Abstract
We conducted integrated transcriptomics network analyses of miRNA and mRNA interactions in human myometrium to identify novel molecular candidates potentially involved in human parturition. Myometrial biopsies were collected from women undergoing primary Cesarean deliveries in well-characterized clinical scenarios: (1) spontaneous term labor (TL, n = 5); (2) term nonlabor (TNL, n = 5); (3) spontaneous preterm birth (PTB) with histologic chorioamnionitis (PTB-HCA, n = 5); and (4) indicated PTB nonlabor (PTB-NL, n = 5). RNAs were profiled using RNA sequencing, and miRNA-target interaction networks were mined for key discriminatory subnetworks. Forty miRNAs differed between TL and TNL myometrium, while seven miRNAs differed between PTB-HCA vs. PTB-NL specimens; six of these were cross-validated using quantitative PCR. Based on the combined sequencing data, unsupervised clustering revealed two nonoverlapping cohorts that differed primarily by absence or presence of uterine quiescence, rather than gestational age or original clinical cohort. The intersection of differentially expressed miRNAs and their targets predicted 22 subnetworks with enriched representation of miR-146b-5p, miR-223-3p, and miR-150-5p among miRNAs, and of myocyte enhancer factor-2C (MEF2C) among mRNAs. Of four known MEF2 transcription factors, decreased MEF2A and MEF2C expression in women with uterine nonquiescence was observed in the sequencing data, and validated in a second cohort by quantitative PCR. Immunohistochemistry localized MEF2A and MEF2C to myometrial smooth muscle cells and confirmed decreased abundance with labor. Collectively, these results suggest altered MEF2 expression may represent a previously unrecognized process through which miRNAs contribute to the phenotypic switch from quiescence to labor in human myometrium.
Keywords: microRNA, transcriptomics, parturition, preterm birth, myometrium, MEF2 transcription factors
Summary Sentence
Integrated miRNA–mRNA study in human myometrium.
Introduction
Throughout most of human pregnancy, the uterus, and especially the myometrium, must accommodate the myometrial wall expansion necessary for feto-placental growth while maintaining functional quiescence. At term, parturition requires the onset of forceful, coordinated contractions that culminate in the delivery of the fetus. Functional genomics studies to date have revealed that transition of the pregnant myometrium to an actively contractile state requires a complex and highly regulated change in the expression of genes encoding factors that affect myometrial structure, contractility, and signaling supported the long-standing consensus that inflammation plays a key role in human parturition [1–11]. Myometrial transcriptome studies have further demonstrated that spontaneous preterm birth (PTB) has numerous features in common with term labor (TL), suggesting shared mechanisms from the mid-second trimester onward [3, 11]. However, as genes that initiate TL appeared to differ from those initiating PTB, it was proposed that although diverse pathways may trigger PTB and TL, they eventually converge into a final common effector pathway characteristic to the human laboring myometrium [9].
Recent advances in RNA-sequencing (RNA-seq) technology have provided new opportunities to integrate changes in protein-coding mRNAs with regulating signals such as those driven by noncoding RNAs and alternative RNA splicing events [1]. Our group recently applied this technology, along with systems-level computational approaches, to facilitate a better understanding of the changes in the decidual and placental compartments in the setting of PTB [12]. Mature microRNAs (miRNAs) are small (∼22 nucleotides in length) noncoding RNAs that regulate protein expression at the post-transcriptional level through interaction with target mRNAs [13]. While most of the studies portray miRNAs as transcriptional repressors of target mRNA, increasing evidence suggests that miRNAs oscillate between inhibition and stimulation in response to specific cellular contexts [14]. Nearly 2600 unique mature human miRNAs have been cataloged in the most recent update of the miRBase 21 database [15]. Since each miRNA has the capacity to potentially regulate hundreds of genes, as a class, miRNAs can exert broad influence on global protein expression.
The role of several miRNA families in the governance of myometrial contractility has been recently reviewed [16]. A combined miRNA and mRNA microarray analysis of mouse uterus demonstrated that miR-199a-3p and miR-214 were significantly decreased in coordination with several mRNAs and transcription factors that are known to be involved in loss of quiescence and initiation of uterine contractions [17]. However, in the context of the human myometrium, an unbiased approach to measure the extent to which global changes in miRNA intersect with changes in their mRNA targets has so far not been undertaken. Herein, we applied integrated RNA-seq of miRNA and mRNA along with various computational algorithms to uncover novel molecular pathways potentially
responsible for the transition of the human myometrium from a quiescent to a contractile laboring state in both term and PTB.
Materials and methods
Subjects and tissue collection
Myometrial biopsy samples were collected from 20 women with singleton gestations undergoing primary Cesarean deliveries in four settings which defined their clinical phenotype as either: (1) spontaneous onset of TL (n = 5, gestational age [GA] median [range]: 39 [38–40] weeks), (2) term nonlabor (TNL, n = 5, GA: 39 [39–39] weeks), (3) spontaneous onset of PTB in the context of histologic chorioamnionitis (PTB-HCA, GA; n = 5, 29 [27–31]; (4) provider-initiated PTB in the absence of labor (PTB-NL, n = 5, GA: 2 [25–32] weeks). An additional set of seven term women (TNL, n = 4; and TL, n = 3) provided tissues solely for immunohistochemistry (IHC) validation, while an additional set of 25 term and preterm women provided tissues solely for quantitative PCR (qPCR) validation (as indicated in text and figure legends). By study design, HCA was diagnosed by a clinical pathologist, and was present in all cases in the PTB-HCA group, but absent in the PTB-NL group. Among cases in the PTB-HCA group, the spontaneous onset of PTB was defined clinically, either through preterm premature rupture of membranes (PPROM), cervical shortening and dilation followed by onset of irregular uterine contractions, or onset of uterine contractions followed by cervical change. All PTB-NL cases were provider-initiated PTBs in the context of severe preeclampsia/eclampsia. All term Cesarean deliveries were performed via an incision on the lower uterine segment. Among preterm cases, 60% (6/10) of uterine incisions were vertical. This study was approved by the Human Investigation Committee of Yale University and the Institutional Review Board of the Ohio State University. All women provided written informed consent.
All patients included in the primary study cohort (n = 20) had a singleton fetus and had requested epidural or combined spinal-epidural analgesia prior to primary Cesarean delivery. Inclusion criteria for term and preterm laboring women required regular myometrial contractions and ≥4 cm cervical dilation. At term, TL women met the criteria for labor dystocia (failure to descend or progress), despite adequate uterine contractility. In four of the five TL patients, uterine contractility was assessed using an intrauterine pressure device. Exclusion criteria included nonsingleton pregnancies, suspected fetal macrosomia ( >4500 g by clinical or ultrasound evaluation), abnormalities of placentation (low-lying placenta, placental abruption), uterine structural abnormalities, and a prior uterine scar. Nonlaboring women at term delivered via elective, primary low segment Cesarean section (LTCS). The indications for LTCS in this group included breech presentation, funic presentation, and suspected anatomic cephalopelvic disproportion.
Following the primary study, a validation cohort was selected to confirm and extend major study findings. This cohort comprised women at term (n = 9) and preterm (n = 16) delivered by Cesarean section, in which the absence or presence of uterine quiescence was determined clinically by tocodynometric or intrauterine pressure catheter measurements prior to surgery (uterine quiescence group, n = 15; uterine nonquiescence group, n = 10).
Myometrial biopsies collection
All myometrial biopsy specimens were collected with surgical scissors immediately after delivery of the fetus and placenta. They consisted of a full-thickness strip of myometrial tissue, excluding decidua, from the upper lip of the incision in LTCS cases (n = 14 in primary cohort, n = 14 in validation cohort), or the side at the midpoint of the incision in classical incisions (n = 6 in primary cohort, n = 11 in validation cohort). A portion of the tissue biopsy was snap-frozen in liquid nitrogen and stored at –80°C. The remainder was fixed in 10% buffered formaldehyde (Fisher Chemical, Fairlawn, NJ) and paraffin-embedded.
