaret L.; Wing S. S. Divergent N-terminal sequences target an inducible testis deubiquitinating enzyme to distinct subcellular structures. Mol. Cell. Biol. 20:6568–6578; 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lin H.; Yin L.; Reid J.; Wilkinson K. D.; Wing S. S. Divergent N-terminal sequences of a deubiquitinating enzyme modulate substrate specificity. J. Biol. Chem. 276:20357–20363; 2001. [DOI] [PubMed] [Google Scholar]
- 21. Meijer H. A.; Thomas A. A. Control of eukaryotic protein synthesis by upstream open reading frames in the 5′-untranslated region of an mRNA. Biochem. J. 367:1–11; 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Miles R. R.; Sluka J. P.; Halladay D. L.; Santerre R. F.; Hale L. V.; Bloem L.; Patanjali S. R.; Galvin R. J.; Ma L.; Hock J. M.; Onyia J. E. Parathyroid hormone (hPTH 1-38) stimulates the expression of UBP41, an ubiquitin-specific protease, in bone. J. Cell. Biochem. 85:229–242; 2002. [DOI] [PubMed] [Google Scholar]
- 23. Papa F.; Hochstrasser M. The yeast DOA4 gene encodes a deubiquitinating enzyme related to a product of the human tre-2 oncogene. Nature 366:313–319; 1993. [DOI] [PubMed] [Google Scholar]
- 24. Park K. C.; Kim J. H.; Choi E. J.; Min S. W.; Rhee S.; Baek S. H.; Chung S. S.; Bang O.; Park D.; Chiba T.; Tanaka K.; Chung C.-H. Antagonistic regulation of myogenesis by two deubiquitinating enzymes, UBP45 and UBP69. Proc. Natl. Acad. Sci. USA 99:9733–9738; 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Penttila T. L.; Yuan L.; Mali P.; Hoog C.; Parvinen M. Haploid gene expression: Temporal onset and storage patterns of 13 novel transcripts during rat and mouse spermiogenesis. Biol. Reprod. 53:499–510; 1995. [DOI] [PubMed] [Google Scholar]
- 26. Thompson J. D.; Higgins D. G.; Gibson T. J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680; 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tobias J. W.; Varshavsky A. Cloning and functional analysis of the ubiquitin-specific protease gene UBP1 of Saccharomyces cerevisiae . J. Biol. Chem. 266:12021–12028; 1991. [PubMed] [Google Scholar]
- 28. Walker W. H.; Delfino D. J.; Habener J. F. RNA processing and the control of spermatogenesis. Front. Horm. Res. 25:34–58; 1999. [DOI] [PubMed] [Google Scholar]
- 29. Walsh K. Coordinate regulation of cell cycle and apoptosis during myogenesis. Prog. Cell Cycle Res. 3:53–58; 1997. [DOI] [PubMed] [Google Scholar]
- 30. Wilkinson K. D. Ubiquitination and deubiquitination: Targeting of proteins for degradation by the proteasome. Semin. Cell Dev. Biol. 11:141–148; 2000. [DOI] [PubMed] [Google Scholar]
- 31. Wilkinson K. D. Regulation of ubiquitin-dependent processes by deubiquitinating enzymes. FASEB J. 11:1245–1256; 1997. [DOI] [PubMed] [Google Scholar]
- 32. Zhu Y.; Carroll M.; Papa F. R.; Hochstrasser M.; D’Andrea A. D. DUB-1, a deubiquitinating enzyme with growth-suppressing activity. Proc. Natl. Acad. Sci. USA 93:3275–3279; 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhu Y.; Lambert K.; Corless C.; Copeland N. G.; Gilbert D. J.; D’Andrea A. D. DUB-2 is a member of a novel family of cytokine-inducible deubiquitinating enzymes. J. Biol. Chem. 272:51–57; 1997. [DOI] [PubMed] [Google Scholar]