Total RNA extraction from tissues
Total RNA was isolated using TRIzol reagent (Life Technologies, Grand Island, NY) according manufacturer's instructions, followed by purification using the RNA Clean-Up and Concentration Kit (Norgen Biotek, Thorold, Ontario, Canada). This protocol was previously validated for extraction of both long RNA and miRNA from the same tissue samples [12].
RNA sequencing
Long total RNA-seq libraries (inclusive of mRNAs and noncoding RNAs) were constructed using the TruSeq Stranded Total RNA Sample Prep Kit with Ribo-Zero Gold (Illumina, San Diego, CA). Small RNA-seq libraries were generated using the NEBNext Multiplex Small RNA Library Prep Set for Illumina (New England Biolabs, Ipswich, MA), according to the manufacturer's protocols. Sequencing was performed using the Illumina HiSeq 2500 system and HiSeq version 3 sequencing reagents to generate 50 bp single-end reads. Image analysis, base calling, and error estimation was performed using Illumina Analysis Pipeline in the HiSeq Control Software version 2.2.38.
Long RNA-seq and miRNA-seq data analysis
The analysis strategy for sequencing data was previously reported [12, 18]. In brief, the quality of all sequenced reads was assessed using FastQC and adaptor, and quality trimming was achieved using Trim Galore version 0.3.2. (both available at http://www.bioinformatics.babraham.ac.uk/index.html). The long RNA reads were mapped to the Homo sapiens UCSC hg38 genome assembly TopHat2 version 2.0.11 [19], and feature counts were generated using HTseq version 0.5.3p3 [20]. The data from the small RNA-seq libraries were mapped to the miRBase21 database [15] using miRExpress version 2.0 [21]. Differential expression was determined statistically using DESeq2 version 1.4.5 [22] with a false discovery rate (FDR) of <0.1 and a linear fold-change cut-off ≥1.5.
MicroRNA target analysis
Experimentally confirmed and predicted miRNA target transcripts were retrieved using the multiMiR R package version 0.99.5 [23]. All experimentally verified target predictions from the miRecords, miRTarBase, and TarBase databases were retained. Potential miRNA–mRNA target interactions were determined by mining the following target prediction databases: DIANA-microT-CDS, ElMMo, MicroCosm, miRanda, miRDB, PicTar, PITA, and TargetScan. Since not all of these databases contained predictions for all of the miRNAs surveyed, we considered only those targets that were present in at least half of the repositories in which miRNA-target interactions were predicted. The Enrichr [24] web server was used to interrogate pathways influenced by miRNA alterations.
Integrated miRNA-target mRNA interaction network analyses
To prioritize myometrial miRNA-target mRNA subnetworks associated with term and preterm labor, we performed an integrative analysis of miRNA and gene expression data based on mRNA abundance. Genes were mapped to the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) protein–protein interaction (PPI) network database [25] with minimum edge confidence set to 0.4 and maximum interactions per node set to 20. Additional nodes were added for differentially expressed miRNAs, with edges representing previously validated miRNA–mRNA target interactions, to form miRNA-PPI networks. These networks were scored using the RNA-seq counts from the term and preterm samples, respectively. The Subnetwork Analysis and Scoring System was used to identify discriminatory small candidate subnetworks [26–28]. The approach used for prioritizing candidate subnetworks did not rely solely on differential gene expression and, as such, mRNA targets and other nodes recruited via PPIs do not necessarily represent transcripts identified as being differentially expressed via the DESeq2 algorithm. Rather, network topology, discretized activity, and an information theoretical metric (the mutual information [MI] score) between clinical phenotypes (PTB-HCA vs. PTB-NL, and TNL vs. TL) was computed as a measure of the ability of a given subnetwork to distinguish between the two phenotypes in each comparison. All possible combinations of two to five genes were scored as possible subnetworks, where subnetwork activity was defined as the aggregate mRNA expression of the subnetwork genes. Gene sets within high scoring subnetworks were connected along all possible shortest paths.
Hierarchical clustering and principal component analysis
Unsupervised hierarchical clustering, dendogram generation, and heatmap plotting were performed using the pheatmap R package version 0.7.7. A complete linkage agglomeration algorithm with Manhattan distance function was applied to normalized, log2-transformed fold change data for statistically significant differentially expressed RNAs. Principal component analysis (PCA) for differentially expressed mRNAs and miRNAs was accomplished using the prcomp function in the base R package. Scatterplots of the principal components (PCs) were generated using SigmaPlot version 12.5 (Systat Software, Chicago, IL). The data presented in this study are accessible through GEO Series accession number GSE99853.
Real-time qPCR
Several mature miRNAs were selected from the sequencing data analysis for cross-validation. The Universal cDNA Synthesis Kit II (Exiqon, Woburn, MA) was used to prepare complementary DNA (cDNA) from isolated total RNA, in tandem with miRNA polyadenylation. Quantitative PCR was performed using the ExiLENT SYBR Green master mix (Exiqon) in the ABI StepOnePlus real-time PCR system (Life Technologies, Carlsbad, CA) according to the manufacturer's recommendations. The following Exiqon locked nucleic acid qPCR primer sets were used for amplification: miR-146b-5p (cat. number 204553), miR-150-5p (cat. number 204660), miR-223-3p (cat. number 205986), miR-888-5p (cat. number 206024), miR-891a-5p (cat. number 204220), miR-892a (cat. number 204092), and miR-892b (cat. number 110497). All miRNAs were normalized to small nucleolar RNA U3 (SNORD38B, cat. number 203901) as an internal loading control [29]. All reactions were performed in duplicate with melting curve analysis following amplification to confirm the presence of single peaks.
myocyte enhancer factor 2A (MEF2A) and myocyte enhancer factor 2C (MEF2C) mRNAs were also cross-validated using qPCR. Reverse transcription for these reactions was performed with Superscript II Reverse Transcriptase (Invitrogen Life Technologies, Carlsbad, CA) using oligo-deoxythymidine primers. The following TaqMan gene expression assays (Thermo Fisher Scientific, Waltham, MA) were used: MEF2A (cat. number 4351372) and MEF2C (cat. number 4331182). The geometric mean of the cycle threshold (CT) values for beta-2-microglobulin (B2M, cat. number 4331182) and ribosomal protein L30 (RPL30, cat. number 4331182) was used as a reference in each reaction. Each 20 μl reaction consisted of 1 μl cDNA (500 ng), 1 μl of TaqMan Gene Expression Assay, 10 μl TaqMan Fast Advanced Master Mix (Thermo Fisher Scientific), and 8 μl of nuclease free water. All reactions were performed in duplicate.
The relative abundance of each miRNA and mRNA was calculated using comparative CT method [30]. Statistical comparisons were conducted using Student t-test on log-transformed data following Shapiro-Wilk normality testing. Correlations between gene expression levels from RNA-seq and qPCR experiments were examined using Pearson's product moment correlation (on log-transformed data).
Immunohistochemistry
Two isoforms of the myocyte enhancer factor-2 (MEF2) family were selected for validation and localization at protein level: MEF2A and MEF2C. IHC was performed on formalin-fixed myometrial specimens using a protocol described previously [31]. IHC was carried out on TL (n = 8) and TNL (n = 9) tissues, of which 10 were the same as those used for RNA-seq experiments. The following antibodies were used: rabbit monoclonal anti-MEF2A (1:100, EP1076Y, Abcam, Cambridge, MA), and rabbit polyclonal anti-MEF2C (1:100, 18290-1-AP, Proteintech, Rosemont, IL) (see Supplemental Table S1). Where indicated, further immunolabeling was performed using mouse monoclonal anti-CD15 also known as fucosyltransferase 4 (FUT4) (1:15, ab53997, Abcam) and mouse monoclonal anti-α-smooth muscle actin (αSMA, also known as actin, alpha 2, smooth muscle, aorta (ACTA2); 1:1000, Thermo Fisher Scientific, Waltham, MA) to identify myeloid lineage cells and smooth muscle cells, respectively. Specificity of staining was confirmed by substituting the primary antibody with species-specific preimmune IgG (not shown). Additional antibody characterization experiments are presented in the Supplemental Materials. Staining intensity of chromogen deposits was evaluated in specific cell types using a semiquantitative 5-point grading scale, with 1 indicating low or absent staining, and 5 indicating intense staining. Statistical comparisons were conducted using the two-tailed Mann–Whitney rank-sum test.
Results
Clinical characteristics of myometrial samples in primary cohort
The characteristics of the cases grouped by clinical phenotype are presented in Table 1. There were no statistically significant differences in GA or birth weight between the two term groups (TL vs. TNL), nor between the two preterm groups (PTB-HCA vs. PTB-NL). Preterm babies were of significantly lower birthweight compared to term neonates (P = 0.002).
Table 1.
Characteristics of the women who provided myometrium samples for RNA sequencing experiments (primary cohort) (n = 20).
| Variable | Term labor (TL, n = 5) | Term nonlabor (TNL, n = 5) | P value (TL vs. TNL) | Preterm birth and HCA (PTB-HCA, n = 5) | Preterm birth nonlabor (PTB-NL, n = 5) | P value (PTB-HCA vs. PTB-NL) | P value (all groups) |
|---|---|---|---|---|---|---|---|
| Clinical characteristics | |||||||
| Maternal age, years† | 31 [23–33] | 28 [25–35] | P = 0.841 | 39 [30–40] | 33 [21–40] | P = 0.548 | P = 0.383 |
| Parity† | 0 [0–0] | 0 [0–1] | P = 0.444 | 2 [1–3] | 0 [0–1] | P = 0.056 | P = 0.022 |
| Gravidity† | 1 [1–2] | 3 [2–4] | P < 0.05 | 4 [2–4] | 1 [1–3] | P = 0.095 | P = 0.038 |
| GA at delivery, wks† | 39 [38–41] | 39 [39–39] | P = 0.690 | 29 [27–31] | 29 [26–32] | P = 0.999 | P = 0.002 |
| PPROM‡ | 0 (0) | 0 (0) | P = 1.000 | 3 (60) | 0 (0) | P < 0.05 | P = 0.014 |
| Clinical chorioamnionitis‡ | 0 (0) | 0 (0) | P = 1.000 | 3 (60) | 0 (0) | P < 0.05 | P = 0.014 |
| Outcome characteristics | |||||||
| Term delivery (≥37 weeks)‡ | 5 (100) | 5 (100) | P = 1.000 | 0 (0) | 0 (0) | P = 1.000 | P < 0.001 |
| PTB at < 34 weeks‡ | 0 (0) | 0 (0) | P = 1.000 | 5 (100) | 5 (100) | P = 1.000 | P < 0.001 |
| HCA, grade† | 0 [0–0] | 0 [0–0] | P = 1.000 | 3 [3–4] | 0 [0–0] | P < 0.01 | P < 0.001 |
| Birth weight, grams† | 3340 [3068–3715] | 2900 [2865–4040] | P = 0.548 | 1205 [1013–1655] | 1100 [573–1,525] | P = 0.548 | P = 0.002 |
†Data presented as median [interquartile range] and analyzed by Mann–Whitney rank-sum test (for two group comparisons) or Kruskal–Wallis ANOVA on Ranks followed by multiple comparisons using Dunn test.
‡Data presented as n (%) and analyzed by Fisher exact or chi-squared test.
Abbreviations: HCA, histological chorioamnionitis; PPROM, preterm premature rupture of membrane; GA, gestational age.
Patterns of myometrial miRNA and mRNA expression in term and PTB
There were 1507 unique mature miRNAs (58% of those currently annotated in miRBase 21) detected in at least one myometrial sample. Overall, abundance of myometrial miRNA spanned five orders of magnitude, with 242 miRNAs (16%) expressed above the median level of 81 counts.
Term myometrium
Relative to TNL, TL was associated with differential expression of 40 mature miRNAs (16 with increased abundance, and 23 with decreased abundance) having, collectively, 3663 unique validated mRNA targets (Supplemental Table S2). Validated targets of these miRNAs were highly enriched for transcripts mapping to pathways involved multiple processes including cardiomyocyte hypertrophy, the DNA damage response, actin cytoskeleton regulation, and proinflammatory signaling pathways.
Preterm myometrium
In the comparison between the PTB-HCA and PTB-NL, there were only seven miRNAs that differed significantly in abundance, and all were increased in the setting of PTB-HCA relative to PTB-NL (Supplemental Table S3). In aggregate, the experimentally verified targetome for these miRNAs comprised 1013 unique transcripts with enrichment for transcripts encoding proteins involved in cardiomyocyte differentiation and hypertrophy, as well as proinflammatory signaling and advanced glycation endproducts (AGE) pathways.
Term vs. preterm myometrium
Of all miRNAs that were differentially expressed in the preterm group, six miRNAs were common within the TL vs. TNL comparison. The miR-146b-5p (toll-like receptor and IL-1 signaling pathways), miR-223-3p (AGE/receptor for AGE) pathways, and miR-223-5p (cardiomyocyte hypertrophy) were increased in both clinical scenarios. The miR-888-5p (nuclear receptors, e.g., progesterone receptor; tumor necrosis factor (TNF)-alpha signaling pathway), miR-891a-5p (estrogen signaling pathway), and miR-892b (apoptosis modulation and signaling pathway) were increased in the setting of PTB-HCA compared to PTB-NL, but exhibited diminished abundance in TL compared to TNL. The remaining miRNA (miR-150-5p; cardiac progenitor differentiation) was uniquely increased in PTL-HCA compared to PTB-NL, with no change between TL and TNL specimens.
Unsupervised clustering analysis
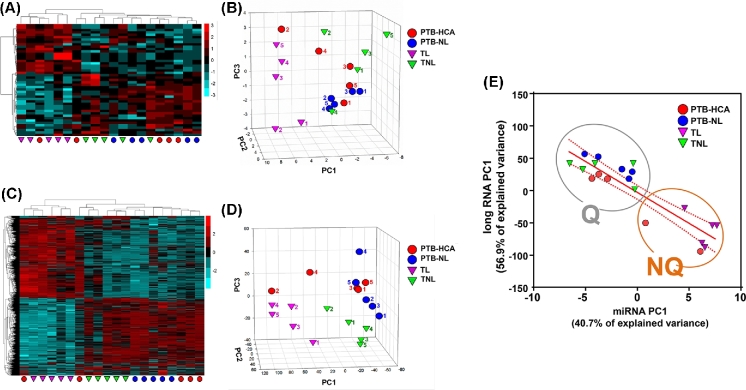
Unsupervised hierarchical and PC clustering of myometrial miRNA expression is shown in Figure 1A and B. The first PC (PC1) explained 40.7% of the variance, while PC2 and PC3 explained 20.9% and 9.4%, respectively. Clustering based on long RNA expression is represented in Figure 1C and D. PC1 explained 56.9% of the variance, while PC2 and PC3 explained only 6.9% and 6.5%, respectively. (The full complement of differentially expressed long RNAs will be reported separately). Both analyses resulted in clear separation of the TL from TNL samples, with less distinction between PTB-HCA and PTB-NL specimens. A significant inverse correlation between the two first PCs (PC1-miRNA vs. PC1-mRNA: r = –0.897, P < 0.001) was suggestive of coregulation between miRNA and long RNA expression patterns transcending the initial clinical groupings. As a result, when tissue samples were clustered using the combination of the two first PCs for each RNA species (Figure 1E), two nonoverlapping molecular clusters were identified: Cluster Q (quiescent, n = 13), which included all TNL and PTB-NL cases together with three of the PT-HCA cases (all of which were from cases presenting with advanced cervical dilation in the absence of uterine contractions); and Cluster NQ (nonquiescent, n = 7), which included all TL cases along with the remaining two PTB-HCA cases, both of which were from cases that exhibited forceful uterine contractions while in hospital. Demographic comparisons for these data-driven clusters are presented in Supplemental Table S4.
Figure 1.
Differentially expressed miRNA and long RNA transcripts in myometrial specimens in term and preterm labor. (A) Heat map of differentially expressed miRNA transcripts comparing: (1) term labor (TL) to term birth in absence of labor (TNL); and (2) spontaneous preterm birth (PTB) with clinical and/or histologic chorioamnionitis (PTB-HCA) to PTB without labor (PTB-NL). (B) Three-dimensional scatterplot of PCA results for miRNA-seq data. (C) Heat map of differentially expressed long RNA transcripts (TL vs. TNL, and PTB-HCA vs. PTB-NL). (D) PCA results for the long RNA-seq data. Note that the TL samples (purple triangles) clustered distinctly from TNL (green triangles) and PTB-NL (blue circles) specimens, whereas the PTB-HCA samples (red circles) clustered heterogeneously among the laboring and nonlaboring specimens. (E) Scatterplot and linear regression analysis comparing the first PC of the miRNA-seq dataset with that of the long RNA-seq data. Two nonoverlapping molecular clusters were identified: Cluster Q (quiescent, n = 13), and Cluster NQ (nonquiescent, n = 7), which corresponded with clinical records identifying whether or not strong uterine contractions had been documented prior to Cesarean delivery.
Identification of regulatory subnetworks through integration of miRNA-mRNA target networks with PPI analyses
Term and preterm miRNA-PPI networks were constructed from the miRNA identified in the RNA-seq dataset mapped to the library of experimentally validated mRNA in the databases. The collection of miRNA-mRNA binding pairs potentially active in term and preterm tissues is shown in Supplemental Figure S1A. No subnetworks were significantly enriched between the TL and TNL samples when the term miRNA-PPI network was scored with the associated transcriptome data in our RNA-seq dataset. In contrast, 22 subnetworks emerged as potentially relevant when scoring the preterm miRNA-PPI network with its associated transcriptome data (Supplemental Figure S1B and S1C; Supplemental Table S5). Three miRNAs populated these subnetworks: miR-146b-5p (present in 17 of 22 subnetworks), miR-223-3p (present in 12 subnetworks), and miR-150-5p (present in eight subnetworks). Four of these subnetworks were characterized by the inclusion of the MEF2C (myocyte-enhancer factor 2C) transcript, which encodes a transcription factor having a pivotal role in myogenesis and cardiac reprogramming/regeneration [32].
PCR cross-validation of differentially expressed miRNAs
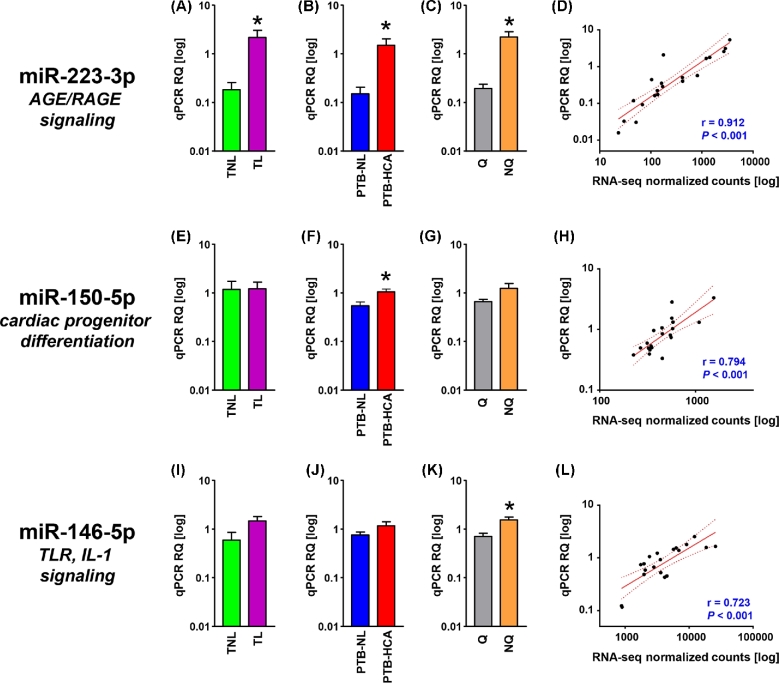
We first selected for cross-validation of the three miRNAs represented in the subnetworks in Supplemental Figure S1 (miR-223-3p, miR-150b-5p, miR-146b-5p). Additionally, miR-888-5p, miR-891a-5p, miR-892b were selected given the differential expression between HCA-PTB and PTB-NL specimens (Supplemental Table S3), and their membership to a primate-specific miRNA cluster residing within a ∼34-kb region on the minus strand of chromosome X (Xq27.3) [33]. Lastly, miR-892a was added given its membership to the same cluster and differential expression in the TL vs. TNL comparison (Supplemental Table S2). The results of the PCR cross-validation are shown in Figures 2 and 3. Expression levels of miR-223-3p were significantly elevated both in TL (P = 0.009, Figure 2A) and in PTB-HCA (P = 0.009, Figure 2B) as compared to TNL and PTB-NL, respectively. The difference remained significant when the tissues were grouped by contractile phenotype (P < 0.001, Figure 2C). There was a significant correlation between miRNA abundance measured by qPCR and RNA-seq (P < 0.001, Figure 2D). miR-150-5p was confirmed to remain unchanged between TL vs. TNL (P = 0.854, Figure 2E), but significantly increased in PTB-HCA vs. PTB-NL (P = 0.021, Figure 2F) myometrium. There was no significant difference in miR-150-5p with quiescence (P = 0.196, Figure 2G), but its qPCR levels correlated with the RNA-seq data (P < 0.001, Figure 2H). For miR-146-5p, only the comparison by contractile phenotype (P = 0.016, Figure 2K) and the correlation with the RNA-seq dataset were significant (P < 0.001, Figure 2L), but comparisons by clinical groups did not reach significance (TL vs. TNL; P = 0.069, Figure 2I; and PTB-HCA vs. PTB-NL: P = 0.247).
Figure 2.
Cross-validation of miRNA-seq data represented in the miRNA-PPI subnetworks. (A–C) Relative quantification (RQ) of real-time quantitative PCR (qPCR) data for mature miR-223-3p in term myometrial specimens (TNL, term nonlabor; TL, term labor; A), preterm myometrial specimens (PTB-NL, preterm birth nonlabor; PTB-HCA, preterm birth with histologic chorioamnionitis; B), and specimens grouped by molecular clustering (Q, quiescent phenotype; NQ, nonquiescent phenotype; C). (D) Scatterplot and linear regression analysis (line of best fit and 95% confidence interval) for miR-223-3p comparing qPCR results with normalized RNA-seq feature counts. (E–H) RQ (grouped as described above) and linear regression analysis (qPCR vs. normalized RNA-seq) for miR-150–5p. (I–L) RQ for mature Q (grouped as described above) and linear regression analysis (qPCR vs. normalized RNA-seq) for miR-146-5p. Data in bar graphs comprise mean ± SEM. Asterisks indicate P < 0.05 by Student t-test.
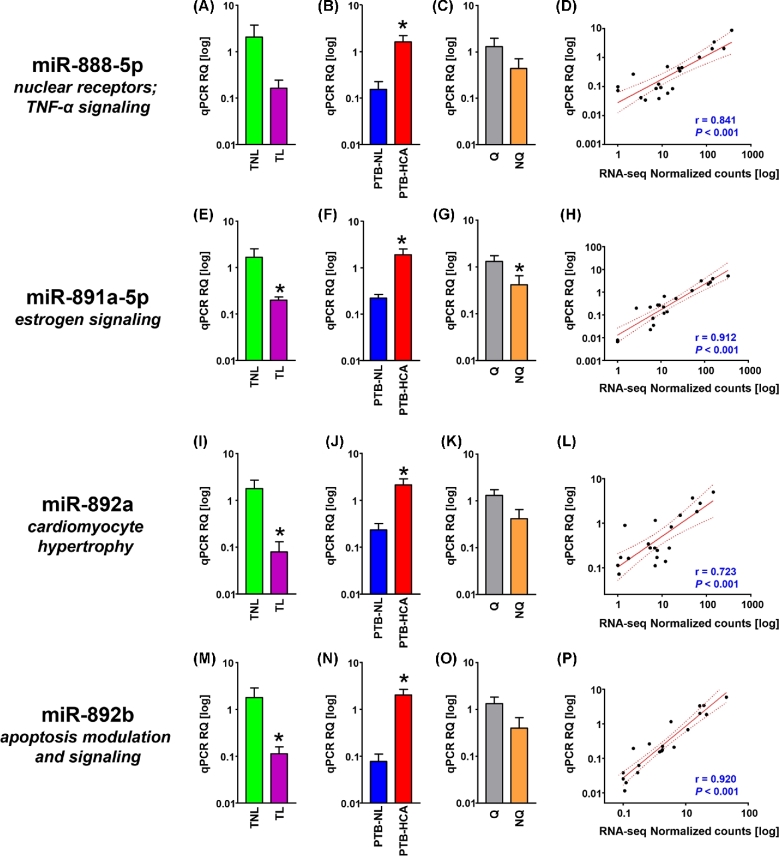
Figure 3.
Cross-validation of miRNA-seq data for members of the primate-specific, X-linked miR-888 cluster. (A–C) Relative quantification (RQ) of real-time quantitative PCR (qPCR) data for mature miR-888-5p in term myometrial specimens (TNL, term nonlabor; TL, term labor; A), preterm myometrial specimens (PTB-NL, preterm birth nonlabor; PTB-HCA, preterm birth with histologic chorioamnionitis; B), and specimens grouped by molecular clustering (Q, quiescent phenotype; NQ, nonquiescent phenotype; C). (D) Scatterplot and linear regression analysis (line of best fit and 95% confidence interval) for miR-888-5p comparing qPCR results with normalized RNA-seq feature counts. (E–H) RQ (grouped as described above) and linear regression analysis (qPCR vs. normalized RNA-seq) for miR-891a-5p. (I–L) RQ (grouped as described above) and linear regression analysis (qPCR vs. normalized RNA-seq) for miR-892a. (M–P) RQ (grouped as described above) and linear regression analysis (qPCR vs. normalized RNA-seq) for miR-892b. Data in bar graphs represent mean ± SEM. Asterisks indicate P < 0.05 by Student t-test.
The qPCR expression levels of the four selected members from the miR-888 cluster are shown in Figure 3 and confirmed the observation from the RNA-seq dataset of increased levels of expression in the PTB-HCA group compared to the PTB-NL group (miR-888-5p: P = 0.007, Figure 3B; miR-891a-5p: P = 0.022, Figure 3F; miR-892a: P = 0.029, Figure 3J; miR-892b: P = 0.002, Figure 3N). The decrease in the TL vs. TNL comparison was confirmed for miR-891a-5p (P = 0.011, Figure 3E), miR-892a (P = 0.028, Figure 3I), miR-892b (P = 0.002, Figure 3N). In quiescent tissues, statistical significance was reached only for miR-891a-5p (P = 0.038, Figure 3G). All correlations between RNA-seq and qPCR levels were significant at P < 0.001.
Localization and differential expression of MEF2 family transcription factors in quiescent vs. nonquiescent myometrium
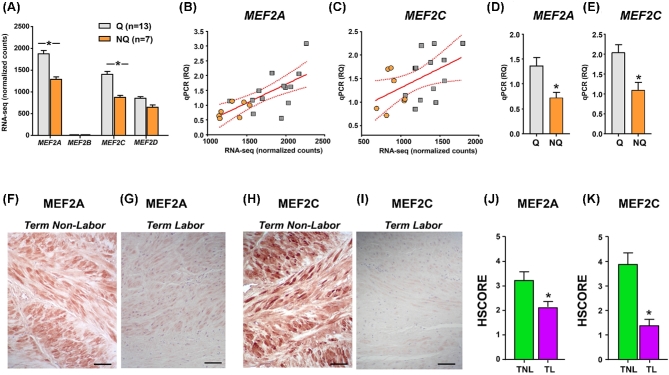
As MEF2C had been prioritized by our integrated miRNA-PPI network as molecular candidate for uterine quiescence, we mined the RNA-seq dataset for presence and expression levels of other members of the MEF2 protein family. We found that the pregnant myometrium at term expressed MEF2A, MEF2C, and MEF2D, but only very low counts were detected for MEF2B (Figure 4A). Both MEF2A and MEF2C transcripts were decreased in nonquiescent compared to quiescent tissues (MEF2A, FDR corrected P < 0.003; and MEF2C, FDR corrected P < 0.001). We cross-validated these expression changes in the primary cohort samples using qPCR, finding significant associations between the original RNA-seq expression levels and the relative quantification (RQ) levels (r = 0.683, P < 0.001 for MEF2A, Figure 4B; r = .478, P < 0,04 for MEF2C, Figure 4C). Using an independent cohort of 25 additional specimens (see Supplemental Table S6 for corresponding demographics information) stratified by the presence or absence of uterine quiescence by clinical contraction monitoring, we confirmed decreased abundance of MEF2A (Figure 4D) and MEF2C (Figure 4E) in nonquiescent relative to quiescent myometrial tissues.
Figure 4.
Expression of MEF2 family members in term myometrium in the absence or presence of uterine quiescence. (A) Bar graphs (mean ± SEM) showing the expression of transcripts encoding MEF2 family members based on normalized RNA-seq feature counts grouped by molecular clustering (Q, quiescent phenotype; NQ, nonquiescent phenotype) in the primary cohort. Asterisks indicate FDR-corrected P values < 0.05. (B, C) Scatterplots and linear regression analysis (line of best fit and 95% confidence interval) comparing the qPCR RQ scores with normalized RNA-seq feature counts for MEF2A and MEF2C in the myometrial samples from the primary cohort. Orange circles denote NQ samples, while gray squares indicate Q samples. Both relationships were statistically significant (P < 0.05). (D, E) Bar graphs (mean ± SEM) showing RQ of qPCR for MEF2A and MEF2C in a validation cohort of 25 additional myometrial samples: Q (n = 15), NQ (n = 10). Asterisks indicate P < 0.05 by Student t-test. (F–I) Representative photomicrographs are shown for term nonlabor and term labor myometrial samples labeled with antibodies directed against MEF2A (F, G) and MEF2C (H, I). Scale bars = 100 μm [56]. (J, K) Bar graphs (mean ± SEM) summarize the histologic staining intensity scores (HSCORE) for term nonlabor (TNL, n = 9, inclusive of four additional samples) and term labor (TL, n = 8, inclusive of three additional samples) myometrial tissues labeled with antibodies directed against MEF2A (G) and MEF2C (H). Asterisks denote P < 0.05 by the Mann–Whitney rank-sum test.
Using IHC, we identified strong immunoreactivity of both MEF2A (Figure 4F) and MEF2C (Figure 4H) in biopsies of women in the TNL group, and we confirmed the overall diminished expression in TL tissues for MEF2A (Figure 4G) and MEF2C (Figure 4I). In further experiments, we found that the overall labeling pattern for the MEF2 transcription factors was similar to that αSMA, consistent with predominant expression in myometrial smooth muscle cells, rather than CD15-positive myeloid cells (Supplemental Figure S3). The results of the IHC scoring for both MEF2A and MEF2C are presented in Figures 4J and K.
Discussion
Here, we surveyed global miRNA expression in myometrial specimens from term laboring and nonlaboring pregnancies, and from preterm pregnancies in the absence of presence of histologic chorioamnionitis. Of the 1507 unique mature miRNAs detected in one or more myometrial samples, we found that 40 were differentially expressed in TL vs. TNL specimens, while seven miRNAs differentiated the PTB-HCA from PTB-NL specimens. Surveying the experimentally verified targets for these differentially expressed miRNAs revealed their potential involvement in a number of pathways important for the maintenance of myocyte integrity (e.g., regulation of the actin cytoskeleton, myometrial contractility, and hypertrophy), as well as pathways governing aspects of the acute inflammatory response (e.g., interleukin, toll-like receptor, and tumor necrosis alpha signaling).
The onset of labor is a complex process characterized by multiple clinical and molecular alterations at the level of the myometrium. Based on the number of differentially expressed miRNAs, our analysis supports the notion that initiation of TL involves participation of a wider array of signals. Clustering of myometrial miRNA expression patterns allowed for identification of miRNA repertoires characterizing the quiescent vs. the nonquiescent myometrium. Interestingly, the molecular signatures of two of the PTB-HCA myometrial samples differed from the remaining specimens (quiescent vs. nonquiescent), despite similar phenotypic classification based on clinical grounds alone. Despite some notable exceptions, successful discovery of molecular signature tests has largely been hampered by limited reproducibility and variability [34]. An expression-based molecular signature capable of differentiating between the laboring and nonlaboring states would be of great practical use, because recognition of women at high risk for PTB is frequently erroneous based on clinical presentation alone [35]. Further work will be necessary to determine whether molecular signatures such as those presently observed in the myometrium may be reflected in other compartments, particularly the maternal circulation.
To prioritize miRNA-target interactions, we constructed miRNA-PPI topological networks, coupled with subnetwork prioritization based on RNA-seq expression data scoring. This in silico analysis identified 22 subnetworks by MI scoring, all of which being associated with one or more of three miRNAs: miR-223-3p, miR-146b-5p, and miR-150-5p. MiR-223, which displayed highly increased expression in both clinical contexts in our study (TL vs. TNL, and PTB-HCA vs. PTB-NL), was previously identified as being increased in human cervical and myometrial tissue following TL or oxytocin administration, respectively, and was found to be elevated in chorioamniotic membranes and villous trophoblast in the presence of inflammation [12, 36–38]. MiR-223 is highly expressed in leukocytes of the myeloid lineage [39], and may, in part, be a marker for inflammatory cell infiltration in tissues, such as that associated with laboring myometrium [40]. By targeting the expression of nuclear factor-kappa B (NF-κB), reactive oxygen species, and AGEs (endogenous mediators of inflammation), miR-223 generally functions to suppress inflammation [41, 42].
MiR-150, an “inflammamiR” associated with hematopoiesis and innate immunity [43, 44], exhibited increased expression only in the context of PTB-HCA. Given our findings, and its absent elevation in the contractile phenotype as identified by PCA, we infer that this miRNA functions in relationship with inflammation or infection. MiR-146b was significantly increased in the setting of the nonquiescent phenotype, in line with the previous data reporting its elevation in the myometrium of women with either spontaneous or oxytocin-augmentation labor [37]. Known targets for miR-146b include members of the toll-like receptor (i.e., CD40 molecule, interferon regulatory factor 5 (IRF5), toll like receptor 4 (TLR4) and IL-1 (i.e., conserved helix-loop-helix ubiquitous kinase (CHUK), interleukin 1 receptor type 1 (IL1R1), interleukin 1 receptor associated kinase 1 (IRAK1)) signaling pathways, implying its involvement in labor-associated inflammatory responses.
The presence of significant coordinated expression changes in a primate-specific, X-linked multicopy miRNA gene family [33], to the best of our knowledge, has not been previously reported in the context of human parturition. This family of miRNAs is highly expressed in reproductive tissues [33, 45, 46], leading to speculation that functionalization of this locus along the primate lineage [33] might have improved reproductive fitness and hence, provided a survival advantage. Given that known and predicted mRNA targets for these miRNAs are involved in estrogen and progesterone signaling, as well as pathways relevant to myometrial contractility (e.g., calcium regulation, contraction and relaxation, and cardiac hypertrophy), we chose to explore these miRNAs in greater detail. Using qPCR, we found significantly decreased expression for all but miR-888 in the TL vs. TNL comparison. Furthermore, we found significantly increased expression for miR-888, miR-891a, miR-892a, and miR-892b in the PTB-HCA vs. PTB-NL comparison. However, only miR-891 was statistically significantly decreased in nonquiescent tissues. Further analysis of this miRNA cluster in human myometrial tissues will likely require careful consideration of covariates such as GA, presence or absence of uterine contractions, and intrauterine inflammation, in addition to larger cohorts.
In an initial integrated miRNA-PPI subnetwork analysis that included the current dataset, MEF2C was found to be one of several transcription factors among prioritized subnetworks [47]. MEF2C was also identified as a candidate PTB-associated locus using a similar approach (PPI subnetwork analysis), but with networks constructed using high-ranking single nucleotide polymorphisms abstracted from a meta-analysis of genome-wide association studies, and scoring based on publically available myometrial transcriptomics datasets [48]. In the current study, we found that in addition to MEF2C, MEF2A also exhibited significantly reduced abundance in nonquiescent myometrial samples at the transcript and also at the protein level.
The factors leading to decreases in myometrial MEF2A and MEF2C in relationship with onset of uterine contractions are yet unknown, although, based on this study, miRNAs may play a significant role. Specifically, MEF2C was found to be an experimentally validated target of miR-223-3p [49]. In our study, miR-223-3p displayed increased abundance in myometrial laboring term and preterm tissues, as well as the nonquiescent phenotype. Our interaction analysis did not prioritize MEF2A, possibly due to the limited number of miRNA–mRNA interactions currently validated; however, this second MEF2 family member was identified as a predicted target of two other miRNAs that were differentially regulated: miR-148a-5p (increased in the TL vs. TNL) and miR-330-3p (decreased in the TL vs. TNL comparison) (Supplemental Table S2). As such, it is possible that regulatory interactions between these miRNAs and their MEF2 family targets contribute to the observed decreased expression of these proteins following onset of term contractions. In human aortic vascular smooth muscle cells, miR-223 upregulation reduced MEF2C expression [44]. Our current analysis cannot exclude the existence of other novel downstream targets of miRNA regulation which cannot be yet predicted given the paucity of validated miRNA–mRNA interactions in various databases. The present data have been deposited in a public data repository for future interrogation by the broader scientific community.
Although originally identified as a protein enhancer of muscle-specific gene expression [50], MEF2 transcription factor paralogs are known to contribute to the development and maintenance of a diverse assortment of tissues, including neuronal, immunological, and hematopoeitic, among others [32, 51]. Other proposed roles include ion transport, contractility, structural maintenance, and metabolism across multiple muscle lineages [32]. Recent observations in mammalian smooth muscle cells and cardiomyocytes demonstrate that MEF2 paralogs contribute to the promotion of myocyte hypertrophy, and transitions between the divergent contractile and synthetic myocyte phenotypes (the latter representing a dedifferentiated, less contractile state) [52–54]. A functional role for MEF2 transcription factors in the uterine myometrium is supported by gel mobility supershift assays, which demonstrated binding of several family members (MEF2A, -B, and -D) to the promoter of a smooth muscle actin gene (ACTG2) in nuclear extracts from nonpegnant samples [55]. It should be noted that while the majority of MEF2A and MEF2C protein expression localized to smooth muscle cells by IHC (Supplemental Figure S3), we cannot exclude the possibility that changes in other MEF2-expressing cells, including immune cells, might also have contributed to the tissue-level transcriptional changes we observed. Nevertheless, given that myometrial contractions are typically associated with an increased complement of immune cells (particularly innate immune cells) [56], this ultimately would tend to bias the observed contraction-associated decreases in MEF2A and MEF2C expression toward the null.
These prior reports, when coupled with our present observations, invite the speculation that MEF2A and MEF2C might facilitate maintenance of a quiescent, hypertrophic myometrial phenotype throughout most of gestation, and that withdrawal from these transcriptions factors might facilitate the onset of uterine contractility. Additionally, there is evidence that MEF2C may suppress inflammation and reduce nuclear factor-kappa B (NF-κB) activity in endothelial cells [57]; given that NF-κB is considered to be a master regulator of labor-associated inflammation in the myometrium [58], it is possible that MEF2 transcription factors might also promote uterine quiescence by opposing proinflammatory signaling. While we acknowledge that the current study was limited by the relatively small number of samples, we were able to query using simultaneous miRNA and long RNA sequencing, our finding of decreased MEF2A and MEF2C expression in the setting of uterine nonquiescence is strengthened by repeating the observation in independent cohort of myometrial samples. Nevertheless, it remains possible that the changes in gene expression profiles might have been influenced by demographic factors unrelated to the absence or presence of uterine quiescence. Further work will be required to determine the broader significance of the current findings in the context of human labor.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge Dr Pearlly Yan of the Nucleic Acid Shared Resource (The Ohio State University Comprehensive Cancer Center, Columbus, OH) for technical support and helpful discussions regarding data analysis. The authors thank Taryn L. Summerfield and Guomao Zhao for excellent technical assistance. The authors are indebted to the High Performance Computing Facility at the Research Institute at Nationwide Children's Hospital for providing computational infrastructure and resources necessary for efficient data storage and processing, and to Dr Ali Snedden for computational support. Portions of this were presented in abstract form at the 36th Annual Pregnancy Meeting of the Society for Maternal-Fetal Medicine (February 1–6, 2016, Atlanta, GA, USA) and at the 63rd Annual Scientific Meeting of the Society for Reproductive Investigation (March 16–19, 2016, Montreal, QC, Canada). We would like to thank Dr Mark B. Landon for assigning departmental funds for completion of this work.
Notes
Edited by Dr. Romana Nowak, PhD, University of Illinois Urbana-Champaign.
Footnotes
Grant Support: The coauthors were supported in part by funds from National Institutes of Health/Eunice Kennedy Shriver National Institute of Child Health and Human Development (HD084628) and by the March of Dimes through the March of Dimes Prematurity Research Center Ohio Collaborative. The sequencing data for this project was supported by internal funds from the Division of Maternal Fetal Medicine at The Ohio State College of Medicine, and from The Center for Perinatal Research in The Research Institute at Nationwide Children's Hospital. The funding sources had no role in study design, data analysis, writing of the report, or decision to submit for publication.
The data presented in this study are accessible through GEO Series accession number GSE99853.
Supplementary data
Supplementary data are available at BIOLRE online.
References
- 1. Aguan K, Carvajal JA, Thompson LP, Weiner CP. Application of a functional genomics approach to identify differentially expressed genes in human myometrium during pregnancy and labour. Mol Hum Reprod 2000; 6(12):1141–1145. [DOI] [PubMed] [Google Scholar]
- 2. Bethin KE, Nagai Y, Sladek R, Asada M, Sadovsky Y, Hudson TJ, Muglia LJ. Microarray analysis of uterine gene expression in mouse and human pregnancy. Mol Endocrinol 2003; 17(8):1454–1469. [DOI] [PubMed] [Google Scholar]
- 3. Charpigny G, Leroy MJ, Breuiller-Fouche M, Tanfin Z, Mhaouty-Kodja S, Robin P, Leiber D, Cohen-Tannoudji J, Cabrol D, Barberis C, Germain G. A functional genomic study to identify differential gene expression in the preterm and term human myometrium. Biol Reprod 2003; 68(6):2289–2296. [DOI] [PubMed] [Google Scholar]
- 4. Havelock JC, Keller P, Muleba N, Mayhew BA, Casey BM, Rainey WE, Word RA. Human myometrial gene expression before and during parturition. Biol Reprod 2005; 72(3):707–719. [DOI] [PubMed] [Google Scholar]
- 5. Esplin MS, Fausett MB, Peltier MR, Hamblin S, Silver RM, Branch DW, Adashi EY, Whiting D. The use of cDNA microarray to identify differentially expressed labor-associated genes within the human myometrium during labor. Am J Obstet Gynecol 2005; 193(2):404–413. [DOI] [PubMed] [Google Scholar]
- 6. Bukowski R, Hankins GD, Saade GR, Anderson GD, Thornton S. Labor-associated gene expression in the human uterine fundus, lower segment, and cervix. PLoS Med 2006; 3(6):e169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bollapragada S, Youssef R, Jordan F, Greer I, Norman J, Nelson S. Term labor is associated with a core inflammatory response in human fetal membranes, myometrium, and cervix. Am J Obstet Gynecol 2009; 200:104.e1–104.e11. [DOI] [PubMed] [Google Scholar]
- 8. Mittal P, Romero R, Tarca AL, Gonzalez J, Draghici S, Xu Y, Dong Z, Nhan-Chang CL, Chaiworapongsa T, Lye S, Kusanovic JP, Lipovich L et al. . Characterization of the myometrial transcriptome and biological pathways of spontaneous human labor at term. J Perinat Med 2010; 38(6):617–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Weiner CP, Mason CW, Dong Y, Buhimschi IA, Swaan PW, Buhimschi CS. Human effector/initiator gene sets that regulate myometrial contractility during term and preterm labor. Am J Obstet Gynecol 2010; 202(5):474.e1–474.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chan YW, van den Berg HA, Moore JD, Quenby S, Blanks AM. Assessment of myometrial transcriptome changes associated with spontaneous human labour by high-throughput RNA-seq. Exp Physiol 2014; 99(3):510–524. [DOI] [PubMed] [Google Scholar]
- 11. Breuiller-Fouche M, Germain G. Gene and protein expression in the myometrium in pregnancy and labor. Reproduction 2006; 131(5):837–850. [DOI] [PubMed] [Google Scholar]
- 12. Ackerman WE, Buhimschi IA, Eidem HR, Rinker DC, Rokas A, Rood K, Zhao G, Summerfield TL, Landon MB, Buhimschi CS. Comprehensive RNA profiling of villous trophoblast and decidua basalis in pregnancies complicated by preterm birth following intra-amniotic infection. Placenta 2016; 44:23–33. [DOI] [PubMed] [Google Scholar]
- 13. Bartel DP. MicroRNAs. Cell 2004; 116(2):281–297. [DOI] [PubMed] [Google Scholar]
- 14. Valinezhad Orang A, Safaralizadeh R, Kazemzadeh-Bavili M. Mechanisms of miRNA-mediated gene regulation from common downregulation to mRNA-specific upregulation. Int J Genomics 2014; 2014:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res 2014; 42(D1):D68–D73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Renthal NE, Williams KC, Mendelson CR. MicroRNAs–mediators of myometrial contractility during pregnancy and labour. Nat Rev Endocrinol 2013; 9(7):391–401. [DOI] [PubMed] [Google Scholar]
- 17. Williams KC, Renthal NE, Gerard RD, Mendelson CR. The microRNA (miR)-199a/214 cluster mediates opposing effects of progesterone and estrogen on uterine contractility during pregnancy and labor. Mol Endocrinol 2012; 26(11):1857–1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Eidem HR, Rinker DC, Ackerman WE, Buhimschi IA, Buhimschi CS, Dunn-Fletcher C, Kallapur SG, Pavlicev M, Muglia LJ, Abbot P, Rokas A. Comparing human and macaque placental transcriptomes to disentangle preterm birth pathology from gestational age effects. Placenta 2016; 41:74–82. [DOI] [PubMed] [Google Scholar]
- 19. Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 2013; 14(4):R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Anders S, Pyl PT, Huber W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 2015; 31(2):166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang WC, Lin FM, Chang WC, Lin KY, Huang HD, Lin NS. miRExpress: analyzing high-throughput sequencing data for profiling microRNA expression. BMC Bioinformatics 2009; 10(1):328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014; 15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ru Y, Kechris KJ, Tabakoff B, Hoffman P, Radcliffe RA, Bowler R, Mahaffey S, Rossi S, Calin GA, Bemis L, Theodorescu D. The multiMiR R package and database: integration of microRNA-target interactions along with their disease and drug associations. Nucleic Acids Res 2014; 42(17):e133–e133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, Clark NR, Ma’ayan A. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 2013; 14(1):128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, Kuhn M, Bork P et al. . STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 2015; 43(D1):D447–D452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nibbe RK, Markowitz S, Myeroff L, Ewing R, Chance MR. Discovery and scoring of protein interaction subnetworks discriminative of late stage human colon cancer. Mol Cell Proteomics 2009; 8(4):827–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Liu Y, Patel S, Nibbe R, Maxwell S, Chowdhury SA, Koyuturk M, Zhu X, Larkin EK, Buxbaum SG, Punjabi NM, Gharib SA, Redline S et al. . Systems biology analyses of gene expression and genome wide association study data in obstructive sleep apnea. Pac Symp Biocomput 2011; 16:14–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Liu Y, Koyuturk M, Maxwell S, Zhao Z, Chance MR. Integrative analysis of common neurodegenerative diseases using gene association, interaction networks and mRNA expression data. AMIA Jt Summits Transl Sci Proc 2012; 2012:62–71. [PMC free article] [PubMed] [Google Scholar]
- 29. Sauer E, Madea B, Courts C. An evidence based strategy for normalization of quantitative PCR data from miRNA expression analysis in forensically relevant body fluids. Forensic Sci Int Genet 2014; 11:174–181. [DOI] [PubMed] [Google Scholar]
- 30. Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method. Nat Protoc 2008; 3(6):1101–1108. [DOI] [PubMed] [Google Scholar]
- 31. Dulay AT, Buhimschi CS, Zhao G, Oliver EA, Abdel-Razeq SS, Shook LL, Bahtiyar MO, Buhimschi IA. Amniotic fluid soluble myeloid differentiation-2 (sMD-2) as regulator of intra-amniotic inflammation in infection-induced preterm birth. Am J Reprod Immunol 2015; 73(6):507–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Potthoff MJ, Olson EN. MEF2: a central regulator of diverse developmental programs. Development 2007; 134(23):4131–4140. [DOI] [PubMed] [Google Scholar]
- 33. Li J, Liu Y, Dong D, Zhang Z. Evolution of an X-linked primate-specific micro RNA cluster. Mol Biol Evol 2010; 27(3):671–683. [DOI] [PubMed] [Google Scholar]
- 34. van’t Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT, Schreiber GJ, Kerkhoven RM et al. . Gene expression profiling predicts clinical outcome of breast cancer. Nature 2002; 415(6871):530–536. [DOI] [PubMed] [Google Scholar]
- 35. Barros FC, Papageorghiou AT, Victora CG, Noble JA, Pang R, Iams J, Cheikh Ismail L, Goldenberg RL, Lambert A, Kramer MS, Carvalho M, Conde-Agudelo A et al. . The distribution of clinical phenotypes of preterm birth syndrome. JAMA Pediatr 2015; 169(3):220–229. [DOI] [PubMed] [Google Scholar]
- 36. Hassan SS, Romero R, Pineles B, Tarca AL, Montenegro D, Erez O, Mittal P, Kusanovic JP, Mazaki-Tovi S, Espinoza J, Nhan-Chang CL, Draghici S et al. . MicroRNA expression profiling of the human uterine cervix after term labor and delivery. Am J Obstet Gynecol 2010; 202(1):80.e1–80.e8. [DOI] [PubMed] [Google Scholar]
- 37. Cook JR, MacIntyre DA, Samara E, Kim SH, Singh N, Johnson MR, Bennett PR, Terzidou V. Exogenous oxytocin modulates human myometrial microRNAs. Am J Obstet Gynecol 2015; 213(1):65.e1–65.e9. [DOI] [PubMed] [Google Scholar]
- 38. Montenegro D, Romero R, Pineles BL, Tarca AL, Kim YM, Draghici S, Kusanovic JP, Kim JS, Erez O, Mazaki-Tovi S, Hassan S, Espinoza J et al. . Differential expression of microRNAs with progression of gestation and inflammation in the human chorioamniotic membranes. Am J Obstet Gynecol 2007; 197(3):289.e1–289.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, Lin C, Socci ND et al. . A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 2007; 129(7):1401–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Thomson AJ, Telfer JF, Young A, Campbell S, Stewart CJ, Cameron IT, Greer IA, Norman JE. Leukocytes infiltrate the myometrium during human parturition: further evidence that labour is an inflammatory process. Hum Reprod 1999; 14(1):229–236. [PubMed] [Google Scholar]
- 41. Haneklaus M, Gerlic M, O’Neill LA, Masters SL. miR-223: infection, inflammation and cancer. J Intern Med 2013; 274(3):215–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wang Z, Li H, Guo R, Wang Q, Zhang D. Antioxidants inhibit advanced glycosylation end-product-induced apoptosis by downregulation of miR-223 in human adipose tissue-derived stem cells. Sci Rep 2016; 6(1):23021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. de Gonzalo-Calvo D Davalos A, Montero A, Garcia-Gonzalez A, Tyshkovska I, Gonzalez-Medina A, Soares SM, Martinez-Camblor P, Casas-Agustench P, Rabadan M, Diaz-Martinez AE, Ubeda N et al. . Circulating inflammatory miRNA signature in response to different doses of aerobic exercise. J Appl Physiol 1985; 2015(119):124–134. [DOI] [PubMed] [Google Scholar]
- 44. He Y, Jiang X, Chen J. The role of miR-150 in normal and malignant hematopoiesis. Oncogene 2014; 33:3887–3893. [DOI] [PubMed] [Google Scholar]
- 45. Belleannee C, Calvo E, Thimon V, Cyr DG, Legare C, Garneau L, Sullivan R. Role of microRNAs in controlling gene expression in different segments of the human epididymis. PLoS One 2012; 7:e34996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hovey AM, Devor EJ, Breheny PJ, Mott SL, Dai D, Thiel KW, Leslie KK. miR-888: a novel cancer-testis antigen that targets the progesterone receptor in endometrial cancer. Transl Oncol 2015; 8:85–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ackerman W, Buhimschi IA, Summerfield T, Zhao G, Jing H, Buhimschi CS. Human myometrial microRNA expression following term and preterm labor. Reprod Sci 2016; 23:300A. [Google Scholar]
- 48. Brubaker D, Liu Y, Wang J, Tan H, Zhang G, Jacobsson B, Muglia L, Mesiano S, Chance MR. Finding lost genes in GWAS via integrative-omics analysis reveals novel sub-networks associated with preterm birth. Hum Mol Genet 2016; 25:5254–5264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rangrez AY, M’Baya-Moutoula E, Metzinger-Le Meuth V, Henaut L, Djelouat MS, Benchitrit J, Massy ZA, Metzinger L. Inorganic phosphate accelerates the migration of vascular smooth muscle cells: evidence for the involvement of miR-223. PLoS One 2012; 7:e47807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Gossett LA, Kelvin DJ, Sternberg EA, Olson EN. A new myocyte-specific enhancer-binding factor that recognizes a conserved element associated with multiple muscle-specific genes. Mol Cell Biol 1989; 9:5022–5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Pon JR, Marra MA. MEF2 transcription factors: developmental regulators and emerging cancer genes. Oncotarget 2016; 7:2297–2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhang T, Kohlhaas M, Backs J, Mishra S, Phillips W, Dybkova N, Chang S, Ling H, Bers DM, Maier LS, Olson EN, Brown JH. CaMKIIdelta isoforms differentially affect calcium handling but similarly regulate HDAC/MEF2 transcriptional responses. J Biol Chem 2007; 282:35078–35087. [DOI] [PubMed] [Google Scholar]
- 53. Ginnan R, Sun LY, Schwarz JJ, Singer HA. MEF2 is regulated by CaMKIIdelta2 and a HDAC4-HDAC5 heterodimer in vascular smooth muscle cells. Biochem J 2012; 444:105–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Li C, Vu K, Hazelgrove K, Kuemmerle JF. Increased IGF-IEc expression and mechano-growth factor production in intestinal muscle of fibrostenotic Crohn's disease and smooth muscle hypertrophy. Am J Physiol Gastrointest Liver Physiol 2015; 309:G888–G899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Phiel CJ, Gabbeta V, Parsons LM, Rothblat D, Harvey RP, McHugh KM. Differential binding of an SRF/NK-2/MEF2 transcription factor complex in normal versus neoplastic smooth muscle tissues. J Biol Chem 2001; 276:34637–34650. [DOI] [PubMed] [Google Scholar]
- 56. Gomez-Lopez N, StLouis D, Lehr MA, Sanchez-Rodriguez EN, Arenas-Hernandez M. Immune cells in term and preterm labor. Cell Mol Immunol 2014; 11:571–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Xu Z, Yoshida T, Wu L, Maiti D, Cebotaru L, Duh EJ. Transcription factor MEF2C suppresses endothelial cell inflammation via regulation of NF-kappaB and KLF2. J Cell Physiol 2015; 230:1310–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Mendelson CR. Minireview: fetal-maternal hormonal signaling in pregnancy and labor. Mol Endocrinol 2009; 23:947–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.